Abstract
Free full text

Convergent multi-miRNA Targeting of ApoE Drives LRP1/LRP8-Dependent Melanoma Metastasis and Angiogenesis
Associated Data
SUMMARY
Through in-vivo selection of human cancer cell populations, we uncover a convergent and cooperative miRNA network that drives melanoma metastasis. We identify miR-1908, miR-199a-5p, and miR-199a-3p as endogenous promoters of metastatic invasion, angiogenesis, and colonization in melanoma. These miRNAs convergently target Apolipoprotein E (ApoE) and the heat-shock factor DNAJA4. Cancer-secreted ApoE suppresses invasion and metastatic endothelial recruitment (MER) by engaging melanoma-cell LRP1 and endothelial-cell LRP8 receptors, respectively–while DNAJA4 promotes ApoE expression. Expression levels of these miRNAs and ApoE correlate with human metastatic progression outcomes. Treatment of cells with locked nucleic acids (LNAs) targeting these miRNAs inhibits metastasis to multiple organs, while therapeutic delivery of these LNAs strongly suppresses melanoma metastasis. We thus identify miRNAs with dual cell-intrinsic/cell-extrinsic roles in cancer, reveal convergent cooperativity in a metastatic miRNA network, identify ApoE as an anti-angiogenic and metastasis-suppressive factor, and uncover multiple prognostic miRNAs with synergistic combinatorial therapeutic potential in melanoma.
INTRODUCTION
Metastatic progression requires that sets of effector proteins involved in common cellular phenotypes be coherently expressed (Png et al., 2011; Gupta and Massagué, 2006; Hanahan and Weinberg, 2011; Talmadge and Fidler, 2010). Such concerted expression states are apparent in gene expression profiles of primary breast cancers that metastasize (Wang et al., 2005) as well as profiles of human cancer cell clones that display enhanced metastatic activity (Kang et al., 2003; Minn et al., 2005). In recent years, post-transcriptional regulation has emerged as a pervasive and robust mode of concerted expression-state and phenotype-level control. The most studied class of post-transcriptional regulators with metastatic regulatory activity are small non-coding RNAs (miRNAs) (He and Hannon, 2004; Bartel, 2009; Filipowicz et al., 2008). Metastasis suppressor miRNAs (Tavazoie et al., 2008) and promoter miRNAs (Ma et al., 2007; Huang et al., 2008) were originally discovered in breast cancer. Subsequent studies revealed many more miRNAs with regulatory roles in the tumorigenesis and metastasis of other cancer types (Hurst et al., 2009). In many cases, the expression levels of such miRNAs in human cancer samples have supported their experimental roles in metastasis. Thus, deregulated miRNA expression and function (Calin and Croce, 2006; Lujambio and Lowe, 2012; Poliseno et al., 2010) appear to be a pervasive feature of human cancer. Clues regarding the robust control exerted by specific miRNAs on metastatic progression emerged from early work showing that concerted targeting of multiple metastasis genes by a single metastasis suppressor miRNA was responsible for the dramatic metastasis suppression effects seen (Tavazoie et al., 2008). Such divergent gene targeting by an individual miRNA has emerged as a defining feature of these regulators.
By applying a systematic, in vivo selection-based approach, we have identified a set of miRNAs that are deregulated in independent metastatic lines derived from multiple patients with melanoma—a highly prevalent cancer with increasing incidence. These miRNAs convergently target the metabolic gene ApoE and the heat-shock protein DNAJA4 and comprise a cooperative miRNA network that maximally silences ApoE signaling. Cancer cell-secreted ApoE inhibits metastatic invasion and endothelial recruitment through its actions on distinct ApoE receptors on melanoma and endothelial cells. These miRNAs are prognostic of clinical metastasis, while their therapeutic inhibition displays in vivo efficacy. The lack of effective therapies in melanoma for metastasis prevention (Garbe et al., 2011) requires novel approaches. Our unbiased and systematic approach has allowed us to discover key non-coding and coding genes involved in melanoma progression and offers novel avenues for both identifying patients at high risk for melanoma metastasis and rationally treating them.
RESULTS
Endogenous miR-1908, miR-199a-3p, and miR-199a-5p Promote Human Melanoma Metastasis
In order to identify miRNA regulators of melanoma metastasis, we utilized in vivo selection (Pollack and Fidler, 1982) with the pigmented MeWo and non-pigmented A375 human melanoma cell lines to generate multiple second (LM2) and third-generation (LM3) lung metastatic derivatives that colonized the lung more efficiently than their respective parental populations (Figures 1A and S1A). Hybridization-based small RNA profiling of 894 mature miRNAs followed by quantitative stem-loop PCR (qRT-PCR) validation revealed four miRNAs (miR-214, miR-199a-5p, miR-1908, and miR-199a-3p) to be upregulated in multiple A375 and MeWo metastatic derivatives relative to their respective parental lines (Figures 1B–C and S1B). The significant induction of these miRNAs suggested potential roles for them in metastatic progression. Indeed, over-expression of the precursor for miR-199a-3p and miR-199a-5p (over-expressed concomitantly as the miR-199a hairpin) or miR-1908 was sufficient to robustly increase lung metastatic colonization by the poorly metastatic MeWo parental cells (Figures 1D and S1C; 9.64-fold (miR-1908); 8.62-fold (miR-199a)), while miR-214 over-expression did not significantly affect metastasis. We next asked if endogenous forms of these miRNAs promote metastasis. Individual inhibition of miR-1908, miR-199a-3p, or miR-199a-5p in the metastatic MeWo-LM2.3 cells (henceforth called MeWo-LM2), as well as in the independent metastatic line A375-LM3.2 (henceforth called A375-LM3), through a miR-Zip-based stable silencing approach significantly suppressed metastatic colonization (Figures 1E–F), establishing these three miRNAs as endogenous promoters of metastasis in melanoma. Notably, both endogenous and exogenous miR-199a and miR-1908 increased the number of metastatic nodules formed (Figures S1D–E), consistent with a role for these miRNAs in metastatic initiation.

(A) Bioluminescence imaging plot of lung metastatic colonization by 4×104 parental MeWo or MeWo-LM2 cells. Lungs were extracted at day 72 and H&E stained. n=4–5. (B–C) Expression levels of miR-214, miR-199a-5p, miR-1908, and miR-199a-3p, determined by qRT-PCR, in multiple independent MeWo-LM2 (B) and A375-LM2/LM3 (C) metastatic derivatives and their respective parental melanoma lines. n=3. (D–E) Bioluminescence quantification and representative H&E-stained lungs corresponding to lung colonization by 4×104 parental MeWo cells over-expressing the precursor for miR-199a (giving rise to both miR-199a-3p and miR-199a-5p), miR-1908, miR-214, or expressing a control hairpin (shCTRL) (D) and 4×104 MeWo-LM2 cells expressing a short hairpin (miR-Zip) inhibiting miR-1908 (m1908 knock-down (KD)), miR-199a-3p (m199a3p KD), miR-199a-5p (m199a5p KD), or a control sequence (shCTRL) (E). n=5–8. (F) Lung colonization by 2×105 A375-LM3 metastatic derivatives with miR-199a-3p KD, miR-199a-5p KD, miR-1908 KD, or a control KD quantified at day 42 by bioluminescence imaging. n=5–8. (G) Expression levels of miR-199a-3p, miR-199a-5p, and miR-1908, determined by qRT-PCR, in non-metastatic (n=38) and metastatic (n=33) primary melanoma skin lesions from the MSKCC patient cohort. All data are represented as mean ± SEM. *p<0.05, **p<0.01, ***p<0.001. See also Figure S1.
Given the robust functional roles of miR-1908, miR-199a-3p, and miR-199a-5p in promoting human melanoma metastasis in a mouse model, we examined whether the expression levels of these miRNAs correlate with the metastatic relapse likelihood of human melanoma. To this end, 71 surgically resected primary melanoma skin lesions were analyzed in a blinded manner for miRNA expression through qRT-PCR. Consistent with our functional studies, all three miRNAs were significantly upregulated in primary melanomas that had metastasized relative to those that had not (Figure 1G), suggesting that induced expression of these miRNAs in primary lesions is an early event predictive of melanoma cancer progression.
miR-1908, miR-199a-3p, and miR-199a-5p Promote Metastatic Invasion, Endothelial Recruitment, and Angiogenesis
We next sought to determine the cellular mechanisms underlying miRNA regulation of metastasis. Importantly, over-expression of each miRNA reduced cell proliferation (Figure S2A) and did not increase tumor growth (Figure 2A), indicating that the pro-metastatic effects of miR-1908 and miR-199a are not secondary to tumor growth promotion or enhanced proliferation.
