Abstract
Free full text

IRF-8 Controls Melanoma Progression by Regulating the Cross Talk between Cancer and Immune Cells within the Tumor Microenvironment1,2
Abstract
The transcription factor interferon regulatory factor-8 (IRF-8) is crucial for myeloid cell development and immune response and also acts as a tumor suppressor gene. Here, we analyzed the role of IRF-8 in the cross talk between melanoma cells and tumor-infiltrating leukocytes. B16-F10 melanoma cells transplanted into IRF-8-deficient (IRF-8-/-) mice grow more rapidly, leading to higher numbers of lung metastasis, with respect to control animals. These events correlated with reduced dendritic cell and T cell infiltration, accumulation of myeloid-derived suppressor cells and a chemokine/chemokine receptor expression profile within the tumor microenvironment supporting tumor growth, angiogenesis, and metastasis. Noticeably, primary tumors developing in IRF-8-/- mice displayed a clear-cut inhibition of IRF-8 expression in melanoma cells. Injection of the demethylating agent 5-aza-2′-deoxycytidine into melanoma-bearing IRF-8-/- animals induced intratumoral IRF-8 expression and resulted in the re-establishment of a chemokine/ chemokine receptor pattern favoring leukocyte infiltration and melanoma growth arrest. Importantly, intrinsic IRF-8 expression was progressively down-modulated during melanoma growth in mice and in human metastatic melanoma cells with respect to primary tumors. Lastly, IRF-8 expression in melanoma cells was directly modulated by soluble factors, among which interleukin-27 (IL-27), released by immune cells from tumor-bearing mice. Collectively, these results underscore a key role of IRF-8 in the cross talk between melanoma and immune cells, thus revealing its critical function within the tumor microenvironment in regulating melanoma progression and invasiveness.
Introduction
Cancer progression and invasiveness are complex multistep phenomena involving continuous interactions between host and cancer cells, preferentially occurring within the tumor microenvironment [1]. Melanoma is a very aggressive tumor with a high metastatic potential, known to be highly resistant to conventional chemotherapy, immunotherapy, and targeted therapy [2]. Melanoma cells can secrete immunomodulatory factors that edit an intratumoral milieu that suppresses immunosurveillance mechanisms, thus enabling tumor progression [3]. During melanoma carcinogenesis, the activity of immune infiltrates, such as dendritic cells (DCs) and CD4+ and CD8+ T lymphocytes, is crucially affected by the prevalence of immunosuppressive signals within the tumor microenvironment and by the accumulation of suppressive immune populations, such as myeloid-derived suppressor cells (MDSCs) or regulatory T cells (Tregs) [4]. However, melanoma cells secrete a number of chemokines that promote tumor angiogenesis [5]. In the recent past, chemokines have also emerged as major determinants of melanoma metastasis [6]. Although it is generally accepted that local interactions between resident and infiltrating cells play a key role in melanoma development, the fine mechanisms generating the continuous alterations of the tumor microenvironment sustaining cancer progression need to be further elucidated.
The transcription factor interferon regulatory factor-8 (IRF-8), a member of the IRF family, plays a dual role in antitumor response by modulating, on the one hand, immune responses and, on the other hand, cell growth and differentiation of various tumor cells [7]. IRF-8 expression governs myeloid cell developmental program, and mice deficient for this factor (IRF-8-/-) display substantial defects in monocyte and DC differentiation and activity, being devoid of plasmacytoid DCs (pDCs) and exhibiting functional alterations of CD8α+ and CD8α- DCs [8–10]. Noteworthy, IRF-8 plays a direct role in tumor development, as it confers resistance to apoptosis in myeloid populations and its deficiency supports the development of a chronic myelogenous leukemia (CML)-like syndrome in mice [11,12]. Of interest, the role of IRF-8 in solid tumor cell biology has also emerged [7,13]. Loss of IRF-8 has been frequently detected in a large collection of primary carcinomas and the suppression of its function has been correlated to enhanced metastatic potential of sarcoma cells [14,15]. These findings have defined IRF-8 as a tumor suppressor gene, although the exact mechanisms by which it operates as well as its role in melanoma biology remain elusive. By using IRF-8-/- mice transplanted with B16-F10 melanoma cells, we here report a previously unrecognized role of IRF-8 in regulating melanoma progression and metastatic process through the active control of cancer and immune cell cross talk within the tumor microenvironment.
Materials and Methods
Cell Lines
B16-F10 murine melanoma cell line and the human cell line Colo-38 were purchased, respectively, from American Type Culture Collection-LGC (ATCC-LGC, Milan, Italy; CRL-6475) and Cell Lines Service (CLS, Eppelheim, Germany; 300151). The cell line M-14 was part of the NC160 cell lines and was obtained from Dr Susan Holbeck (National Cancer Institute). The cell lines Alo-39, PES-41, PES-43, and PES-47 were generated from metastatic melanoma lesions by Dr Lombardi in the Laboratory of Immunology, “G. Pascale Foundation” (Naples, Italy) [16]. Ovalbumin (OVA)-expressing B16 melanoma cells (B16.OVA) were kindly provided by Dr Laurence Zitvogel (Institut Gustave Roussy, Villejuif, France). Parental B16-F0 melanoma cells were a generous gift from Dr Elena Pagani (Istituto Dermopatico dell'Immacolata, Rome, Italy). The cells were routinely tested for morphology, growth curve, and absence of mycoplasma and passaged for no more than three times from thawing.
Mice and In Vivo Treatments
IRF-8-/- were generated as described [11] and backcrossed on a C57BL/6 background. C57BL/6 IRF-8-/- and wild-type (WT) mice were housed in the animal facility at the Istituto Superiore di Sanità (Rome, Italy) and manipulated in accordance with the local Ethical Committee guidelines. B16-F10 or B16.OVA melanoma cells were injected subcutaneously (s.c.; 0.8 x 106). Where indicated, mice were injected intraperitoneally (i.p.) with 250 µg/kg of 5-aza-2′-deoxycytidine (5-Aza-dC; Sigma, St Louis, MO) dissolved in 200 µlof phosphate-buffered saline (PBS). The control group consisted of mice injected with 200 µl of PBS. For induction of experimental pulmonary metastasis, mice were injected intravenously with 1.5 x 106 B16-F10 cells and sacrificed 5 days later for counts of metastatic foci in lungs.
Histologic Analysis
Tumors were excised with scissors and the inner part of the lesions was taken. Explanted melanoma tissues were fixed with 4% para-formaldehyde for 24 hours, then embedded in paraffin, sectioned at 5-µm thickness, and stained with hematoxylin and eosin (H&E).
