Abstract
Free full text

Influenza neuraminidase inhibitors: antiviral action and mechanisms of resistance
Abstract
Please cite this paper as: McKimm‐Breschkin (2012) Influenza neuraminidase inhibitors: Antiviral action and mechanisms of resistance. Influenza and Other Respiratory Viruses 7(Suppl. 1), 25–36.
There are two major classes of antivirals available for the treatment and prevention of influenza, the M2 inhibitors and the neuraminidase inhibitors (NAIs). The M2 inhibitors are cheap, but they are only effective against influenza A viruses, and resistance arises rapidly. The current influenza A H3N2 and pandemic A(H1N1)pdm09 viruses are already resistant to the M2 inhibitors as are many H5N1 viruses. There are four NAIs licensed in some parts of the world, zanamivir, oseltamivir, peramivir, and a long‐acting NAI, laninamivir. This review focuses on resistance to the NAIs. Because of differences in their chemistry and subtle differences in NA structures, resistance can be both NAI‐ and subtype specific. This results in different drug resistance profiles, for example, the H274Y mutation confers resistance to oseltamivir and peramivir, but not to zanamivir, and only in N1 NAs. Mutations at E119, D198, I222, R292, and N294 can also reduce NAI sensitivity. In the winter of 2007–2008, an oseltamivir‐resistant seasonal influenza A(H1N1) strain with an H274Y mutation emerged in the northern hemisphere and spread rapidly around the world. In contrast to earlier evidence of such resistant viruses being unfit, this mutant virus remained fully transmissible and pathogenic and became the major seasonal A(H1N1) virus globally within a year. This resistant A(H1N1) virus was displaced by the sensitive A(H1N1)pdm09 virus. Approximately 0·5–1·0% of community A(H1N1)pdm09 isolates are currently resistant to oseltamivir. It is now apparent that variation in non‐active site amino acids can affect the fitness of the enzyme and compensate for mutations that confer high‐level oseltamivir resistance resulting in minimal impact on enzyme function.
Introduction
Influenza virions have three surface proteins, the hemagglutinin (HA), neuraminidase (NA), and M2 protein. The HA binds to terminal sialic acids on cellular receptors, after which the virus is endocytosed. The low pH (5·5–6·0) of the endosome activates the M2 proton channel in the influenza A virus membrane to allow acid to enter the virus, prior to HA‐mediated fusion, triggering the release of the virus ribonucleoprotein (RNP). After replication, the NA of progeny virions cleaves sialic acids from the cell receptors and from the HA and NA which are also glycosylated, to release progeny virions from the cell surface and prevent self aggregation. There are two major classes of antivirals licensed for the treatment and prevention of influenza, the M2 inhibitors and the NA inhibitors (NAIs). By blocking the M2 proton channel, the M2 inhibitors prevent release of the virus RNP for migration to the nucleus of the cell. The NA inhibitors (NAIs) prevent release of newly formed virions from the cell surface.
The M2 inhibitors, amantadine and rimantadine, only act on influenza A viruses. Although influenza B viruses have a BM2 protein which is analogous to the M2 protein in influenza A, this is not sensitive to the M2 inhibitors. The M2 inhibitors have two potential binding sites on the M2 protein: a high‐affinity site in the ion channel pore and a second low‐affinity site on the lipid face of the pore. 1 The two most common mutations V27A and S31N are in the ion channel pore, confirming this as the pharmacologically relevant site.
However, although the M2 inhibitors are cheap and have been around for almost 50 years,
2
their use for the treatment of influenza has been limited, in part because resistant viruses emerge rapidly in treated patients, and in a single passage in tissue culture.
3
The pandemic A(H1N1)pdm09 virus was already resistant
4
, and the majority of seasonal A(H3N2) viruses have been resistant since the mid‐2000s, with an increase in oseltamivir‐resistant seasonal H1N1 viruses also observed.
5
,
6
Many of the H5N1 strains circulating in Southeast Asia, especially in Vietnam and Thailand, are also resistant to M2 inhibitors.
7
,
8
Due to resistance, the usefulness of these drugs is currently limited. This review therefore focuses on the most recently developed NAIs.
years,
2
their use for the treatment of influenza has been limited, in part because resistant viruses emerge rapidly in treated patients, and in a single passage in tissue culture.
3
The pandemic A(H1N1)pdm09 virus was already resistant
4
, and the majority of seasonal A(H3N2) viruses have been resistant since the mid‐2000s, with an increase in oseltamivir‐resistant seasonal H1N1 viruses also observed.
5
,
6
Many of the H5N1 strains circulating in Southeast Asia, especially in Vietnam and Thailand, are also resistant to M2 inhibitors.
7
,
8
Due to resistance, the usefulness of these drugs is currently limited. This review therefore focuses on the most recently developed NAIs.
Licensed neuraminidase inhibitors
There are two NAIs licensed globally for the treatment and prevention of influenza. Relenza (zanamivir) was the first in this class
9
followed by Tamiflu (oseltamivir).
10
Zanamivir was developed based on two key findings. Firstly, the transition‐state analog 2,3‐dehydro‐2‐deoxy‐N‐acetylneuraminic acid (DANA) was known to be a weak inhibitor of the NA. Secondly, the structure of the sialic acid substrate in complex with the enzyme active site revealed an empty negatively charged pocket in the region of the C4 on the sugar ring. This suggested that substitution of the C4‐OH with a larger basic residue might lead to higher affinity binding.
11
A single substitution of the C4‐OH with a 4‐guanidino group enhanced binding more than 10 000‐fold over DANA (Figure 1). Zanamivir is administered by oral inhalation as it is not absorbed. Oseltamivir was subsequently designed based on knowledge from zanamivir. While based on DANA, it has a cyclohexene ring with two substitutions compared with DANA. It has a C4 amino group and a bulky hydrophobic pentyl ether side chain in place of the glycerol side chain. It is administered as the prodrug oseltamivir phosphate and converted by hepatic esterases to the active compound oseltamivir carboxylate.
000‐fold over DANA (Figure 1). Zanamivir is administered by oral inhalation as it is not absorbed. Oseltamivir was subsequently designed based on knowledge from zanamivir. While based on DANA, it has a cyclohexene ring with two substitutions compared with DANA. It has a C4 amino group and a bulky hydrophobic pentyl ether side chain in place of the glycerol side chain. It is administered as the prodrug oseltamivir phosphate and converted by hepatic esterases to the active compound oseltamivir carboxylate.
