Abstract
Free full text

Comparative Genomic Analysis of East Asian and Non-Asian Helicobacter pylori Strains Identifies Rapidly Evolving Genes
Associated Data
Abstract
Helicobacter pylori infection is a risk factor for the development of gastric adenocarcinoma, a disease that has a high incidence in East Asia. Genes that are highly divergent in East Asian H. pylori strains compared to non-Asian strains are predicted to encode proteins that differ in functional activity and could represent novel determinants of virulence. To identify such proteins, we undertook a comparative analysis of sixteen H. pylori genomes, selected equally from strains classified as East Asian or non-Asian. As expected, the deduced sequences of two known virulence determinants (CagA and VacA) are highly divergent, with 77% and 87% mean amino acid sequence identities between East Asian and non-Asian groups, respectively. In total, we identified 57 protein sequences that are highly divergent between East Asian and non-Asian strains, but relatively conserved within East Asian strains. The most highly represented functional groups are hypothetical proteins, cell envelope proteins and proteins involved in DNA metabolism. Among the divergent genes with known or predicted functions, population genetic analyses indicate that 86% exhibit evidence of positive selection. McDonald-Kreitman tests further indicate that about one third of these highly divergent genes, including cagA and vacA, are under diversifying selection. We conclude that, similar to cagA and vacA, most of the divergent genes identified in this study evolved under positive selection, and represent candidate factors that may account for the disproportionately high incidence of gastric cancer associated with East Asian H. pylori strains. Moreover, these divergent genes represent robust biomarkers that can be used to differentiate East Asian and non-Asian H. pylori strains.
Introduction
Over half of the world’s human population is persistently colonized with Helicobacter pylori, a Gram-negative bacterium that inhabits the human stomach. H. pylori infection is an important risk factor for gastric adenocarcinoma, peptic ulcer disease, and gastric mucosa-associated lymphoid tissue (MALT) lymphoma [1], [2], [3], [4]. Gastric adenocarcinoma is the second leading cause of cancer-related death worldwide [5], [6], [7], [8], [9]. The incidence of this malignancy varies globally, and is particularly high in several parts of East Asia [5], [7], [8].
There is a high level of genetic diversity among H. pylori strains from unrelated humans, which has been attributed to an elevated mutation rate and a high rate of intraspecies genetic recombination [10], [11], [12]. Multiple populations and subpopulations of H. pylori with distinct geographic distributions have been recognized, based on multilocus sequence typing (MLST) analysis of conserved housekeeping genes [13], [14], [15]. Genetic diversity in H. pylori decreases with geographic distance from eastern Africa, a finding that is consistent with an African origin of H. pylori [16], [17], [18].
Considerable effort has been devoted to analyzing two important H. pylori virulence factors, CagA and VacA, which each exhibit a high level of intraspecies genetic diversity. VacA is a secreted pore-forming toxin that causes multiple alterations in human cells, including cell vacuolation, apoptosis, and inhibition of T-cell activation and proliferation [19]. CagA alters numerous signaling pathways, many of which are associated with malignant transformation of cells [20], [21]. Thus, CagA has been termed a “bacterial oncoprotein” [20], [22]. Within gastric epithelial cells, CagA undergoes tyrosine-phosphorylation at sites known as EPIYA motifs (EPIYA-A, B, C and D), and such phosphorylation is required for many of its actions on host cells [20], [21].
The cagA gene is located within a ~40 kb chromosomal region known as the cag pathogenicity island (cag PAI), which also contains genes encoding components of a type IV secretion system that translocates CagA into gastric epithelial cells [23], [24]. Some H. pylori genomes contain an intact cag PAI, some strains contain a partial cag PAI, and others lack the cag PAI [23]. All strains contain vacA, but there is variation among strains in levels of vacA expression and VacA activity [19]. Based on observed similarities in the phylogenies of VacA and CagA in large numbers of strains, it has been suggested that these functionally interacting proteins have co-evolved in a manner that facilitates H. pylori colonization of the human stomach [18], [25]. Strains containing the cag PAI and expressing active forms of VacA are associated with a higher risk of gastric disease than are strains that lack these features [26], [27], [28].
To account for the high incidence of gastric cancer in East Asia, one hypothesis is that H. pylori strains from East Asia are more virulent or more frequently produce specific oncogenic factors than do strains from other parts of the world with lower rates of gastric cancer. Several lines of evidence support this hypothesis. Specifically, most H. pylori strains from East Asia contain the cag PAI and produce active forms of VacA [29]. Moreover, the CagA and VacA sequences found in many East Asian H. pylori strains are phylogenetically distinct from corresponding sequences found in non-Asian strains [18], [25]. East Asian strains typically produce a form of CagA that contains a tyrosine phosphorylation motif known as EPIYA-D, whereas non-Asian forms of CagA typically contain an EPIYA-C tyrosine phosphorylation motif [20], [21]. CagA proteins that contain an EPIYA-D motif have been associated with increased activity in vitro compared to other forms of CagA [30], [31]. East Asian strains of H. pylori often contain vacA alleles with a distinct set of 5′ polymorphisms known as type s1c and a form of the vacA mid-region that is highly divergent compared to vacA mid-regions found in non-East Asian strains [18], [32], [33]. Sequence differences in AlpA/B adhesins of East Asian strains compared to non-Asian strains have been associated with differences in functional activity, including variations in intracellular signaling [34]. The results of several studies suggest there may be many other functionally important differences when comparing East Asian and non-Asian strains [35], [36], [37]. Although several previous studies have analyzed diversity in East Asian strains of H. pylori compared to non-Asian strains, most of these studies were limited by the availability of only a small number of whole genome sequences or inclusion of a restricted number of genes in the analysis.