Analysis of Immune Cell Infiltrate in Melanoma
For fluorescence-activated cell sorting (FACS) analysis, melanoma explants were cut into small fragments using curved scissors and then digested in type III collagenase-containing medium (1 mg/ml; Worthington Biochemicals, Lakewood, NJ) for 30 minutes at room temperature in agitation, followed by EDTA (0.1 M, pH 7.2) for additional 5 minutes. The homogenate was then passed through a cell strainer and the resulting cell suspension was treated with lysis buffer (140 mM NH4Cl, 17 mM Tris HCl, pH 7.2) to eliminate red blood cells. Cells were then stained with peridinin-chlorophyll-protein complex (PerCP) anti-CD45, allophycocyanin anti-CD11c, fluorescein isothiocyanate (FITC) anti-CD8a, PerCP or biotin anti-CD4, phycoerythrin (PE) anti-CD357/GITR, PE anti-CD124/interleukin-4 (IL-4R), FITC anti-Foxp3, biotin anti-CD45R/B220, biotin anti-CD40, biotin antiCD80, biotin anti-CD86, biotin anti-I-A/I-E (all from BD Pharmingen, San Diego, CA), allophycocyanin anti-CD25, allophycocyanin anti-Gr-1, and FITC anti-CD11b (Miltenyi Biotec, Bergisch Gladbach, Germany). Biotinylated antibodies were detected by streptavidin PE (BD Pharmingen). The CD45 labeling allowed discriminating between leukocytes (CD45+ cells) and tumor tissue cells, composed of stromal cells and B16 melanoma cells. T lymphocytes were detected as CD4+ or CD8+ CD45+CD11c-. Total tumor-infiltrating DCs were defined as CD45+CD11c+I-A+, plasmacytoid DCs as CD45+CD11lowB220+ CD11b-, MDSCs as CD45+CD11b+Gr-1+CD124/IL-4R+, and Tregs as CD4+CD25+CD357/GITR+Foxp3+. For confocal laser scanning microscopy (CLSM) analysis, melanoma tissues grown in WT and IRF-8-/- mice were snap-frozen, embedded in optimal cutting temperature compound (Tissue-Tek), and stored at -80°C. Frozen tumor sections (5 µm tick) were then obtained and fixed in cold acetone. Sections were stained with FITC anti-CD11c antibody combined with biotin anti- I-Ad/I-Ed for detection of DCs or with biotin anti-mPDCA1 (Miltenyi Biotec) for detection of pDCs. T lymphocytes were detected by labeling with biotin anti-CD3 antibody in combination with FITC-conjugated anti-CD4 or anti-CD8. Biotin-conjugated monoclonal antibodies were detected with streptavidin-Alexa Fluor 594 (Invitrogen, Carlsbad, CA). Nuclei were visualized with 4′,6-diaminidino-2-phenylindole (DAPI) (1 µg/ml). CLSM observations were performed with a Leica TCS SP2 AOBS apparatus, using 405-, 488-, 594-nm excitation spectral laser lines appropriately tuned by acousto-optical tunable filter. Signals from different fluorescent probes were taken in sequential scan settings, and co-localization was detected in yellow.
Proliferation Assays
Tumor-infiltrating leukocytes from B16.OVA-bearing mice were isolated by magnetic cell sorting using anti-CD45 microbeads (Miltenyi Biotec). CD45+ cells (>80% pure; 3 x 105) and spleen cells (3 x 106) from B16.OVA-bearing mice were seeded in 96-well round-bottomed plates in the presence or absence of graded amounts of OVA protein or of major histocompatibility complex (MHC) class I-restricted SIINFEKEL peptide of OVA (1 µM). Cell cultures were incubated in complete medium for 3 days at 37°C in 5% CO2 and then pulsed with 3H-thymidine (1 µCi/well) for 16 hours. Incorporation of 3H-thymidine was analyzed by liquid scintillation counting.
Methylation-specific Polymerase Chain Reaction
Genomic DNA was purified from tumor cells by using the salting out method [17]. Sodium bisulfite treatment of genomic DNA to convert unmethylated cytosine to thymidine was carried out using CpGenome Universal DNA Modification Kit (Chemicon, Temecula, CA) according to the manufacturer's instructions. Methylation-sensitive polymerase chain reaction (PCR) primers were designed by using the Methyl Primer Express software. The PCR sequences, targeting a CpG island within the IRF-8 promoter region (Figure W1), are the following: unmethylated forward primer, 5′-AGGATTATTGAGTTTGAAAGAGTG-3′; unmethylated reverse primer, 5′-TTTTCCTACTACTATCACATAAAAA-3′;methylatedforward primer,5′-ATTATCGAGTTTGAAAGAGCG-3′; methylated reverse primer 5′-TCCTACTACTATCGCGTAAAAA-3′. These primers amplify a 179-bp sequence inside the mentioned CpG island.
Quantitative Reverse Transcription-PCR
Total RNA was extracted from tumor tissue by using TRIzol reagent (Invitrogen). Messenger RNA was reverse transcribed by means of Verso cDNA Kit (Thermo Scientific, Waltham, MA). Quantitative reverse transcription-PCR (qRT-PCR) was performed using Sensimix Plus SYBR Kit containing the fluorescent dye SYBR Green (Quantace, Watford, United Kingdom). Forward and reverse primers (Table W1) were purchased from Primm (Milan, Italy). The conditions of real-time PCR reaction were given as follows: 15 seconds at 95°C, 30 seconds at 60°C, and 45 seconds at 72°C (45 cycles). PCR products were continuously measured by means of an ABI 7500 Real-Time PCR System (Applied Biosystems, Carlsbad, CA). Quality and specificity of amplicons in each sample was detected by dissociation curve analysis. Triplicates were performed for each experimental point. For quantization, threshold cycle (CT) values were determined by the Sequence Detection System software (Applied Biosystems), and ΔCT was obtained by subtracting CT of reference gene, β-actin, to CT of target gene. Gene expression was presented as relative amount of mRNA normalized to β-actin and was calculated as 2-ΔCt, a modification of the 2-ΔΔCt method [18].
Protein Extraction and Western Blot Analysis
Proteins were extracted from melanoma bulks at different tumor progression stages or from B16-F10 cells, untreated or treated with interferon-γ (IFN-γ, 100 ng/ml) as positive control. In addition, proteins were also purified from melanoma lesions grown in IRF-8-/- mice treated with PBS or 5-Aza-dC. Briefly, 10 x 106 tumor cells were incubated for 30 minutes on ice in lysis buffer [50 mM Tris HCl (pH 8.0), 120 mM NaCl, 0.25% NP-40, 0.1% sodium dodecyl sulfate; Sigma] containing the protease inhibitors PMSF, aprotinin, leupeptin, and pepstatin (Roche, Mannheim, Germany), each at a final concentration of 10 ng/ml, and 1 mM DTT (Sigma). A quantity of 30 µg of each protein sample was loaded onto a 10% sodium dodecyl sulfate-polyacrylamide gel electrophoresis gel. Following separation, proteins were blotted onto a nitrocellulose membrane (Whatman). Membranes were blocked with 5% nonfat dry milk in PBS-0.5% Tween 20 and then probed with rabbit anti-mouse IRF-8 polyclonal antibody (Santa Cruz Biotechnology, Santa Cruz, CA) or anti-mouse β-tubulin monoclonal antibody (Sigma), followed by HRP-conjugated anti-rabbit IgG antibody or anti-mouse IgG antibody (Amersham Pharmacia Biotech, Uppsala, Sweden). Immunoreactive protein bands were detected by using the ECL Detection Kit (Amersham Pharmacia Biotech).