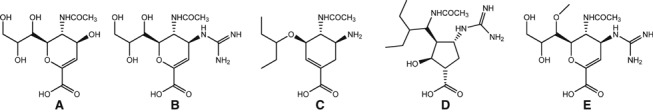
 Chemical structures of the neuraminidase inhibitors (A) DANA, (B) zanamivir, (C) oseltamivir carboxylate, (D) peramivir, and (E) laninamivir. Structures are oriented to demonstrate the differences relative to DANA – the C4‐guanidinium group on zanamivir, peramivir, and laninamivir, the C4‐amino group on oseltamivir, and the pentyl side chains on oseltamivir and peramivir.
Chemical structures of the neuraminidase inhibitors (A) DANA, (B) zanamivir, (C) oseltamivir carboxylate, (D) peramivir, and (E) laninamivir. Structures are oriented to demonstrate the differences relative to DANA – the C4‐guanidinium group on zanamivir, peramivir, and laninamivir, the C4‐amino group on oseltamivir, and the pentyl side chains on oseltamivir and peramivir.
Peramivir has subsequently been developed
12
and is now licensed in Japan and for emergency use in some other countries while undergoing further clinical trials. It is based on DANA, but has a cyclopentane ring and features of both zanamivir and oseltamivir, the C4‐guanidino substitution and a hydrophobic side chain, respectively. It is only effective if administered intravenously. A fourth compound, laninamivir (Inavir), based on zanamivir with a 7‐OCH3 substitution, is a long‐acting NAI
13
and is administered by oral inhalation as a single 40 mg dose of the laninamivir octanoate prodrug. Laninamivir has been licensed in Japan and is undergoing clinical trials in other countries.
mg dose of the laninamivir octanoate prodrug. Laninamivir has been licensed in Japan and is undergoing clinical trials in other countries.
The NAIs prevent release and spread of progeny virions by blocking NA function. The sensitivity of the NA enzyme to the NAIs is evaluated in an in vitro enzyme inhibition assay, using either a fluorescent
14
or chemiluminescent substrate.
15
The IC50 is defined as the concentration inhibiting 50% of the enzyme activity compared with the uninhibited control. Decreased sensitivity due to a mutation in the NA is identified by an elevated IC50.
16
,
17
Sensitivities vary in different laboratories due to subtle differences in assay methodology, but in general influenza A(H3N2) viruses are slightly more sensitive to oseltamivir than N1 subtype viruses. Conversely, N1 subtype viruses are slightly more sensitive to zanamivir than to N2 subtype viruses. IC50s are generally <5 nm for both drugs for N1 and N2 subtypes. Influenza B viruses have slightly higher IC50s for zanamivir, but they are still <10
nm for both drugs for N1 and N2 subtypes. Influenza B viruses have slightly higher IC50s for zanamivir, but they are still <10 nm.
16
,
18
In contrast, influenza B viruses have 10–20‐fold higher IC50s for oseltamivir compared with influenza A viruses.
16
,
17
,
18
nm.
16
,
18
In contrast, influenza B viruses have 10–20‐fold higher IC50s for oseltamivir compared with influenza A viruses.
16
,
17
,
18
Oseltamivir is taken orally twice daily, with a dose of 75 mg for adults. The levels of oseltamivir in plasma are estimated to be in the range from 400 to 1200
mg for adults. The levels of oseltamivir in plasma are estimated to be in the range from 400 to 1200 nm
19
,
20
and in saliva to be <5% of plasma levels.
21
Thus, levels in the upper respiratory tract may be significantly lower than 100
nm
19
,
20
and in saliva to be <5% of plasma levels.
21
Thus, levels in the upper respiratory tract may be significantly lower than 100 nm. This may only be 20–50 times the IC50s for influenza A strains and 2–5‐fold higher than the IC50s for wild‐type influenza B strains.
nm. This may only be 20–50 times the IC50s for influenza A strains and 2–5‐fold higher than the IC50s for wild‐type influenza B strains.
Zanamivir dosing is 10 mg inhaled twice daily, delivering high levels to the upper respiratory tract, estimated to be up to 10
mg inhaled twice daily, delivering high levels to the upper respiratory tract, estimated to be up to 10 000
000 nm.
22
,
23
This would be up to 5000‐fold higher than the average IC50s for influenza A viruses.
nm.
22
,
23
This would be up to 5000‐fold higher than the average IC50s for influenza A viruses.
Emergence of resistance
In early studies, resistance to oseltamivir emerged both in challenge studies and in naturally acquired infections, with resistant virus isolated from 1 to 4% of oseltamivir‐treated adult patients. 24 , 25 , 26 Subsequently, resistant viruses have been isolated from patients after treatment or prophylaxis with oseltamivir, or with no apparent history of drug exposure. Due to differences in the chemical structures of the inhibitors, many of the mutations do not confer reduced sensitivity to all the NAIs. Additionally, despite high conservation of residues in the active site, there are mutations which confer resistance in only one subtype, for example, H274Y (H275Y in N1 numbering) confers oseltamivir resistance only in N1, E119V, and R292K confer high‐level oseltamivir resistance only in N2. (Note: There are subtle differences in the lengths of the sequences of different NAs; however, traditionally NA numbering is based on alignment to the N2 NA, which is used throughout unless otherwise specified.) Although a resistant influenza B virus with an R152K mutation conferring reduced sensitivity to all NAIs was isolated from an immunocompromised child on zanamivir therapy, 27 no resistant virus has yet been isolated from an immunocompetent patient treated with zanamivir.
In early studies, resistant viruses could be isolated from 4 to 8% of oseltamivir‐treated pediatric patients, possibly due to prolonged virus shedding in children.
26
,
28
However, three post‐release studies of oseltamivir‐treated children have demonstrated much higher frequencies of resistant viruses. The first two studies were conducted in Japan, where weight‐based dosing of 2 mg/kg of oseltamivir is used for children. In one study, in which the viruses were primarily H1N1, oseltamivir‐resistant H274Y viruses were isolated from 7 of 43 children (16%).
29
The H274Y mutation decreases sensitivity to both oseltamivir and peramivir, but not to zanamivir. In the second study, predominantly of H3N2 viruses, resistant viruses with E119V (2), R292K (6), or N294S (1) mutations were isolated from 9 of 50 children (18%).
30
The first two mutations have only been seen clinically in N2 subtype viruses, with E119V and N294S conferring reduced sensitivity specifically to oseltamivir. The N294S mutation has also been seen in the N1 subtype (N295S in N1 numbering) conferring a mild reduction in zanamivir sensitivity, but a greater reduction in oseltamivir sensitivity.
