Abstract
Free full text

Sortase-mediated modification of αDEC205 affords optimization of antigen presentation and immunization against a set of viral epitopes
Abstract
A monoclonal antibody against the C-type lectin DEC205 (αDEC205) is an effective vehicle for delivery of antigens to dendritic cells through creation of covalent αDEC205–antigen adducts. These adducts can induce antigen-specific T-cell immune responses or tolerance. We exploit the transpeptidase activity of sortase to install modified peptides and protein-sized antigens onto the heavy chain of αDEC205, including linkers that contain nonnatural amino acids. We demonstrate stoichiometric site-specific labeling on a scale not easily achievable by genetic fusions (49 distinct fusions in this report). We conjugated a biotinylated version of a class I MHC-restricted epitope to unlabeled αDEC205 and monitored epitope generation upon binding of the adduct to dendritic cells. Our results show transfer of αDEC205 heavy chain to the cytoplasm, followed by proteasomal degradation. Introduction of a labile dipeptide linker at the N terminus of a T-cell epitope improves proteasome-dependent class I MHC-restricted peptide cross-presentation when delivered by αDEC205 in vitro and in vivo. We also conjugated αDEC205 with a linker-optimized peptide library of known CD8 T-cell epitopes from the mouse γ-herpes virus 68. Animals immunized with such conjugates displayed a 10-fold reduction in viral load.
A monoclonal antibody against the C-type lectin DEC205, αDEC205, recognizes DEC205, a ~250-kDa surface protein on dendritic cells (DCs) (1, 2). Attachment of an antigenic moiety to αDEC205, either through chemical conjugation (3) or as a genetic fusion (4), allows its targeted delivery to DCs in vivo, and elicits potent immune responses even with very small amounts of the αDEC205 adduct. This response includes CD4 and CD8 T cells specific for the antigen attached to the antibody (5). Equipping αDEC205 with a suitable payload, as well as similar modifications of other antibodies directed against yet other surface proteins on professional antigen presenting cells (6), holds promise as a purely protein-based vaccine strategy that does not require replicating or infectious agents.
Most chemical conjugation methods one might consider for modification of αDEC205 lack precision, as they predominantly target exposed lysine or cysteine residues. Even the introduction of a single suitably exposed cysteine by mutagenesis, although it affords good site-specificity of labeling, still limits the range of possibilities for covalent attachment of peptides and proteins. Fusion of αDEC205 and the payload of interest can be attained genetically, but expression and purification of each new individual adduct needs to be explored and optimized. These limitations impelled us to develop an alternative method to install payloads of interest onto αDEC205 in a more flexible manner. An efficient and straightforward chemoenzymatic alternative with the use of sortase A (7) should allow not only the facile installation of a large diversity of antigens, but also an analysis of the mechanisms that underlie the desirable immunogenic properties of αDEC205 adducts, with particular emphasis on the mode of attachment and sequences flanking the antigen. This approach is unique in that it allows the use of nonnatural amino acids as linkers, as well as the selective introduction of chemical modifications on the payload, while leaving the αDEC205 portion otherwise unmodified.
Sortase A from Staphylococcus aureus enables the covalent linkage of proteins that contain a suitably exposed LPXTG motif to probes composed of N-terminal oligoglycines, modified with a cargo of choice: peptide, protein, nucleic acid, (glyco)lipid, or any other entity that can be provided in linkage to an oligoglycine peptide (8, 9). We show that the introduction of an LPXTG motif at the C terminus of the heavy chain of αDEC205 allows the attachment of a T-cell epitope, a fluorescent, or a biotinylated cargo of choice in a stoichiometric manner. This procedure, referred to as sortagging, affords delivery of any T-cell epitope or traceable payload to DEC205+ DCs in vitro and in vivo. Importantly, it allows the installation of a peptide that can be labeled independently from the protein to which it is attached, an essential attribute for investigating T-cell epitope processing.
We show that conjugation of peptides or a protein, such as GFP, to αDEC205 is achieved with efficiencies approximating 90%. The conjugated antibody was readily separated from the sortase enzyme and unincorporated probes, thus allowing rapid processing of numerous samples in parallel. We used sortagging to install a biotinylated class I MHC-restricted epitope on αDEC205 and unravel the sequence of events that leads to the generation of the epitope upon binding to DEC205. We investigated the factors that influence the presentation by DCs of a peptide conjugated to αDEC205 and show that the introduction of labile dipeptide linkers at the N terminus of a class I MHC-restricted epitope sortagged onto αDEC205 strongly affects the in vivo CD8 immune response upon immunization of mice with conjugated αDEC205 by favoring the generation of the final epitope in a proteasome-dependent manner. We used these findings to design and conjugate to αDEC205 a complex set of peptides corresponding to 19 known epitopes of mouse γ-herpes virus (MHV-68). Immunization with αDEC205 sortagged to the MHV-68 epitope set reduced viral burden upon subsequent infection with live MHV-68. Our study thus addresses the mechanism that underlies antibody-mediated targeting of antigens to DCs and exploits these findings to elicit a CD8 T-cell response that helps curtail a herpesvirus infection.
Results
Sortagging of αDEC205 with (Modified) Peptides or GFP.
We modified the DNA construct encoding the heavy chain of αDEC205 (2, 4) to introduce an LPETG motif, required for sortase-mediated installation of payloads of interest, followed by a histidine tag (His6; Fig. 1). The modified antibody was expressed in CHO cells and purified from the culture media. The sortase reaction was then performed under native conditions in physiological buffers, with no collateral chemical damage inflicted on αDEC205 or its cargo. We started by coupling peptides (containing an HA or a biotin tag) to αDEC205 and monitored the kinetics of the reaction by immunoblotting against both His6 (input αDEC205 and sortase) and HA or biotin (desired product) tags (Fig. 2A). After 4 h of incubation, 90% of input αDEC205 was conjugated, and, after 20 h, the reaction was nearly complete, as judged from the loss of the His6 tag (Fig. 2A). Gel filtration chromatography or purification by using protein G-coupled beads allowed complete separation of sortagged αDEC205 from sortase A (Fig. S1). αDEC205 could be conjugated to a protein-sized probe with similar efficiency. Fig. 2B shows the result of a sortase reaction in which we used GFP equipped with a five-glycine N-terminal extension as the nucleophile. Incubation of αDEC205 with this modified GFP and sortase allowed its conjugation to the αDEC205 heavy chain in excellent yield (>90%, Fig. 2B, Upper). Sortagging of αDEC205 with the biotinylated peptide or GFP neither inhibited antibody binding nor compromised GFP fluorescence (Fig. S2). The sortase method thus allows conjugation of (biotinylated) peptides or proteins with excellent efficiency and without compromising the binding specificity of αDEC205.