Cytokines
Recombinant murine IL-3, IL-6, IL-10 (ProSpec, East Brunswick, NJ), and interleukin-27 (IL-27; Cell Signaling Technology, Beverly, MA) were used at 50 ng/ml.
Transwell Co-cultures of B16-F10 Melanoma/Immune Cells
Spleen cells (2 x 106) from naïve or melanoma-bearing WT and IRF-8-/- mice, at 19 days after tumor implant, were plated on the upper compartment of 0.4-µmporesizetranswell plates (Costar) in 0.1 ml of RPMI complete medium. B16-F10 melanoma cells (0.3 x 106 in 0.6 ml) were placed on the lower chamber. Co-cultures were incubated for 24 hours at 37°C and 5% CO2, after which B16 cells were harvested and total RNA was extracted. Samples were then assayed for IRF-8 expression by qRT-PCR. Supernatants from co-cultures were harvested and tested for cytokine release using RayBio mouse cytokine antibody array membranes (Raybiotech, Norcross, GA) following the manufacturer's instructions. Quantitative analysis of protein expression was performed by using ImageJ software. Levels of IL-27 were determined by ELISA (eBioscience, San Diego, CA).
Statistical Analysis
Levels of significance for comparison between samples were determined by the Student's t test. P values minor to .05 were considered statistically significant.
Results
IRF-8 Host Deficiency Supports Tumor Growth and Metastatic Potential of Melanoma
To investigate the role of IRF-8 in melanoma growth control, we transplanted B16-F10 melanoma cells into IRF-8-/- and WT mice. As shown in Figure 1A, melanoma grew significantly faster in IRF-8-/- with respect to WT hosts. These observations paralleled with a decreased survival time of tumor-bearing IRF-8-/- mice (16.8 ± 4.2 days; Figure 1B) compared to that observed in the WT counterparts (30.6 ± 8.9 days, Figure 1B). Because B16-F10 cells are also endowed with the ability to metastasize lung tissue when injected intravenously into mice [19], we asked whether IRF-8 host deficiency could affect melanoma cell metastasization. Remarkably, we found a large amount of metastatic foci in lungs of IRF-8-/- mice with respect to the WT counterparts as early as 5 days after injection of B16 cells (Figure 1C). These results indicate that IRF-8 deficiency promotes both melanoma growth and metastatic process.
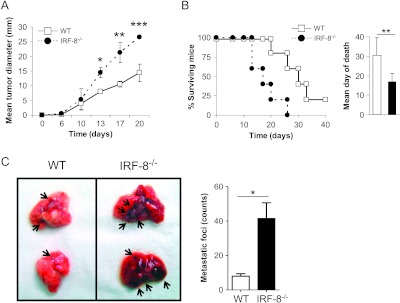
Melanoma progression and metastasis development are highly increased in IRF-8-/- mice. (A) WT and IRF-8-/- mice were injected s.c. with 0.8 x 106 B16-F10 melanoma cells and tumor size was measured. Data represent the mean tumor diameter ± SD. One representative experiment of seven is shown. (B) Kaplan-Meyer plot representing percent of surviving mice. Surviving mice after 40 days (all with tumor) were humanely killed. Histograms depict mean day of death (n = 6–10 mice) ± SD. (C) WT and IRF-8-/- mice were injected intravenously with B16-F10 cells (1.5 x 106) and sacrificed 5 days later. (Left) Photograph of representative isolated lungs showing metastatic foci (indicated by black arrows). (Right) Counts of lung metastatic foci. Histograms represent the mean number of metastatic foci per lung (n = 10 mice) ± SD. One representative experiment of two is shown. *P < .05, **P < .01, ***P < .001.
Impaired Immune Cell Infiltration in Melanoma-bearing IRF-8-/- Mice
We asked whether the inability to control melanoma growth in IRF-8-/- mice could reflect a defect in immune cell infiltration within the tumor burden. Staining of tumor tissue sections with H&E revealed the presence of little dense spots corresponding to leukocyte nuclei in melanoma from IRF-8-/- mice (IRF-8-melanoma) with respect to that from WT animals (WT-melanoma; Figure 2A, red arrows). Of note, a consistent number of blood vessels indicative of a major vascularization was observed in IRF-8-melanoma sections compared to WT ones (Figure 2A, yellow arrows). In addition, significantly higher expression of the angiogenic markers vascular endothelial growth factor A (VEGF-A), VEGF-B, and VEGF-R2 was found in IRF-8-melanoma, with respect to WT-melanoma (Figure W2). Flow cytometry analysis of tumor infiltrates at day 19 post-implantation (p.i.) evidenced a 3.5-fold lower percentage of CD45+ infiltrating leukocytes in IRF-8-melanoma compared to WT-melanoma (Figure 2B; P < .001). Furthermore, CLSM of leukocyte populations revealed little infiltration of DCs and none of pDCs in IRF-8-melanoma sections, with respect to the WT counterparts (Figure 2C). Notably, the few DCs detected in IRF-8-melanoma displayed an immature phenotype, revealed by lack of MHC-II co-expression (Figure 2C). CD4+ and CD8+ T cells were also barely detectable in IRF-8-melanoma, while these cells were present in WT-melanoma (Figure 2C).
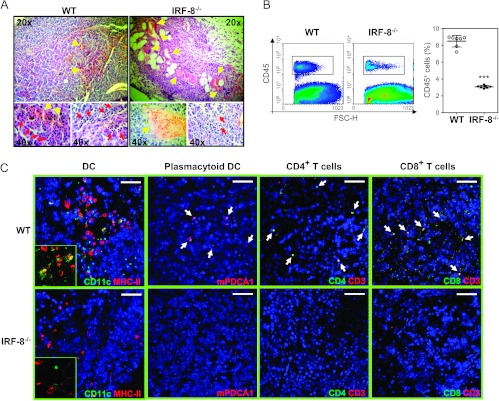
Defective leukocyte infiltration in melanoma-bearing IRF-8-/- mice. (A) H&E staining of frozen melanoma sections from IRF-8-/- and WT mice at early stage (12-mm mean diameter). Yellow arrows depict blood vessels; red arrows indicate immune infiltrates. (B) FACS analysis of total infiltrating CD45+ leukocytes. Scatter plots indicate mean percent values of each experiment ± SD. ***P < .001. (C) CLSM analysis of immune infiltrates in IRF-8-melanoma and WT-melanoma sections. DCs are represented by co-localization (yellow) of CD11c and MHC-II expression; pDCs (white arrows) are depicted by mPDCA1 expression; CD4 and CD8 T cells are shown by coexpression of CD3 and CD4 or CD8 markers, respectively (white arrows). Inserts represent high magnification portions of the fields displayed. Bars correspond to 100 µm. One representative experiment of two is presented.