31
,
32
mg/kg of oseltamivir is used for children. In one study, in which the viruses were primarily H1N1, oseltamivir‐resistant H274Y viruses were isolated from 7 of 43 children (16%).
29
The H274Y mutation decreases sensitivity to both oseltamivir and peramivir, but not to zanamivir. In the second study, predominantly of H3N2 viruses, resistant viruses with E119V (2), R292K (6), or N294S (1) mutations were isolated from 9 of 50 children (18%).
30
The first two mutations have only been seen clinically in N2 subtype viruses, with E119V and N294S conferring reduced sensitivity specifically to oseltamivir. The N294S mutation has also been seen in the N1 subtype (N295S in N1 numbering) conferring a mild reduction in zanamivir sensitivity, but a greater reduction in oseltamivir sensitivity.
31
,
32
There were concerns that the weight‐based dosing used in Japan delivered suboptimal concentrations of oseltamivir, facilitating selection of resistant viruses. Hence, another trial was carried out in the United Kingdom, in which tiered weight‐based dosing of oseltamivir was used. 33 Despite small numbers of patients, significant resistance was seen. Three of 11 patients (27%) infected with H1N1 viruses shed H274Y‐resistant virus and one of 34 patients (3%) infected with H3N2 viruses shed an R292K‐resistant virus. None of 19 patients infected with influenza B shed a resistant virus.
Due to concerns over the high level of oseltamivir resistance seen in the Japanese pediatric studies, a 3‐year study was carried out in Japan to monitor for the emergence of resistant virus after zanamivir therapy. A total of 273 children were enrolled over three influenza seasons from 2006 to 2009.
34
All children enrolled were <15 years of age, and were influenza positive by a rapid diagnostic assay and culture positive by throat swab. Samples from pre‐ and post‐treatment were tested for resistant virus by RT‐PCR and in enzyme assays of cultured virus. Three viruses from two subjects infected with H1N1 viruses showed virus with reduced sensitivity prior to treatment. One virus with a N70S mutation in the NA showed a 46‐fold increase in IC50 for zanamivir. Two viruses from one subject had a Q136K NA mutation, which showed a 300‐fold increase in IC50 for zanamivir, but this mutation was only detected after culture and not in the primary sample. Hence, despite more patients than the oseltamivir pediatric trials, no emergence of resistance was seen in 273 zanamivir‐treated children.
years of age, and were influenza positive by a rapid diagnostic assay and culture positive by throat swab. Samples from pre‐ and post‐treatment were tested for resistant virus by RT‐PCR and in enzyme assays of cultured virus. Three viruses from two subjects infected with H1N1 viruses showed virus with reduced sensitivity prior to treatment. One virus with a N70S mutation in the NA showed a 46‐fold increase in IC50 for zanamivir. Two viruses from one subject had a Q136K NA mutation, which showed a 300‐fold increase in IC50 for zanamivir, but this mutation was only detected after culture and not in the primary sample. Hence, despite more patients than the oseltamivir pediatric trials, no emergence of resistance was seen in 273 zanamivir‐treated children.
Seasonal H1N1
Early results demonstrated that mutations in the NA which conferred reduced NAI sensitivity also impacted on the function of the NA such that resistant viruses were compromised in their fitness and unlikely to be transmitted.
35
,
36
However, in late 2007, several seasonal H1N1 viruses with an H274Y mutation were isolated in Norway. There was a minimal use of oseltamivir in Norway, and none of the patients had a history of drug exposure. Subsequent testing revealed 183 of 272 isolates (67%) bore this mutation. This virus was clearly fit, and transmissible and within weeks resistant viruses were detected in North America, Europe, and Asia.
37
,
38
,
39
The resistant viruses continued to spread to the southern hemisphere,
40
ultimately displacing the sensitive virus. More than 90% of H1N1 isolates were resistant by 2008–2009, with IC50s in the fluorescent assay generally in the 500–1000 nm range. It appears that permissive mutations had evolved that enabled the NA to tolerate the H274Y mutation, maintaining fitness of the enzyme.
41
,
42
,
43
,
44
The substitutions include R193G, R221Q, V233M, and D343N (R194G, R222Q, V234M, and D344N in N1 numbering). While H274Y is the primary mutation seen in N1 viruses, a seasonal H1N1 virus with an I222V mutation, conferring reduced susceptibility to oseltamivir, was also detected in surveillance of community isolates from untreated patients.
45
nm range. It appears that permissive mutations had evolved that enabled the NA to tolerate the H274Y mutation, maintaining fitness of the enzyme.
41
,
42
,
43
,
44
The substitutions include R193G, R221Q, V233M, and D343N (R194G, R222Q, V234M, and D344N in N1 numbering). While H274Y is the primary mutation seen in N1 viruses, a seasonal H1N1 virus with an I222V mutation, conferring reduced susceptibility to oseltamivir, was also detected in surveillance of community isolates from untreated patients.
45
Pandemic influenza A(H1N1)pdm09
The sudden emergence and spread of the swine‐derived influenza virus from Mexico led to the displacement of the oseltamivir‐resistant seasonal H1N1 virus by the new A(H1N1)pdm09 virus. However, given the awareness of oseltamivir resistance, much closer monitoring has been carried out, by both phenotypic testing by enzyme assay and sequencing. Viruses with the H274Y mutation have been detected in patients after treatment or prophylaxis with oseltamivir or peramivir
46
,
47
,
48
and also in untreated patients. Resistance has been detected as early as 48 hours post‐treatment.
46
hours post‐treatment.
46
One of the earliest reports of resistance was in summer campers in the USA undergoing oseltamivir prophylaxis. Resistant virus was isolated from the second patient several days after contact with the first. In addition to the H274Y mutation, both viruses had a common I222V (I223V in N1 numbering) mutation, suggesting possible human–human spread.
49
Another case was reported after prophylaxis of a family contact of an infected patient.
50
As the dose of oseltamivir for prophylaxis is only half that for therapy (75 mg once daily), administering subtherapeutic doses when virus replication has already begun could increase the selection of resistant A(H1N1)pdm09 virus.
mg once daily), administering subtherapeutic doses when virus replication has already begun could increase the selection of resistant A(H1N1)pdm09 virus.