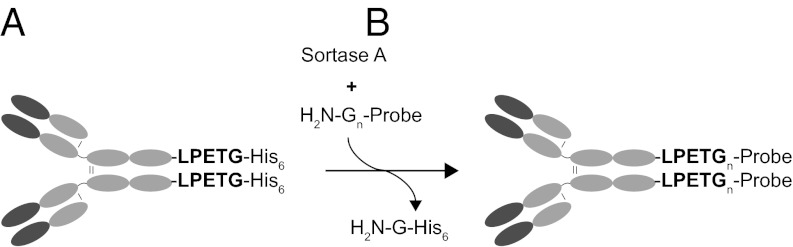
Site-specific labeling of αDEC205 by sortase-mediated transpeptidation. (A) Schematic representation of the modified αDEC205 antibody. The cDNA construct encoding the heavy chain of αDEC205 was modified to introduce an LPETG motif, followed by a histidine tag (His6) at the C terminus of the protein. (B) Sortase-mediated transpeptidation mechanism. Sortase A recognizes the LPETG motif, cleaving the peptide bond between the threonine and glycine and resulting in a thioacyl intermediate. A modified oligoglycine nucleophile then attacks the thioacyl intermediate to yield the transpeptidation product with the probe in amide linkage to the target protein. The final reaction product consists of αDEC205 bearing the newly attached C-terminal probe on the heavy chain. The efficiency of the reaction can be conveniently monitored by loss of the His6 tag.
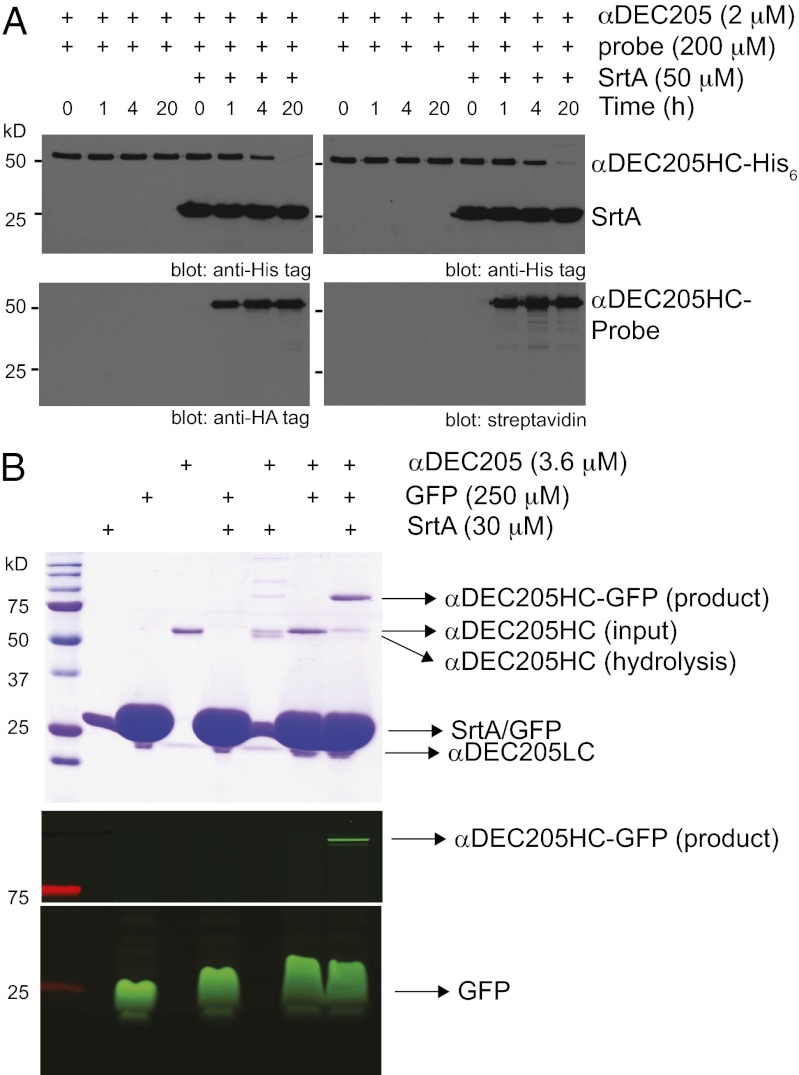
Covalent attachment of HA-, biotin-containing peptides, and GFP to αDEC205 by sortase. (A) αDEC205 was incubated with an oligoglycine probe containing an HA tag (Left) or an oligoglycine probe modified with a biotin group (Right) in the presence or absence of sortase A for 0, 1, 4, or 20 h at 37 °C. The reaction was monitored by immunoblotting against His6 (Upper), HA tag or by blotting with streptavidin–HRP (Lower). (B) Site-specific attachment of GFP to the C terminus of αDEC205. The reaction was monitored by Coomassie staining (Upper) and fluorescence imaging (Lower). αDEC205HC, heavy chain of the αDEC205 antibody; αDEC205LC, light chain of the αDEC205 antibody; SrtA, sortase A from S. aureus.
Insertion of Dipeptide Linkers at N Terminus of T-Cell Epitopes Sortagged to αDEC205 Improves Presentation of Class I MHC-Restricted but Not Class II MHC-Restricted Epitopes in Vitro.
