Abstract
Free full text

Cloning and Regulation of Cholesterol 7α-Hydroxylase, the Rate-limiting Enzyme in Bile Acid Biosynthesis*
Abstract
The rate-limiting step in bile acid biosynthesis is catalyzed by the microsomal cytochrome P-450 cholesterol 7α-hydroxylase (7α-hydroxylase). The expression of this enzyme is subject to feedback regulation by sterols and is thought to be coordinately regulated with enzymes in the cholesterol supply pathways, including the low density lipoprotein receptor and 3-hydroxy-3-methylglutaryl-coenzyme A reductase and synthase. Here we report the purification of rat 7α-hydroxylase and the determination of a partial amino acid sequence. Oligonucleotides derived from peptide sequence were used to clone a full-length cDNA encoding 7α-hydroxylase. DNA sequence analysis of the cDNA revealed a 7α-hydroxylase protein of 503 amino acids with a predicted molecular weight of 56,890 which represents a novel family of cytochrome P-450 enzymes. Transfection of a 7α-hydroxylase cDNA into simian COS cells resulted in the synthesis of a functional enzyme whose activity was stimulated in vitro by the addition of rat microsomal cytochrome P-450 reductase protein. RNA blot hybridization experiments indicated that the mRNA for 7α-hydroxylase is found only in the liver. The levels of this mRNA increased when bile acids were depleted by dietary cholestyramine and decreased when bile acids were consumed. Dietary cholesterol led to an increase in 7α-hydroxylase mRNA levels. The enzymatic activity of 7α-hydroxylase paralleled the observed changes in mRNA levels. These results suggest that bile acids and sterols are able to alter the transcription of the 7α-hydroxylase gene and that this control explains the previously observed feedback regulation of bile acid synthesis.
Cholesterol homeostasis in mammals represents a delicate balance between pathways of supply and catabolism. In the liver, the chief organ of cholesterol metabolism (1), supply is accomplished by a receptor-mediated pathway (2) and by the de novo synthesis of cholesterol from acetate precursors (3). A single pathway of catabolism involves the conversion of cholesterol into hydrophilic bile acids that are subsequently excreted from the body via the bile and intestine (4). Although the regulatory mechanisms underlying the expression of key enzymes in the cholesterol supply pathways are beginning to be understood (5–8), little is currently known about similar molecular mechanisms in cholesterol catabolism or about the coordinate regulation of the three pathways.
Bile acids play two opposing roles in the maintenance of cholesterol homeostasis. In one role, they are the end products of cholesterol catabolism, and their biosynthesis and excretion serve to decrease the levels of cholesterol in the liver. In a second role, their presence in the intestine facilitates the solubilization of dietary fats and cholesterol and the subsequent uptake of these essential nutrients (4). In this manner, bile acids increase whole body cholesterol levels. These observations suggest that the production of bile acids in the liver must be tightly regulated in order to balance these two antagonistic functions.
The synthesis of bile acids is carried out by at least 10 enzymes in the liver which hydroxylate the four-ring structure of cholesterol and shorten and oxidize the side chain (9). In the human and rat, the major products thus formed are the primary bile acids, cholic acid and chenodeoxycholic acid (Fig. 1). The rate-limiting step in this pathway involves the introduction of a hydroxyl moiety at the 7 position of cholesterol (10) and is catalyzed by cholesterol 7α-hydroxylase (7α-hydroxylase,1, Fig. 1), a microsomal enzyme that is a member of the cytochrome P-450 family. Once formed, 7α-hydroxycholesterol is rapidly converted into a primary bile acid, as few of the subsequent enzymatic steps appear to be regulated (9). The activity of 7α-hydroxylase is subject to end product repression by bile acids in the enterohepatic circulation (10). Thus, in animals maintained on a diet containing bile acids, the level of 7α-hydroxylase activity is reduced (10). Conversely, animals fed drugs such as cholestyramine, which enhance the excretion of bile acids (11), increase their hepatic levels of this enzyme and hence the production of bile acids (10). Because of the complexity of the enzyme assay and the lack of an independent measure of 7α-hydroxylase protein, the changes in enzyme activity have not yet been shown unequivocally to result from changes in the amount of enzyme as opposed to changes in its activity. Moreover, nothing is known about the regulation of the 7α-hydroxylase mRNA.

The primary bile acids cholic acid and chenodeoxycholic acid are synthesized from cholesterol in the liver via the actions of 10 or more different enzymes (9). Cholesterol 7α-hydroxylase converts cholesterol into 7α-hydroxycholesterol and constitutes the rate-limiting step in the pathway.
Recently, a preliminary report describing the purification and cloning of the rat 7α-hydroxylase was published by Okuda and colleagues (12). In the current studies, we confirm and extend this work by reporting the purification of rat 7α-hydroxylase and the subsequent determination of a partial protein sequence. These biochemical tools were then used to isolate cDNA clones that in turn allowed the determination of the structure of the protein and the expression of the enzyme in transfected COS cells. Finally, the cDNA clone was used as a probe in RNA-blotting experiments to demonstrate feedback regulation of the 7α-hydroxylase mRNA.
EXPERIMENTAL PROCEDURES
Materials
Male Sprague-Dawley rats (250–300 g) were purchased from Sasco (Omaha, NE) and maintained on a normal chow diet and a 12-h light/12-h dark cycle for at least 7 days prior to being placed on the indicated diets. Aminohexyl-Sepharose 4B resin, DEAE-Sepharose CL-6B resin, protein G-Sepharose 4B resin, cyanogen bromide-activated Sepharose 4B resin, 2′,5′-ADP-Sepharose 4B resin, and bacteriophage M13 replicative form DNAs were purchased from Pharmacia LKB Biotechnology Inc. Hydroxylapatite (Bio-Gel HTP) was obtained from Bio-Rad, and CF1 cellulose powder was purchased from Whatman. Preabsorbent multichannel thin layer chromatography (TLC) plates were obtained from J. T. Baker Chemical Co. Silica Gel 60 TLC plates (no. 5748-7) were obtained from Merck. Cholic acid was purchased from United States Biochemical and crystallized three times with ethanol prior to use. NADPH, chenodeoxycholic acid, Lubrol (type PX), and Triton X-100 were obtained from Sigma. Granular cholestyramine (Colestid) was purchased from the Upjohn Co. Ethyl acetate, methylene chloride, and toluene were from Aldrich Chemical Co. Protein concentration determinations were accomplished using kits from Bio-Rad and the micro BCA protein assay kit from Pierce. Perkin-Elmer Cetus was the source of Taq polymerase. Radioisotopes were purchased from Du Pont-New England Nuclear. Steroid standards were obtained from Steraloids, Inc. Glycerol, competent Escherichia coli cells, and a cDNA synthesis cloning kit were purchased from GIBCO. Enzymes used in cloning and DNA sequencing were obtained from New England BioLabs, United States Bio-chemicals, and GIBCO. A rat liver cDNA library was constructed by methods described previously (13). Nylon filters (Biotrans) for hybridization experiments were obtained from ICN Pharmaceutical, Irvine, CA. Oligonucleotides were synthesized on an Applied Biosystems model 380A DNA synthesizer. DNA sequence analysis was carried out with an Applied Biosystems model 370A DNA sequencer employing fluorescently labeled primers. COS-M6 cells (SV40-trans-formed monkey kidney cells) were a kind gift of Arnold J. Berk (Dept. of Microbiology, University of California, Los Angeles). NADPH-cytochrome P-450 reductase was purified from the microsomes of phenobarbital-induced Sprague-Dawley male rats as described by Yasukochi and Masters (14).