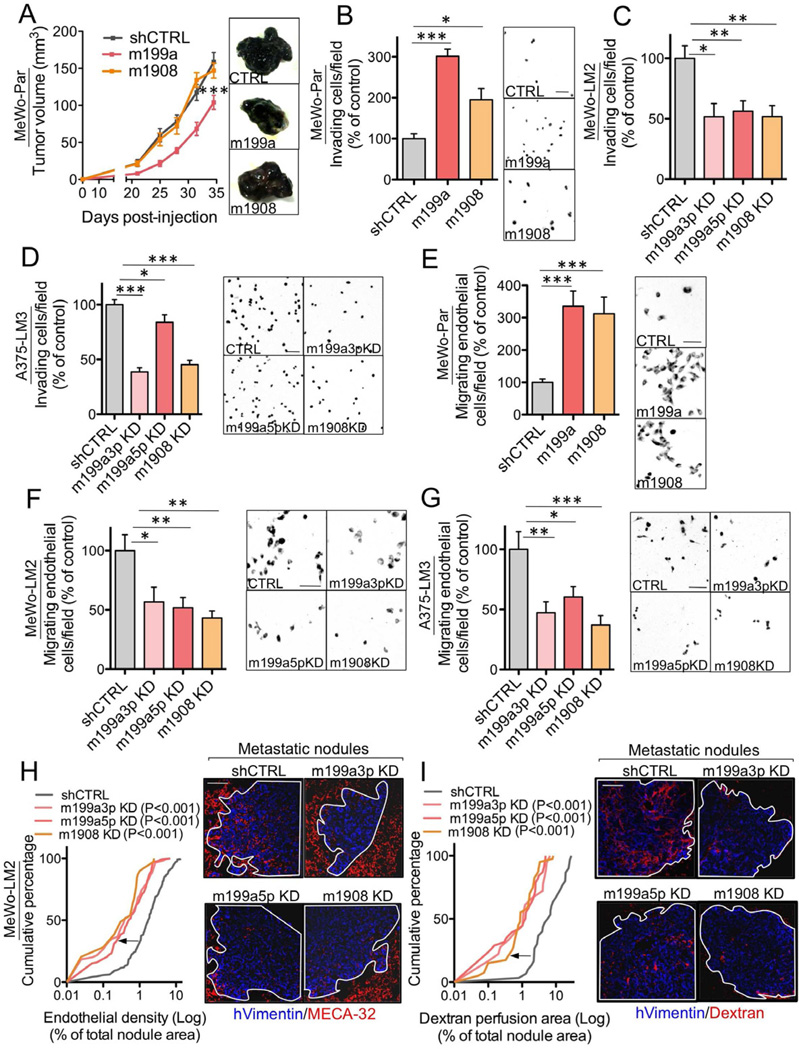
(A) Primary tumor growth by 1×106 MeWo cells over-expressing the miR-199a or miR-1908 precursor hairpins or expressing an shCTRL following subcutaneous injection into mice. n=4–6.(B) Matrigel invasion by 1×105 MeWo cells over-expressing miR-199a or miR-1908 or expressing an shCTRL was quantified by counting the number of cells that invaded into the basal side of matrigel-coated trans-well inserts after 24 hours. n=7. (C–D) Matrigel invasion by MeWo-LM2 (C) and A375-LM3 (D) cells with miR-199a-3p KD, miR-199a-5p KD, miR-1908 KD, or a control KD. n=6–8. (E) Trans-well recruitment of 1×105 human umbilical vein endothelial cells (HUVEC) by 5×104 MeWo cells over-expressing miR-199a or miR-1908 or control MeWo cells. Endothelial recruitment capacity was measured by counting the number of HUVECs that migrated to the basal side of each trans-well insert after 16 hours. n=7. (F–G) Endothelial recruitment by MeWo-LM2 (F) and A375-LM3 (G) cells inhibited for miR-199a-3p, miR-199a-5p, miR-1908, or a control sequence. n=6–10. (H–I) Cumulative fraction plots depicting the endothelial cell (H) or dextran perfusion (I) density distributions for metastatic nodules formed by 2×105 MeWo-LM2 cells with miR-199a-3p KD, miR-199a-5p KD, miR-1908 KD, or a control KD. Lung sections were double-stained for vimentin and MECA-32 (H) or vimentin and intravenously injected dextran (I). n=211, 30, 138, and 39 nodules for control KD, m199a3p KD, m199a5p KD, and m1908 KD, respectively (H); n=30, 18, 38, and 40 nodules for control KD, m199a3p KD, m199a5p KD, and m1908 KD, respectively (I). All data are represented as mean ± SEM. Scale bar, 100 µm. See also Figure S2
We next asked whether these miRNAs regulate cell invasion, a key metastatic phenotype. Over-expression of either miR-199a or miR-1908 enhanced the ability of parental MeWo cells to invade through matrigel (Figure 2B), while metastatic LM2 cells, which express higher levels of these miRNAs, displayed significantly greater matrigel invasion capacity (Figure S2B). Conversely, individual inhibition of miR-199a-3p, miR-199a-5p, or miR-1908 decreased the invasive capacity of MeWo-LM2 (Figure 2C) and A375-LM3 (Figure 2D) metastatic melanoma cell derivatives.
Given the robust effects of these miRNAs on metastatic progression, we examined whether they regulate additional pro-metastatic phenotypes. While over-expression of miR-199a or miR-1908 did not modulate melanoma cell adhesion to endothelial cells (Figure S2C), resistance to anoikis (Figure S2D), survival during serum starvation (Figure S2E), or colony formation (Figure S2F), over-expression of each miRNA dramatically enhanced the ability of parental MeWo cells to recruit endothelial cells in trans-well endothelial recruitment assays (Figure 2E). Consistent with this, metastatic MeWo-LM2 cells were more efficient at recruiting endothelial cells relative to their parental line (Figure S2G). Furthermore, inhibition of miR-199a-3p, miR-199a-5p, or miR-1908 in the metastatic MeWo-LM2 (Figure 2F) as well as A375-LM3 (Figure 2G) cells suppressed endothelial recruitment.
To determine whether endogenous miR-199a-3p, miR-199a-5p, and miR-1908 mediate endothelial recruitment by metastatic cells in vivo, we examined metastatic endothelial density in lung nodules formed by melanoma cells over-expressing or knocked-down for each miRNA. Consistent with the robust miRNA-dependent endothelial recruitment phenotype, each of the three miRNAs was both required and sufficient for enhanced metastatic nodule endothelial content (Figures 2H and S2H–I). Remarkably, individual inhibition of each miRNA robustly suppressed metastatic nodule perfusion by roughly 5-fold following dextran injection (Figures 2I and S2J), indicating that each of these miRNAs also promotes functional metastatic angiogenesis. Our findings reveal miR-199a-3p, miR-199a-5p, and miR-1908 as necessary and sufficient for enhanced invasion, metastatic endothelial recruitment (MER), and angiogenesis during melanoma progression.
miR-1908, miR-199a-3p, and miR-199a-5p Convergently and Cooperatively Target ApoE and DNAJA4
We next employed a systematic and global approach to identify the direct molecular targets of these miRNAs. Since miR-1908, miR-199a-3p, and miR-199a-5p mediate the same sets of in vitro and in vivo phenotypes, and since miR-199a-5p and miR-199a-3p arise from the same precursor hairpin, we hypothesized that the pro-metastatic phenotypes of these miRNAs may emerge through silencing of common target genes. We thus performed transcriptomic profiling of melanoma cells in the context of both loss- and gain-of-function for each miRNA (Figure S3A). This analysis, followed by qRT-PCR validation, revealed two genes (ApoE and DNAJA4) to be repressed by both exogenous and endogenous miR-199a and miR-1908 and also present at lower levels in the metastatic LM2 derivatives—which display elevated levels of these miRNAs (Figures 3A and S3B–D).
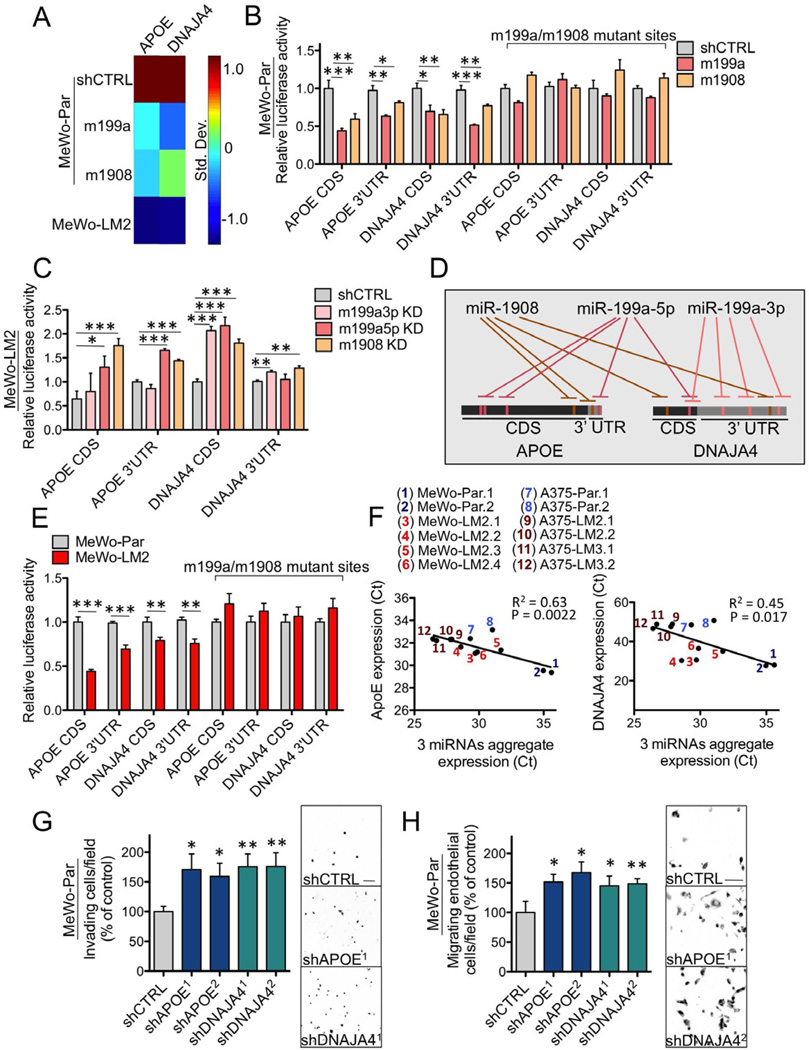
(A) Heat-map depicting the mRNA expression levels of ApoE and DNAJA4, measured by qRT-PCR, in MeWo parental cells expressing an shCTRL or over-expressing miR-199a or miR-1908 and in MeWo-LM2 cells. Color-map illustrates standard deviations change from the mean for each gene’s expression. (B–C) Heterologous luciferase reporter assays measuring the expression of wild type or miRNA target site mutant ApoE and DNAJA4 CDS’s and 3’UTR’s in MeWo cells over-expressing miR-199a or miR-1908 or in control MeWo cells (B) and in MeWo-LM2 cells with miR-199a-3p KD, miR-199a-5p KD, miR-1908 KD, or a control KD (C). n=3–4. (D) Schematic of an experimentally derived model depicting the convergent targeting of the CDS’s and 3’UTR’s of ApoE and DNAJA4 by miR-199a-3p, miR-199a-5p, and miR-1908. (E) Luciferase activity of wild type and miRNA target site mutant ApoE and DNAJA4 3’UTR/CDS luciferase constructs transfected into MeWo parental and MeWo-LM2 cells. n=4. (F) Linear regression analyses of the expression levels of each gene (ApoE and DNAJA4) and the aggregate expression levels of the three miRNAs (average of the individual miRNA expression values) in multiple MeWo and A375 metastatic derivatives and their parental lines. n=12. (G–H) Trans-well matrigel invasion (G) and endothelial recruitment (H) by parental MeWo cells expressing short hairpin RNAs (shRNAs) targeting ApoE (shAPOE), DNAJA4 (shDNAJA4), or a control sequence (shCTRL). n=6–8. All data are represented as mean ± SEM. Scale bar, 100 µm. See also Figure S3.