αDEC205 has been used extensively as a vehicle for the delivery of T-cell epitopes to antigen-presenting cells as a means of evoking a T-cell immune response or to induce tolerance (5, 6, 10). We sortagged a single batch of αDEC205 with a larger set of peptides to explore the importance of the mode of linkage of a T-cell epitope to αDEC205. We measured presentation by DCs as assayed by activation of T cells of the appropriate specificity. We chose the ovalbumin peptide OVA323-339 (ISQAVHAAHAEINEAGR) recognized by CD4 T cells from OTII transgenic mice (11, 12) to explore the influence of the flanking residues on presentation via class II MHC. We synthesized a set of sortase-compatible peptides containing three N-terminal glycines followed by the T-cell epitope, flanked by residues contained in intact ovalbumin or preceded by dipeptide linkers that have been used in antibody–drug conjugates (Table S1) (13, 14). We measured the capacity of DCs exposed to αDEC205 sortagged with OVA323-339 peptide variants to induce proliferation of carboxyfluorescein succinimidyl ester (CFSE)-labeled CD4 T cells from ovalbumin-specific OTII T-cell receptor (TCR) transgenic mice. Exposure of DCs to αDEC205 sortagged with OVA323-339 peptide induced robust proliferation, compared with incubation with αDEC205-HA. All OVA323-339 peptide variants sortagged onto αDEC205 showed a similar dose–response curve in this assay (Fig. 3A). We conclude that the flanking residues of this class II MHC-restricted epitope do not affect the capacity of the αDEC205–peptide adducts to activate CD4 OTII T cells.
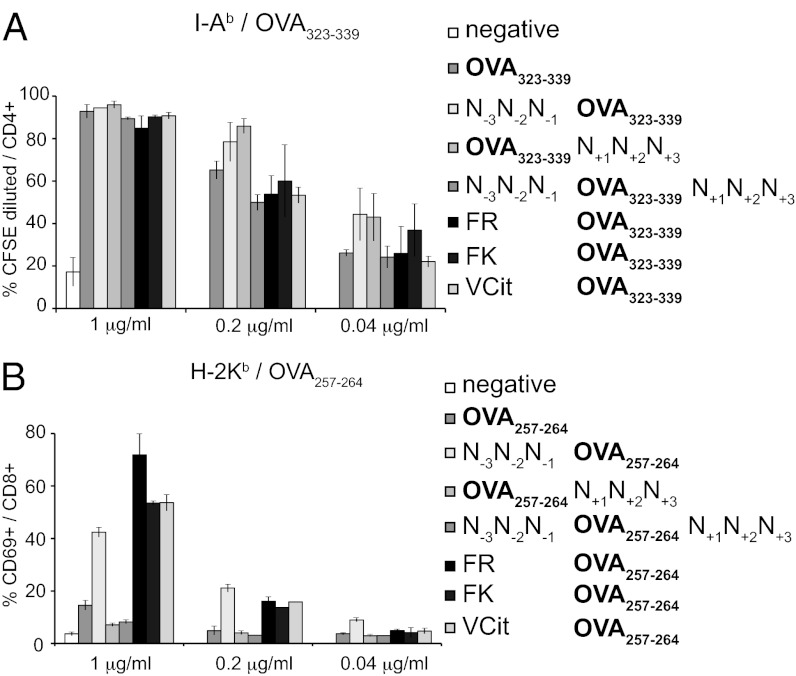
In vitro stimulation of antigen-specific CD4 and CD8 T cells by DCs incubated with sortagged αDEC205. (A) DCs were incubated in vitro with αDEC205 sortagged with HA tag (negative) or OVA323-339–containing probes (Table S1) at various concentrations, washed, and incubated together with CFSE-labeled splenocytes from OTII transgenic mice at day 0. The histogram shows the percentage of CFSE-diluted OTII CD4 T cells at day 4 for each concentration of αDEC205 adduct used. (B) BMDCs were incubated in vitro with medium alone (negative) or αDEC205 sortagged with OVA257-264–containing probes (Table S1) at various concentrations, washed, and incubated together with splenocytes from OTI transgenic mice. The histogram shows the percentage of CD69-positive CD8 OTI T cells for each concentration of αDEC205 adduct used after 16 h. Errors bars show SDs of three independent measurements; representative of three independent experiments.
αDEC205 can induce CD8 T-cell stimulation by cross-presentation in vitro and in vivo (3). As cross-presentation involves steps in epitope processing and routing that likely include the cytoplasm, the exact manner of attachment of an epitope sortagged onto αDEC205 potentially affects its presentation via class I MHC. We synthesized a set of sortase-compatible peptides (Table S1) derived from the OVA257-264 (SIINFEKL) peptide, an H-2Kb restricted epitope recognized by CD8 T cells from OTI mice (15). Exposure of bone marrow-derived DCs (BMDCs) to αDEC205 sortagged with SIINFEKL activated OTI CD8 T cells, based on the up-regulation of CD69 (Fig. 3B). We observed striking differences in the ability of αDEC205 sortagged with different peptides to activate T cells (Fig. 3B). BMDCs exposed to αDEC205 sortagged with OVA257-264 preceded by dipeptide linkers stimulated approximately five times more OTI cells than BMDCs exposed to αDEC205 sortagged with most of the other peptides (Fig. 3B). We investigated two additional class I MHC-restricted epitopes: the H-2Ld restricted Rop7161-169 epitope (IPAAAGRFF) from Toxoplasma gondii (16), recognized by Rop7-specific transnuclear mice (17) and the H-2Db restricted Gp33-41 epitope (KAVYNFATC) from lymphocytic choriomeningitis virus, recognized by P14 TCR transgenic mice (18), to assess the generality of these findings. BMDCs exposed to αDEC205 sortagged with the relevant peptides preceded by a dipeptide linker were significantly more potent in stimulating antigen-specific CD8 T cell than constructs that lacked them (Fig. S3). The ability of DCs to stimulate antigen-specific CD8 T cells upon incubation with sortagged αDEC205 thus improves upon introduction of a dipeptide linker preceding the epitope, as shown for three unrelated epitopes, presented by three different class I MHC products.
Insertion of Dipeptide Linkers at N Terminus of T Epitopes Sortagged to αDEC205 Dramatically Increases CD8 Immune Response upon Immunization with αDEC205 Adducts.