There are numerous reports of the emergence of resistant viruses among immunocompromised patients undergoing oseltamivir treatment or prophylaxis, which is not unexpected due to the longer periods of therapy. 51 , 52 , 53 , 54 , 55 There are also reports of transmission of resistant viruses in hospitalized settings among immunocompromised patients. 56 , 57
Of more concern is the isolation and transmission of viruses in the community among patients with no history of drug use. A traveler from the USA, identified as feverish upon entry into Hong Kong, was one of the first patients from which resistant virus was isolated with no known exposure to oseltamivir. 58 The nasopharyngeal specimen contained a mixture of wild‐type (47%) and H274Y‐resistant (53%) viruses. After culture in MDCK cells, the sample contained 98% H274Y virus. Hence, the mutation did not compromise replication in vitro. H274Y‐resistant virus was also detected in 7 of 10 untreated students traveling on a train in Vietnam. 59 More recently, a community cluster of 29 patients infected with a H274Y virus has been identified in Australia, 28 of whom had no known drug exposure, 60 thus demonstrating the fitness and transmissibility of the H274Y A(H1N1)pdm09 virus. There are various reports of the assessment of fitness of resistant A(H1N1)pdm09 viruses in vitro and in animal models. Some report no compromise in fitness, 61 , 62 while others demonstrate reduced fitness. 63 , 64
In addition to the early report of an I222V (I223V in N1 numbering) mutation,
49
I222R mutations have been reported. I222R variants emerged in two immunocompromised patients, one treated sequentially with oseltamivir than zanamivir and a second treated with oseltamivir.
65
,
66
One virus had both I222R and H274Y mutations. There is also a recent report of isolation of an I222R variant from a third patient with no history of drug exposure.
67
The single I222R mutation conferred reduced sensitivity to oseltamivir, zanamivir, and peramivir with 45‐, 10‐, and 7‐fold reductions, respectively, in sensitivity in the NA enzyme inhibition assay.
66
The dual I222R/H274Y mutation further increased the IC50 by 10–90‐fold over that due to the H274Y mutation alone to approximately 10 000
000 nm for both oseltamivir and peramivir. While this dual mutation also reduced sensitivity to zanamivir by 10‐fold compared with the single H274Y mutation, the IC50 was still <10
nm for both oseltamivir and peramivir. While this dual mutation also reduced sensitivity to zanamivir by 10‐fold compared with the single H274Y mutation, the IC50 was still <10 nm.
nm.
The WHO maintained a Web site to record the numbers of resistant A(H1N1)pdm09 viruses reported to it.
68
By October 2011, after which there have been no further additions, there were only 605 cases of oseltamivir resistance reported globally. This is clearly an underrepresentation of the true number of cases of resistant virus because many countries reported an incidence of 0·5–1% of oseltamivir‐resistant isolates in their community surveillance.
69
,
70
,
71
This equates to 5000–10 000‐resistant viruses per 1‐million cases of infection. As it was estimated that there were around 60‐million cases in the USA,
72
there could be many thousands of cases of oseltamivir resistance in the USA alone. Although the WHO figures are misleading in terms of the total number of resistant isolates, the WHO site provided useful statistics on patient history and oseltamivir exposure. Of the total cases, 28% of the resistant isolates in 2009–2010 were from immunocompromised patients. Of the 72% immunocompetent patients, 63% had some exposure to oseltamivir, but 37% had no history of any exposure to drug or drug‐treated individuals. In 2010–2011, there was an increase in the frequency of resistance detection in the USA from 0·5 to 1% of isolates, but additionally 74% of patients had no exposure to oseltamivir versus only 11% in the 2009–2010 season, indicating greater transmission of oseltamivir‐resistant virus in the community.
71
000‐resistant viruses per 1‐million cases of infection. As it was estimated that there were around 60‐million cases in the USA,
72
there could be many thousands of cases of oseltamivir resistance in the USA alone. Although the WHO figures are misleading in terms of the total number of resistant isolates, the WHO site provided useful statistics on patient history and oseltamivir exposure. Of the total cases, 28% of the resistant isolates in 2009–2010 were from immunocompromised patients. Of the 72% immunocompetent patients, 63% had some exposure to oseltamivir, but 37% had no history of any exposure to drug or drug‐treated individuals. In 2010–2011, there was an increase in the frequency of resistance detection in the USA from 0·5 to 1% of isolates, but additionally 74% of patients had no exposure to oseltamivir versus only 11% in the 2009–2010 season, indicating greater transmission of oseltamivir‐resistant virus in the community.
71
H3N2 viruses
While oseltamivir‐resistant H3N2 viruses were isolated in the pediatric studies described above, the incidence of detection of resistant H3N2 viruses either after drug exposure or among community acquired infections remains low.
16
,
18
,
26
,
45
E119V and R292K have been the two most common mutations detected, with N294S also reported in the pediatric study. The E119V mutation confers resistance only to oseltamivir, with a 20 to several hundredfold increase in IC50 depending on whether tested in the chemiluminescent or fluorescent enzyme inhibition assay, respectively.
16
,
17
,
18
,
73
,
74
,
75
The R292K mutation confers a very high level of resistance to oseltamivir, usually in the μm range, intermediate resistance to peramivir, and a smaller reduction in sensitivity to zanamivir, usually with an IC50 <50
μm range, intermediate resistance to peramivir, and a smaller reduction in sensitivity to zanamivir, usually with an IC50 <50 nm.
17
,
74
,
76
,
77
,
78
The N294S also reduces oseltamivir sensitivity by several hundredfold.
30
Viruses with an I222V mutation have been isolated from untreated patients
45
and an oseltamivir‐treated immunocompromised child. The isolate in the latter case also had an E119V mutation, I222V further enhancing oseltamivir resistance while remaining sensitive to zanamivir.
79
The I222V mutation also appeared to partially restore the impaired viral fitness conferred by the E119V mutation.
80
A novel E119I mutation was detected in a virus from an oseltamivir‐treated immunocompromised child, which reduced susceptibility to oseltamivir by several hundredfold, more than the E119V mutation. It also reduced peramivir sensitivity by several hundredfold, but zanamivir sensitivity by only 5‐fold.
75
Isolation of a naturally occurring Q136K mutant from Myanmar, with a 30–60‐fold reduction in zanamivir sensitivity, has also been described.
81
nm.
17
,
74
,
76
,
77
,
78
The N294S also reduces oseltamivir sensitivity by several hundredfold.
30
Viruses with an I222V mutation have been isolated from untreated patients
45
and an oseltamivir‐treated immunocompromised child. The isolate in the latter case also had an E119V mutation, I222V further enhancing oseltamivir resistance while remaining sensitive to zanamivir.
79
The I222V mutation also appeared to partially restore the impaired viral fitness conferred by the E119V mutation.