To determine whether ApoE and DNAJA4 are directly targeted by miR-1908, miR-199a-3p, and miR-199a-5p, we examined the effects of each miRNA on the expression of its putative targets through heterologous luciferase reporter assays. Interestingly, both miR-199a and miR-1908 individually repressed the expression levels of the 3’ untranslated regions (UTR) and coding sequences (CDS) of both ApoE and DNAJA4. Consistent with direct targeting, mutating the miRNA complementary sequences on each target abrogated miRNA-mediated regulation (Figure 3B). In a direct test of endogenous targeting, individual miRNA inhibition in metastatic LM2 cells led to an increased target-driven luciferase activity (Figure 3C) that was abrogated upon mutating the miRNA target sites (Figure S3E), revealing ApoE to be directly targeted by miR-1908 and miR-199a-5p and DNAJA4 to be directly targeted by all three miRNAs (Figure 3D). Importantly, the CDS’s and 3’UTR’s of both genes were expressed at lower levels in highly metastatic MeWo-LM2 cells, which exhibit physiologically higher endogenous levels of the three regulatory miRNAs (Figure 3E). These findings establish ApoE and DNAJA4 as direct targets of these miRNAs in melanoma. In agreement with this, we found a significant anti-correlation between the endogenous levels of both ApoE and DNAJA4 and the aggregate expression levels of the three miRNAs across our collection of parental and metastatic lines (Figure 3F). Consistent with the phenotypes of their miRNA regulators, both ApoE and DNAJA4 were required and sufficient for suppression of cell invasion and endothelial recruitment–phenotypes that drive metastatic progression (Figures 3G–H and S3F–I).
ApoE and DNAJA4 Mediate miR-199a- and miR-1908-Dependent Metastatic Invasion, Endothelial Recruitment, and Colonization
To determine whether ApoE and DNAJA4 are the direct biological effectors downstream of miR-199a and miR-1908, we examined whether these two target genes are epistatic to each miRNA. Consistent with direct and epistatic miRNA/target gene interactions, knock-down of ApoE or DNAJA4 in the setting of miRNA inhibition (Figures S4A–D) significantly occluded the suppressed invasion (Figures 4A and 4C) and endothelial recruitment phenotypes (Figures 4B and 4D) seen upon silencing of each miRNA and fully rescued the dramatically suppressed metastatic colonization phenotype resulting from miRNA inhibition (Figures 4E–F, and S4E). Conversely, over-expression of the coding regions of ApoE or DNAJA4 in cells over-expressing miR-1908 (Figures 4G–H and S4F) or miR-199a (Figures S4G–I) was sufficient to suppress cell invasion and endothelial recruitment, as well as miRNA-dependent metastatic colonization (Figure S4J). Importantly, downregulation of ApoE and DNAJA4 was also required for miRNA-induced enhancement of cell invasion and endothelial recruitment by the highly metastatic A375-LM3 line (Figures 4I–J and S4K). Notably, endogenous ApoE and DNAJA4 were also found to suppress metastatic endothelial recruitment in vivo (Figure 4K). These effects of ApoE and DNAJA4 were both epistatic to and dependent on the miRNAs. Our findings implicate ApoE and DNAJA4 as direct downstream effectors of miRNA-dependent metastatic invasion, colonization, and endothelial recruitment phenotypes in melanoma.

(A–D) MeWo-LM2 cells expressing a control shRNA or shRNAs targeting ApoE or DNAJA4 in the context of miR-1908 KD (A–B) or miR-199a-5p KD (C–D) were subjected to the trans-well invasion (A and C) and endothelial recruitment (B and D) assays. n=6–8. (E–F) Bioluminescence imaging plots and H&E-stained lungs corresponding to lung metastasis by 1×105 MeWo-LM2 cells expressing a control shRNA or shRNAs targeting ApoE or DNAJA4 in the setting of miR-1908 KD (E) or miR-199a-5p KD (F). n=5. (G–H) MeWo cells over-expressing the coding regions of ApoE or DNAJA4 or expressing a control vector in the context of miR-1908 over-expression were analyzed for the invasion (G) and endothelial recruitment (H) phenotypes. (I–J) A375-LM3 cells expressing a control shRNA or shRNAs targeting ApoE and DNAJA4 were transfected with a cocktail of LNAs targeting miR-199a-3p, miR-199a-5p, and miR-1908 or a control LNA and subjected to the invasion (I) and endothelial recruitment (J) assays. n=4. (K) Cumulative fraction plots of metastatic nodule endothelial content distributions corresponding to nodules formed by 1×105 MeWo-LM2 cells knocked down for ApoE, DNAJA4, or a control sequence in the setting of miR-1908 KD. Thresholded images depicting MECA-32 signal are shown in lower panels. n=39 nodules (shCTRL); n=97 (shAPOE1); n=38 (shAPOE2); n=200 (shDNAJA41); n=19 (shDNAJA42). All data are represented as mean ± SEM. Scale bar, 100 µm. See also Figure S4.
Melanoma Cell-Secreted ApoE Suppresses Invasion and Endothelial Recruitment
ApoE is a secreted factor. As such, we wondered whether melanoma cell-secreted ApoE could suppress invasion and endothelial recruitment. Consistent with this, poorly metastatic parental cells displayed higher levels of secreted ApoE than highly metastatic LM2 cells (Figure 5A). Secreted ApoE levels were also significantly suppressed by endogenous and exogenous miR-199a and miR-1908 (Figures 5B and S5A). Next, inhibiting extracellular ApoE through the use of a neutralizing antibody (1D7) that recognizes its receptor-binding domain enhanced both cell invasion (Figure 5C) and endothelial recruitment (Figure 5D) by parental MeWo cells. Conversely, addition of recombinant human ApoE protein significantly suppressed invasion and endothelial recruitment by metastatic LM2 cells (Figure 5E). Importantly, antibody-mediated neutralization of extracellular ApoE significantly abrogated the suppressed invasion and endothelial recruitment phenotypes seen with inhibition of each miRNA (Figures S5B–C)— consistent with secreted ApoE being downstream of miR-199a and miR-1908. Notably, recombinant ApoE addition did not affect melanoma cell or endothelial cell proliferation (Figures S5D–E) or survival in serum starvation conditions (Figures S5F–G), indicating that suppression of invasion or endothelial recruitment by ApoE is not secondary to decreased proliferation or impaired survival. Our findings reveal melanoma cell-secreted ApoE as a necessary and sufficient suppressor of miRNA-dependent invasion and endothelial recruitment phenotypes in melanoma.