To identify candidate genes that could underlie the disproportionately high incidence of gastric cancer associated with East Asian strains, we set out to systematically compare East Asian H. pylori genomes with non-Asian genomes and identify rapidly evolving genes. By performing comparative genomic and phylogenetic analyses of 16 whole genome sequences (from eight East Asian and eight non-Asian H. pylori strains), we report the following key results: (i) 57 proteins, including CagA and VacA, are highly divergent in East Asian H. pylori strains compared to non-Asian strains, but relatively conserved within East Asian strains. (ii) The most highly represented functional groups of divergent proteins are hypothetical proteins, cell envelope proteins and proteins involved in DNA metabolism. (iii) These highly divergent genes exhibit significantly higher Ka/Ks ratios than control housekeeping genes, suggesting that the highly divergent genes experience more positive selection. (iv) Finally, diversifying selection has driven the divergence of about one third of the highly divergent genes, including cagA and vacA, and these genes exhibit sequence signatures of a reduction in effective population size, as measured by the mean nucleotide diversity of synonymous sites (πs). We propose that, similar to CagA and VacA, these proteins represent a panel of candidates that may contribute to H. pylori virulence and may account for the high incidence of gastric cancer associated with East Asian H. pylori strains. Moreover, these divergent genes represent robust biomarkers that can be used to differentiate East Asian and non-Asian H. pylori strains.
Materials and Methods
Selection of H. pylori Strains for Comparative Analysis
To identify strains for inclusion in this study, we evaluated all complete or nearly complete genome sequences that were available in Genbank at the time when the study was initiated. To assign these H. pylori strains to previously described populations and subpopulations, we used multilocus sequence typing (MLST) analysis [13]. Partial nucleotide sequences of 7 conserved housekeeping genes (atpA, efp, mutY, ppa, trpC, yphC, and ureI) from each strain were concatenated and aligned to corresponding loci from 445 reference strains contained in a MLST database (http://pubmlst.org/helicobacter) using the Muscle algorithm within MEGA5 [38]. Phylogenetic relationships were analyzed using MEGA5 with the Kimura 2-parameter model of nucleotide substitution, neighbor-joining clustering, and 10,000 bootstrap replicates. This led to the identification of 8 strains that were classified as East Asian (hspEAsian) (F16, F30, F32, F57, 35A, 51, 52 and 98-10) [35], [37], [39]. Six of these strains (F16, F30, F32, F57, 98-10, and 35A) were originally isolated from patients in Japan, and two were from Korea (51 and 52). We selected the same number of strains that were distantly related to East Asian strains and classified as non-Asian (either hpEurope or hpAfrica1), based on MLST analysis (26695, J99, HPAG1, G27, P12, B8, B38, 908) [40], [41], [42], [43], [44], [45], [46], [47].
Identification and Classification of Genes Encoding Highly Divergent Proteins
Whole genome sequences were retrieved from Genbank and protein sequences were extracted using Bioperl [48]. As a first step to identify predicted gene products that are highly divergent in East Asian strains when compared to non-Asian strains, we conducted Blast Score Ratio (BSR) analysis [49]. This approach allows for comparisons among 3 strains (2 query strains against a single reference strain). To evaluate protein sequence similarity, two Blast score ratios are calculated, based on comparison of a query sequence (BS Q1 or BS Q2) to a reference sequence (BS Ref). Thus, BSR1 =
= (BS Q1)/(BS Ref) and BSR2
(BS Q1)/(BS Ref) and BSR2 =
= (BS Q2)/(BS Ref). In this manner, all scores are normalized in the range of 0 to 1 [49]. For example, if a perfect match is found between a protein in the reference strain and a protein in a query strain, this corresponds to a BSR of 1.0. BSR analysis was performed using comparisons of each of the 8 non-Asian strains (used as Query strains) with the 8 East Asian strains (used as reference strains). For analysis of each East Asian strain, we selected proteins that yielded 0.4≤BSR≤0.93 in comparisons with at least 6 of the 8 non-Asian strains. The lower threshold value (BSR
(BS Q2)/(BS Ref). In this manner, all scores are normalized in the range of 0 to 1 [49]. For example, if a perfect match is found between a protein in the reference strain and a protein in a query strain, this corresponds to a BSR of 1.0. BSR analysis was performed using comparisons of each of the 8 non-Asian strains (used as Query strains) with the 8 East Asian strains (used as reference strains). For analysis of each East Asian strain, we selected proteins that yielded 0.4≤BSR≤0.93 in comparisons with at least 6 of the 8 non-Asian strains. The lower threshold value (BSR =
= 0.4) represents approximately 30% amino acid identity over approximately 30% of the peptide length, a commonly used threshold for peptide similarity [49]. The upper threshold value (BSR≤0.93) was chosen empirically in a manner so that we would detect proteins such as VacA (mean BSR
0.4) represents approximately 30% amino acid identity over approximately 30% of the peptide length, a commonly used threshold for peptide similarity [49]. The upper threshold value (BSR≤0.93) was chosen empirically in a manner so that we would detect proteins such as VacA (mean BSR =
= 0.93, calculated by averaging all of the BSR values resulting from all comparisons of East Asian and non-Asian strains), which although less divergent than CagA, is known to be divergent when comparing East Asian and non-Asian strains [18]. This approach, involving analysis of about 1500 proteins from each strain, led to the identification of 1140 candidate divergent proteins. Subsequently, we sought to refine this list by identifying protein sequences that were relatively conserved within the East Asian population, as might be expected if the corresponding genes arose from a process involving positive selection, and excluding genes that exhibited a very high rate of overall sequence divergence that was unrelated to geographic origin of strains. To do this, we performed comparisons among the 8 East Asian strains, and for each strain, we selected proteins that yielded BSR≥0.90 in comparisons with at least 6 other East Asian strains. This led to the identification of 159 candidate divergent proteins that were selected for further analysis.