Because IRF-8-/- and WT mice display different tumor growth kinetics, we addressed whether the extent of immune cell infiltration in the two strains was associated with the tumor burden. Thus, we explanted melanoma from tumor-bearing IRF-8-/- and WT at three stages of progression, when tumor size was 10 to 12 mm (early), 20 mm (medium), or 28 mm (late). At early and medium stages, CD4+ T lymphocytes were present at comparable levels, whereas CD8+ T cells were slightly decreased in IRF-8-melanoma with respect to the WT counterparts (Figure 3A). At late stages of tumor progression, the percentages of both CD4+ and CD8+ T cells dropped dramatically in IRF-8-melanoma, whereas they remained constantly present in WT-melanoma (Figure 3A). Moreover, few CD11c+ DCs infiltrated IRF-8-melanoma ever since early stages, whereas these cells were found at all stages in WT-melanoma bulks (Figure 3B). As expected, pDCs (CD11clowB220+) were almost completely absent in IRF-8-melanoma, being IRF-8-/- mice devoid of this DC subset (Figure 3C) [8]. In contrast, pDCs were detected in WT-melanoma at all stages of tumor progression (Figure 3C). Tumor-infiltrating DCs from WT host, but not those from IRF-8-/- animals, displayed a mature phenotype, shown by expression of co-stimulatory molecules at the three stages of tumor progression (Figure 3D). In contrast to what was observed with effect or immune cells, we found much larger numbers of infiltrating MDSCs within IRF-8-melanoma with respect to WT-melanoma (Figure 4A). Higher frequencies of MDSCs were also observed in the spleens of melanoma-bearing IRF-8-/- with respect to WT mice, thus supporting a systemic suppressive environment in these animals (Figure 4B). Tregs were found slightly decreased in melanoma-bearing IRF-8-/- mice compared to the WT counterparts, both in tumor bulk and spleen (Figure W3). To evaluate whether the suppressive immune compartment in IRF-8-/- mice resulted in impaired antitumor response, we transplanted IRF-8-/- and WT mice with B16.OVA cells and analyzed the response of CD45+ tumor-infiltrating leukocytes and spleen cells to OVA recall in vitro. Strikingly, no proliferative response was observed with either tumor-infiltrating CD45+ (Figure 4C) or spleen cells (Figure 4D) from IRF-8-/- mice to OVA stimulation. In contrast, WT immune cells from both tumor and spleen exhibited significant proliferation in response to OVA (Figure 4, C and D). Thus, IRF-8 host deficiency determines defective tumor infiltration of T lymphocytes and mature DCs but high frequencies of MDSCs in melanoma-bearing mice that result in a defective antitumor response, thus suggesting a role for these immune cells in controlling melanoma growth.
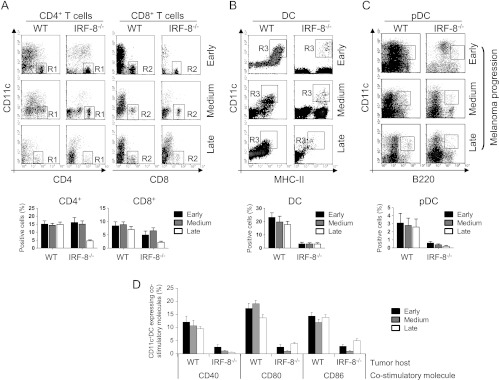
T lymphocytes and DC trafficking is impaired in IRF-8-melanoma lesions during tumor progression. Melanoma lesions from WT and IRF-8-/- mice were excised at various stages of growth. (A) FACS analysis of infiltrating CD4+ and CD8+ T lymphocytes gated on total leukocytes (CD45+) as CD11c-CD4+ (R1 region) and CD11c-CD8+ (R2 region) T cells, respectively. (B) Representative analysis of infiltrating DCs. R3 region represents CD11c+MHC-II+DC gated on CD45+ cells. (C) Representative analysis of infiltrating pDCs. R4 region represents CD11clowB220+ pDC gated on CD45+ cells. Histograms below each panel depict mean percentage values of the various infiltrating immune cell populations among total CD45+ cells in each group (n = 18 mice) ± SD. (D) Phenotype of DCs. Data represent the percentage of CD45+CD11c+MHC-II+ DC expressing the indicated co-stimulatory molecules (mean values ± SD). One representative experiment of three is shown.
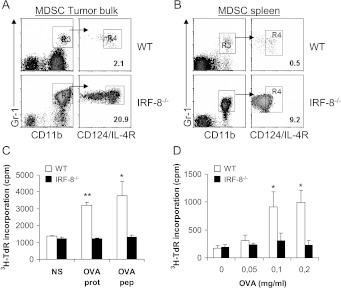
Increased frequency of MDSC and impaired T cell response in tumor and spleen of melanoma-bearing IRF-8-/- mice. (A, B) Melanoma-bearing IRF-8-/- and WT mice were sacrificed at early tumor stage (12-mm mean diameter). FACS analysis of MDSCs was performed in tumor bulks (A) and spleens (B). Left-side dot plots show cell population CD45+ gated; right-side plots show the population gated as indicated by the arrow. Values depicted refer to percent of positive cells over total CD45+ leukocytes. One representative experiment of three is shown. (C, D) IRF-8-/- and WT mice transplanted with B16.OVA tumors were sacrificed after 17 days and tumors and spleens were harvested. (C) CD45+ tumor-infiltrating leukocytes were cultured for 4 days with 1 µM SIINFEKL OVA peptide, 0.2 mg/ml OVA protein, or medium alone. Proliferative response was measured by pulsing with 3H-thimydine for the last 16 hours of culture. Data are expressed as mean cpm ± SD of culture triplicates. (D) Proliferative response of spleen cells after incubation with graded amounts of OVA protein. 3H-thimydine incorporation was measured on the fourth day of culture. Data are expressed as mean cpm ± SD of culture triplicates. One representative of two is shown. *P < .05; **P < .01 versus unstimulated.