Animals Diets
Rats maintained on an induction diet were fed ad libitum normal rat chow supplemented with 2% cholestyramine (w/ w) for 7 days. Animals were killed between the 7th and 8th h of the dark period. A cholesterol diet was prepared by first dissolving cholesterol (1 g/100 g of chow) in corn oil (10 ml/100 g of chow) followed by emulsification with normal chow. Rats were maintained on this diet for 7 days prior to killing between the 6th and 7th h of the light period. Suppression diets consisted of either normal chow supplemented with 0.2% chenodeoxycholic acid (w/w) given for a 14-day period (see Fig. 3) or normal chow supplemented with either 1% chenodeoxycholic acid (w/w) or 1% cholic acid (w/w) given for 7 days (see Fig. 7). Rats maintained on suppression diets were killed between the 6th and 7th h of the light period. Rats maintained on normal chow diets were killed at the light/dark interface.
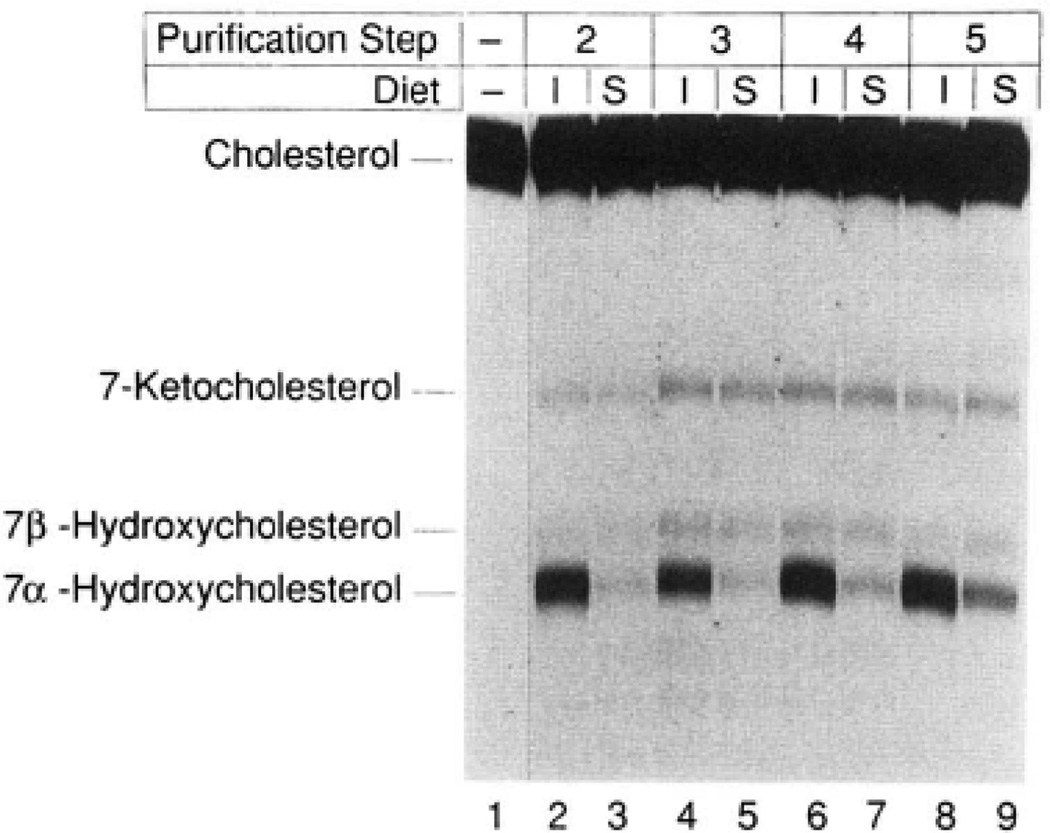
7α-Hydroxylase was simultaneously purified from groups of 100 animals maintained for 2 weeks on rat chow supplemented with 2% cholestyramine (induced, I) or 0.2% chenodeoxycholic acid (suppressed, S) as described under “Experimental Procedures.” An equal amount of protein derived from the two groups at different purification steps was assayed for 7α-hydroxylase activity by thin layer chromatography. The purification steps correspond to those of Table I, Approximately 500 µg of protein from step 2 (lanes 2 and 3), 7 µg from step 3 (lanes 4 and 5), 6 µg from step 4 (lanes 6 and 7), and 1 µg from step 5 (lanes 8 and 9) were assayed for 5 min at 37 °C in a reaction containing 25 µM [14C] cholesterol and 1000 units of cytochrome P-450 reductase. Lane 1 contained only the starting isotope and was not subjected to solvent evaporation. The chromatogram was exposed to Kodak XAR-5 film for a period of 44 h at −70 °C. The identities of the observed sterols are indicated on the left and were determined by comparison with authentic standards. The 7-keto and 7β-hydroxylated forms of cholesterol represent spontaneous oxidation products derived from cholesterol during solvent evaporation in the workup of the various reactions.