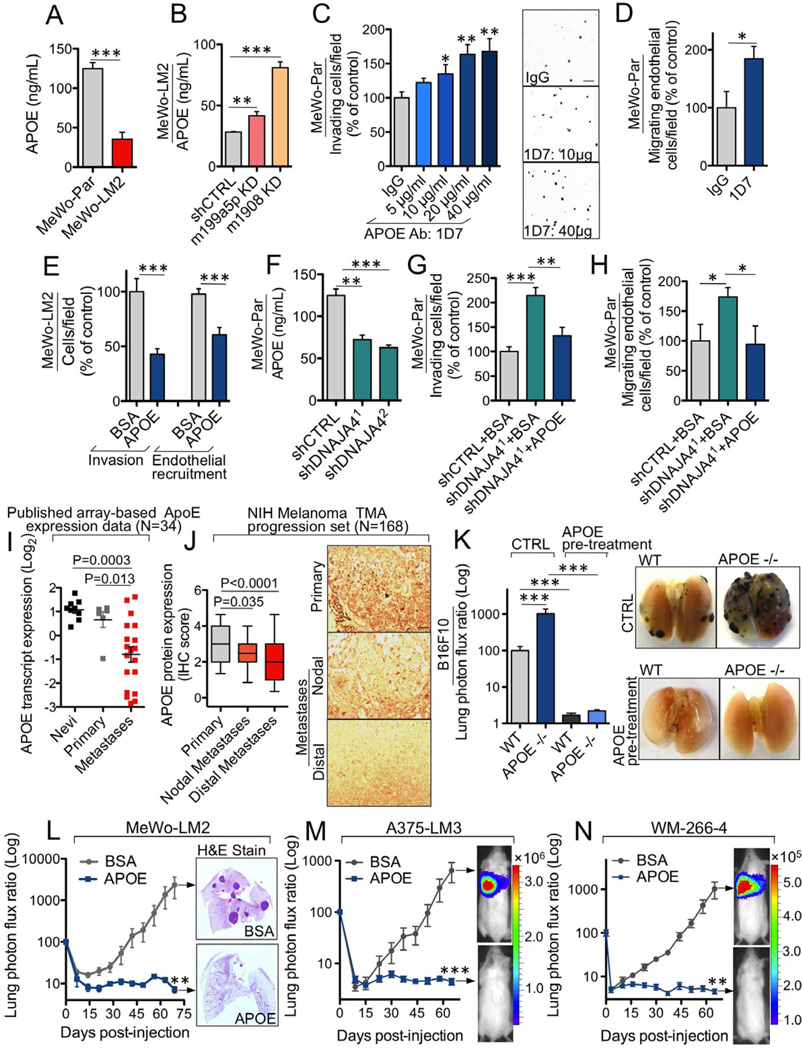
(A–B) Extracellular ApoE levels, quantified by ELISA, in conditioned media from MeWo parental and LM2 cells (A) and in MeWo-LM2 cells silenced for miR-199a-5p, miR-1908, or a control sequence (B). n=3. (C) Matrigel invasion by MeWo cells in response to the ApoE-neutralizing antibody 1D7 (5, 10, 20, or 40 µg/mL) or IgG control (40 µg/mL). n=4–6. (D) Endothelial recruitment by MeWo cells in the presence of 1D7 or control IgG antibody at 40 µg/mL. n=4. (E) The invasion and endothelial recruitment phenotypes were assessed in MeWo-LM2 cells in response to bovine serum albumin (BSA) or recombinant ApoE3 added to the cell media at 100 µM. n=7–10. (F) Extracellular ApoE levels, quantified by ELISA, in conditioned media from MeWo cells expressing shRNAs targeting DNAJA4 or an shCTRL. n=3. (G–H) MeWo cells with shRNA-induced silencing of DNAJA4 were analyzed for the matrigel invasion (G) and endothelial recruitment (H) phenotypes in the presence of either BSA or recombinant ApoE3 at 100 µM. n=4. (I) Array-based ApoE transcript expression levels in nevi (n=9), primary melanomas (n=6), and distal melanoma metastases (n=19). (J) ApoE protein expression determined by blinded immunohistochemical analysis in a tissue microarray (TMA) melanoma progression set (NIH) comprised of primary melanoma lesions (n=66), nodal (n=36), and distal (n=66) melanoma metastases. (K) Bioluminescence quantification and representative ex vivo lung images corresponding to lung metastasis 19 days post-injection of 5×104 B16F10 mouse melanoma cells pre-treated with recombinant ApoE3 (100 µg/mL) or control for 24 hours and intravenously injected into ApoE genetically null or wild type mice. n=12–22 (control) and n=5–7 (ApoE pre-treatment). (L–N) Bioluminescence imaging plots of lung colonization by 4×104 MeWo-LM2 cells (L), 2×105 A375-LM3 cells (M), and 1.5×105 WM-266-4 cells (N) pre-treated with ApoE3 or BSA at 100 µg/mL for 24 hours prior to injection. n=5–8. All data are represented as mean ± SEM. Scale bar, 100 µm. See also Figure S5.
We next wished to investigate the mechanism by which DNAJA4, a poorly characterized heat-shock protein, mediates endothelial recruitment and invasion. Given the phenotypic commonalities displayed by ApoE and DNAJA4, we hypothesized that DNAJA4 may play a regulatory role and enhance ApoE levels. Indeed, knock-down of DNAJA4 reduced both ApoE transcript levels (Figure S5H) as well as secreted ApoE levels (Figure 5F), while DNAJA4 over-expression substantially elevated ApoE transcript expression (Figure S5I). Consistent with DNAJA4 acting upstream of ApoE, addition of recombinant ApoE abrogated the enhanced cell invasion and endothelial recruitment phenotypes resulting from DNAJA4 knock-down (Figures 5G–H), while antibody-mediated neutralization of ApoE significantly occluded the suppression of invasion and endothelial recruitment resulting from DNAJA4 over-expression (Figures S5J–K). These findings reveal DNAJA4 to suppress melanoma invasion and endothelial recruitment through positive regulation of ApoE expression, leading to its increased extracellular levels.
The regulatory convergence of three metastasis-promoting miRNAs and the DNAJA4 gene onto ApoE motivated us to determine whether ApoE expression correlates with human melanoma progression. To this end, we analyzed published array-based expression data for ApoE (Haqq et al., 2005) in nevi, primary, and metastatic lesions. Consistent with a metastasis-suppressive role, ApoE transcript levels were significantly lower in distal organ metastases relative to primary melanoma and nevi lesions (Figure 5I). We also examined ApoE protein expression in a large (N = 168) human tissue-microarray melanoma progression panel. Blinded immunohistochemical analysis revealed that ApoE protein levels were significantly lower in lymph node metastases (P = 0.035) and even further downregulated in distal melanoma metastases (P < 0.0001) relative to primary melanomas (Figure 5J).
Genetic inactivation of ApoE promotes metastasis, while ApoE pre-treatment blocks metastatic initiation
Given the robust suppression of metastatic phenotypes exerted by ApoE and its clinical association with melanoma progression, we next investigated the impact of genetic deletion of systemic ApoE on melanoma progression in an immunocompetent mouse model of melanoma metastasis. Consistent with a major suppressive role for extracellular ApoE in metastasis, B16F10 mouse melanoma cells injected into the circulation exhibited a 10-fold increase in metastatic colonization in ApoE genetically null mice compared to their wild type littermates. Importantly, the metastatic capacity of B16F10 cells was dramatically abrogated by ApoE cell pre-treatment in both wild type (60-fold inhibition) and ApoE-null (460-fold inhibition) genetic contexts (Figure 5K). These findings establish both systemic and cancer-secreted ApoE as a robust suppressor of metastasis by human and mouse melanoma cells.
We next wondered whether enhanced ApoE signaling in melanoma cells could have therapeutic efficacy in preventing melanoma metastasis. We thus pre-incubated melanoma cells with recombinant ApoE or BSA for 24 hours prior to injection into mice. Notably, ApoE pre-treatment robustly suppressed metastatic colonization by the highly aggressive MeWo-LM2 cells by over 300-fold (Figure 5L). Remarkably, ApoE pre-incubation abrogated the metastatic capacity of four additional human melanoma cell lines: A375-LM3 (145-fold, Figure 5M), WM-266-4 (80-fold, Figure 5N), HT-144 (119-fold, Figure S5L), and A2058 (85-fold, Figure S5M).
Extracellular ApoE Divergently Targets Melanoma Cell LRP1 and Endothelial Cell LRP8 Receptors
We next investigated the molecular mechanisms by which ApoE suppresses metastasis. In order to identify the ApoE receptor(s) that mediate(s) invasion, we knocked down all four known ApoE receptors (VLDLR, LRP1, LRP8, and LDLR) in melanoma cells. Interestingly, knock-down of LRP1, but not the other ApoE receptors, abolished ApoE-induced suppression of invasion (Figures 6A and S6A), while LRP1 knock-down in the setting of miRNA silencing rescued the suppressed invasion phenotype resulting from miRNA inhibition (Figures 6B and S6B) and significantly enhanced in vivo metastatic colonization by MeWo-LM2 cells depleted for miR-1908 (Figures 6C and S6C). These findings reveal LRP1 to be a downstream mediator of miRNA/ApoE-dependent melanoma invasion and metastatic colonization phenotypes.