80
A novel E119I mutation was detected in a virus from an oseltamivir‐treated immunocompromised child, which reduced susceptibility to oseltamivir by several hundredfold, more than the E119V mutation. It also reduced peramivir sensitivity by several hundredfold, but zanamivir sensitivity by only 5‐fold.
75
Isolation of a naturally occurring Q136K mutant from Myanmar, with a 30–60‐fold reduction in zanamivir sensitivity, has also been described.
81
Influenza B viruses
Although influenza B viruses do not cause pandemics, because they have no animal reservoir, they often cause a significant proportion of seasonal influenza. Oseltamivir has also been reported to have lower clinical efficacy in children infected with influenza B compared with influenza A,
82
consistent with observations of higher IC50s in enzyme assays.
16
,
17
,
18
,
45
Viruses with mutations conferring oseltamivir resistance were detected in 1·4% (1/74) of oseltamivir‐treated pediatric patients (G402S), but also in 1·7% (7/422) of untreated patients. Mutations detected in isolates from this study and other untreated patients include D198N, D198E, I222T, and S250G (N2 numbering).
45
,
83
,
84
The IC50s for NAs with the mutations were 50 nm for zanamivir and 250
nm for zanamivir and 250 nm for oseltamivir for G402S and D198N, 25
nm for oseltamivir for G402S and D198N, 25 nm for zanamivir and 450–500
nm for zanamivir and 450–500 nm for oseltamivir for I222T, and 190
nm for oseltamivir for I222T, and 190 nm for zanamivir and 50
nm for zanamivir and 50 nm (equivalent to wild type) for oseltamivir for the S250G mutation. Infection with these resistant isolates may have been acquired through exposure to treated contacts, but others had no known contact. There is also a recent report of a cluster of 14 cases of influenza B viruses with an I222V mutation conferring reduced susceptibility to both oseltamivir and peramivir.
85
Although the incidence of resistance in influenza B is low, it appears from both the Japanese and US studies that resistant influenza B viruses are fit and transmissible.
86
nm (equivalent to wild type) for oseltamivir for the S250G mutation. Infection with these resistant isolates may have been acquired through exposure to treated contacts, but others had no known contact. There is also a recent report of a cluster of 14 cases of influenza B viruses with an I222V mutation conferring reduced susceptibility to both oseltamivir and peramivir.
85
Although the incidence of resistance in influenza B is low, it appears from both the Japanese and US studies that resistant influenza B viruses are fit and transmissible.
86
H5N1 viruses
Like the seasonal and pandemic A(H1N1)pdm09 strains, H274Y mutations have been seen in H5N1 isolates from infected patients treated with oseltamivir. 32 , 87 One of these patients contained a mixed population of wild‐type virus and viruses with H274Y or N294S mutations. 32 The N294S mutation also appears to have emerged spontaneously in H5N1 viruses in Egypt 31 and confers a reduction in oseltamivir sensitivity of 57–138‐fold.
There are ongoing concerns that H5N1 viruses may yet cause a pandemic; hence, surveillance of avian viruses has been carried out using molecular techniques to identify known mutations conferring resistance, as well as phenotypic testing by the enzyme inhibition assay. Each method has its advantages and disadvantages. Sequencing can detect mixtures of wild‐type and known mutant NAs. In contrast, screening for an alteration in enzyme sensitivity in the NAI assay will detect both known and novel mutations; however, the resistant virus population needs to be in excess over the wild‐type population to detect a shift in sensitivity.
17
Sequence analyses of the NAs of H5N1 viruses have revealed mixed populations of wild‐type and H274Y viruses in samples from chickens, ducks, and swans, and virus with a N294S mutation was detected in ducks.
88
Enzyme inhibition assays demonstrated that clade 2 viruses from Indonesia have a naturally occurring 15–30‐fold lower sensitivity to oseltamivir in vitro compared with both clade 1 viruses from Vietnam and human H1N1 viruses.
16
,
17
,
89
We suggested this could be attributed to the H252Y difference between clade 1 and clade 2 NAs.
89
Subsequent mutagenesis confirmed that Y252 does reduce oseltamivir sensitivity,
90
,
91
while zanamivir sensitivity is not affected. Reduced oseltamivir sensitivity in vitro was shown to correspond to reduced sensitivity in animal models.
92
,
93
Several I222T/V/M mutants have also been identified in H5N1 isolates. While mutations in I222 reduced oseltamivir sensitivity by only a few fold in clade 1 H5N1 viruses,
94
it would appear that they act synergistically with the H252Y in the clade 2 viruses, because IC50s for I222V/T mutants were in the range of 40–70 nm and for I222M mutants >250
nm and for I222M mutants >250 nm.
95
Screening 29 clade 2·3·2 isolates from the Republic of Laos identified three with reduced NAI sensitivity due to different mutations. V116A reduced sensitivity to oseltamivir by 18‐fold and to zanamivir by 10‐fold, I222L contributed to a 77‐fold reduction in susceptibility to oseltamivir, and S246N reduced the sensitivity to oseltamivir by 24‐fold.
96
This emphasizes the importance of ongoing surveillance in the avian populations.
nm.
95
Screening 29 clade 2·3·2 isolates from the Republic of Laos identified three with reduced NAI sensitivity due to different mutations. V116A reduced sensitivity to oseltamivir by 18‐fold and to zanamivir by 10‐fold, I222L contributed to a 77‐fold reduction in susceptibility to oseltamivir, and S246N reduced the sensitivity to oseltamivir by 24‐fold.
96
This emphasizes the importance of ongoing surveillance in the avian populations.
Structural and functional insights into the impacts of mutations
Knowledge obtained from the structure of the NA with sialic acid bound in the active site was used to design zanamivir. 11 We have subsequently used structural analysis to understand the mechanisms of resistance to the NAIs. The first structure of an oseltamivir‐resistant NA was published in 1998, even prior to the introduction of the drugs into clinical practice, and provided important insights into the relationship between the chemical structure of the inhibitors and the likelihood of resistance emerging. 97 We demonstrated that high‐affinity binding of oseltamivir required reorientation of E276 (E277 in N1 numbering) to create a pocket to accommodate the bulky hydrophobic side chain (Figure 2). The R292K mutation prevented the rotation of E276, leading to the loss of high‐affinity binding. We therefore developed our minimalist hypothesis of drug design that the closer the ligand is to the natural substrate, the less likely resistance is to emerge. Zanamivir has only a single difference compared with the natural DANA ligand and does not need any structural change in the NA to bind (Figure 1). Therefore, mutations conferring resistance to zanamivir would most likely impact on substrate binding, compromising the fitness of the NA. Conversely, the more differences from the natural ligand, the more potential targets for resistance. Hence, we predicted resistance was more likely to arise to oseltamivir, as has now been seen. Peramivir has both the C4‐guanidino group like zanamivir and a hydrophobic side chain like oseltamivir (Figure 1). We showed structurally that it also requires reorientation of E276 (Figure 2). 77 We have demonstrated by structural and functional studies that cross‐resistance to peramivir can be caused by mutations conferring resistance to either zanamivir or oseltamivir. 77 Laninamivir is similar to zanamivir, and hence, it is expected that resistance to laninamivir is also less likely to arise.