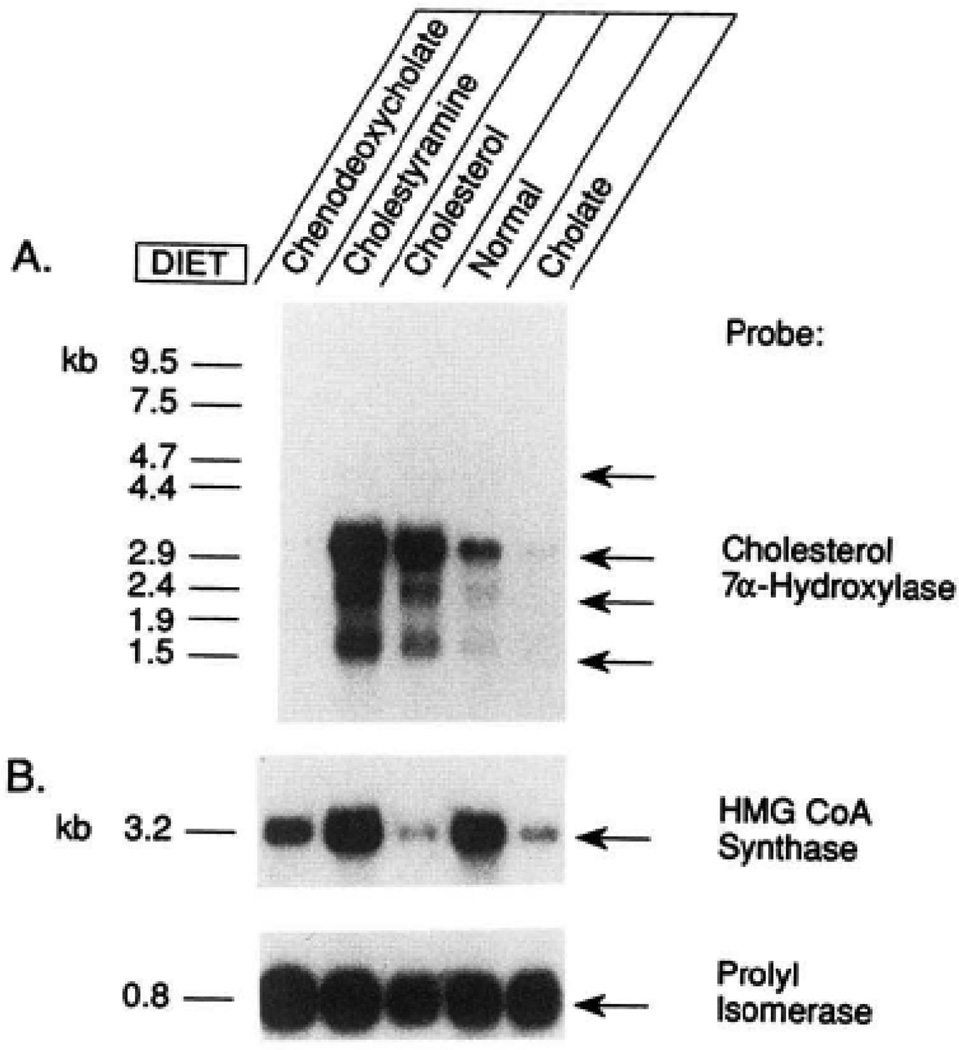
Polyadenylated RNA was isolated from the livers of animals maintained on the indicated diets, and 10 µg aliquots were subjected to blot analysis as described in the legend to Fig. 6. The filter was initially probed with 32P-labeled cDNAs for 7α-hydroxylase (panel A) and subsequently stripped and reprobed with cDNAs for the HMG-CoA synthase (plasmid p53K-3, Ref. 33) and cis-trans-prolyl isomerase mRNAs (panel B). The positions to which RNA standards of known size migrated in an adjacent lane are indicated on the left of panel A. These same standards were used to estimate the sizes of the HMG-CoA synthase and cis-trans-prolyl isomerase mRNAs shown in panel B.
Enzyme Assay
Cholesterol 7α-hydroxylase activity was assayed in the presence of 5–10 µg of [14C]cholesterol (60 Ci/mmol) in a 0.5-ml volume containing 50 mM Tris acetate (pH 7.5), 20% glycerol, 1 mM EDTA, 2 mM dithiothreitol (DTT), 0.06% Triton X-100, 2 mM NADPH, and 1000 units of purified NADPH-cytochrome P-450 reductase. Reactions were carried out for 5–20 min at 37 °C followed by extraction with either methylene chloride (purified fractions) or Folch (chloroform:methanol, 2:1, COS cell experiments). The organic phase was dried under an N2 stream at 50 °C, resuspended in 100 µl of acetone, and subjected to thin layer chromatography in ethyl acetate/toluene (3:2, v/v). Autoradiography was used to identify areas on the plates corresponding to the various metabolites. These areas were scraped, and radioactivity was quantitated in a liquid scintillation counter. The identities of the products were determined by comparison with the RF values of known standards.
Preparation of Liver Microsomes
Freshly isolated rat livers were washed in ice-cold sucrose buffer (250 mM sucrose, 1 mM EDTA, 10 mM Tris-Cl, pH 7.4) and immediately homogenized in a Potter-Elvehjem Teflon glass homogenizer with 4 volumes of the same buffer/g of liver. All operations were carried out at 0–4 °C. The homogenate was filtered through Miracloth (Calbiochem) and centrifuged for 20 min at 7,000 × g. The supernatant was filtered again through Miracloth and centrifuged 70 min at 106,000 × g. The microsomal pellet was rehomogenized in 100 mM potassium pyrophosphate, pH 7.4, and recentrifuged at 106,000 × g for 70 min. This pellet, referred to as washed liver microsomes, was resuspended in a minimal volume of sucrose buffer, divided into multiple aliquots, quick-frozen in liquid N2, and stored at −70 °C.
Purification of Cholesterol 7α-Hydroxylase
Aliquots of washed microsomes were thawed and resuspended in 100 mM Tris-Cl (pH 7.5), 0.1 mM DTT, and 20% glycerol at a protein concentration of 6 mg ml−1. Solubilization of membrane proteins was accomplished by the slow addition of cholic acid to a final concentration of 1.8%. After stirring for 30–60 min at 4 °C, the solubilized proteins were adjusted to 8% (v/v) polyethylene glycol by the addition of a solution of 50% polyethylene glycol, and precipitated proteins were separated by centrifugation (20 min, 16,000 × g) and discarded. The polyethylene glycol concentration of the supernatant was then raised to 17%, and the resultant pellet containing 7α-hydroxylase activity was resolubilized in 100 mM potassium phosphate (pH 7.4), 20% glycerol, 0.1 mM EDTA, and 0.7% cholic acid. The final protein concentration of this solution was adjusted to 7 mg/ml. Material from approximately 75 rats (4,000 mg of protein) was applied to two 2.5 × 40-cm aminohexyl-Sepharose 4B columns previously equilibrated with 100 mM potassium phosphate (pH 7.4), 20% glycerol, 0.1 mM EDTA, 0.1 mM DTT, and 0.5% cholic acid. The columns were then washed with this same buffer until the absorbance at 280 nm was less than 0.05. Elution of 7α-hydroxylase was carried out with a buffer containing 100 mM potassium phosphate, pH 7.4, 20% glycerol, 0.1 mM EDTA, 0.1 mM DTT, 0.4% cholic acid, and 0.06% Lubrol PX. Fractions of 20-ml were collected until the absorbance at 280 nm was less than 0.2 and the absorbance at 416 nm was less than 0.05. Fractions were assayed for 7α-hydroxylase activity as described above and pooled accordingly.
The active pool was adjusted such that the final buffer components were 20 mM potassium phosphate (pH 7.4), 20% glycerol, 0.1 mM DTT, 0.2% cholic acid, and 0.2% Lubrol PX. A1/15 volume of packed hydroxylapatite, preequilibrated in the same buffer, was added to the pool, and the resulting slurry was allowed to stir for 1 h at 4 °C. A 1/ 30 volume of packed preequilibrated Whatman CF1 celluose powder was then added, and the mixture was transferred to a glass column (2.5 × 40 cm). After packing, the column was washed with the starting buffer containing 50 mM potassium phosphate (pH 7.4) in place of 20 mM potassium phosphate until the absorbance at 280 nm was less than 0.1. 7α-Hydroxylase was eluted by washing with buffer containing 180 mM potassium phosphate (pH 7.4). Active fractions were pooled and dialyzed overnight against 20 mM Tris acetate (pH 7.5), 20% glycerol, 0.1 mM EDTA, 0.1 mM DTT, and 0.4% Lubrol PX.