Establishment of a Chemokine/Chemokine Receptor Pattern Supporting Tumor Growth in IRF-8-melanoma
Next, we investigated the intratumoral expression by resident and infiltrating cells of selected chemokines and chemokine receptors involved in immune cell trafficking, tumor angiogenesis, and metastasis [20–22]. To this end, we sorted CD45+ cells, representative of the whole infiltrating leukocyte population and CD45- cells, representative of the bulk of melanoma cells, and stromal resident cells from IRF-8-melanoma and WT-melanoma. WT-melanoma showed elevated levels of CCL19 and CCL21 in the CD45- fraction and of their receptor CCR7 in the CD45+ fraction (Figure 5). In contrast, this axis, known to recruit DCs and T cells [22], was completely inhibited in IRF-8- melanoma (Figure 5). Furthermore, the DCs and T cell-attracting chemokines CCL20, CCL5, CCL27, andCXCL10werehighly expressed in both CD45- and CD45+ fractions of WT-melanoma, while they were significantly suppressed in IRF-8-melanoma (Figure 5) [23–25]. Of note, CCR5, the receptor for CCL5 known to be associated with CD8+ T cell cytotoxicity in response to antigen presentation by DCs [26], was significantly more expressed in the CD45+ fraction of WT-melanoma with respect to the IRF-8 counterpart (Figure 5). With regard to chemokine and chemokine receptors involved in metastasis and angiogenesis, we found CCR10, which promotes escape from host immunosurveillance and tumor growth [27,28], expressed at high levels selectively in the CD45- fraction of IRF-8-melanoma (Figure 5). Moreover, the angiostatic axis CXCL10/CXCR3 [29,30] was significantly inhibited in IRF-8-melanoma but highly activated in WT-melanoma (Figure 5). Conversely, the angiogenic CXCL1 and CX3CL1 chemokines [31] were highly expressed in the CD45- fraction of IRF-8-melanoma (Figure 5). Together, these data suggest a close correlation between host IRF-8 expression and a pattern of chemokines/chemokine receptors that control immune cell infiltration and tumor progression.
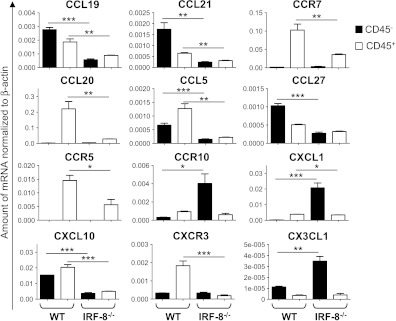
Altered chemokine and chemokine receptor expression in IRF-8-melanoma lesions. Melanoma was excised from WT and IRF-8-/- mice (n = 6) at medium stage (20-mm mean diameter); cell suspensions were sorted into CD45+ and CD45- cell fractions and RNA was purified. qRT-PCR for the indicated chemokines or chemokine receptors was carried out. Histograms represent mean mRNA expression normalized to β-actin in each sample run in triplicate (mean values ± SD). One representative experiment of three is shown. *P < .05, **P < .01, ***P < .001.
IRF-8 Intratumoral Expression Is Suppressed in IRF-8-melanoma and in Human Metastatic Melanoma
As IRF-8 acts as a tumor suppressor gene in different types of tumors, we asked whether IRF-8 host deficiency could also influence the intrinsic expression of IRF-8 in melanoma. To this end, we analyzed the intratumoral expression of this transcription factor in IRF-8-melanoma and WT-melanoma at early, medium, and late stages of tumor progression. Strikingly, no IRF-8 expression, either mRNA (Figure 6A) or protein (Figure 6B), was observed in IRF-8-melanoma at the three stages of growth analyzed. In contrast, WT-melanoma displayed high IRF-8 expression at early stage, progressively decreasing at medium to late stages, indicating an inverse correlation between IRF-8 expression and melanoma progression in immunocompetent mice (Figure 6, A and B). Importantly, B16-F10 cells expressed basal levels of IRF-8 before injection, meaning that this factor is upregulated when melanoma cells are transplanted into WT mice but not into IRF-8-/- hosts (Figure 6A). We verified IRF-8 expression in sorted CD45+ cells and CD45- cells. Remarkably, IRF-8 expression was strongly suppressed in the CD45- fraction of IRF-8-melanoma, whereas substantial mRNA levels were observed in the same fraction of WT-melanoma (Figure 6C). As expected, no IRF-8 mRNA expression was found in the CD45+ fraction from IRF-8-melanoma, whereas it was detected at very high levels in the same fraction from WT-melanoma (Figure 6C).
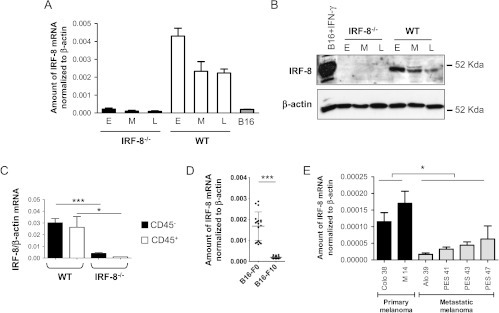
IRF-8 expression is inhibited in IRF-8-melanoma and human metastatic melanoma. (A) IRF-8 mRNA expression in melanoma bulk from WT and IRF-8-/- mice at various stages of growth and in B16-F10 cells before transplantation. Histograms represent mean expression values normalized to β-actin in each group (18 mice) ± SD. (B) IRF-8 protein expression in melanoma lysates from WT and IRF-8-/- hosts. β-Tubulin was used as normalization control for IRF-8 expression (E, early; M, medium; L, late stage of tumor growth). IFN-γ-treated B16 cells were used as positive control. One representative experiment of two is shown. (C) IRF-8 mRNA expression in sorted CD45+ and CD45- fractions from melanoma-bearing mice at medium stage (20-mm mean diameter). Histograms represent mean mRNA expression normalized to β-actin in each sample run in triplicate (± SD). One representative experiment of three is shown. (D) Expression levels of IRF-8 mRNA in non-metastatic (B16-F0, n = 18) or metastatic (B16-F10, n = 18) murine melanoma cells. Bars in the plot represent mean values of IRF-8 expression normalized to β-actin in the indicated group. (E) Expression levels of IRF-8 mRNA in the indicated human primary and metastatic melanoma cells. Histograms represent mean values of IRF-8 mRNA expression normalized to β-actin. *P < .05; ***P < .001.
To further determine whether down-regulation of IRF-8 expression in B16 melanoma correlates with tumor growth and metastasis, we analyzed the expression levels of IRF-8 in the non-metastatic B16-F0 melanoma cell line [32] compared to that in metastatic B16-F10 cells. Consistently, B16-F0 cells exhibited significantly higher expression of IRF-8 with respect to B16-F10 cells (Figure 6D). We asked whether also in human melanoma cells with diverse grade of malignancy IRF-8 was differentially expressed. To this end, we analyzed IRF-8 mRNA levels in Colo-38 and M14, originated by primary tumors [33], and Alo-39, PES-41, PES-43, and PES-47, generated from metastatic lesions [16]. Of interest, the metastatic melanoma cell lines exhibited significantly lower expression of IRF-8 with respect to the primary melanomas (P < .05, Figure 6E). Together, these observations strongly suggest a strict correlation between intrinsic IRF-8 expression and the metastatic phenotype of melanoma cells.