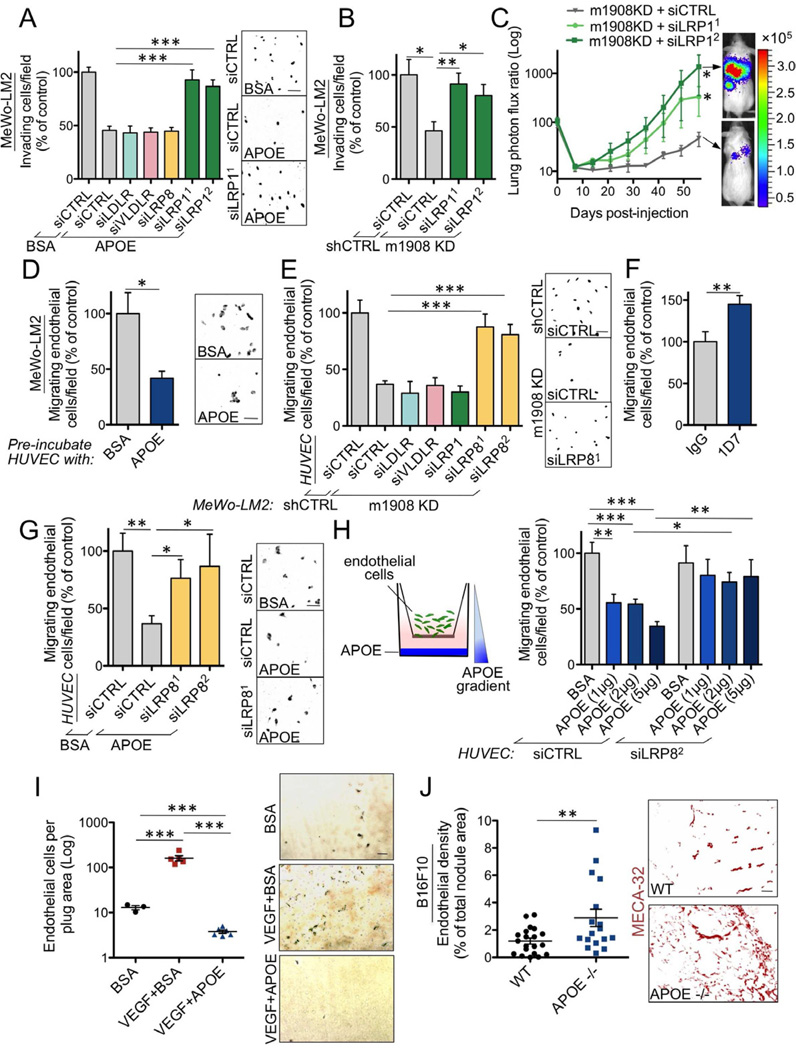
(A) Trans-well invasion by MeWo-LM2 cells transfected with short interfering RNAs (siRNAs) targeting LDLR, VLDLR, LRP8, LRP1, or a control siRNA (siCTRL) in the presence of either BSA (100 µM) or recombinant ApoE3 (100 µM). n=4–7. (B) MeWo-LM2 cells with miR-1908 KD or a control KD were transfected with siRNAs targeting LRP1 or siCTRL and subjected to the matrigel invasion assay. n=4. (C) Lung colonization by 1×105 MeWo-LM2 cells transfected with siRNAs targeting LRP1 or siCTRL in the setting of miR-1908 KD. n=5. (D) Trans-well recruitment of endothelial cells pre-incubated with BSA or recombinant ApoE3 at 100 µM for 24 hours by MeWo-LM2 cells. n=3–4. (E) Recruitment of HUVECs transfected with siRNAs targeting LDLR, VLDLR, LRP1, LRP8, or siCTRL by MeWo-LM2 cells with miR-1908 KD or control KD. n=4–12. (F) Trans-well migration by HUVECs in the presence of IgG or 1D7 antibody at 40 µg/mL. n=6–8. (G) Trans-well migration by endothelial cells transfected with siRNAs targeting LRP8 or siCTRL in the presence of BSA or recombinant ApoE3 at 100 µM. n=6–7. (H) Trans-well migration along an ApoE matrigel gradient by endothelial cells transfected with siRNAs targeting LRP8 or siCTRL. n=6–8. (I) Endothelial recruitment into subcutaneously implanted matrigel plugs containing BSA (10 µg/mL), VEGF (400 ng/mL) + BSA (10 µg/mL), or VEGF (400 ng/mL) + ApoE3 (10 µg/mL). The number of MECA-32 positive endothelial cells was quantified after three days by immunohistochemical detection. n=3–6. (J) Endothelial cell content, determined by staining for MECA-32, within lung metastatic nodules formed 19 days following injection of 5×104 B16F10 cells into wild type or ApoE genetically null mice. n=17–20. All data are represented as mean ± SEM. Scale bar, 100 µm. See also Figure S6.
While ApoE’s invasion phenotype reflects its cell-autonomous effect on melanoma cells, the endothelial recruitment phenotype suggests a non-cell-autonomous role of cancer-expressed ApoE on endothelial cells. Consistent with this, pre-treatment of endothelial cells with ApoE significantly reduced their ability to migrate towards highly metastatic cancer cells (Figure 6D). We next wished to identify the ApoE receptor(s) on endothelial cells that mediate(s) the endothelial recruitment phenotype. Interestingly, unlike for cancer cell invasion, knock-down of endothelial LRP8 receptors selectively and significantly abrogated the suppressed endothelial recruitment phenotype induced by miRNA silencing (Figures 6E and S6D–F). These findings are consistent with the LRP8 receptor being the endothelial mediator of miRNA/ApoE-dependent effects on endothelial recruitment. We next investigated whether ApoE might also regulate general endothelial migration in a cancer cell-free system. Indeed, antibody neutralization of ApoE, which is present in endothelial cell media, significantly enhanced endothelial migration (Figure 6F), while recombinant ApoE was sufficient to inhibit endothelial migration in a trans-well assay (Figure 6G) and a gradient-based migration assay (Figure 6H) in an endothelial cell LRP8 receptor-dependent manner. Importantly, ApoE dramatically suppressed by more than 40-fold VEGF-induced in vivo endothelial recruitment into subcutaneous matrigel plugs (Figure 6I). Consistent with the requirement and sufficiency of ApoE for suppressing endothelial migration, genetically null ApoE mice displayed higher blood vessel densities within their lung metastatic nodules formed by B16F10 mouse melanoma cells compared to their wild type littermates (Figure 6J), implicating systemic ApoE as a suppressor of metastatic angiogenesis.
miR-199a-3p, miR-199a-5p, and miR-1908 as Robust Prognostic and Therapeutic Targets in Melanoma Metastasis
To examine whether the metastasis promoter miRNAs we describe herein could serve as clinical predictors of metastatic outcomes, we quantified their expression levels in a blinded manner by qRT-PCR in a cohort of human melanoma samples obtained from patients at MSKCC. Patients whose primary melanoma lesions expressed higher (greater than the median for the population) levels of miR-199a-3p, miR-199a-5p, or miR-1908 exhibited significantly shorter metastasis-free survival times than patients whose primary melanomas expressed lower levels of each of these miRNAs (Figures 7A–C, P = 0.0032 for miR-199a-3p, P = 0.0034 for miR-199a-5p, and P = 0.027 for miR-1908). Importantly, the aggregate expression level of the three miRNAs displayed the strongest prognostic capacity in stratifying patients at high risk from those at very low risk for metastatic relapse (Figure 7D, P < 0.001). These clinical findings are indicative of functional cooperativity between the three miRNAs in the regulation of cancer progression and suggest utility for these molecules as clinical prognostic biomarkers of melanoma metastasis. Our identification of miR-199a in melanoma is consistent with a previous clinical association study that revealed increased miR-199a levels to correlate with uveal melanoma progression as well (Worley et al., 2008).