The dialyzed fraction (~40 mg of protein) was applied to a DEAE-Sepharose CL-6B column (2.5 × 10 cm) previously equilibrated with dialysis buffer. 7α-Hydroxylase activity was eluted with 50 mM sodium acetate, 20 mM Tris acetate (pH 7.5), 20% glycerol, 0.1 mM EDTA, 0.1 mM DTT, and 0.4% Lubrol PX. Active fractions were pooled, quick-frozen in liquid N2, and stored at −70 °C.
The final purification step involved immunoaffinity chromatography using a monoclonal antibody (2B4) raised against a major contaminant in the DEAE-Sepharose-purified material. This antibody was initially identified in an unsuccessful screen for anti-7α-hydroxylase antibodies.2 For purification, 5 mg of 2B4 antibody was coupled to 1 ml of cyanogen bromide-activated Sepharose 4B using a protocol supplied by the manufacturer. A column of antibody-Sepharose (1.0 × 5.0 cm) was poured and equilibrated in a buffer containing 50 mM sodium acetate, 20 mM Tris acetate (pH 7.5), 20% glycerol, 0.1 mM EDTA, 1 mM DTT, and 0.4% Lubrol PX. DEAE-Sepharose fractions (0.5 mg of protein/ml of resin) were then passed through the column at a flow rate of 0.5 ml/min. 7α-Hydroxylase-containing fractions in the flow-through were identified by thin layer chromatography assay, pooled, and stored at −70 °C.
Protein Sequence Analysis
Approximately 500 pmol of step 6 material (see Table I) was subjected to sodium dodecyl sulfate (SDS)-polyacrylamide gel electrophoresis and transferred to polyvinylidene difluoride membranes for amino-terminal sequence analysis on an Applied Biosystems model 470A sequenator. Internal peptide sequence was obtained after transfer to nitrocellulose paper and solid phase tryptic digestion (15). Peptides were separated by reverse phase HPLC on a Brownlee RP300 (2.1 × 100-mm) C8 column in 0.1% (v/ v) trifluoroacetic acid with a gradient of 0–50% acetonitrile for 100 min at a flow rate of 50 ml/min. Some peptides were resolved further on a Brownlee RP300 (2.1 × 100-mm) C8 column in 0.1% (w/v) ammonium acetate with a gradient of 0–50% acetonitrile. Individual peaks were collected manually on 1-cm discs of Whatman GF/C paper prior to sequence analysis.
Table I
Microsomes were prepared from 75 rats maintained for 7 days on a 2% cholestyramine diet and subjected to the indicated purification steps as detailed under “Experimental Procedures.”
| Step | Fraction | Protein | Specific activitya | Total activity | Purification | Recovery |
|---|---|---|---|---|---|---|
| mg | nmol/min/mg | nmol/min | -fold | % | ||
| 1 | Microsomes | 16,000 | 0.065 | 1,040 | 1.0 | 100 |
| 2 | 8–17% PEGb | 8,148 | 0.18 | 1,466 | 2.7 | 100 |
| 3 | AH-Sepharosec | 240 | 5.2 | 1,248 | 80 | 85 |
| 4 | Hydroxylapatite | 84 | 10.0 | 840 | 154 | 57 |
| 5 | DEAE-Sepharose | 11 | 35.2 | 387 | 542 | 26 |
| 6 | 2B4 Affinity column | 6 | 43.0 | 258 | 662 | 18 |
cDNA Cloning
A size-fractionated cDNA library in bacteriophage λgt10 was constructed from liver mRNA isolated from cholestyramine-fed animals and screened with 32P-labeled oligonucleotides (16) corresponding to peptides 1, 2, 6, and 7 of Table II by standard methods (17). Hybridization-positive plaques were picked and subjected to polymerase chain reaction analysis to size the harbored cDNA insert (18). As described in the text a partial cDNA was initially isolated and subsequently used to isolate cDNAs that spanned the entire 7α-hydroxylase mRNA. DNA sequence analysis of both strands of the cDNA was carried out as described by Sanger et al. (19) or Smith et al. (20). RNA blotting was carried out as described previously (21).
Table II
The Mr 50,300 protein was isolated by SDS-polyacrylamide gel electrophoresis, transferred to nitrocellulose membranes, and digested in filtero with trypsin. The resulting peptides were resolved by HPLC and subjected to sequence analysis as described under “Experimental Procedures.” A blank in the sequence of a given peptide indicates an amino acid residue for which an unambiguous identification could not be made.
| Peptide | Amino acid sequence |
|---|---|
| 1 | TAKEDFTLHLEDGSYNIR |
| 2 | AGLGILPPLHDIEFK |
| 3 | LSSASLNI |
| 4 | DDMIALYPQLMHLDPEIYPDP |
| 5 | FGSNPLEFL |
| 6 | —SEEVSGALQSAVQEL |
| 7 | SIDPNDGNTTENINNTFTK |
Construction of 7α-Hydroxylase Expression Vector
A plasmid vector capable of expressing 7α-hydroxylase in mammalian cells was constructed via standard methods of genetic engineering. A DNA fragment corresponding to nucleotides 1–2172 of Fig. 5 was ligated into the EcoRI site of the pCMV2 eukaryotic expression vector (22). The desired recombinant plasmid (p7α-OHase) was characterized by Southern blotting and restriction mapping and banded twice in CsCl2 density gradients prior to transfection analysis.

Amino acids are numbered above the sequence, and nucleotides are numbered on the right. A dot is placed under every 10th nucleotide. Protein sequence determined from the amino terminus and from seven tryptic peptides is underlined. The protein sequence deduced from the cDNA matched that determined by Edman degradation (Fig. 4 and Table II) with the exception of the 14th residue of tryptic peptide 6, which was valine in the peptide and glycine (residue 323) in the cDNA. A cysteine residue found in all cytochrome P-450 enzymes is circled at position 444. An Alu sequence in the 3′-untranslated region is overlined.
Transfection of COS-M6 Cells
Stock cultures were maintained, grown, and transfected as described previously (23). To assay transfected cells for 7α-hydroxylase activity, microsomes were prepared as follows. Sixty h post-transfection, cells were washed with ice-cold phosphate-buffered saline, harvested with a rubber policeman, and homogenized in 0.5 M sucrose, 10 mM Tris-Cl (pH 7.5), 1.0 mM EDTA, 1 mM phenylmethylsulfonyl fluoride, 0.04 units/ml aprotinin, 0.1 mM EGTA, and 0.1 µg/ml leupeptin. The homogenate was diluted to 0.25M sucrose, layered on a 0.5 M sucrose cushion in the above buffer, and centrifuged at 10,000 × g for 10 min. Microsomes were subsequently isolated from the supernatant by a 206,000 × g centrifugation for 30 min. 7α-Hydroxylase enzyme activity in the purified microsomes was assayed as described above.