Our results suggest that residues preceding a class I MHC-restricted epitope critically influence success of antigen presentation upon αDEC205-mediated delivery. To investigate whether the insertion of dipeptide linker would improve CD8 T-cell immune response in vivo, we monitored expansion and IFN-γ production of OTI T cells upon immunization with αDEC205-G3-SIINFEKL vs. αDEC205-G3-FR-SIINFEKL. Immunization of mice with 0.75 μg of αDEC205-G3-SIINFEKL induced a modest increase in the percentage of OTI T cells in the spleen compared with unimmunized mice (Fig. 4 A and C). In contrast, immunization of mice with 0.75 μg of αDEC205-G3-FR-SIINFEKL induced a significant increase in the percentage of OTI T cells in the spleen (Fig. 4 A and C). These results correlate with an increase in the number of proliferating cells in mice immunized with αDEC205-G3-FR-SIINFEKL compared with mice immunized with αDEC205-G3-SIINFEKL (Fig. 4B). There were two times more IFN-γ–producing OTI T cells in the spleen of mice immunized with αDEC205-G3-FR-SIINFEKL compared with mice immunized with αDEC205-G3-SIINFEKL (Fig. 4D). Proper linker design is therefore important to ensure delivery of T-cell epitopes: even minor modifications may have an outsized impact on the desired immune response.
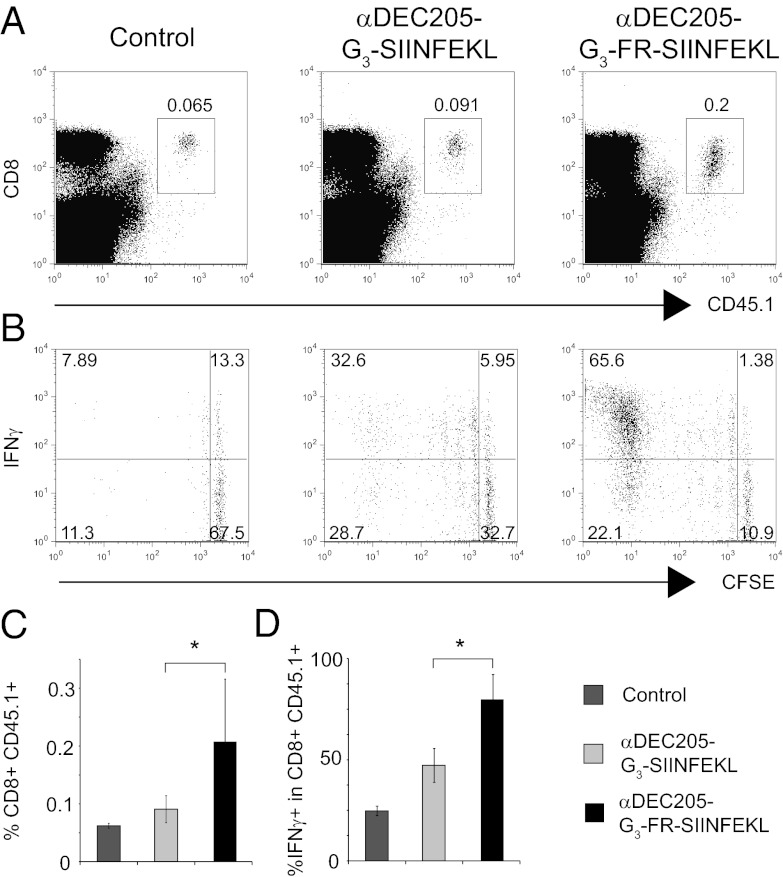
OTI T-cell proliferation and activation in mice immunized with αDEC205 sortagged with G3-SIINFEKL or G3-FR-SIINFEKL. CFSE-labeled CD8 T cells from CD45.1 OTI transgenic mice were transferred i.v. into CD45.2 C57/BL6 mice. At 24 h after cell transfer, mice were immunized with 0.75 μg of αDEC205 sortagged with G3-SIINFEKL or G3-FR-SIINFEKL together with 25 μg of αCD40 and 50 μg of Poly I:C or left untreated. Transferred T-cell expansion and activation was measured 7 d after immunization. (A) CD8 vs. CD45.1 staining of splenocytes from control mice (Left), mice immunized with αDEC205-G3-SIINFEKL or αDEC205-G3-FR-SIINFEKL (Middle and Right, respectively). (B) Dot plots show CFSE vs. IFN-γ staining on donor OTI T cells from the spleen of control mice and mice immunized with αDEC205-G3-SIINFEKL or αDEC205-G3-FR-SIINFEKL upon in vitro stimulation with SIINFEKL peptide. (C) Histogram shows percentage of transferred OTI T cells in the spleen of control mice and mice immunized with αDEC205-G3-SIINFEKL or αDEC205-G3-FR-SIINFEKL, respectively. (D) Histogram shows percentage of IFN-γ positive cells upon in vitro stimulation with SIINFEKL peptide among donor OTI T cells from the spleen of control mice or mice immunized with αDEC205-G3-SIINFEKL or αDEC205-G3-FR-SIINFEKL, respectively. Errors bars show SDs; control, n = 2; αDEC205-G3-SIINFEKL immunized, n = 4; αDEC205-G3-FR-SIINFEKL immunized, n = 4 (*P < 0.05, Student t test); representative of three independent experiments.
Presence of Dipeptide Linker Favors Generation of Proteasome-Dependent Class I MHC-Restricted Epitope.
Our results show that insertion of an FR motif immediately upstream of the antigen epitope attached to αDEC205 significantly increases the ability of DCs to stimulate antigen-specific CD8 T cells. Different routes of antigen presentation have been described, involving transfer of the antigen to the cytoplasm or antigen processing in vacuoles with subsequent loading onto class I MHC (19). To investigate the role of cathepsins and proteasomes in the generation of the epitopes from αDEC205 adducts, we treated DCs with the cathepsin inhibitor Z-FA-FMK or with the proteasome inhibitor ZL3VS and addressed their ability to stimulate OTI T cells upon incubation with αDEC205-G3-SIINFEKL. Neither Z-FA-FMK nor ZL3VS inhibited the ability of SIINFEKL-loaded BMDCs to stimulate OTI T cells (Fig. 5A). However, the ability of ZL3VS-treated BMDCs to stimulate OTI T cells upon αDEC205-G3-SIINFEKL incubation was fivefold less than seen for control BMDCs (Fig. 5B). In addition, we observed a small but reproducible increase in the ability of Z-FA-FMK–treated BMDCs to stimulate OTI T cells upon incubation with αDEC205-G3-SIINFEKL (Fig. 5B). These data suggest that the proteasome contributes to the generation of DEC205-dependent class I MHC-restricted epitopes. Therefore, αDEC205-G3-SIINFEKL or a fragment derived from it must reach the cytoplasm.