0.93, calculated by averaging all of the BSR values resulting from all comparisons of East Asian and non-Asian strains), which although less divergent than CagA, is known to be divergent when comparing East Asian and non-Asian strains [18]. This approach, involving analysis of about 1500 proteins from each strain, led to the identification of 1140 candidate divergent proteins. Subsequently, we sought to refine this list by identifying protein sequences that were relatively conserved within the East Asian population, as might be expected if the corresponding genes arose from a process involving positive selection, and excluding genes that exhibited a very high rate of overall sequence divergence that was unrelated to geographic origin of strains. To do this, we performed comparisons among the 8 East Asian strains, and for each strain, we selected proteins that yielded BSR≥0.90 in comparisons with at least 6 other East Asian strains. This led to the identification of 159 candidate divergent proteins that were selected for further analysis.
As a complementary analytical approach, we further analyzed the predicted protein sequences encoded by the 8 East Asian and 8 non-Asian strains of H. pylori using nWayComp analysis, which allows for the comparison of protein sequences among multiple strains at the whole-genome level [50]. nWayComp analysis compares DNA or protein sequences, searches for homologous sequences among multiple strains, and identifies genes or proteins that are either unique to a particular strain or are encoded in multiple strains. For each set of orthologous sequences, we generated a table of maximum size n×n, where n =
= 16, which displayed amino acid sequence identities among the analyzed sequences. Mean percent amino acid identities were calculated based on all possible comparisons of East Asian sequences with orthologous non-Asian sequences. Sequences were excluded if there were marked differences in peptide lengths (when compared to orthologous sequences in other strains) or in cases in which proteins had been incorrectly identified as orthologues. This manual curation resulted in a reduction in the number of highly divergent gene products from 159 to 57. To further examine the divergent gene products selected with BSR and nWayComp analyses, neighbor-joining trees were constructed for each of the 57 proteins, using the program Geneious (Drummond AJ, Ashton B, Buxton S, Cheung M, Cooper A, Heled J, Kearse M, Moir R, Stones-Havas S, Sturrock S, Thierer T, Wilson A (2010) Geneious v5.1, available from http://www.geneious.com). Trees were inspected to determine whether East Asian sequences clustered together or whether East Asian and non-Asian sequences were intermingled. We also performed Bayesian analyses of a subset of trees to ensure that the neighbor joining inference methods were accurate [51], [52]. ProtTest model selection and Bayesian inference generally recapitulated the patterns observed in neighbor joining trees, and consistently revealed clustering of East Asian sequences. Predicted main functional classes and sub-functional classes for each of 57 divergent sequences were assigned based on previous classifications (J. Craig Venter Institute Comprehensive Microbial Resource database).
16, which displayed amino acid sequence identities among the analyzed sequences. Mean percent amino acid identities were calculated based on all possible comparisons of East Asian sequences with orthologous non-Asian sequences. Sequences were excluded if there were marked differences in peptide lengths (when compared to orthologous sequences in other strains) or in cases in which proteins had been incorrectly identified as orthologues. This manual curation resulted in a reduction in the number of highly divergent gene products from 159 to 57. To further examine the divergent gene products selected with BSR and nWayComp analyses, neighbor-joining trees were constructed for each of the 57 proteins, using the program Geneious (Drummond AJ, Ashton B, Buxton S, Cheung M, Cooper A, Heled J, Kearse M, Moir R, Stones-Havas S, Sturrock S, Thierer T, Wilson A (2010) Geneious v5.1, available from http://www.geneious.com). Trees were inspected to determine whether East Asian sequences clustered together or whether East Asian and non-Asian sequences were intermingled. We also performed Bayesian analyses of a subset of trees to ensure that the neighbor joining inference methods were accurate [51], [52]. ProtTest model selection and Bayesian inference generally recapitulated the patterns observed in neighbor joining trees, and consistently revealed clustering of East Asian sequences. Predicted main functional classes and sub-functional classes for each of 57 divergent sequences were assigned based on previous classifications (J. Craig Venter Institute Comprehensive Microbial Resource database).