RESULTS
Purification of 7α-Hydroxylase
To increase the level of enzyme in the liver, rats were maintained for 1 week on a normal chow diet supplemented with 2% cholestyramine and then killed in the middle of the dark period of a 12-h light/ dark cycle. Cholestyramine increases 7α-hydroxylase activity by binding bile acids in the intestine and preventing their reutilization via the enterohepatic circulation (11). In compensation, the liver boosts the production of bile acids by increasing the synthesis of 7α-hydroxylase. Similarly, the production of this enzyme is maximized during the nocturnal feeding phase of the rat (10).
Washed microsomes were prepared and used as starting material for the isolation of the enzyme (Table I). The purification protocol was a modification of that described by Andersson et al. (24). Two key steps were employed in the purification. The first was chromatography on aminohexyl-Sepharose 4B, a mixed hydrophobic/ion exchange resin on which 7α-hydroxylase could be separated from the bulk of other members of the cytochrome P-450 enzyme family (26). The second was immunoaffinity chromatography using a monoclonal antibody directed against a major contaminant that co-chromatographed with 7α-hydroxylase. After the immunoaffinity chromatography step, an enzyme preparation was obtained in which 7α-hydroxylase activity had been enriched 660-fold with an overall yield of 18% (Table I). Electrophoresis of this highly purified material on SDS-polyacrylamide gels yielded four major protein bands with molecular weights in the 40,000–50,000 range (Fig. 2, lane 6).
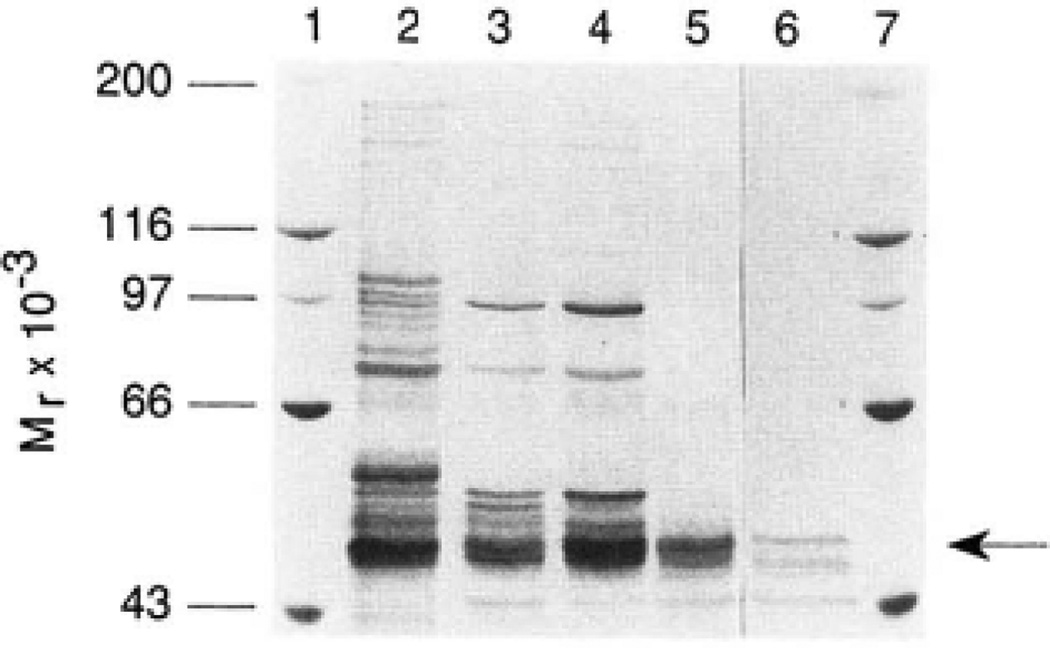
20 µg of rat microsomal 8–17% polyethylene glycol precipitate (lane 2), 2.5 µg of aminohexyl-Sepharose fraction (lane 3), 5.0 µg of hydroxy1apatite fraction (lane 4), 1.5 µg of DEAE-Sepharose fraction (lane 5), and 1.0 µg of postmonoclonal antibody 2B4 fraction (lane 6) were electrophoresed on a 7% SDS-polyacrylamide gel and subsequently visualized by silver staining. The molecular weights of protein standards (lanes 1 and 7) are indicated on the left of the stained gel. The 7α-hydroxylase protein is indicated by an arrow on the right.
Despite further chromatography attempts on more than a dozen different resins, we were unsuccessful in obtaining a more highly enriched preparation of 7α-hydroxylase. In an attempt to identify which of the four major bands in the most highly purified material corresponded to 7α-hydroxylase, advantage was taken of the regulated expression of the enzyme. To this end, 7α-hydroxylase was simultaneously purified from microsomes derived from animals maintained on an induction diet (2% cholestyramine) and from microsomes from a similar number of animals maintained on a suppression diet (0.2% chenodeoxycholic acid). As shown in Pig. 3, a starting differential of 10-fold in 7α-hydroxylase activity was maintained throughout five steps of the purification scheme. When an equal mass of protein derived at purification step 5 (Table I) from the induced and suppressed microsomes was electrophoresed on SDS-polyacrylamide gels, only one protein was seen to vary in accordance with the difference in 7α-hydroxylase activity between the two preparations (data not shown). The regulated protein (apparent Mr 50,300) corresponded to the largest of the four polypeptides visualized in Fig. 2, lane 6. These results suggested that this protein corresponded to 7α-hydroxylase.
In an attempt to identify 7α-hydroxylase further, we next took advantage of the observation that a majority of cytochrome P-450 enzymes do not have blocked amino termini and thus yield protein sequence data when subjected to Ed-man degradation. In addition, despite being members of a superfamily of proteins with hydrophobic amino termini, the sequences of cytochrome P-450s in this region are sufficiently different to distinguish the various members (25). To this end, material from step 6 of the purification (Table I) obtained from animals on an induction diet was electrophoresed, transferred to Immobilon membranes, and the four proteins were subjected individually to sequence analysis on an Applied Biosystems model 470A sequenator. The results obtained are shown in Fig. 4. The largest protein with an Mr of 50,300 corresponded to the cholestyramine-regulated protein and had a hydrophobic amino-terminal sequence that was characteristic of, but different from, all other cytochrome P-450 sequences reported to date (25). The amino-terminal sequences of the Mr 46,600 and 45,400 proteins revealed them to be identical to the rat cytochrome P-450g (27) and cytochrome P-450a (28) enzymes, respectively. Cytochrome P-450g has been shown to catalyze the hydroxylation of testosterone at carbon atoms 6, 15, and an unknown additional position (29), whereas cytochrome P-450a catalyzes hydroxylation at the 7 position of testosterone (28). Consistent with these observations, step 6 material (Table I) also demonstrated these catalytic activities (data not shown). The Mr 42,100 protein was somewhat small to be a eukaryotic cytochrome P-450, and consistent with this notion, the amino-terminal sequence of the protein contained a number of glycine residues (Fig. 4) that are generally underepresented in the amino-terminal sequences of cytochrome P-450s (25).