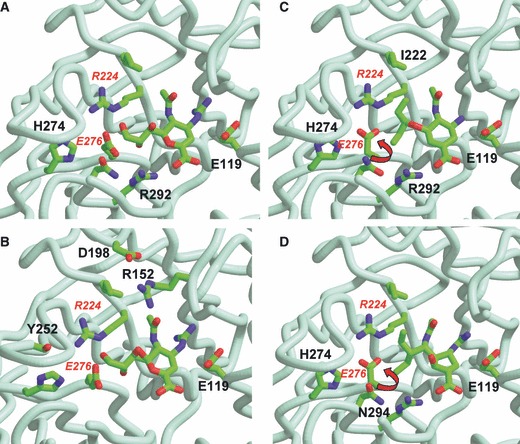
 Structures of the N9 NA with bound inhibitors (A,B) zanamivir (PDB NNC), (C) oseltamivir (PDB QWK), (D) peramivir (PDB IL7F). To minimize overcrowding, only some amino acids which affect NAI binding are labeled in each figure. (B) is slightly rotated from (A) to show Y252 situated behind H274. Residue 252 is Y in N9, but the H252Y difference between clade 1 and clade 2 avian H5N1 NAs reduces binding of oseltamivir. Arrows show rotation of E276 to form a salt bridge to R224, creating the hydrophobic pocket to accommodate pentyl side chains of oseltamivir and peramivir.
Structures of the N9 NA with bound inhibitors (A,B) zanamivir (PDB NNC), (C) oseltamivir (PDB QWK), (D) peramivir (PDB IL7F). To minimize overcrowding, only some amino acids which affect NAI binding are labeled in each figure. (B) is slightly rotated from (A) to show Y252 situated behind H274. Residue 252 is Y in N9, but the H252Y difference between clade 1 and clade 2 avian H5N1 NAs reduces binding of oseltamivir. Arrows show rotation of E276 to form a salt bridge to R224, creating the hydrophobic pocket to accommodate pentyl side chains of oseltamivir and peramivir.
Many mutations conferring reduced sensitivity to oseltamivir cluster around the hydrophobic pocket, impairing E276 rotation (Figure 2). Structural analysis of the H274Y mutant NA confirmed that, like our earlier R292K findings, this also prevents full rotation of E276. 90 The structure of the N1 NA from wild‐type H5N1 virus also did not show full rotation of the E276. 98 Interestingly, although this was a clade 1 NA, the authors had mutated the H252 to Y252, which prevented full E276 rotation, correlating with reduced sensitivity of the clade 2 viruses which have Y252. 89
In the N1 NA, the N294S mutation weakens the hydrogen bond interaction of Y344 with the carboxylate common to sialic acid and all NAIs, leading to a small reduction in substrate and zanamivir binding. 44 The S294 also forms a hydrogen bond with the carboxylate of E276 perturbing the hydrophobic pocket in which the oseltamivir side chain binds, leading to a further reduction in oseltamivir binding.
Reports of I222 (I223 in N1 numbering) mutations associated with reduced NAI sensitivity are appearing more frequently. Unlike other mutations, I222 mutations are not type or subtype specific and confer reduced sensitivity on N1, N2, and B NAs. They have emerged either spontaneously or after oseltamivir treatment. Mutations I222T have been seen in influenza B NA, 45 , 83 , 99 I222V/M/T/R in N1, 45 , 49 , 65 , 66 , 67 , 94 and I222V in N2. 45 , 79 I222 is located near the N‐acetyl and glycerol side chains of zanamivir or the pentyl ether group of oseltamivir (Figure 2). It is also near R224, which is important in the formation of a salt link to E276, involved in the formation of the hydrophobic pocket for oseltamivir binding. Mutations at I222 would lead to the loss of interactions between the side chain of I222 and the pentyl ether group of oseltamivir, thus having a greater impact on binding of oseltamivir than of zanamivir.
E119 is critical for interactions either with the C4‐guanidino group of zanamivir and peramivir or with the C4‐amino group of oseltamivir. E119G/A/D‐resistant mutants have only been generated in laboratory studies. 94 , 100 , 101 , 102 While the E119A/D confers reduced sensitivity to all NAIs, E119G confers high‐level resistance only to zanamivir and peramivir. 77 However, the E119G also makes the NA unstable. 102 , 103 , 104 In contrast, viruses with E119V/I have been detected in both oseltamivir‐treated and untreated patients, indicating they are fit and transmissible. E119V confers high‐level resistance only to oseltamivir. 18 , 30 , 75 , 79 , 105 , 106 We have shown structurally that the E119G mutation causes resistance to zanamivir due to the loss of interactions of the carboxylate side chain with the C4‐guanidinium group, as well as to alterations in the solvent structure, because a water molecule occupies the position previously occupied by the carboxylate side chain. 101 Resistance of the E119G mutant NA to peramivir is not as high as to zanamivir. This may be because zanamivir has a stronger interaction with E119 due to the formation of a weak ionic hydrogen bond. 77 The structural basis for the selectivity of E119V for oseltamivir has not yet been elucidated.
Our structures of wild‐type influenza B and D198E (D197 in B NA numbering) NAs provided insight into the reduced binding of not only this mutant, but also wild‐type influenza B NAs to oseltamivir. 107 We showed that the D198E mutation affected the interaction of R152 (R150 in B numbering) with the N‐acetyl group on the ligand, thus conferring reduced sensitivity to all NAIs. However, we also showed that while E276 rotated on binding peramivir in both the wild‐type and E198 NAs, the full rotation did not occur on binding oseltamivir, providing a structural explanation for the reduced sensitivity of the influenza B NA to oseltamivir.
The NAIs are described as being time‐dependent or slow‐binding inhibitors. Hence, in the enzyme inhibition assay used to determine drug sensitivity, virus and inhibitor are pre‐incubated to facilitate maximum occupancy of the enzyme active site. Loss of slow binding often occurs with NA mutations conferring drug resistance.