Induction of IRF-8 Expression in Melanoma Cells Reactivates Immune Cell Infiltration and Stimulates Tumor Regression
We investigated whether the induction of IRF-8 into IRF-8-melanoma could influence chemokine/chemokine receptor profiles, immune cell infiltration, and tumor progression. To this end, we took advantage of 5-Aza-dC, a DNA methyltransferase inhibitor known to upregulate IRF-8 expression in malignant cells by epigenetic mechanisms [34]. A single injection of 5-Aza-dC in tumor-bearing IRF-8-/- mice induced IRF-8 expression in cancer cells as early as 24 hours p.i. (Figure 7A). 5-Aza-dC-induced IRF-8 expression was associated to demethylation of a CpG island inside the IRF-8 promoter (Figures 7B and W1). In contrast, IRF-8 expression in WT-melanoma was not affected by treatment with 5-Aza-dC in accordance with the unmethylated status of melanoma DNA in both 5-Aza-dC-treated and untreated WT mice (Figure W4). We then investigated whether 5-Aza-dC could influence tumor infiltration in IRF-8-/- mice. Noteworthy, IRF-8-melanoma from 5-Aza-dC-treated mice displayed large infiltration of T lymphocytes, CD4+ and CD8+ (Figure 7C), and DCs (Figure 7D). Moreover, following 5-Aza-dC treatment, the axes CCL21/CCR7 and CCL5/CCR5 were fully reactivated in IRF-8-melanoma (Figure 7E). Likewise, the expression of CCL20, CCL27, and CXCL10 was strongly upregulated in melanoma of these mice (Figure 7E). In addition, 5-AzadC induced up-regulation of the angiostatic axis CXCL10/CXCR3 in the CD45+ fraction and down-regulation of the angiogenic chemokine CXCL1 and of CCR10 in the CD45- fraction (Figure 7E). Of note, 5-Aza-dC promoted a significant expression of CCR10 selectively in the CD45+ fraction of IRF-8-melanoma, in accordance with high levels of its ligand CCL27, thus suggesting the activation of this T cell inflammation axis following drug treatment [35] (Figure 7E). All these changes correlated with a significant tumor regression in 5-Aza-dC-injected IRF-8-/- mice (Figure 7F). Forty-eight hours after 5-Aza-dC administration, melanoma resumed its growth propensity, which could then again be blocked by a new 5-Aza-dC injection (Figure 7F). Overall, these observations indicate a strict correlation between IRF-8 expression in melanoma cells and the establishment of appropriate chemokines and chemokine receptor profiles at the tumor site that control immune infiltration and tumor growth.
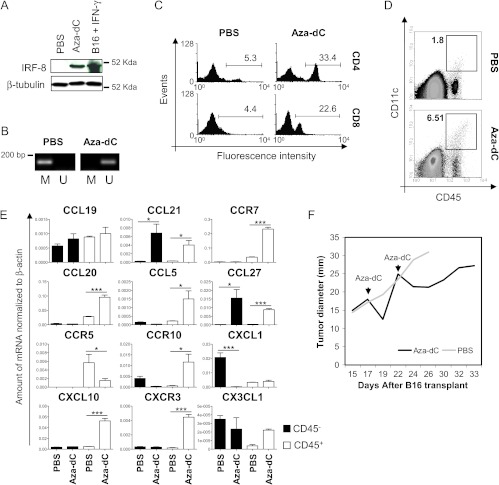
Induction of intratumoral IRF-8 expression by 5-Aza-dC reactivates immune cell infiltration and leads to melanoma regression in IRF-8-/- hosts. Melanoma-bearing IRF-8-/- mice (n = 8) were injected i.p. with 5-Aza-dC or PBS at 17 days after tumor implant. (A) Western blot analysis of intratumoral IRF-8 expression (24 hours p.i.) and of IFN-γ-treated B16 (positive control). (B) DNA methylation assay specific for IRF-8 promoter region in IRF-8-melanoma at the indicated experimental conditions (24 h p.i.). M, methylation primers; U, unmethylation primers. (C) FACS analysis of infiltrating CD4+ and CD8+ T cells. Numbers in plots represent the percentage of CD4+ or CD8+ T cells among CD45+-gated leukocytes. (D) FACS analysis of DCs. Numbers in dot plots represent the percentage of CD45+CD11c+ cells in tumor bulks. (E) Sorted CD45+ and CD45- fractions from melanoma explants were subjected to qRT-PCR analysis for the indicated chemokine or chemokine receptors. Histograms represent the relative mRNA amount normalized to β-actin in samples run in triplicate (mean values ± SD). *P < .05, ***P < .001. (F) Mean tumor diameter of mice treated with 5-Aza-dC or PBS (n = 5–8). One representative experiment of three is shown.
Immune Factors Secreted by Splenocytes of Melanoma-bearing Mice Affect IRF-8 Expression in B16-F10 Cells
To investigate the interplay between cancer and immune cells, we assessed whether IRF-8 expression in melanoma cells could be directly modulated by the release of soluble mediators from the host immune system. Thus, we co-cultured splenocytes from melanoma-bearing IRF-8-/- or WT mice (day 19) in transwell plates with B16-F10 melanoma cells and analyzed mRNA IRF-8 expression in B16 cells after 24 hours. As shown in Figure 8A, IRF-8 expression in B16 cells was significantly upregulated by splenocytes from tumor-bearing WT mice with respect to B16 cultured in medium alone. In contrast, soluble factors released from IRF-8-/- splenocytes failed to upregulate IRF-8 expression in B16 melanoma cells (Figure 8A). Of interest, IRF-8 expression was not induced when B16 cells were cultured with splenocytes from naïve mice, either WT or IRF8-/-, indicating that the immune cell-secreted factors were induced in response to melanoma implant in mice (Figure 8A). To seek for candidate soluble mediators responsible for IRF-8 up-regulation, we measured an array of cytokines and growth factors in supernatants from splenocytes/B16 cell co-cultures (Figure W5). Among the 96 soluble factors tested, we found three cytokines differentially expressed selectively in splenocytes from tumor-bearing WT mice, with respect to tumor-bearing IRF-8-/-, naïve WT, and naïve IRF-8-/- mice (Figure W6). These factors were IL-3, IL-6, and IL-10 (Figure 8B). Specifically, the protein levels of IL-3 and IL-6 were up-modulated, whereas IL-10 was down-modulated selectively in co-cultures containing splenocytes of tumor-bearing WT mice with respect to the other conditions (Figure 8B). Because IL-27 was shown to regulate IRF-8 expression in B16 melanoma cells [36], we also tested the levels of this cytokine in the co-cultures and found higher levels in supernatants from splenocytes of tumor-bearing WT mice with respect to the IRF-8-/- counterpart (Figure 8C). We then examined the ability of IL-3, IL-6, IL-10, and IL-27 to directly modulate IRF-8 expression in B16 melanoma and found that only IL-27 was able alone to induce IRF-8 in B16 cells (Figure 8D). Of note, a synergistic effect was observed when IL-27 was combined with IL-6 (Figure 8D). Addition of IL-10 did not significantly inhibit IL-27/IL-6 combined action and IL-3 did not affect IRF-8 expression (Figure 8D). To establish a correlation between intratumoral IRF-8 expression and IL-27 and IL-6 in vivo, we analyzed the expression of the two cytokines in the CD45+ and CD45- fractions of IRF-8-melanoma and WT-melanoma. Remarkably, both cytokines were significantly more expressed in the CD45+ fraction of WT-melanoma than in the IRF-8-/- counterpart (Figure 8E). Together, these data demonstrate that immune cells are endowed with the ability to control intratumoral IRF-8 expression through the secretion of soluble mediators, among which IL-27 and IL-6, and imply IRF-8 as a potential determinant for the cross talk between immune cells and melanoma.