Analysis of Mean Nucleotide Diversity and Positive Selection
For analysis of nucleotide diversity and positive selection, sequences of orthologous genes were aligned using Muscle in Geneious, version 5.4.5. Hypervariable regions and insertions/deletions (indels) were manually removed. Nucleotide divergence at non-synonymous and synonymous sites (Ka and Ks, respectively, with Jukes and Cantor correction) and silent site diversity (πs) was calculated for each set of orthologous sequences with the program DnaSP (http://www.ub.edu/dnasp). Sequences from East Asian strains were compared with corresponding sequences from non-Asian strains.
The McDonald-Kreitman test (http://mkt.uab.es/mkt/) for positive selection [53] was performed with the exclusion of low-frequency variants less than or equal to 15% to reduce artifacts associated with detecting adaptive evolution. The neutrality index (NI) was calculated as follows: NI =
= (Pn/Ps)/(Dn/Ds), where P is polymorphic within the population, D is divergence or fixed difference between populations, n is nonsynonymous, and s is synonymous.
(Pn/Ps)/(Dn/Ds), where P is polymorphic within the population, D is divergence or fixed difference between populations, n is nonsynonymous, and s is synonymous.
Results
MLST Analysis of H. pylori Strains
In an analysis of H. pylori strains for which complete or nearly-complete genome sequences were available in Genbank when this study was initiated, we identified eight strains that were classified as East Asian (hspEAsia), based on MLST analysis (Fig. 1). We selected the same number of genome sequences from strains that were classified as non-Asian, based on MLST analysis. Six of the latter strains were classified as hpEurope and two were classified as hspWAfrica (Fig. 1). The assignment of the 16 strains to these population groups is consistent with previous analyses [23].
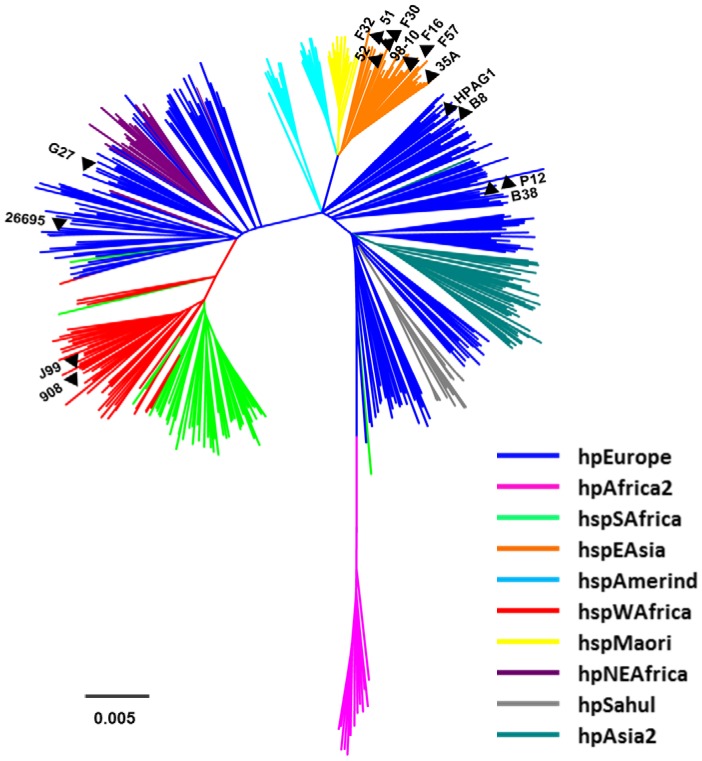
Nucleotide sequences of 7 conserved housekeeping genes (atpA, efp, mutY, ppa, trpC, ureI, and yphC) from 16 strains of H. pylori were concatenated and compared to corresponding loci from 445 reference strains (see Methods). Eight strains (98-10, 35A, 51, 52, F16, F30, F32 and F57) were classified as hspEAsia, six strains (26695, HPAG1, G27, P12, B8 and B38) were classified as hpEurope and two strains (J99 and 908) were classified as hspWAfrica.
Identification of Highly Divergent Alleles in East Asian Strains
The sequences of two virulence factors, CagA and VacA, are known to be highly divergent in East Asian strains compared to non-Asian strains [18], [25]. Consistent with expectations, all eight East Asian strains in the current study (98-10, 35A, F16, F30, F32, F57, 51 and 52) encode CagA proteins with an EPIYA-D motif, whereas 7 non-Asian strains (26695, J99, HPAG1, G27, P12, B8 and 908) encode CagA with an EPIYA-C motif (Table 1). The eight non-Asian strain (B38) does not contain the cag pathogenicity island, and therefore, this strain does not contain cagA. Seven out of the eight East Asian strains contain an s1c vacA allele, whereas the s1c genotype was not identified in any of the non-Asian strains. These features of CagA and VacA conform to the profiles that are predicted based on the MLST classification of the 16 strains.