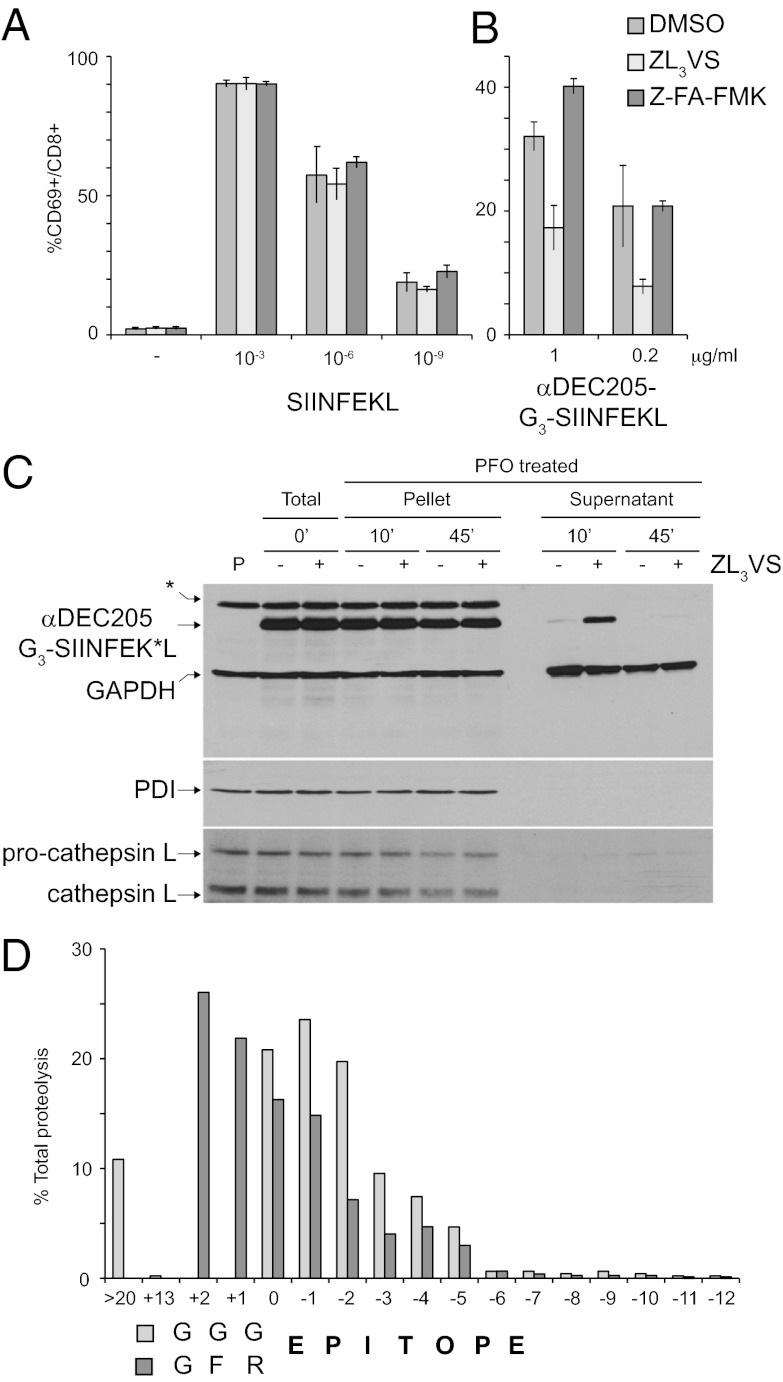
Proteasome-dependent generation of αDEC205-conjugated class I MHC-restricted epitopes. DMSO-, ZL3VS-, or Z-FA-FMK–treated BMDCs were incubated in vitro with medium alone (negative), with free SIINFEKL peptide (A), or with αDEC205-G3-SIINFEKL (B) at various concentrations, washed, and incubated together with splenocytes from OTI transgenic mice. Histogram shows percentage of CD69-positive CD8 OTI T cells for each concentration of αDEC205 adduct used after 16 h. Errors bars show SDs of four independent measurements; representative of three independent experiments. (C) DMSO- or ZL3VS-treated BMDCs were incubated with αDEC205-G3-SIINFEK*L for 10 min on ice. The cells were washed (total 0′) and incubated for 10 and 45 min, at 37 °C, 5% CO2. Cells were perfringolysin O-treated to separate organelle (i.e., pellet) and cytoplasmic (i.e., supernatant) fractions. All fractions were analyzed by SDS/PAGE under reducing conditions and immunoblotted with streptavidin–HRP. The blot was subsequently probed with anti-PDI (luminal ER-resident protein), anti-cathepsin L (soluble lysosomal enzyme), and anti-GAPDH (soluble cytosolic protein) antibodies as loading and fractionation controls. An anti-mouse HRP (GAPDH) and anti-rabbit HRP (PDI and cathepsin L) were used as secondary antibodies. Five equivalents of supernatant were loaded to facilitate visualization. Lane P, cells not incubated with αDEC205-G3-SIINFEK*L. Background biotinylated protein band is marked with an asterisk. Representative of six independent experiments. (D) Proteasomal cleavage site prediction of the C terminus of αDEC205 sortagged in silico with 235 H-2K, H-2D, or H-2L–restricted T-cell epitopes modified with GGG or GGGFK linkers. Histogram shows percentage of total digestions releasing a peptide at the indicated position. Null score represents final epitope liberation; positive score represents cleavage at the N terminus of the epitope; and negative score represents digestion in the epitope.