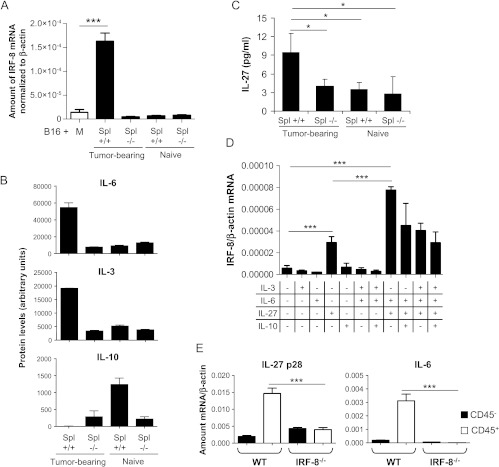
IRF-8 expression in B16 melanoma cells is modulated by immune cell-derived soluble factors. (A) Spleen cells of melanoma-bearing or naïve WT and IRF-8-/- mice (n = 6) were co-cultured with B16 cells in a 0.4-µm pore size transwell culture system. After 24 hours, B16 cells were collected, total RNA was purified, and qRT-PCR for IRF-8 mRNA was performed. Histograms represent the mean values of IRF-8 mRNA expression of triplicate samples normalized to β-actin ± SD. Spl, spleen cells; M, medium alone. One representative experiment of two is shown. (B) Cytokines from supernatants of spleen cell-B16 melanoma co-cultures, as in A, were measured using a commercial protein array and quantified using ImageJ software. Values are expressed in arbitrary units. (C) IL-27 release in the co-cultures, as measured by ELISA. (D) IRF-8 mRNA expression in B16 melanoma cells after 24 hours culture with the indicated cytokines. Histograms represent the mean values of IRF-8 mRNA expression of triplicate samples normalized to β-actin ± SD. One representative experiment of two is shown. (E) IL-27 p28 and IL-6 mRNA expression in sorted CD45+ and CD45- fractions from melanoma-bearing mice at medium stage (20-mm mean diameter). Histograms represent mean mRNA expression normalized to β-actin in each sample run in triplicate (mean values ± SD). One representative experiment of three is shown. *P < .05, ***P < .001.
Discussion
In this study, we report the dual role of IRF-8 as a crucial immune regulator and as a tumor suppressor factor regulating melanoma progression and metastasis development. IRF-8 is a major determinant of myeloid cell development and function [37,38]. This has been clearly demonstrated in IRF-8-/- mice, which display an altered hematopoiesis, with impaired macrophage and DC compartments [11]. Our laboratory has shown that IRF-8-/- mice are selectively devoid of pDCs and exhibit reduced frequencies and function of CD8α+ DCs, which are critical for the induction of protective T cell immunity against many pathogens and tumors [8–10]. Here, we report that the uncontrolled melanoma growth and the high rate of metastases in IRF-8-/- mice strongly correlate with the severely defective immunosurveillance in these mice due to the effects of IRF-8 deficiency on the immune system. This is indicated by our observations that IRF-8-/- mice display poor homing of DCs and T cells at the tumor site, whereas they show large numbers of MDSCs in both tumor tissue and spleen. Increased numbers of MDSCs in spleen and tumor tissue have been described in various murine tumor models [39]. MDSCs are known to suppress the infiltration and activities of T cells at the tumor site and to inhibit DC maturation, thus hampering the onset of effector antitumor responses [40,41]. Accordingly, the few DCs infiltrating IRF-8-melanoma exhibited an immature phenotype and, as expected, lacked totally the pDC component. As a result of this immunosuppressive environment, T cell-mediated antitumor immune responses were completely abrogated in IRF-8-/- mice, both at the tumor site and at systemic level. In keeping with this, at late stage of tumor development, IRF-8-melanoma contained very few infiltrating CD4+ and CD8+ T cells, crucial effectors of the antitumor immune response against melanoma [42]. On the contrary, WT-melanoma showed a considerable infiltration of T cells and mature DCs, including a significant fraction of pDCs, which associated with induced T cell response. Correlation between the presence of intratumoral mature DCs and favorable clinical prognosis has been established for a variety of human solid tumors, including melanoma [43,44]. Furthermore, pDC recruitment in melanoma lesions has been reported to be an important correlate for the clinical response triggered by imiquimod, confirming a crucial role for these cells in the antitumor response against melanoma [45].
At the tumor site, a tight cross talk between various types of cells occurs, affecting angiogenesis, cancer metastatic potential, and intratumoral immune trafficking through a complex network of cytokines, chemokines, and growth factors [5]. In this scenario, cancer cells modulate the expression of chemokines and chemokine receptors favoring or inhibiting the homing of immune cells that dictate the course of tumor progression [31]. Our data clearly reveal a close correlation between the extent of tumor infiltration and the chemokine expression profiles in IRF-8-melanoma. Indeed, the impaired expression of the chemokine receptor CCR7 and its two ligands, CCL19 and CCL21, driving the recruitment of mature DCs and CD8+ T cells [46], strongly correlate both with the scarce frequency of these cells and with the immature phenotype of the few available DCs found in IRF-8-melanoma. Moreover, the absence of infiltrating pDCs in IRF-8-melanoma correlates with minimal levels of intratumoral CCL20, shown to participate to pDC recruitment from the blood to the tumor in melanoma patients [24], and of CCR5/CCL5, driving antitumor immunity through the activation of DCs and CD8+ T cells [25]. Noteworthy, these expression profiles were completely reversed in WT-melanoma, suggesting a potential key role of IRF-8-mediated DC activity in the induction of melanoma intratumoral immunity. The expression pattern of chemokine and chemokine receptors in IRF-8-melanoma is also distinctive of a tumor microenvironment favoring angiogenesis and metastasis development and correlates with increased vascularization and expression of angiogenic factors in these tumors. Indeed, the inhibition of the angiostatic axis CXCL10/CXCR3, the high levels of the angiogenic CXCL1 and CX3CL1, and the activation of the CCL27/CCR10 axis indicate a milieu supporting neoplastic cell growth, metastatic process, and immune escape [30,31,47–49].
Remarkably, IRF-8 host deficiency resulted in the inhibition of IRF-8 expression in the transplanted B16 melanoma. Early investigations revealed that IRF-8 deficiency is associated with the development of a CML-like syndrome in mice as well as with the expression of BCRABL in CML patients with poor prognosis [12,50]. In addition, recent reports have identified IRF-8 as a crucial determinant in solid tumor cell biology and defined this transcription factor as a tumor suppressor gene [7,14,51,52]. The suppression of IRF-8 function was also found to correlate with enhanced metastatic potential of cancer cells in a mouse model of mammary carcinoma [13,15]. Accordingly, we found that melanoma transplanted in immunocompetent mice exhibits high IRF-8 expression at early stage of development, progressively declining with tumor growth, indicating that repression of this transcription factor is a hallmark of melanoma malignant phenotype. This assumption is further sustained by our observation that metastatic B16-F10 melanoma expresses lower levels of IRF-8 than non-metastatic parental B16-F0 cells and that in human melanoma cell lines IRF-8 expression is substantially reduced in metastatic with respect to primary melanoma cells. In this regard, silencing of IRF-8 by DNA methylation or other epigenetic mechanisms has been associated with the malignant phenotype of many hematological and solid cancer cells [34,52].