Table 1
| Strain | cag PAI | CagA type (EPIYA) | VacA type |
| 98-10 | + | EPIYA-D | s1c/i1/m1 |
| 35A | + | EPIYA-D | s1c/i1/m1 |
| F16 | + | EPIYA-D | s1c/i1/m1 |
| F30 | + | EPIYA-D | s1c/i1/m1 |
| F32 | + | EPIYA-D | s1c/i1/m1 |
| F57 | + | EPIYA-D | s1a/i1/m1 |
| 51 | + | EPIYA-D | s1c/i1/m1 |
| 52 | + | EPIYA-D | s1c/i1/m1a |
| 26695 | + | EPIYA-C | s1a/i1/m1 |
| J99 | + | EPIYA-C | s1a/i1/m1 |
| HPAG1 | + | EPIYA-C | s1a/i1/m1 |
| G27 | + | EPIYA-C | s1b/i1/m1 |
| P12 | + | EPIYA-C | s1a/i1/m1 |
| B8 | + | EPIYA-C | s1a/i2/m2a |
| 908 | + | EPIYA-C | s1b/i1/m1 |
| B38 | − | Not applicableb | s2/i2/m2 |
Neighbor-joining tree analyses confirmed that CagA and VacA were highly divergent in East Asian strains of H. pylori compared to non-Asian strains (Fig. S1). Similar analysis of concatenated housekeeping gene sequences revealed that East Asian strains were distinguishable from non-Asian strains, but the level of divergence among housekeeping genes was much lower than observed for CagA and VacA (compare Fig. S1C with Fig. S1A, B). When comparing 7 housekeeping genes from East Asian and non-Asian strains of H. pylori, the mean amino acid identity (based on all possible comparisons of orthologous sequences) was 96%. In contrast, the mean amino acid identity of CagA sequences between groups was 77%, and the mean amino acid identity of VacA sequences between groups was 87%. Therefore, when East Asian strains are compared to non-Asian strains, there is a much higher level of divergence in CagA and VacA than in the products of housekeeping genes.
To identify other proteins encoded by East Asian strains that might be highly divergent compared to those encoded by non-Asian strains, we compared the protein sequences of eight East Asian H. pylori strains and eight non-Asian strains, using Blast Score Ratio and nWayComp analyses (described in Methods). We identified 57 predicted gene products, including CagA and VacA, that were highly divergent between East Asian and non-Asian strains and relatively conserved within the East Asian group (Table 2). Analysis of these proteins indicated that the intergroup differences in amino acid identities ranged from 71%–91% (Table 3). As shown in Table 2, the 57 divergent proteins were grouped based upon their predicted main functional class (J. Craig Venter Institute Comprehensive Microbial Resource database). The most highly represented groups were hypothetical proteins, cell envelope proteins, and proteins involved in DNA metabolism (Table 2).
Table 2
| Main rolea | Subrole | Gene IDb | Annotationc,d |
| Cell envelope | Other | HP0009 | outer membrane protein HopZ (omp1) |
| Cell envelope | Other | HP0025 | outer membrane protein HopD (omp2) |
| Cell envelope | Other | HP1243 | outer membrane protein BabA (omp28) |
| Cell envelope | Other | HP0373 | outer membrane protein HomC/HomD |
| Cell envelope | Other | NAe | outer membrane protein HomB |
| Cell envelope | Other | HP0725 | outer membrane protein SabA/HopP (omp17) |
| Cell envelope | Other | HP0923 | outer membrane protein HopK (omp12) |
| Cell envelope | Other | HP0229 | outer membrane protein HopA (omp6) |
| Cell envelope | Other | HP1157 | outer membrane protein HopL (omp26) |
| Cell envelope | Other | HP0609/0610 | vacuolating cytotoxin (VacA)-like protein |
| Cell envelope | Other | HP0922 | vacuolating cytotoxin (VacA)-like protein |
| Cell envelope | Other | HP0492 | HpaA-like protein |
| Cell envelope | Biosynthesis and degradation of surface polysaccharides and lipopolysaccharides | HP0651 | alpha-(1,3)-fucosyltransferase |
| Cell envelope | Biosynthesis and degradation of surface polysaccharides and lipopolysaccharides | HP0159 | lipopolysaccharide 1,2-glucosyltransferase (RfaJ) |
| Cell envelope | Biosynthesis and degradation of murein sacculus and peptidoglycan | HP0160 | cysteine-rich protein D/beta-lactamase HcpD |
| Cellular processes | Pathogenesis | HP0547 | cytotoxin associated protein A (CagA) |
| Cellular processes | Toxin production and resistance | HP0887 | vacuolating cytotoxin A (VacA) |