To address whether αDEC205 traffics to the cytoplasm with its cargo, we monitored the fate of αDEC205 sortagged to G3-SIINFEK*L (a peptide modified to contain a single biotin on the side chain of its lysine residue) in BMDCS—treated with DMSO or ZL3VS—by immunoblot using streptavidin–horseradish peroxidase (HRP). Arrival of αDEC205-G3-SIINFEK*L in the cytoplasm was assessed by subcellular fractionation using perfringolysin O, a toxin that selectively perforates the plasma membrane and allows retrieval of the cytoplasmic fraction after a simple centrifugation step (20). Inclusion of the proteasome inhibitor ZL3VS does not affect binding of αDEC205-G3-SIINFEK*L to BMDCs (Fig. 5C). After 10 min of incubation, we detected a cytoplasmic fraction of αDEC205-G3-SIINFEK*L. In ZL3VS-treated BMDCs, the level of the cytoplasmic pool of αDEC205-G3-SIINFEK*L was increased twofold compared with control BMDCs (8.1 ± 3.1% and 4.1 ± 3.8%, respectively, as percentage of the total fraction). We monitored the distribution of cathepsin L, protein disulfide isomerase (PDI), and GAPDH (Fig. 5C) to demonstrate the efficacy of this fractionation protocol and to establish that subcellular organellar integrity had been maintained. After 45 min, the cytoplasmic fraction of αDEC205-G3-SIINFEK*L was barely detectable, but the organellar fraction (i.e., pellet) remained stable.
Altogether, our results suggest that a fraction of αDEC205 is internalized by DCs and reaches the cytoplasm, and that the proteasome is key to generate the class I MHC-restricted–αDEC205-conjugated epitope. Thus, the presence of an FR linker must favor proteasome-dependent generation of the (extended) epitope. To address this, we sortagged in silico a total of 235 known H-2K–, H-2D–, and H-2L–restricted T-cell epitopes through extension with a GGG or a GGGFR motif and analyzed the predicted digestion products at the C terminus of the resultant αDEC205 adducts by using a proteasomal cleavage prediction algorithm (21). In the presence of the FR motif, the program predicts release of the epitope extended by zero, one, or two N-terminal residues in more than 60% of the cases (Fig. 5D). In sharp contrast, in the absence of the FR linker, in almost 70% of the cases, proteolysis is predicted to destroy the epitope (Fig. 5D).
Protection Against MHV-68 Infection upon Immunization with αDEC205 Sortagged with Virus Epitope Library.
MHV-68 is a rodent pathogen (22) used as a model for the human gammaherpesviruses Kaposi sarcoma-associated herpesvirus (HHV8) and EBV (HHV4) (23). We individually sortagged 19 known class I MHC-restricted epitopes (Table S1, probe 29–47) covering 14 ORFs of MHV-68 and containing a N-terminal GGGFR motif onto αDEC205 (αDEC205-G3-FR-MHV-68) (24). C57/BL6 mice were immunized with 20 μg of total αDEC205-G3-FR-MHV-68 (~1 μg of αDEC205 adduct per epitope) in combination with Poly I:C and αCD40, or animals received free MHV-68 peptides together with Poly I:C and αCD40 at 14 and 7 d before intranasal infection with MHV-68. Immunization of mice with αDEC205-G3-FR-MHV-68 resulted in a reduction of lung viral titers by 10-fold upon subsequent challenge with MHV-68 (Fig. 6), whereas administration of free peptides was without discernible effect.
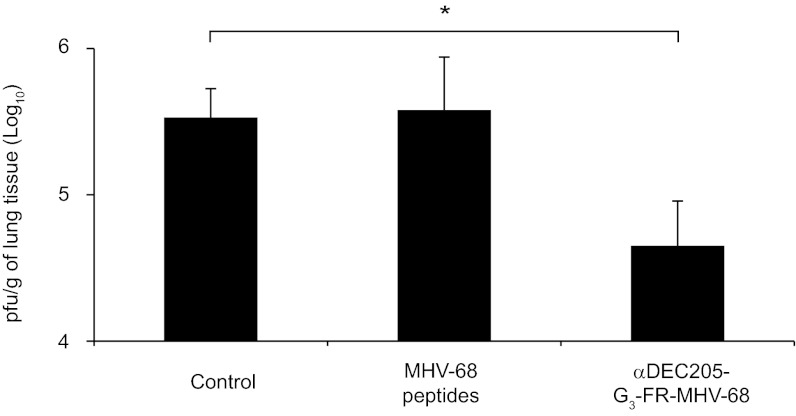
Viral titers in mice immunized with free MHV-68 peptides or αDEC205 sortagged with MHV-68 epitope library. The MHV-68 epitope library (Table S1, probe 29–47) was sortagged to αDEC205, and the desired product was purified as described in Fig. S1A. C57/BL6 mice were immunized with 20 μg of αDEC205-G3-FR-MHV-68 mix together with 25 μg of αCD40 and 50 μg of Poly I:C or an equimolar quantity of free MHV-68 peptide mix together with 25 μg of αCD40 and 50 μg of Poly I:C at 14 and 7 d before intranasal infection with 105 pfu of MHV-68. Histograms show pfu per gram of lung tissue in control mice (n = 8), mice immunized with free MHV-68 peptide (n = 7), or mice immunized with αDEC205-G3-FR-MHV-68 (n = 8) 6 d after infection. Errors bars show SDs (*P < 0.05, Student t test); representative of two independent experiments.
Discussion
Sortase-mediated modification of antibodies is flexible and robust. It offers considerable advantages vs. genetic fusions and chemical crosslinking methods. First, unlike crosslinking approaches, it allows stoichiometric and site-specific installation of probes at the C terminus of the heavy chain of the antibody in a manner that does not perturb antigen binding. Second, it enables rapid conjugation of a single batch of antibody with numerous different peptides. In this study alone, 49 different αDEC205 adducts were synthesized, each of which would have required a separate genetic construct and transfection if generated by conventional cloning and expression methods. Third, we report here the site-specific conjugation of peptides containing (modified) residues that are not template-encoded (e.g., citrulline, biotinylated lysine) and hence cannot be inserted through genetic fusion. Fourth, sortagging allowed us to install a traceable peptide (i.e., epitope) on αDEC205 and monitor its fate during antibody processing. This procedure can be extended to the installation of isotopically labeled peptide if needed. Fifth, it affords a practical alternative to genetic fusions for the installation of protein-sized fusion partners that may affect the conformation and secretion of one or both fusion partners, as well as heterooligomeric proteins, provided at least one of them has an exposed stretch of glycine residues. The sortase-mediated installation of “click” handles or aldehyde tags further extends the range of possibilities (9, 25). Whether there is an upper limit to the size of incoming nucleophiles that can be used in a sortase-mediated transpeptidation reaction remains to be determined, but we have successfully used polypeptides of as much as ~100 kDa in reactions that effectuate head-to-tail protein–protein fusions (26).