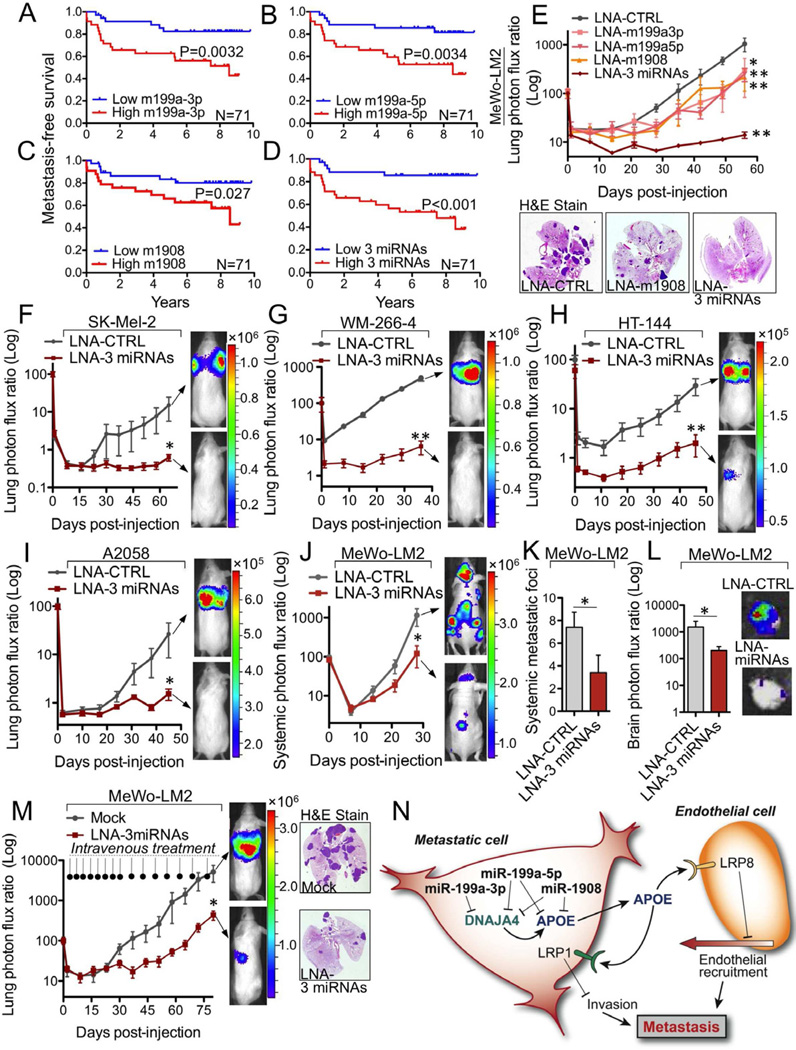
(A–D) Kaplan-Meier curves for the MSKCC cohort (N=71) representing metastasis-free survival of patients as a function of their primary melanoma lesion’s miR-199a-3p (A), miR-199a-5p (B), or miR-1908 (C) expression levels and the aggregate three miRNA expression (sum of the expression values of the individual miRNAs) (D). Patients whose primary tumors’ miRNA expression levels were greater or lower than the median for the population were classified as miRNA expression positive (red) or negative (blue), respectively. (E) Lung metastasis by 1×105 MeWo-LM2 cells transfected with LNAs individually targeting miR-1908, miR-199a-3p, or miR-199a-5p, or a combination of LNAs targeting all three miRNAs (LNA-3 miRNAs), or a control LNA (LNA-CTRL) 48 hours prior to injection. n=5–6. (F–I) Melanoma cells were pre-treated with LNA-CTRL or LNA-3 miRNAs. After 48 hours, 5×105 SK-Mel-2 (F), WM-266-4 (G), HT-144 cells (H), or A2058 (I) cells were intravenously injected into mice, and lung colonization was monitored by bioluminescence imaging. n=5–6. (J) Systemic metastasis by 1×105 MeWo-LM2 cells transfected with LNA-CTRL or LNA-3 miRNAs 48 hours prior to intracardiac injection into mice. n=5. (K–L) Number of systemic metastatic foci (K) and bioluminescence signal quantification of brain metastasis (L) arising from LNA-CTRL and LNA-3 miRNAs treated MeWo-LM2 cells at day 28 post-intracardiac injection. n=5. (M) Following tail-vein injection of 4×104 MeWo-LM2 cells, the next day mice were intravenously treated with a cocktail of in vivo-optimized LNAs targeting miR-1908, miR-199a-3p, and miR-199a-5p (12.5 mg/kg total dose) or a mock PBS control treatment as indicated. Lung colonization was quantified by bioluminescence imaging, and representative H&E-stained lungs extracted at day 80 are shown. n=5–6. (N) Emerging model of miRNA-dependent regulation of metastatic invasion, endothelial recruitment, and colonization in melanoma through cooperative targeting of ApoE-mediated LRP1/LRP8 receptor binding. See also Figure S7.
In light of the current lack of effective treatment options for the prevention of melanoma metastasis after surgical resection of the primary lesion (Garbe et al., 2011) and the strong prognostic value of these regulatory miRNAs in melanoma metastasis, we decided to therapeutically target them using antisense locked nucleic acids (LNAs) (Elmén et al., 2008). Highly metastatic MeWo-LM2 cells pre-treated with LNA oligonucleotides antisense to each mature miRNA (miR-199a-3p, miR-199a-5p, or miR-1908) exhibited a roughly four-fold decrease in metastatic activity (Figure 7E). Given our clinical evidence for cooperativity among these miRNAs as well as their convergent targeting of ApoE and DNAJA4, we examined the impact of silencing all three miRNAs on metastatic progression. Co-transfection of LNAs targeting all three miRNAs suppressed metastatic colonization by over seventy-fold, revealing dramatic synergy and cooperativity between endogenous miR-199a-3p, miR-199a-5p, and miR-1908 (Figure 7E). Importantly, inhibition of these miRNAs with triple LNA pre-treatment did not decrease in vitro proliferation (Figure S7A), indicating that the metastasis suppression phenotype is not secondary to impaired proliferation. Remarkably, combinatorial LNA-mediated miRNA inhibition robustly inhibited lung colonization by the independent SK-Mel-2 (24-fold, Figure 7F), WM-266-4 (77-fold, Figure 7G), HT-144 (15-fold, Figure 7H), A2058 (17-fold, Figure 7I), and A375-LM3 (3-fold, Figure S7B) human melanoma cell lines, as well as micrometastasis formation by the SK-Mel-28 human melanoma cell line (Figures S7C–D). These findings indicate that combinatorial targeting of the three miRNAs exhibits robust therapeutic potential across a large variety of melanotic and amelanotic human melanoma lines of diverse BRAF and NRAS mutational statuses. Additionally, intracardiac injection of highly metastatic MeWo-LM2 cells pre-treated with a cocktail of LNAs targeting these miRNAs suppressed systemic metastasis to multiple distal organs (Figures 7J – K) such as the brain (Figure 7L) and bone (Figure S7E). We next examined the therapeutic efficacy of systemically administered in vivo-optimized LNAs in melanoma metastasis prevention. To this end, highly metastatic MeWo-LM2 cells were injected into mice. The following day, mice were intravenously treated with LNAs targeting miR-199a-3p, miR-199a-5p, and miR-1908 at a low dose (12.5 mg/kg total) on a bi-weekly basis for four weeks followed by weekly dosing for seven weeks. Notably, combinatorial LNA treatment reduced lung colonization by more than 10-fold (Figure 7M, P = 0.039) without causing weight loss (Figure S7F). Importantly, therapeutic LNA-mediated miRNA targeting was confirmed by qRT-PCR analysis of lung metastatic nodules (Figure S7G) and mouse cardiac and liver tissues (Figures S7H–I). Taken together, our findings reveal a novel miRNA-dependent regulatory network that converges onto ApoE signaling to control cell-autonomous and non-cell-autonomous features of melanoma metastatic initiation and progression (Figure 7N).
DISCUSSION
The complexity of cancer requires the application of systems-level and integrative approaches. Using a systematic global approach, we have uncovered a cooperative network of miRNAs that are i) upregulated in highly metastatic human melanoma cells, ii) required and sufficient for optimal metastatic colonization and angiogenesis in melanoma, iii) pathologic predictors of human melanoma metastatic relapse, and iv) potential therapeutic targets across a variety of human melanoma cell lines. Through a transcriptomic-based and biologically guided target identification approach, we have found miR-1908, miR-199a-3p, and miR-199a-5p to convergently target the heat-shock factor DNAJA4 and the metabolic gene ApoE. Our identification of ApoE as a gene negatively regulated by three metastasis promoter miRNAs, positively regulated by a metastasis suppressor gene (DNAJA4), and silenced in clinical metastasis samples highlights the significance of this gene as a key novel suppressor of melanoma metastatic progression. ApoE mediates its effects by targeting two distinct yet homologous receptors on two diverse cell types. ApoE acting on melanoma cell LRP1 receptors inhibits melanoma invasion, while its action on endothelial cell LRP8 receptors suppresses endothelial migration. Our findings also reveal LRP1 and LRP8 to be novel endogenous suppressors of pro-metastatic phenotypes in melanoma. Our results from loss-of-function, gain-of-function, epistasis, clinical correlation, and in vivo selection-based expression analyses give rise to a model wherein three miRNAs convergently target a metastasis suppressor network to limit ApoE secretion, thus suppressing ApoE engagement of melanoma LRP1 receptors and endothelial LRP8 receptors (Figure 7N). The cooperative suppression of these inhibitory pathways by these miRNAs provides melanoma cells with maximal invasive and endothelial recruitment capacity. While we have identified LRP1 as a mediator of the ApoE response on melanoma cells, it remains possible that additional ApoE receptors may mediate ApoE responses in melanoma cells from distinct patients.
We reveal combined molecular, genetic, and in vivo evidence for a required and sufficient role for melanoma cell-secreted and systemic ApoE in the suppression of melanoma metastatic progression. The ability of recombinant ApoE to inhibit these pro-metastatic phenotypes, as well as the enhanced melanoma invasion and endothelial recruitment phenotypes resulting from ApoE neutralization by an antibody that targets the ApoE protein suggest that the ApoE molecule itself, rather than ApoE-associated lipids in lipoprotein particles, is the key mediator of these phenotypes.
Our molecular and in vivo studies reveal a role for endogenous cancer-derived ApoE in the modulation of endothelial migration and cancer angiogenesis through engagement of the endothelial LRP8 receptor. This non-cell-autonomous endothelial recruitment phenotype mediated by ApoE suggests that ApoE may also modulate metastatic angiogenesis and tumor angiogenesis in other cancer types. ApoE is a polymorphic molecule with well-established roles in cardiovascular and neurodegenerative disorders. Its three major variants—ApoE2, ApoE3, and ApoE4—display varying prevalences in the human population, with ApoE3 being the most common variant. The three isoforms differ at residues 112 and 158 in the N-terminal region, which contains the receptor-binding domain (Hatters et al., 2006). Given that the MeWo and A375 cell lines analyzed in our study are homozygous for the ApoE3 allele and given the ability of recombinant ApoE3 to inhibit melanoma metastatic phenotypes, our findings are consistent with ApoE3 being sufficient and required for the suppression of melanoma metastatic progression.