| Cellular processes | Chemotaxis and motility | HP0906 | flagellar hook-length control protein |
| DNA metabolism | DNA replication, recombination, and repair | HP1553 | recombination protein RecB/helicase |
| DNA metabolism | DNA replication, recombination, and repair | HP0661 | ribonuclease H (RnhA) |
| DNA metabolism | DNA replication, recombination, and repair | HP1323 | ribonuclease HII (RnhB) |
| DNA metabolism | Restriction/modification | HP0463 | type I restriction enzyme M protein/HsdM |
| DNA metabolism | Restriction/modification | HP0850 | type I restriction enzyme M protein (HsdM) |
| DNA metabolism | Restriction/modification | HP1354 | type IIG restriction-modification enzyme/adenine specific DNA methyltransferase |
| DNA metabolism | Restriction/modification | HP1371 | type III restriction enzyme R protein |
| Protein fate | Degradation of proteins, peptides, and glycopeptides | HP0806 | metalloprotease |
| Protein fate | Protein and peptide secretion and trafficking | HP1255 | preprotein translocase subunit SecG |
| Protein synthesis | tRNA and rRNA base modification | HP1415 | tRNA delta(2)-isopentenylpyrophosphate transferase (MiaA) |
| Protein synthesis | tRNA aminoacylation | HP1513 | selenocysteine synthase (SelA)/L-seryl-tRNA(Sec)selenium transferase |
| Purines, pyrimidines, nucleosides, and nucleotides | Purine ribonucleotide biosynthesis | HP1530 | purine nucleoside phosphorylase (PunB) |
| Transcription | RNA processing | HP0640 | poly(A) polymerase (PapS) |
| Unknown function | General | HP0322 | poly E-rich protein |
| Hypothetical | Conserved | HP0728 | tRNA(Ile)-lysidine synthase (TilS) |
| Hypothetical | Conserved | HP0729 | probable ATP/GTP binding protein |
| Hypothetical | Conserved | HP1250 | bacterial SH3 domain protein |
| Hypothetical | Conserved | HP0852 | excinuclease ATPase subunit |
| Hypothetical | Conserved | HP1265 | NADH-ubiquinone oxidoreductase chain F (NuoF) |
| Hypothetical | Conserved | HP0721 | hypothetical protein |
| Hypothetical | Conserved | HP0636 | hypothetical protein |
| Hypothetical | Conserved | HP1579 | hypothetical protein |
| Hypothetical | Conserved | HP0861 | hypothetical protein |
| Hypothetical | Conserved | HP0384 | hypothetical protein |
| Hypothetical | Conserved | HP0635 | hypothetical protein |
| Hypothetical | Conserved | HP0897 | hypothetical protein |
| Hypothetical | Conserved | HP0398 | hypothetical protein |
| Hypothetical | Conserved | HP0629 | hypothetical protein |
| Hypothetical | Conserved | HP0973 | hypothetical protein |
| Hypothetical | Conserved | HP0167 | hypothetical protein |
| Hypothetical | Conserved | HP0120 | hypothetical protein |
| Hypothetical | Conserved | HP0583 | hypothetical protein |
| Hypothetical | Conserved | HP0119 | hypothetical protein |
| Hypothetical | Conserved | HP0681 | hypothetical protein |
| Hypothetical | Conserved | HP1321 | hypothetical protein |
| Hypothetical | Conserved | HP0833 | hypothetical protein |
| Hypothetical | Conserved | HP0338 | hypothetical protein |
| Hypothetical | Conserved | HP0061 | hypothetical protein |
| Hypothetical | Conserved | HP1322 | hypothetical protein |
| Control Group | |||
| Energy metabolism | ATP-proton motive force interconversion | HP1134 | ATP synthase F0F1 subunit alpha (AtpA) |
| Protein synthesis | Translation factors | HP0177 | elongation factor P (Efp) |
| DNA metabolism | DNA replication, recombination, and repair | HP0142 | A/G-specific adenine glycosylase (MutY) |
| Central intermediary metabolism | Phosphorus compounds | HP0620 | inorganic pyrophosphatase (Ppa) |
| Tryptophan biosynthesis | Aromatic amino acid family | HP1279 | anthranilate isomerase (TrpC) |
| Central intermediary metabolism | Other | HP0071 | urease accessory protein (UreI) |
| Unknown function | General | HP0834 | GTP-binding protein (YphC) |
Table 3
| Annotation | Gene ID (26695) | Mean % aa identity (EA vs. Non-EA)b | πa-EA | πa-Non EA | πs-EA | πs-Non EA | Ka/Ks (EA-NEA)c |