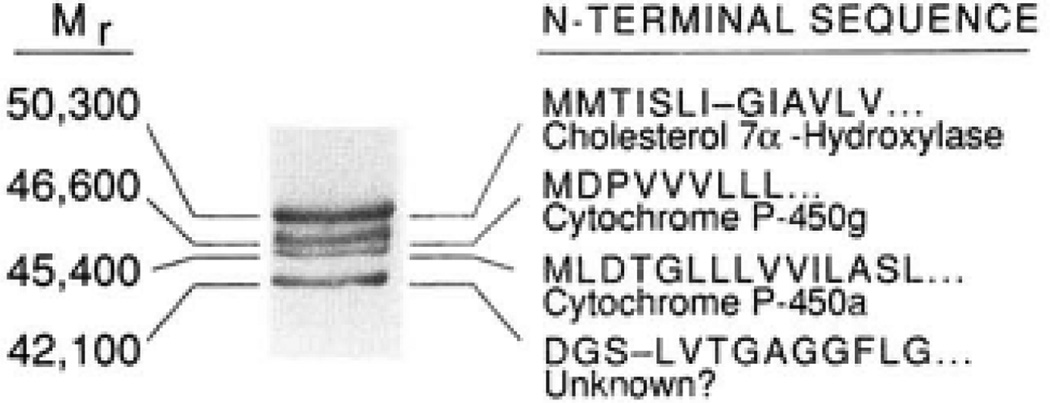
A Coomassie Blue-stained gel of 20 µg of step 6-purified material (Table I) is shown together with the amino-terminal sequences determined from each of the four major proteins remaining at this stage of the purification. The calculated molecular weight of each protein is shown on the left. The identities of cytochrome P-450a and cytochrome P-450g were determined by comparison of their deduced amino-terminal sequences with those in the National Biomedical Research Foundation protein data base and by enzyme assay (see “Results”).
The results of the regulation experiment shown in Fig. 3 and the unique hydrophobic amino-terminal sequence determined for the Mr 50,300 polypeptide of Fig. 4 strongly suggested that this protein was 7α-hydroxylase. To aid in the isolation of cDNA clones, the sequences of seven internal tryptic peptides from this protein were subsequently determined (Table II).
cDNA Cloning of 7α-Hydroxylase
To isolate cDNA clones corresponding to the rat 7α-hydroxylase mRNA, size-fractionated cDNA libraries constructed from liver mRNA isolated from cholestyramine-fed animals were screened with multiple oligonucleotide probes derived from the peptide sequences of the Mr 50,300 protein. After screening 8 × 104 clones, a cDNA was isolated which hybridized with three oligonucleotide probes. The amino acid sequence deduced from the DNA sequence of this clone revealed an open translation reading frame of 135 residues which included peptides 1, 2, and 4 (Table II) that were used to design the hybridizing oligonucleotides. Comparison with other cytochrome P-450 sequences indicated that this cDNA encoded the carboxyl-terminal end of a member of this family of proteins. cDNAs spanning the complete 7α-hydroxylase mRNA were subsequently identified using hybridization probes derived from the 5′ end of the initial cDNA clone.
Structure of 7α-Hydroxylase
The nucleotide sequence and the predicted amino acid sequence of 7α-hydroxylase derived from five overlapping cDNA clones are shown in Fig. 5. The deduced amino acid sequence begins with the unusual occurrence of 2 methionine residues and proceeds for a total of 503 residues. As indicated by the underlines in Fig. 5, the 2 methionines were present in the mature 7α-hydroxylase protein as well as in the cDNA sequence. The first 6 amino acids of the 7α-hydroxylase cDNA including the two methionines (Met-Met-Thr-Ile-Ser-Leu) bear little resemblance to a previously published aminotermmal sequence (Met-Phe-Glu-Val/Ile-Ser-Leu) for 7α-hydroxylase derived from an apparently impure sample (30). The remainder of the protein sequence contains many of the hallmarks that identify a microsomal cytochrome P-450, including an overall hydrophobic nature and a conserved cysteine residue at position 444. The mRNA for 7α-hydroxylase has an unusually long 3′-untranslated region of over 2 kb (Fig. 5). Included within this region is a single copy of the Alu family of middle repetitive DNAs (nucleotides 2195–2313, Fig. 5).
Expression of 7α-Hydroxylase cDNA in COS Cells
To confirm that the sequence shown in Fig. 4 encoded a functional 7α-hydroxylase enzyme, we expressed the cDNA in simian COS-M6 cells. A DNA fragment corresponding to nucleotides 1–2172 of Fig. 5 was ligated into the pCMV2 eukaryotic expression vector and transfected into COS cells using a DEAE-dextran protocol. Enzyme activity could not be detected in transfected whole cells. However, as shown in Table III, 7α-hydroxylase activity was readily measured in microsomes prepared from COS cells transfected with the 7α-hydroxylase cDNA. This activity was stimulated approximately 18-fold by the addition of purified rat microsomal cytochrome P-450 reductase (experiment 2, Table III). No 7α-hydroxylase activity could be detected in mock-transfected COS cells. In experiments not shown, polyclonal antibodies raised against the purified 7α-hydroxylase cross-reacted with an Mr 50,000 protein that was present in COS cells transfected with the 7α-hydroxylase cDNA but absent from the mock-transfected cells.
Table III
Expression of 7α-hydroxylase in transfected COS-M6 cells
| Experiment | Plasmid | Microsomal protein | Cytochrome P-450 reductase | Conversiona | Specific activity |
|---|---|---|---|---|---|
| µg | % | Pmol/min/mg protein | |||
| 1 | Vector alone | 250 | + | <0.1 | <5.0 |
| 500 | + | <0.1 | <2.5 | ||
| 7α-Hydroxylase cDNA | 250 | + | 2.5 | 130 | |
| 500 | + | 4.9 | 127 | ||
| 2 | Vector alone | 250 | + | <0.1 | <5.0 |
| 500 | + | <0.1 | <2.5 | ||
| 7α-Hydroxylase cDNA | 250 | + | 3.7 | 192 | |
| 500 | − | 0.4 | 10 | ||
| 500 | + | 7.2 | 187 |
Expression and Regulation of Cholesterol 7α-Hydroxylase
To determine the tissue distribution of 7α-hydroxylase in the rat, polyadenylated RNA was prepared from eight organs and four different regions of the brain and subjected to blot analysis using hybridization probes derived from the coding region of the 7α-hydroxylase cDNA. As indicated in Fig. 6, only the liver expressed 7α-hydroxylase mRNA. In this tissue, three species of mRNA were detected, including a prominent 3.6-kb mRNA and two less abundant mRNAs of 2.4 and 1.7 kb (Fig. 5A). With longer periods of autoradiography, an additional band of 4.7 kb was also detected (see below). As a control, the mRNA encoding a cis-trans-prolyl isomerase (31) was present in most lanes. Certain tissues did not express prolyl isomerase, and in these tissues we used β-actin mRNA as a control (32; data not shown).