Despite recent promising advances such as the BRAF V600E inhibitor Vemurafenib (Chapman et al., 2011; Sosman et al., 2012) and the immunomodulatory antibody ipilimumab (Hodi et al., 2010; Sharma et al., 2011) that have extended the median survival times for subsets of patients diagnosed with metastatic (stage IV) melanoma, there are currently no effective therapies for the prevention of melanoma metastasis in the adjuvant setting, with interferon therapy increasing overall survival rates at 5 years by a meager 3% based on meta-analyses—while phase III trial data demonstration of a significant survival benefit is still outstanding (Garbe et al., 2011). The substantial enhancement of melanoma metastatic progression in the context of genetic ablation of systemic ApoE, as well as the dramatic abrogation of metastatic capacity by ApoE pre-treatment, suggest that modulating ApoE levels may have significant therapeutic implications for melanoma—a disease that claims approximately 48,000 lives a year globally (Lucas et al., 2006). Therapeutic approaches aimed at pharmacological induction of endogenous ApoE levels could potentially reduce melanoma mortality rates by decreasing metastatic relapse incidence.
Our establishment of in vivo selection models of melanotic and amelanotic melanoma metastasis has allowed us to identify the cellular phenotypes displayed by highly metastatic melanoma cells. Our work reveals that, in addition to enhanced invasiveness, the capacity of melanoma cells to recruit endothelial cells in vitro and in vivo is significantly enhanced in highly metastatic melanoma cells. Additionally, we find that three major post-transcriptional regulators of metastasis strongly regulate endothelial recruitment. We further show that the downstream signaling pathway modulated by these miRNAs also regulates endothelial recruitment. These findings reveal endothelial recruitment to be a defining feature of metastatic melanoma cells. Metastatic breast cancer derivatives obtained from multiple patients were also recently reported to recruit endothelial cells more efficiently than their parental lines by silencing the metastasis suppressor miRNA miR-126, thereby suppressing metastatic endothelial recruitment (MER) (Png et al., 2011). These findings as a whole highlight enhanced MER capacity as a key attribute of metastatic cells from multiple epithelial cancer types and suggest a novel and significant role for cancer-endothelial interactions in metastatic initiation and progression that complements their established conventional roles in primary tumor growth and perfusion (Carmeliet and Jain, 2000).
The ability of miR-199a-3p, miR-199a-5p, and miR-1908 to individually predict metastasis-free survival in melanoma patients indicates the significance of each miRNA as a clinical predictor of melanoma cancer progression. Importantly, the robust capacity of the three miRNA aggregate signature to separate patients at high risk from those at very low risk for metastatic relapse reveals both the cooperativity of these miRNAs as well as their clinical potential as melanoma biomarkers (Sawyers, 2008) for identifying the subset of patients that might benefit from experimental therapies such as miRNA inhibition therapy. Therapeutic miRNA targeting has gained momentum through the use of in vivo LNAs that have been shown to antagonize miRNAs in mice and primates (Elmén et al., 2008) and are currently being tested in human clinical trials. The powerful prognostic capacity of the three miRNAs, the proof-of-principle demonstration of robust metastasis suppression by combinatorial LNA-mediated miRNA targeting across multiple independent human melanoma cell lines, as well as the metastasis inhibitory effect of therapeutically delivered in vivo-optimized LNAs targeting these miRNAs motivate future clinical studies aimed at determining the therapeutic potential of combinatorially targeting these pro-metastatic and pro-angiogenic miRNAs in patients at high risk for melanoma metastasis—an outcome currently lacking effective chemotherapeutic options.
EXPERIMENTAL PROCEDURES
Animal Studies
All mouse experiments were conducted in agreement with a protocol approved by the Institutional Animal Care and Use Committee (IACUC) at The Rockefeller University. 6–8-week old sex-matched NOD scid, NOD scid gamma, athymic nu/nu, and C57Bl6 mice (Jackson Labs) were used. In vivo selection and metastasis assays were performed as previously described (Pollack and Fidler, 1982; Tavazoie et al., 2008). For ApoE pre-incubation metastasis experiments, cells were pre-treated with recombinant human ApoE3 (BioVision) or BSA (Sigma-Aldrich) at 100 µg/mL for 24 hours prior to intravenous injection into mice. For LNA pre-treatment metastasis assays, cells were transfected with LNAs (Exiqon) at a final concentration of 50 nM and injected into mice 48 hours later. In the LNA therapy experiment, MeWo-LM2 cells were injected tail-vein into mice, which were intravenously administered in vivo-optimized LNAs (Exiqon) antisense to miR-199a-3p, miR-199a-5p, and miR-1908 at a total dose of 12.5 mg/kg delivered in 0.1 mL of PBS bi-weekly for 4 weeks and then once weekly for 7 weeks. See Extended Experimental Procedures.
Cell Culture
Various cell lines were obtained from ATCC and maintained in standard conditions. Cell line information, miRNA and gene knock-down/over-expression studies in cell lines, and in vitro functional assays are described in Extended Experimental Procedures.
Microarray Hybridization
In order to identify miRNAs deregulated across highly metastatic derivatives, small RNAs were enriched from total RNA derived from MeWo and A375 cell lines and profiled by LC Sciences. For identification of miRNA gene targets, total RNA from MeWo cell lines was labeled and hybridized onto Illumina HT-12 v3 Expression BeadChip arrays by The Rockefeller University genomics core facility. Microarray-based expression findings were validated by qRT-PCR using TaqMan miRNA expression assays (Applied Biosystems) or SYBR® Green-based detection of gene expression (Invitrogen). See Extended Experimental Procedures.
Analysis of miRNA Expression in Human Melanoma Skin Lesions
All human clinical samples were obtained, processed, and analyzed in accordance with IRB guidelines. Total RNA was extracted from paraffin-embedded tissue sections of primary melanoma skin lesions previously resected from patients at MSKCC, and specific miRNA expression levels were analyzed in a blinded manner using TaqMan miRNA Assays. See Extended Experimental Procedures.
ApoE ELISA
Conditioned cancer cell media was prepared by incubating 70%-confluent cells in 0.2% FBS serum starvation DMEM-based media for 24 hours. ApoE levels in conditioned media were quantified using the APOE ELISA kit (Innovative Research).
Histochemistry
For gross macroscopic metastatic nodule visualization, 5-µm-thick lung tissue sections were H&E stained. For metastatic endothelial content and perfusion analyses, lung sections were double-stained with antibodies against MECA-32 (Developmental Studies Hybridoma Bank, University of Iowa) and human vimentin (Vector Laboratories) or biotinylated dextran (Vector Laboratories) and vimentin, respectively. ApoE protein expression in the melanoma tissue microarray progression set (NIH) was detected by staining with D6E10 anti-ApoE antibody (Abcam). See Extended Experimental Procedures.
Data Analysis
The Kolmogorov-Smirnov test was used to determine significance of differences in metastatic nodule endothelial content and dextran perfusion cumulative distributions. The prognostic power of the miRNAs to predict metastatic outcomes was tested for significance using the Mantel-Cox log-rank test. The one-way Mann-Whitney t-test was used to determine significance of non-Gaussian bioluminescence measurements. For all other comparisons, the one-tailed student’s t-test was used. The following designations apply to all figures: *p<0.05, **p<0.01, ***p<0.001; while p≤0.05 were deemed statistically significant.
ACKNOWLEDGEMENTS
We thank the members of the Tavazoie Laboratory for thoughtful comments on previous versions of this manuscript and Masoud Tavazoie for insightful discussions regarding melanoma. ApoE null mice were generously provided by Jan Breslow. We thank Jordana Ray-Kirton for tremendous help with clinical sample procurement and Yves Marcel for kindly providing ApoE neutralization antibody. S.F.T. was supported by the Rita Allen and the Elizabeth and Vincent Mayer foundations and a DOD Era of Hope Scholar award. This work was also generously supported by the Melanoma Research Alliance and the Melanoma Research Foundation.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
ACCESSION NUMBERS
The data from the miRNA gain- and loss-of-function microarray experiments are deposited at GEO under the accession number GSE35668.
SUPPLEMENTAL INFORMATION
Supplemental information includes Extended Experimental Procedures and seven figures.