| HopZ (omp1) | HP0009 | 73.61 | 0.069 | 0.039 | 0.228 | 0.186 | 0.264 |
| HopD (omp2) | HP0025 | 88.96 | 0.015 | 0.042 | 0.097 | 0.212 | 0.224 |
| BabA (omp28) | HP1243 | 87.65 | 0.050 | 0.053 | 0.186 | 0.266 | 0.226 |
| HomC/HomD | HP0373 | 80.21 | 0.012 | 0.068 | 0.073 | 0.272 | 0.293 |
| HomB | NAd | 86.77 | 0.053 | 0.048 | 0.195 | 0.236 | 0.220 |
| SabA/HopP/(omp17) | HP0725 | 82.78 | 0.070 | 0.039 | 0.171 | 0.183 | 0.299 |
| HopK (omp12) | HP0923 | 89.23 | 0.021 | 0.040 | 0.100 | 0.205 | 0.197 |
| HopA (omp6) | HP0229 | 89.24 | 0.040 | 0.043 | 0.116 | 0.170 | 0.259 |
| HopL (omp26) | HP1157 | 89.58 | 0.032 | 0.036 | 0.132 | 0.202 | 0.210 |
| VacA-like protein | HP0609/0610 | 91.09 | 0.024 | 0.037 | 0.144 | 0.251 | 0.162 |
| VacA-like protein | HP0922 | 89.93 | 0.020 | 0.027 | 0.083 | 0.167 | 0.197 |
| HpaA-like protein | HP0492 | 71.03 | 0.014 | 0.038 | 0.053 | 0.128 | 0.403 |
| alpha-(1,3)-fucosyltransferase | HP0651 | 81.68 | 0.023 | 0.057 | 0.174 | 0.294 | 0.218 |
| lipopolysaccharide 1,2-glucosyltransferase (rfaJ) | HP0159 | 86.99 | 0.020 | 0.057 | 0.063 | 0.193 | 0.331 |
| cysteine-rich protein D/beta-lactamase (hcpD) | HP0160 | 89.61 | 0.022 | 0.035 | 0.088 | 0.141 | 0.320 |
| cytotoxin associated protein A (cagA) | HP0547 | 77.74 | 0.018 | 0.054 | 0.067 | 0.116 | 0.414 |
| vacuolating cytotoxin A (vacA) | HP0887 | 87.39 | 0.013 | 0.057 | 0.097 | 0.203 | 0.260 |
| flagellar hook-length control protein | HP0906 | 85.76 | 0.030 | 0.049 | 0.108 | 0.178 | 0.279 |
| recombination protein RecB/helicase | HP1553 | 89.52 | 0.020 | 0.032 | 0.102 | 0.101 | 0.229 |
| ribonuclease H (rnhA) | HP0661 | 79.16 | 0.020 | 0.020 | 0.149 | 0.076 | 0.206 |
| ribonuclease HII (rnhB) | HP1323 | 87.90 | 0.035 | 0.042 | 0.165 | 0.173 | 0.222 |
| type I restriction enzyme M protein (hsdM) | HP0463 | 89.23 | 0.025 | 0.037 | 0.091 | 0.147 | 0.250 |
| type I restriction enzyme M protein (hsdM) | HP0850 | 87.75 | 0.026 | 0.044 | 0.131 | 0.208 | 0.213 |
| type IIG restriction-modification enzyme | HP1354 | 82.75 | 0.031 | 0.072 | 0.098 | 0.242 | 0.321 |
| type III restriction enzyme R protein | HP1371 | 83.71 | 0.031 | 0.036 | 0.093 | 0.155 | 0.264 |
| metalloprotease | HP0806 | 87.69 | 0.020 | 0.047 | 0.090 | 0.231 | 0.251 |
| preprotein translocase subunit secG | HP1255 | 89.15 | 0.012 | 0.024 | 0.087 | 0.169 | 0.179 |
| tRNA delta(2)-isopentenylpyrophosphate transferase (miaA) | HP1415 | 80.77 | 0.020 | 0.055 | 0.100 | 0.195 | 0.268 |
| selenocysteine synthase (SelA)/L-seryl-tRNA(Sec) selenium transferase | HP1513 | 89.50 | 0.024 | 0.038 | 0.093 | 0.195 | 0.221 |
| purine nucleoside phosphorylase (punB) | HP1530 | 89.03 | 0.016 | 0.037 | 0.091 | 0.200 | 0.223 |
| poly(A) polymerase (papS) | HP0640 | 89.11 | 0.021 | 0.034 | 0.106 | 0.187 | 0.190 |
| poly E-rich protein | HP0322 | 72.27 | 0.027 | 0.048 | 0.105 | 0.189 | 0.243 |
| tRNA(Ile)-lysidine synthase | HP0728 | 89.96 | 0.017 | 0.034 | 0.078 | 0.150 | 0.236 |
| probable ATP/GTP binding protein | HP0729 | 88.15 | 0.035 | 0.031 | 0.151 | 0.158 | 0.190 |
| bacterial SH3 domain protein | HP1250 | 77.58 | 0.038 | 0.056 | 0.122 | 0.131 | 0.445 |
| Excinuclease ATPase subunit | HP0852 | 83.85 | 0.038 | 0.053 | 0.100 | 0.194 | 0.284 |
| NADH-ubiquinone oxidoreductase chain F | HP1265 | 88.97 | 0.018 | 0.038 | 0.078 | 0.186 | 0.218 |
| Control group | |||||||
| ATP synthase F0F1 subunit alpha (atpA) | HP1134 | 98.00 | 0.003 | 0.003 | 0.076 | 0.105 | 0.027 |
| elongation factor P (efp) | HP0177 | 98.00 | 0.002 | 0.003 | 0.107 | 0.159 | 0.020 |
| A/G-specific adenine glycosylase (mutY) | HP0142 | 94.00 | 0.011 | 0.026 | 0.095 | 0.223 | 0.114 |
| inorganic pyrophosphatase (ppa) | HP0620 | 96.00 | 0.004 | 0.005 | 0.060 | 0.123 | 0.092 |
| anthranilate isomerase (trpC) | HP1279 | 94.00 | 0.020 | 0.032 | 0.088 | 0.188 | 0.173 |
| urease accessory protein (ureI) | HP0071 | 97.00 | 0.001 | 0.007 | 0.047 | 0.103 | 0.061 |
| GTP-binding protein (yphC) | HP0834 | 96.00 | 0.008 | 0.018 | 0.078 | 0.154 | 0.096 |