101
,
108
,
109
,
110
,
111
We have recently developed a real‐time IC50 kinetics assay,
107
,
112
which compares the rate of inhibitor binding with and without pre‐incubation (Figure 3). This simple phenotypic assay allows the easy identification of slow and fast binding of multiple inhibitors and multiple viruses without requiring purified virus or NA, or a detailed knowledge of enzyme kinetics. By following the changes in IC50, each 10 minutes for 60
minutes for 60 minutes after addition of substrate, we demonstrated that pre‐incubation only enhances binding to wild‐type slow‐binding NAs, resulting in lower IC50s. Loss of slow binding to resistant NAs is shown by a minimal change in IC50 with or without pre‐incubation over the 60
minutes after addition of substrate, we demonstrated that pre‐incubation only enhances binding to wild‐type slow‐binding NAs, resulting in lower IC50s. Loss of slow binding to resistant NAs is shown by a minimal change in IC50 with or without pre‐incubation over the 60 minutes reaction. Figure 3 shows the differences in binding of oseltamivir and zanamivir to wild‐type and resistant NAs. Drugs to which the NAs remain sensitive are still slow binding. Subsequently, we have demonstrated that reduced NAI sensitivity of a virus with a Y155H NA mutation
45
,
113
was due to even slower binding than to the wild‐type NA. IC50 kinetics assays also suggest there are differences in the dissociation rates of the NAIs after pre‐incubation with inhibitors. Oseltamivir appears to dissociate faster than zanamivir. We are developing a new solid‐phase dissociation assay, which confirms these trends.
114
minutes reaction. Figure 3 shows the differences in binding of oseltamivir and zanamivir to wild‐type and resistant NAs. Drugs to which the NAs remain sensitive are still slow binding. Subsequently, we have demonstrated that reduced NAI sensitivity of a virus with a Y155H NA mutation
45
,
113
was due to even slower binding than to the wild‐type NA. IC50 kinetics assays also suggest there are differences in the dissociation rates of the NAIs after pre‐incubation with inhibitors. Oseltamivir appears to dissociate faster than zanamivir. We are developing a new solid‐phase dissociation assay, which confirms these trends.
114
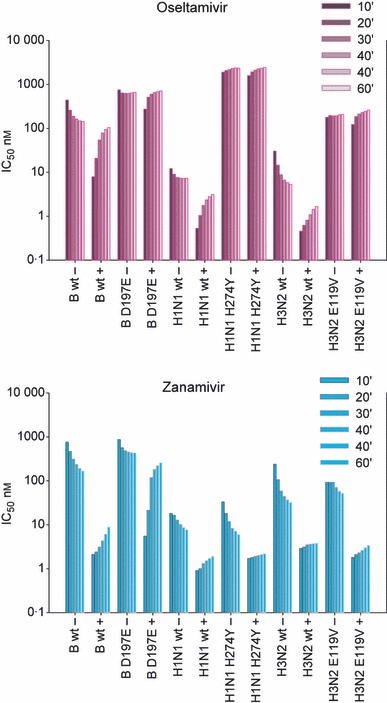
 Graphs of IC50 kinetics show how changes in IC50 during the enzyme inhibition assay can identify slow and fast binding inhibitors. IC50s are compared either with pre‐incubation with inhibitors (+) or with no pre‐incubation, where virus, inhibitor, and substrate are added simultaneously (−). Pre‐incubation enhances binding, leading to lower IC50s for slow‐binding inhibitors compared with no pre‐incubation. Where inhibitors are no longer slow binding, there is little difference in the IC50s with or without pre‐incubation, for example, wild‐type B virus with oseltamivir, D198E mutant with both inhibitors, and H274Y and E119V only with oseltamivir. The latter two remain sensitive to zanamivir and still demonstrate slow binding. As substrate competes with the inhibitor in pre‐incubation reactions, there is an increase in IC50. For wild‐type viruses, there is a more rapid dissociation of oseltamivir than that of zanamivir.
Graphs of IC50 kinetics show how changes in IC50 during the enzyme inhibition assay can identify slow and fast binding inhibitors. IC50s are compared either with pre‐incubation with inhibitors (+) or with no pre‐incubation, where virus, inhibitor, and substrate are added simultaneously (−). Pre‐incubation enhances binding, leading to lower IC50s for slow‐binding inhibitors compared with no pre‐incubation. Where inhibitors are no longer slow binding, there is little difference in the IC50s with or without pre‐incubation, for example, wild‐type B virus with oseltamivir, D198E mutant with both inhibitors, and H274Y and E119V only with oseltamivir. The latter two remain sensitive to zanamivir and still demonstrate slow binding. As substrate competes with the inhibitor in pre‐incubation reactions, there is an increase in IC50. For wild‐type viruses, there is a more rapid dissociation of oseltamivir than that of zanamivir.
Conclusions
Resistance to the NAIs can be both drug and virus type or subtype specific. A summary of the effects of mutations referred to in this review is presented in Table 1. A designation of low, medium, or high resistance is based on reported fold differences in sensitivity relative to the wild‐type virus. However, while resistance is often defined as greater than a 10‐fold change in IC50 compared with the wild type, because some viruses have a higher base line IC50, for example, influenza B and clade 2 H5N1 strains, such viruses may be clinically resistant with only a few fold increase in IC50. It may be more appropriate to define resistance in terms of the IC50 and the drug concentrations delivered to the upper respiratory tract. There is also an issue of how the IC50 is measured, because values from the chemiluminescent assay are often lower than those from the fluorescent assay, and our IC50 kinetics experiments demonstrate how the IC50 can change with incubation times in the NAI assays. Hence, there is currently no consensus on a definition of resistance, as it can really only be demonstrated by the lack of response to treatment.