REFERENCES
- Bartel DP. MicroRNAs: target recognition and regulatory functions. Cell. 2009;136:215–233. [Europe PMC free article] [Abstract] [Google Scholar]
- Calin GA, Croce CM. MicroRNA signatures in human cancers. Nat. Rev. Cancer. 2006;6:857–866. [Abstract] [Google Scholar]
- Carmeliet P, Jain RK. Angiogenesis in cancer and other diseases. Nature. 2000;407:249–257. [Abstract] [Google Scholar]
- Chapman PB, Hauschild A, Robert C, Haanen JB, Ascierto P, Larkin J, Dummer R, Garbe C, Testori A, Maio M, et al. Improved survival with vemurafenib in melanoma with BRAF V600E mutation. N Engl J Med. 2011;364:2507–2516. [Europe PMC free article] [Abstract] [Google Scholar]
- Elmén J, Lindow M, Schütz S, Lawrence M, Petri A, Obad S, Lindholm M, Hedtjärn M, Hansen HF, Berger U, et al. LNA-mediated microRNA silencing in non-human primates. Nature. 2008;452:896–899. [Abstract] [Google Scholar]
- Filipowicz W, Bhattacharyya SN, Sonenberg N. Mechanisms of post-transcriptional regulation by microRNAs: are the answers in sight? Nat. Rev. Genet. 2008;9:102–114. [Abstract] [Google Scholar]
- Garbe C, Eigentler TK, Keilholz U, Hauschild A, Kirkwood JM. Systematic review of medical treatment in melanoma: current status and future prospects. Oncologist. 2011;16:5–24. [Europe PMC free article] [Abstract] [Google Scholar]
- Gupta GP, Massagué J. Cancer metastasis: building a framework. Cell. 2006;127:679–695. [Abstract] [Google Scholar]
- Hanahan D, Weinberg RA. Hallmarks of cancer: the next generation. Cell. 2011;144:646–674. [Abstract] [Google Scholar]
- Haqq C, Nosrati M, Sudilovsky D, Crothers J, Khodabakhsh D, Pulliam BL, Federman S, Miller JR, Allen RE, Singer MI, et al. The gene expression signatures of melanoma progression. Proc. Natl. Acad. Sci. U S A. 2005;102:6092–6097. [Europe PMC free article] [Abstract] [Google Scholar]
- He L, Hannon GJ. MicroRNAs: small RNAs with a big role in gene regulation. Nat Rev Genet. 2004;5:522–531. [Abstract] [Google Scholar]
- Hatters DM, Peters-Libeu CA, Weisgraber KH. Apolipoprotein E structure: insights into function. Trends Biochem. Sci. 2006;31:445–454. [Abstract] [Google Scholar]
- Hodi FS, O’Day SJ, McDermott DF, Weber RW, Sosman JA, Haanen JB, Gonzalez R, Robert C, Schadendorf D, Hassel JC, et al. Improved survival with ipilimumab in patients with metastatic melanoma. N Engl J Med. 2010;363:711–723. [Europe PMC free article] [Abstract] [Google Scholar]
- Huang Q, Gumireddy K, Schrier M, le Sage C, Nagel R, Nair S, Egan DA, Li A, Huang G, Klein-Szanto AJ, et al. The microRNAs miR-373 and miR-520c promote tumour invasion and metastasis. Nat. Cell Biol. 2008;10:202–210. [Abstract] [Google Scholar]
- Hurst DR, Edmonds MD, Welch DR. Metastamir: the field of metastasis-regulatory microRNA is spreading. Cancer Res. 2009;69:7495–7498. [Europe PMC free article] [Abstract] [Google Scholar]
- Kang Y, Siegel PM, Shu W, Drobnjak M, Kakonen SM, Cordón-Cardo C, Guise TA, Massagué J. A multigenic program mediating breast cancer metastasis to bone. Cancer Cell. 2003;3:537–549. [Abstract] [Google Scholar]
- Lucas R, McMichael T, Wayne S, Armstrong B World Health Organization. Solar ultraviolet radiation: global burden of disease from solar ultraviolet radiation. Environmental Burden of Disease Series. 2006;13 [Google Scholar]
- Lujambio A, Lowe SW. The microcosmos of cancer. Nature. 2012;482:347–355. [Europe PMC free article] [Abstract] [Google Scholar]
- Ma L, Teruya-Feldstein J, Weinberg RA. Tumour invasion and metastasis initiated by microRNA-10b in breast cancer. Nature. 2007;449:682–688. [Abstract] [Google Scholar]
- Minn AJ, Gupta GP, Siegel PM, Bos PD, Shu W, Giri DD, Viale A, Olshen AB, Gerald WL, Massagué J. Genes that mediate breast cancer metastasis to lung. Nature. 2005;436:518–524. [Europe PMC free article] [Abstract] [Google Scholar]
- Png KJ, Halberg N, Yoshida M, Tavazoie SF. A microRNA regulon that mediates endothelial recruitment and metastasis by cancer cells. Nature. 2011;481:190–194. [Abstract] [Google Scholar]
- Poliseno L, Salmena L, Zhang J, Carver B, Haveman WJ, Pandolfi PP. A coding-independent function of gene and pseudogene mRNAs regulates tumour biology. Nature. 2010;465:1033–1038. [Europe PMC free article] [Abstract] [Google Scholar]
- Pollack VA, Fidler IJ. Use of young nude mice for selection of subpopulations of cells with increased metastatic potential from nonsyngeneic neoplasms. J. Natl. Cancer Inst. 1982;69:137–141. [Abstract] [Google Scholar]
- Sawyers CL. The cancer biomarker problem. Nature. 2008;452:548–552. [Abstract] [Google Scholar]
- Sharma P, Wagner K, Wolchok JD, Allison JP. Novel cancer immunotherapy agents with survival benefit: recent successes and next steps. Nat Rev Cancer. 2011;11:805–812. [Europe PMC free article] [Abstract] [Google Scholar]
- Sosman JA, Kim KB, Schuchter L, Gonzalez R, Pavlick AC, Weber JS, McArthur GA, Hutson TE, Moschos SJ, Flaherty KT, et al. Survival in BRAF V600-mutant advanced melanoma treated with vemurafenib. N Engl J Med. 2012;366:707–714. [Europe PMC free article] [Abstract] [Google Scholar]
- Talmadge JE, Fidler IJ. AACR centennial series: the biology of cancer metastasis: historical perspective. Cancer Res. 2010;70:5649–5669. [Europe PMC free article] [Abstract] [Google Scholar]
- Tavazoie SF, Alarcón C, Oskarsson T, Padua D, Wang Q, Bos PD, Gerald WL, Massagué J. Endogenous human microRNAs that suppress breast cancer metastasis. Nature. 2008;451:147–152. [Europe PMC free article] [Abstract] [Google Scholar]
- Wang Y, Klijn JG, Zhang Y, Sieuwerts AM, Look MP, Yang F, Talantov D, Timmermans M, Meijer-van Gelder ME, Yu J, et al. Gene-expression profiles to predict distant metastasis of lymph-node-negative primary breast cancer. Lancet. 2005;365:671–679. [Abstract] [Google Scholar]
- Worley LA, Long MD, Onken MD, Harbour JW. Micro-RNAs associated with metastasis in uveal melanoma identified by multiplexed microarray profiling. Melanoma Res. 2008;18:184–190. [Abstract] [Google Scholar]
Full text links
Read article at publisher's site: https://doi.org/10.1016/j.cell.2012.10.028
Read article for free, from open access legal sources, via Unpaywall:
http://www.cell.com/article/S0092867412012895/pdf
Citations & impact
Impact metrics
Citations of article over time
Alternative metrics
Smart citations by scite.ai
Explore citation contexts and check if this article has been
supported or disputed.
https://scite.ai/reports/10.1016/j.cell.2012.10.028
Article citations
Epigenetics and Control of Tumor Angiogenesis in Melanoma: An Update with Therapeutic Implications.
Cancers (Basel), 16(16):2843, 14 Aug 2024
Cited by: 0 articles | PMID: 39199614 | PMCID: PMC11352434
Review Free full text in Europe PMC
Lactylated Apolipoprotein C-II Induces Immunotherapy Resistance by Promoting Extracellular Lipolysis.
Adv Sci (Weinh), 11(38):e2406333, 09 Jul 2024
Cited by: 2 articles | PMID: 38981044 | PMCID: PMC11481198
Comprehensive analysis of bulk and single-cell transcriptomic data reveals a novel signature associated with endoplasmic reticulum stress, lipid metabolism, and liver metastasis in pancreatic cancer.
J Transl Med, 22(1):393, 29 Apr 2024
Cited by: 0 articles | PMID: 38685045 | PMCID: PMC11057100
The Analysis of ceRNA Networks and Tumor Microenvironment in Endometrial Cancer.
J Cancer, 15(8):2147-2159, 24 Feb 2024
Cited by: 0 articles | PMID: 38495486 | PMCID: PMC10937290
ApoE maintains neuronal integrity via microRNA and H3K27me3-mediated repression.
iScience, 27(3):109231, 15 Feb 2024
Cited by: 0 articles | PMID: 38439966 | PMCID: PMC10909902
Go to all (247) article citations
Data
Data behind the article
This data has been text mined from the article, or deposited into data resources.
BioStudies: supplemental material and supporting data
GEO - Gene Expression Omnibus
- (1 citation) GEO - GSE35668
Similar Articles
To arrive at the top five similar articles we use a word-weighted algorithm to compare words from the Title and Abstract of each citation.
A microRNA regulon that mediates endothelial recruitment and metastasis by cancer cells.
Nature, 481(7380):190-194, 14 Dec 2011
Cited by: 346 articles | PMID: 22170610
MicroRNA-199a-3p and MicroRNA-199a-5p Take Part to a Redundant Network of Regulation of the NOS (NO Synthase)/NO Pathway in the Endothelium.
Arterioscler Thromb Vasc Biol, 38(10):2345-2357, 01 Oct 2018
Cited by: 31 articles | PMID: 29976767
miR-98 suppresses melanoma metastasis through a negative feedback loop with its target gene IL-6.
Exp Mol Med, 46:e116, 03 Oct 2014
Cited by: 41 articles | PMID: 25277211 | PMCID: PMC4221693
Anti-Angiogenic Effects of Phytochemicals on miRNA Regulating Breast Cancer Progression.
Biomolecules, 10(2):E191, 27 Jan 2020
Cited by: 26 articles | PMID: 32012744 | PMCID: PMC7072640
Review Free full text in Europe PMC
Funding
Funders who supported this work.
NCRR NIH HHS (2)
Grant ID: 5KL2 RR024142-04
Grant ID: KL2 RR024142