The development of a sortase-compatible version of αDEC205 allowed us to optimize αDEC205-mediated antigen presentation in an iterative process, as well as to dissect the route of DEC205-dependent antigen cross-presentation. Although it has been reported that αDEC205-mediated antigen cross-presentation is transporter of antigenic peptides (TAP)-dependent (3), the sequence of steps that lead to class I MHC-restricted epitope generation, especially when the antigen is delivered via specific surface structures such as DEC205, remains largely unknown. The inability of TAP-deficient DCs to cross-present αDEC205-conjugated antigens suggests that transfer of the (extended) epitope from the cytoplasm to the lumen of the ER or endosomal compartments is required (19), or that class I MHC molecules do not reach endolysosomal compartments in TAP-deficient DCs.
Our results show that, within minutes, a minor fraction of intact αDEC205 adduct is detectable in the cytoplasm. Several observations suggest that degradation of αDEC205 adducts involves the proteasome. First, at no time did we detect other streptavidin-reactive material corresponding to a C-terminal proteolytic fragment of αDEC205-G3-SIINFEK*L (Fig. 5C). Second, degradation of the cytoplasmic pool of αDEC205-G3-SIINFEK*L was partially inhibited by the proteasomal inhibitor ZL3VS. Even though a significant amount of αDEC205-G3-SIINFEK*L was present in the organellar (i.e., pellet) fraction at 45 min, far less was detected in the cytoplasm (i.e., supernatant; Fig. 5C). This suggests spatial and/or temporal constraints on the transfer of αDEC205-G3-SIINFEK*L to the cytoplasm. Whether protein-conducting channels of the type implicated in dislocation (27) or disintegration of a vacuolar compartment are involved in αDEC205 transfer will need further investigation.
We demonstrate that the insertion of a dipeptide linker at the N terminus of a class I MHC-restricted epitope increases its presentation by DCs in vitro and in vivo. The insertion of such motifs favors the generation of the final epitope in a proteasome-dependent fashion. We show here that αDEC205 conjugates bearing a defined dipeptide motif are fivefold more potent in inducing CD8 T-cell response upon incubation with DCs. If a strategy analogous to fusion of antigens to αDEC205 were ever considered for clinical applications, a fivefold difference in administered dose is meaningful, from both a manufacturing and a dosing perspective.
The engineering of antibody-based vaccines requires a platform that considers ease of comparison of potency. Conventional conjugation methods are less well suited to investigate this parameter in a high-throughput manner and could not easily have replaced sortagging. The method described here affords conjugation of large numbers of synthetic probes to an antibody substrate in a straightforward and robust fashion. It not only permits immunization against pathogens by using complex sets of peptides (Fig. 6), but, in an analogous manner, should also allow the induction of tolerance toward comparably large sets of epitopes through αDEC205-mediated differentiation of regulatory T cells (10). This might be of particular interest for autoimmune diseases in which self-antigens are still poorly defined. The range and number of different synthetic class I or II MHC-restricted peptides that can be conjugated to αDEC205 is in essence limitless, including peptides with nonnatural amino acids, or modified with entities that cannot be encoded genetically [e.g., CpG oligonucleotides, fluorophores, lipids, (oligo)saccharides]. The ability to evoke a CD8 T-cell response that reduces viral titers, by using a purely protein-based preparation, demonstrates the feasibility of this approach, as demonstrated here for MHV68.
Materials and Methods
αDEC204 Sortagging.
All sortase reactions were performed in 50 mM Tris-Cl, pH 7.5, 150 mM NaCl, 1 mM CaCl2. Unless specified otherwise in the figure legend, reactions were incubated for 4 h at 37 °C, and the reagents were used at the following final concentrations: αDEC205, 1 to 3 μM; nucleophile, 0.1 to 1 mM; and sortase A, 50 to 100 μM. Labeling of αDEC205 with G5GFP was performed at room temperature, overnight as shown in Fig. 2.
In Vitro Assay.
Primary DCs were obtained from C57/BL6 or BALB/c spleen and enriched by using anti-mouse CD11c magnetic beads following the manufacturer’s instructions (cat. 130-052-001; Miltenyi Biotec). BMDCs were obtained by culturing bone marrow cells from C57/BL6 or BALB/c mice during 5 to 7 d in supplemented RPMI 1640 (cat. 11875; Gibco) supplemented with 10% (vol/vol) inactivated FCS (Gibco), β-mercaptoethanol (cat. M7522; Sigma), and penicillin–streptomycin (Invitrogen, cat. 10378-016), plus recombinant GM-CSF and IL-4 (cat. 315-03 and 214-14; Peprotech). DCs or BMDCs were incubated at 1 × 106 cells per milliliter of supplemented RPMI with sortagged αDEC205 or agonist peptide for 45 min at 37 °C and washed three times with PBS solution or supplemented RPMI. A total of 0.3 to 1 × 105 DCs were then seeded in supplemented RPMI in round-bottom 96-well plates (Corning), together with 1 to 2 × 105 CFSE-labeled (final concentration, 5 μM in PBS solution; Sigma) lymph node or spleen cells from OTI rag−/−, OTII, LCMV-specific TCR transgenic mice (18) or Rop7-I (17) transnuclear mouse. After 72 or 16 h, cells were stained with CD4 or CD8α together with CD69 and propidium iodide and analyzed on a FACSCalibur device (BD Biosciences). Flow cytometry results were analyzed by using FlowJo software (TreeStar). When indicated, BMDCs were incubated with 20 μM of proteasome inhibitor ZL3VS for 1.5 h before and during incubation with sortagged αDEC205 or free peptide. When indicated, BMDCs were incubated with 5 μM of Z-FA-FMK (cat. C1480; Sigma) 1.5 h before incubation with sortagged αDEC205 or free peptide and throughout the assay.
In Vivo Experiments.