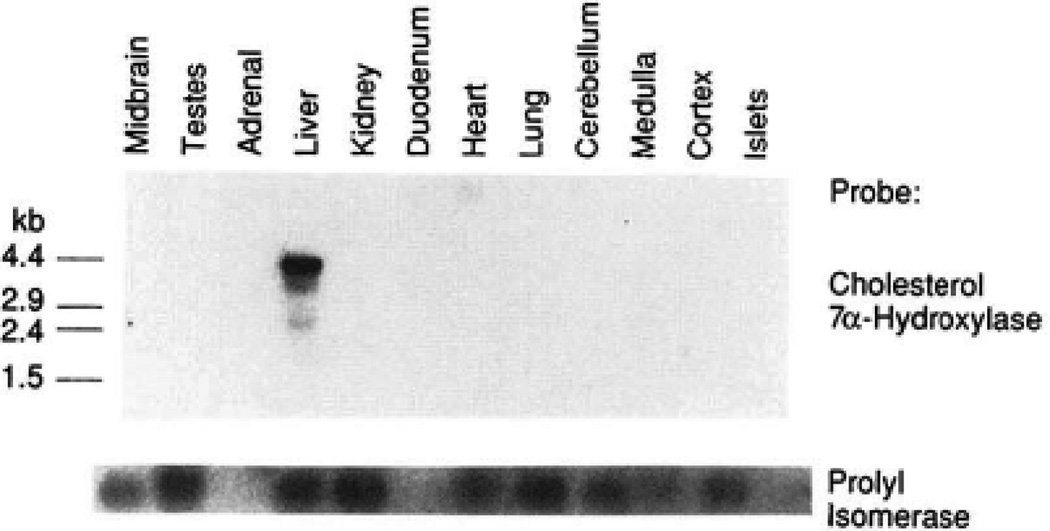
Top, RNA was isolated from the indicated tissue or region of the brain by a guanidinium/CsCl procedure (17). Approximately 5 µg of polyadenylated RNA from each source was denatured with glyoxal and size fractionated by electrophoresis in a 1.5% agarose gel. Following transfer to a nylon membrane, hybridization was carried out with 32P-labeled single-stranded probes (21) derived from the coding region of the 7α-hydroxylase cDNA. The filter was washed as described previously (21) and subjected to autoradiography with intensifying screens at −70 °C for 96 h. Bottom, the blot from panel A was stripped of 32P radioactivity and reprobed with a cDNA corresponding to the cis-trans-prolyl isomerase mRNA. For the tissues that had low levels of this mRNA (adrenal, duodenum, pancreatic islets), previous studies with this same filter have shown that β-actin mRNA was present in these lanes (32).
We next examined the dietary regulation of hepatic 7α-hydroxylase mRNA expression. Rats were fed normal chow or chow supplemented with cholestyramine, bile acids, or cholesterol for periods of 7–10 days. Polyadenylated RNA was prepared from the pooled livers of multiple animals and subjected to blot analysis with probes corresponding to three different mRNAs. One probe was derived from the coding region of 7α-hydroxylase (Fig. 5). A second probe was derived from the cDNA for HMG-CoA synthase, an enzyme in the cholesterol biosynthetic pathway which is subject to negative feedback regulation by dietary cholesterol (33, 34). The third probe was complementary to the cis-trans-prolyl isomerase mRNA.
As indicated in Fig. 7A, the presence of chenodeoxycholic acid (first lane) or cholic acid (last lane) in the diet suppressed mRNA levels for 7α–hydroxylase relative to animals maintained on a normal chow diet (fourth lane). Conversely, cholestyramine (second lane) or cholesterol (third lane) in the diet induced 7α-hydroxylase mRNA levels. The mRNA for HMG-CoA synthase (Fig. 7B) demonstrated a similar pattern of suppression in the presence of dietary bile acids (first and last lanes) and of induction by cholestyramine (second lane). However, in contrast to the results obtained with 7α-hydroxylase, cholesterol suppressed hepatic levels of HMG-CoA synthase mRNA (third lane). Hybridization with the prolyl isomerase probe (Fig. 7B, bottom) indicated that near equal amounts of RNA were present in all lanes.
DISCUSSION
The current paper describes the purification, sequence, cDNA cloning, expression, and regulation at the protein and mRNA levels of rat hepatic 7α-hydroxylase. The structure and expression of this enzyme agree well with that described by Noshiro et al. (12).
The protein sequence of 7α-hydroxylase as deduced by peptide sequencing and from the cDNA (Fig. 5) indicates that this enzyme is a member of the cytochrome P-450 superfamily. Some 35% of the amino acids in the protein have hydrophobic side chains, and the ubiquitously conserved cysteine that is thought to be a protein ligand for the heme group is located at residue 444. Comparison of the sequence with those of other cytochrome P-450 enzymes (35) indicates that the 7α-hydroxylase represents a novel family of cytochrome P-450s (family VII) with a gene nomenclature of CYP73.
The 7α-hydroxylase protein migrated on SDS-polyacrylamide gels as a single band with an Mr of 50,300, well separated from the cytochrome P-450a and cytochrome P-450g species (Fig. 2). This migration is somewhat anomalous as 7α-hydroxylase is only 9 and 13 amino acids larger than the cytochrome P-450a (28) and cytochrome P-450g (27) proteins, respectively. Although well resolved on an SDS-polyacrylamide gel, the enzymes co-chromatographed on a large number of different resins. At the primary structure level, cytochrome P-450a and cytochrome P-450g are 46% identical in sequence and are classified into two subfamilies (IIA and HC, respectively) (35). The sequence of 7α-hydroxylase is only 22% identical to these two proteins and as mentioned above, is classified into a different family (family VII). Despite these differences, however, the chromatographic behavior of these proteins indicates that they must share similar hydrophobic and hydrophilic surface properties.
At least four mRNAs for 7α-hydroxylase are present in the rat liver (Figs. 6 and and7).7). It is not known whether the multiple mRNAs are the products of the same gene or of different members of a closely related gene family. However, all of the cDNA clones isolated in this study were identical in sequence to that reported in Fig. 5, and the four mRNAs demonstrated the same pattern of regulation (Fig. 7). The most abundant mRNA species has a size of approximately 3.6 kilobases (Fig. 7) and has an unusually long 3′-untranslated region of 2002 nucleotides. Contained within this region of the mRNA is a single copy of a repetitive sequence of the rat Alu family (Fig. 5). The Alu family in rodents is composed of DNA repeats that are approximately 130 base pairs in length and that are homologous to the right arm of the bipartite human Alu sequence (36). Although the function of these sequences is at present not known, they are found in a number of mature mRNAs in the rat, including several encoding cytochrome P-450s (26). Interestingly, multiple copies of an Alu repeat are also located in the 3′-untranslated region of the human low density lipoprotein receptor mRNA (37). The regulatory significance if any of this observation remains to be determined.