The observation that IRF-8 expression in B16 melanoma cells is upregulated following transplantation into WT but not IRF-8-/- mice supports the concept that host IRF-8 deficiency abrogates the natural mechanisms of immunosurveillance that normally occur in immunocompetent animals. Of interest, treatment of melanoma-bearing IRF-8-/- mice with the demethylating agent 5-Aza-dC, which induced IRF-8 expression in cancer cells by epigenetic mechanisms, radically reversed the chemokine expression pattern, re-established intratumoral trafficking of DCs and T cells, and led to melanoma growth arrest. Although we cannot rule out the possibility that other factors, especially tumor suppressor genes, may be induced by 5-Aza-dC, our observations suggest a close correlation between intratumoral IRF-8 expression, immune infiltration, and melanoma growth control. Furthermore, these findings indicate that IRF-8 may function as a critical determinant in the cross talk between melanoma and infiltrating cells at the tumor site. In this context, the immune system may release signals that activate or repress IRF-8 in cancer cells that in turn shape melanoma phenotype and affect host immunosurveillance. Indeed, transwell co-culture experiments showed that only immune cells from melanoma-bearing WT mice, but not those from IRF-8-/- animals, released soluble factors capable of inducing IRF-8 expression in B16 cells.
Among the factors responsible for immune cell-cancer cross talk, we identified IL-27 as a candidate cytokine expressed at higher levels in tumor-bearing WT mice and able to induce alone IRF-8 expression in melanoma cells. IL-6, also expressed by WT immune cells, was able to synergize with IL-27 to upregulate intratumoral IRF-8. Therefore, the observation that both IL-27 and IL-6 were significantly up-regulated in the CD45+ fraction of WT-melanoma, with respect to IRF-8-melanoma, suggest that these cytokines may play a coordinate role in controlling intratumoral IRF-8 expression in vivo. Although tumor-released IL-6 has been reported to act as a pro-tumorigenic factor in advanced melanoma, its role in host antitumor response is less clear [53]. Instead, IL-27 has been shown to exert anti-proliferative activity on B16 melanoma through a mechanism involving IRF-1 and IRF-8 up-regulation [36]. Of interest, IRF-8-deficient macrophages were shown to be highly defective in the production of IL-27 [54]. Our results are in line with these reports and suggest the existence of a mutual regulatory pathway linking IRF-8 and IL-27. Hence, IRF-8-/- host deficiency may impair IL-27 production by innate immune cells resulting in failure to upregulate intratumoral IRF-8 in melanoma cancer cells, which in turn acquire a more aggressive phenotype escaping immune surveillance. Whether and which other factors besides IL-27 are involved in IRF-8-driven immunosurveillance against melanoma will need further investigations. Nevertheless, our results unravel a clinically relevant role of IRF-8 in bridging cancer and immune cross talk in the control of melanoma progression and metastatic process within the tumor microenvironment, thus opening new perspectives for the design of innovative therapies for melanoma patients.
Acknowledgments
We thank Laura Abalsamo for genotyping procedure of IRF-8-/- mice and Anna Maria Pacca and Teresa D'Urso for their helpful technical assistance in mice breeding. We are grateful to Anna Riccio for providing RNA from human cell lines. We also thank Maria Buoncervello for providing us with human GAPDH primers.
Footnotes
1This study was funded by the Italian Association for Cancer Research (AIRC) Project No. 11610 and the Italian Ministry of Health Programma Integrato Oncologia 2006 to L.G. and by AIRC Project No. 10720 to F.B.
2This article refers to supplementary materials, which are designated by Table W1 and Figures W1 to W5 and are available online at www.neoplasia.com.
References
Articles from Neoplasia (New York, N.Y.) are provided here courtesy of Neoplasia Press
Citations & impact
Impact metrics
Citations of article over time
Article citations
NEUROD1 predicts better prognosis in pancreatic cancer revealed by a TILs-based prognostic signature.
Front Pharmacol, 13:1025921, 13 Oct 2022
Cited by: 1 article | PMID: 36313290 | PMCID: PMC9612957
IRF8 Regulates Intrinsic Ferroptosis through Repressing p53 Expression to Maintain Tumor Cell Sensitivity to Cytotoxic T Lymphocytes.
Cells, 12(2):310, 13 Jan 2023
Cited by: 4 articles | PMID: 36672246 | PMCID: PMC9856547
IRF8: Mechanism of Action and Health Implications.
Cells, 11(17):2630, 24 Aug 2022
Cited by: 12 articles | PMID: 36078039 | PMCID: PMC9454819
Review Free full text in Europe PMC
Bona Fide Tumor Suppressor Genes Hypermethylated in Melanoma: A Narrative Review.
Int J Mol Sci, 22(19):10674, 01 Oct 2021
Cited by: 2 articles | PMID: 34639015 | PMCID: PMC8508892
Review Free full text in Europe PMC
Single cell RNA sequencing of AML initiating cells reveals RNA-based evolution during disease progression.
Leukemia, 35(10):2799-2812, 09 Jul 2021
Cited by: 36 articles | PMID: 34244611 | PMCID: PMC8807029
Go to all (32) article citations
Data
Data behind the article
This data has been text mined from the article, or deposited into data resources.
BioStudies: supplemental material and supporting data
Similar Articles
To arrive at the top five similar articles we use a word-weighted algorithm to compare words from the Title and Abstract of each citation.
A multidisciplinary study using in vivo tumor models and microfluidic cell-on-chip approach to explore the cross-talk between cancer and immune cells.
J Immunotoxicol, 11(4):337-346, 06 Mar 2014
Cited by: 28 articles | PMID: 24597645
VEGF suppresses T-lymphocyte infiltration in the tumor microenvironment through inhibition of NF-κB-induced endothelial activation.
FASEB J, 29(1):227-238, 31 Oct 2014
Cited by: 110 articles | PMID: 25361735
Host-derived MCP-1 and MIP-1α regulate protective anti-tumor immunity to localized and metastatic B16 melanoma.
Am J Pathol, 180(1):365-374, 26 Oct 2011
Cited by: 39 articles | PMID: 22037251
The good and the bad of chemokines/chemokine receptors in melanoma.
Pigment Cell Melanoma Res, 22(2):175-186, 14 Feb 2009
Cited by: 77 articles | PMID: 19222802 | PMCID: PMC2848967
Review Free full text in Europe PMC