Splenic CD8 T cells from CD45.1 OTI mice were enriched by negative selection using magnetic beads (cat. 130-095-236; Miltenyi Biotec), labeled with CSFE (cat. 21888; Sigma), and transferred i.v. into 7- to 12-wk-old CD45.2 C56/BL6 mice. One day after T cells transfer, mice were immunized with sortagged αDEC205 together with 50 μg of Poly I:C (cat P0913; Sigma) and 25 μg of αCD40 (FGK1.5). At 7 d after immunization, mice were killed, and T-cell proliferation was measured by monitoring CFSE dilution by flow cytometry. For measurements of IFN-γ production, 2 × 106 splenocytes were incubated for 4 h with OVA peptide at 1 mg/mL in 96-well U-bottom plates in complete RPMI medium and in the presence of brefeldin A (GolgiPlug; cat. 555029; BD Biosciences). Cells were then stained by using a fixation/permeabilization kit (cat. 554722; BD Biosciences) according to the manufacturer’s instructions.
Dislocation Assay.
For in vitro dislocation analysis of αDEC205-G3-SIINFEK*L in semiintact cells, 13 × 106 BMDCs were treated with 20 μM ZL3VS dispensed from a DMSO stock solution or with the corresponding volume of DMSO for 45 min at 37 °C, 5% CO2. The cells were then incubated with 35 μg/mL αDEC205-G3-SIINFEK*L on ice for 10 min, washed with ice-cold PBS solution, and resuspended in RPMI media. An aliquot was taken, and the cells were immediately transferred to 37 °C, 5%CO2, for 10 and 45 min. Subcellular fractionation using perfringolysin O was performed as described previously (20).
Immunization and Infection of Mice.
MHV-68 peptide probes were sortagged to αDEC205 in separate reactions as described earlier. Reactions were pooled after 4 h, and the final product was purified by gel filtration chromatography as described in SI Materials and Methods. C57/BL6 mice (age 7–12 wk) were immunized with 20 μg of sortagged αDEC205 or an equimolar quantity of free MHV-68 peptides together with 50 μg of Poly I:C and 25 μg of αCD40 (FGK1.5) 14 and 7 d before intranasal challenge with 105 pfu of MHV-68. Viral titers were measured in lungs 6 d after infection. Infections and viral titer measurements were performed as described elsewhere (28). All described procedures were approved by the Massachusetts Institute of Technology Committee on Animal Care.
Acknowledgments
L.K.S. received a fellowship from the Swiss National Science Foundation and the Swiss Foundation for Grants in Biology and Medicine. This research was further supported by National Institutes of Health Grant AI098159.
Footnotes
The authors declare no conflict of interest.
†This Direct Submission article had a prearranged editor.
This article contains supporting information online at www.pnas.org/lookup/suppl/10.1073/pnas.1214994110/-/DCSupplemental.
References
Articles from Proceedings of the National Academy of Sciences of the United States of America are provided here courtesy of National Academy of Sciences
Full text links
Read article at publisher's site: https://doi.org/10.1073/pnas.1214994110
Read article for free, from open access legal sources, via Unpaywall:
https://www.pnas.org/content/pnas/110/4/1428.full.pdf
Citations & impact
Impact metrics
Citations of article over time
Alternative metrics
Article citations
A Chemical Approach to Assess the Impact of Post-translational Modification on MHC Peptide Binding and Effector Cell Engagement.
ACS Chem Biol, 19(9):1991-2001, 16 Aug 2024
Cited by: 0 articles | PMID: 39150956 | PMCID: PMC11420952
Episomal Vectors for Stable Production of Recombinant Proteins and Engineered Antibodies.
Antibodies (Basel), 13(1):18, 11 Mar 2024
Cited by: 0 articles | PMID: 38534208 | PMCID: PMC10967652
The enzyme activity of sortase A is regulated by phosphorylation in Staphylococcus aureus.
Virulence, 14(1):2171641, 01 Dec 2023
Cited by: 1 article | PMID: 36694285 | PMCID: PMC9928477
Enzymatic Bioconjugation: A Perspective from the Pharmaceutical Industry.
JACS Au, 3(5):1267-1283, 04 May 2023
Cited by: 5 articles | PMID: 37234110 | PMCID: PMC10207132
Review Free full text in Europe PMC
Efficient targeting of NY-ESO-1 tumor antigen to human cDC1s by lymphotactin results in cross-presentation and antigen-specific T cell expansion.
J Immunother Cancer, 10(4):e004309, 01 Apr 2022
Cited by: 5 articles | PMID: 35428705 | PMCID: PMC9014073
Go to all (43) article citations
Data
Data behind the article
This data has been text mined from the article, or deposited into data resources.
BioStudies: supplemental material and supporting data
Similar Articles
To arrive at the top five similar articles we use a word-weighted algorithm to compare words from the Title and Abstract of each citation.
Enhancement of the priming efficacy of DNA vaccines encoding dendritic cell-targeted antigens by synergistic toll-like receptor ligands.
BMC Immunol, 10:43, 03 Aug 2009
Cited by: 27 articles | PMID: 19650904 | PMCID: PMC2731061
Antigen Delivery to DEC205+ Dendritic Cells Induces Immunological Memory and Protective Therapeutic Effects against HPV-Associated Tumors at Different Anatomical Sites.
Int J Biol Sci, 17(11):2944-2956, 13 Jul 2021
Cited by: 10 articles | PMID: 34345218 | PMCID: PMC8326119
Enhanced delivery of exogenous peptides into the class I antigen processing and presentation pathway.
Infect Immun, 70(6):3249-3258, 01 Jun 2002
Cited by: 20 articles | PMID: 12011020 | PMCID: PMC128024
Applications for T-cell epitope queries and tools in the Immune Epitope Database and Analysis Resource.
J Immunol Methods, 374(1-2):62-69, 31 Oct 2010
Cited by: 31 articles | PMID: 21047510 | PMCID: PMC3041860
Review Free full text in Europe PMC
Funding
Funders who supported this work.
NIAID NIH HHS (3)
Grant ID: R01 AI087879
Grant ID: R21 AI098159
Grant ID: AI098159
NIGMS NIH HHS (1)
Grant ID: R01 GM100518