The RNA-blotting results of Figs. 6 and and77 demonstrate that 7α-hydroxylase expression is limited to the liver and subject to feedback regulation by product as well as induction by substrate. These features of 7α-hydroxylase expression underscore the unique and important regulatory role of this microsomal cytochrome P-450 in the bile acid biosynthetic pathway. By way of comparison, the expression of sterol 26-hydroxylase, a mitochondrial cytochrome P-450 that catalyzes oxidations at the 26 position of sterol intermediates in this pathway, is neither liver specific nor subject to feedback regulation in this organ (22).
The mechanism by which dietary bile acids decrease the expression of 7α-hydroxylase mRNA remains to be determined. Given the precedence established for the regulation of other genes involved in cholesterol homeostasis (5–8), the observations reported here suggest the existence of DNA sequences in the promoter of the 7α-hydroxylase gene which respond to bile acids. The proteins that interact with these sequences would presumably sense the levels of bile acids in the enterohepatic circulation and regulate the expression of the gene accordingly. However, the addition of bile acids to the medium of cultured rat hepatocytes did not result in a decrease in bile acid production (38, 39). Similarly, the infusion of bile acids into bile-diverted animals did not down-regulate bile acid biosynthesis (40). Taken together, these results suggest that either an intact enterohepatic circulation is required for the regulation observed here or that bile acids act indirectly to influence 7α-hydroxylase expression (40), perhaps by influencing the total flux of cholesterol across the liver (41).
That the expression of 7α-hydroxylase mRNA can respond to cholesterol is demonstrated by the increase in the levels of this mRNA upon cholesterol feeding (Fig. 7). This response predicts the existence of regulatory elements in the promoter of the gene which respond in a positive fashion to cholesterol. Such elements may or may not be different from the sterol regulatory elements identified in genes such as HMG-CoA synthase and others in the cholesterol supply pathways which respond negatively to cholesterol (Fig. 7 and Refs. 5–7). Similarly, the 7α-hydroxylase promoter may share sequences with certain apolipoprotein genes whose expression is enhanced by dietary cholesterol (42). Clearly, the elucidation of the mechanisms that underlie the regulated expression of 7α-hydroxylase will be a rich area of future study.
Acknowledgments
We thank Henry Danielsson, Bengt Gustavsson, and Kjell Wikvall for invaluable assistance during the early stages of this work; Lyman Bilhartz, John Dietschy, and David Spady for bile acid and dietary advice; William Campbell, Barbara Clark, Don Capra, Elliot Ross, and Michael Waterman for protein purification and sequencing advice; John Leffert for the RNA blot used in Fig. 6; and Michael Brown and Joseph Goldstein for critical reading of the manuscript. Richard Gibson provided able veterinary assistance; and Steven Bauer, Kristine Cala, Daphne Davis, Joan Hsu, Carolyn Moomaw, Daphne Norsworthy, and Edith Womack provided excellent technical assistance.
Footnotes
*This research was supported in part by National Institutes of Health Grant HL 20948, Robert A. Welch Foundation Grant 1-0971, and by the Perot Family Foundation.
The nucleotide sequence(s) reported in this paper has been submitted to the GenBank™/EMBL Data Bank with accession number(s) J05430.
1The abbreviations used are: 7α-hydroxylase, cholesterol 7α-monooxygenase (EC 1.14.13.17); DTT, dithiothreitol; SDS, sodium dodecyl sulfate; kb, kilobase(s); HMG-CoA, 3-hydroxy-3-methylglu-taryl-coenzyme A; HPLC, high performance liquid chromatography.
2S. Andersson, unpublished observations.
3D. W. Nebert, personal communication.
REFERENCES
Full text links
Read article at publisher's site: https://doi.org/10.1016/s0021-9258(19)39056-8
Free to read at intl.jbc.org
http://intl.jbc.org/cgi/content/abstract/265/14/8190
Free after 12 months at intl.jbc.org
http://intl.jbc.org/cgi/reprint/265/14/8190.pdf
Citations & impact
Impact metrics
Citations of article over time
Alternative metrics
Smart citations by scite.ai
Explore citation contexts and check if this article has been
supported or disputed.
https://scite.ai/reports/10.1016/s0021-9258(19)39056-8
Article citations
An overview of the cholesterol metabolism and its proinflammatory role in the development of MASLD.
Hepatol Commun, 8(5):e0434, 02 May 2024
Cited by: 1 article | PMID: 38696365 | PMCID: PMC11068152
Review Free full text in Europe PMC
Macrophage inhibitory cytokine-1 aggravates diet-induced gallstone formation via increased ABCG5/ABCG8 expression.
PLoS One, 18(6):e0287146, 13 Jun 2023
Cited by: 0 articles | PMID: 37310967 | PMCID: PMC10263326
The regulatory role of bile acid microbiota in the progression of liver cirrhosis.
Front Pharmacol, 14:1214685, 21 Jun 2023
Cited by: 1 article | PMID: 37416060 | PMCID: PMC10320161
Review Free full text in Europe PMC
My lifelong dedication to bile acid research.
J Biol Chem, 299(5):104672, 03 Apr 2023
Cited by: 0 articles | PMID: 37019215 | PMCID: PMC10173005
Interaction between gut microbiota and sex hormones and their relation to sexual dimorphism in metabolic diseases.
Biol Sex Differ, 14(1):4, 07 Feb 2023
Cited by: 19 articles | PMID: 36750874 | PMCID: PMC9903633
Review Free full text in Europe PMC
Go to all (190) article citations
Data
Data behind the article
This data has been text mined from the article, or deposited into data resources.
BioStudies: supplemental material and supporting data
Nucleotide Sequences
- (1 citation) ENA - J05430
Similar Articles
To arrive at the top five similar articles we use a word-weighted algorithm to compare words from the Title and Abstract of each citation.
Hepatic P-450 cholesterol 7 alpha-hydroxylase. Regulation in vivo at the protein and mRNA level in response to mevalonate, diurnal rhythm, and bile acid feedback.
J Biol Chem, 265(25):15090-15095, 01 Sep 1990
Cited by: 30 articles | PMID: 2394714
Regulation of cholesterol 7 alpha-hydroxylase in the liver. Cloning, sequencing, and regulation of cholesterol 7 alpha-hydroxylase mRNA.
J Biol Chem, 265(20):12012-12019, 01 Jul 1990
Cited by: 107 articles | PMID: 1694852
Purification of cholesterol 7 alpha-hydroxylase from human and rat liver and production of inhibiting polyclonal antibodies.
J Biol Chem, 265(8):4541-4546, 01 Mar 1990
Cited by: 15 articles | PMID: 2106520
[New bile acid biosynthesis pathways].
Gastroenterol Clin Biol, 25(1):81-92, 01 Jan 2001
Cited by: 2 articles | PMID: 11275621
Review
Funding
Funders who supported this work.
NHLBI NIH HHS (2)
Grant ID: HL 20948
Grant ID: P01 HL020948
NIGMS NIH HHS (1)
Grant ID: GM 12783