Table 1
 Mutations and relative susceptibility to NAIs*
Mutations and relative susceptibility to NAIs*
| Type/subtype | Amino acid substitution** | Oseltamivir | Zanamivir | Peramivir |
|---|---|---|---|---|
| H1N1 | N70S | S | M | ? |
| H1N1 | Q136K | S | H | H |
| H1N1 | Y155H | M | H | H |
| H1N1 | I222V | L | S | S |
| H1N1 | I222M | L | S | S |
| H1N1 | H274Y | H | S | H |
| H1N1pdm(09) | I222V | M | S | ? |
| H1N1pdm(09) | I222R | M | L | L |
| H1N1pdm(09) | H274Y | H | S | H |
| H3N2 | E119A | H | H | H |
| H3N2 | E119D | H | H | H |
| H3N2 | E119G | S | H | M |
| H3N2 | E119V | H | S | S |
| H3N2 | E119I | H | L | H |
| H3N2 | Q136K | S | M | ? |
| H3N2 | I222V | M | S | S |
| H3N2 | R292K | H | M | H |
| H3N2 | N294S | H | S | S |
| B | E119A | H | H | H |
| B | E119D | H | H | H |
| B | E119G | M | M/H | H |
| B | R152K | H | H | H |
| B | D198N | H | M | L |
| B | D198E | M | L | M |
| B | I222V | L | S | L |
| B | I222T | H | M | ? |
| B | S250G | M | H | ? |
| B | G402S | H | M/H | ? |
| H5N1 | V116A | M | M | L |
| H5N1 | I222L | M | S | ? |
| H5N1 | I222V | L/H*** | S | S |
| H5N1 | I222M | M/H*** | S | S |
| H5N1 | I222T | M/H*** | S | S |
| H5N1 | S246N | M | S | ? |
| H5N1 | H252Y | M | S | S |
| H5N1 | H274Y | H | S | H |
| H5N1 | N294S | M/H | L | L |
S, sensitive; L, low <10‐fold change; M, medium 10–50‐fold change; H, High=>50‐fold change compared with wild type.
*A more comprehensive list of both in vitro and in vivo selected isolates is given in Nguyen et al. 115
**N2 numbering.
***Clade 2 isolates have a higher baseline IC50 and these mutations act synergistically, conferring higher IC50s compared to clade 1 viruses.
In contrast to earlier results, we have now seen that viruses with mutations conferring resistance can be fit and transmissible. Resistance is more likely to arise to oseltamivir, due to the structural changes needed for oseltamivir to bind with high affinity. No resistance has yet emerged under standard zanamivir treatment of immunocompetent patients. Resistance to either NAI may also confer reduced sensitivity to peramivir. Hence, diversification of antiviral stockpiles to include zanamivir as well as oseltamivir is an important strategy to minimize the impact of oseltamivir resistance. Development of new inhibitors with different modes of action should also be a priority.
Conflicts of interest
Dr McKimm‐Breschkin has received honoraria and/or travel assistance from GlaxoSmithKline (GSK) and Hoffman La‐Roche for the participation in advisory groups and scientific meetings. She has also received research support from GSK and Biota for studies on resistance to the neuraminidase inhibitors.
Acknowledgements
Parts of this work were funded through CSIRO, GlaxoSmithKline, by a grant from the National Institutes of Health (NIAID RO1A1062721 awarded to JMB), a Pandemic influenza grant 595625 from the NHMRC Australia and Grant ID 77869 from the MRC UK. The funders had no role in data analysis or the preparation of the manuscript.
References
 years of their use.
Antimicrob Agents Chemother
2006; 50:2395–2402.
[Europe PMC free article] [Abstract] [Google Scholar]
years of their use.
Antimicrob Agents Chemother
2006; 50:2395–2402.
[Europe PMC free article] [Abstract] [Google Scholar] hours.
Emerg Infect Dis
2010; 16:1633–1636.
[Europe PMC free article] [Abstract] [Google Scholar]
hours.
Emerg Infect Dis
2010; 16:1633–1636.
[Europe PMC free article] [Abstract] [Google Scholar] year by an immunocompromised child.
Clin Infect Dis
2006; 43:1555–1561.
[Abstract] [Google Scholar]
year by an immunocompromised child.
Clin Infect Dis
2006; 43:1555–1561.
[Abstract] [Google Scholar]Articles from Influenza and Other Respiratory Viruses are provided here courtesy of Wiley
Full text links
Read article at publisher's site: https://doi.org/10.1111/irv.12047
Read article for free, from open access legal sources, via Unpaywall:
https://onlinelibrary.wiley.com/doi/pdfdirect/10.1111/irv.12047
Citations & impact
Impact metrics
Article citations
Genotypic and phenotypic susceptibility of emerging avian influenza A viruses to neuraminidase and cap-dependent endonuclease inhibitors.
Antiviral Res, 229:105959, 08 Jul 2024
Cited by: 0 articles | PMID: 38986873
A pan-respiratory antiviral chemotype targeting a transient host multi-protein complex.
Open Biol, 14(6):230363, 19 Jun 2024
Cited by: 2 articles | PMID: 38889796 | PMCID: PMC11285769
Retracted and republished from: "The current state of research on influenza antiviral drug development: drugs in clinical trial and licensed drugs".
mBio, 15(5):e0017524, 29 Mar 2024
Cited by: 3 articles | PMID: 38551343 | PMCID: PMC11077966
Review Free full text in Europe PMC
N-Glycan Profiles of Neuraminidase from Avian Influenza Viruses.
Viruses, 16(2):190, 26 Jan 2024
Cited by: 0 articles | PMID: 38399967 | PMCID: PMC10893399
An orally active entry inhibitor of influenza A viruses protects mice and synergizes with oseltamivir and baloxavir marboxil.
Sci Adv, 10(8):eadk9004, 23 Feb 2024
Cited by: 1 article | PMID: 38394202 | PMCID: PMC10889430
Go to all (167) article citations
Other citations
Similar Articles
To arrive at the top five similar articles we use a word-weighted algorithm to compare words from the Title and Abstract of each citation.
Neuraminidase inhibitor susceptibility profile of pandemic and seasonal influenza viruses during the 2009-2010 and 2010-2011 influenza seasons in Japan.
Antiviral Res, 99(3):261-269, 17 Jun 2013
Cited by: 23 articles | PMID: 23791870
Neuraminidase inhibitor susceptibility surveillance of influenza viruses circulating worldwide during the 2011 Southern Hemisphere season.
Influenza Other Respir Viruses, 7(5):645-658, 10 Apr 2013
Cited by: 25 articles | PMID: 23575174 | PMCID: PMC5781198
Global update on the susceptibility of human influenza viruses to neuraminidase inhibitors, 2013-2014.
Antiviral Res, 117:27-38, 23 Feb 2015
Cited by: 74 articles | PMID: 25721488 | PMCID: PMC9036627
Laninamivir octanoate: a new long-acting neuraminidase inhibitor for the treatment of influenza.
Expert Rev Anti Infect Ther, 9(10):851-857, 01 Oct 2011
Cited by: 43 articles | PMID: 21973296
Review
Funding
Funders who supported this work.
Medical Research Council
NIAID NIH HHS (1)
Grant ID: R01 AI062721
PHS HHS (1)
Grant ID: R01A1062721