In order to link predicted functions with patterns of molecular evolution, we identified 37 genes that had been assigned to predicted functional groups or for which annotations were available (Table 2), and focused further analyses on these proteins. We identified intact coding sequences for thirteen of the 37 proteins in all 16 strains, and thus, these 13 genes (HP0159, HP0160, HP0229, HP0640, HP0728, HP0806, HP0906, HP0922, HP1255, HP1265, HP1323, HP1415, HP1513) represent a subset of the core genome. The apparent absence of intact coding sequences was most commonly observed in strains 908 and 98-10. There are many possible reasons for why a particular gene sequence might not be identified in an individual strain; these include absence of the gene from the strain, presence of a truncated gene or pseudogene, or failure to detect the gene due to shortcomings in sequencing, assembly or annotation of a genome. To examine relationships among sequences for the sets of divergent gene products, we constructed neighbor-joining phylogenetic trees for each of the 37 predicted proteins. Phylogenetic analysis (prior to removal of outliers) revealed that for 17 of the 37 proteins, all of the East Asian sequences formed a well-defined cluster that was distinct from non-Asian sequences. These include CagA (HP0547), VacA (HP0887), HpaA paralog (HP0492), HopL (HP1157), a VacA-like protein (HP0922), HP0159, HP0160, HP0651, HP0728, HP0906, HP1243, HP1250, HP1255, HP1265, HP1323, HP1415, and HP1553. Representative phylogenetic trees are shown in Fig. 2. For the remaining 20 proteins, at least one sequence did not cluster within the expected East Asian or non-Asian group; these outlier sequences were randomly distributed among the 16 strains analyzed, and may have arisen through recombination. All subsequent analyses of the 37 proteins were performed both with and without the removal of outlier sequences, and the two approaches generally yielded similar results.
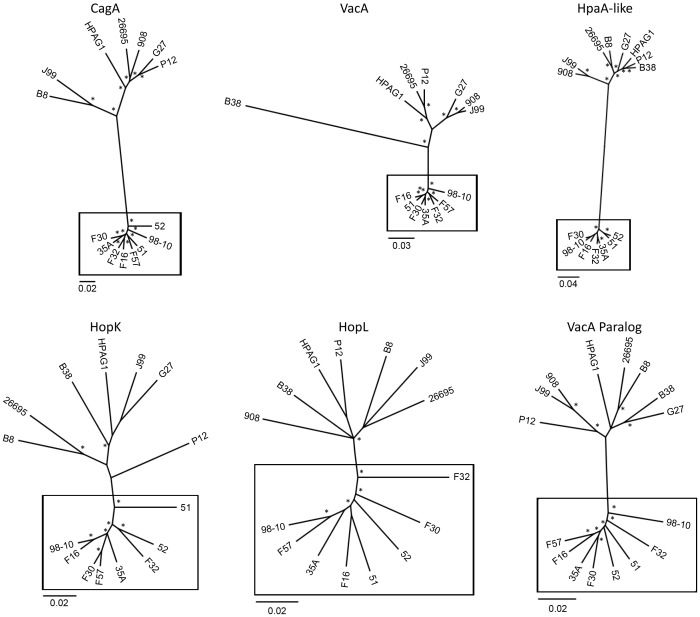
These include CagA (HP0547), VacA (HP0887), HpaA-like protein (HP0492), HopK (HP0923), HopL (HP1157), and a VacA-like protein (HP0922). The best available model of evolution was determined with ProtTest and phylogenies were inferred using MrBayes. Asterisks indicate posterior probabilities greater than 0.75. Sequences from East Asian strains (boxed) are highly divergent when compared to corresponding amino acid sequences from non-Asian strains of H. pylori. Scale bars show number of substitutions per site.
Analysis of Nucleotide Diversity and Positive Selection
To determine if the 37 genes were under positive selection, we calculated gene-wide Ka/Ks ratios, comparing sequences from East Asian strains with corresponding sequences from non-Asian strains (Table 3). As shown in Fig. 3A, Ka/Ks values of highly divergent genes were higher than Ka/Ks values of housekeeping genes. Specifically, the mean Ka/Ks ratio for the group of divergent 37 genes (0.256+0.064) was significantly higher than that of a control group of 7 housekeeping genes (0.083+0.049) (Mann-Whitney U test, p<0.001) (Table 3 and Table S1). Therefore, as a group, the 37 divergent genes are under less purifying selection than the seven housekeeping genes, as expected. Furthermore, the Ka/Ks ratios of the 37 divergent genes were negatively correlated with the level of amino acid similarity (comparing East Asian and non-Asian sequences) (Fig. 3B). These results indicate that sequences with higher protein sequence divergence exhibit less purifying selection.

Gene-wide Ka/Ks ratios were calculated, comparing sequences from East Asian strains with corresponding sequences from non-Asian strains, without the removal of outliers. (A) Distribution of Ka/Ks values. Ka/Ks values of highly divergent genes were significantly higher than Ka/Ks values of housekeeping genes. (B) Simple linear regression analysis comparing Ka/Ks values with mean % amino acid identity values (East Asian vs. non-Asian) for the 37 highly divergent proteins.
The elevated Ka/Ks ratios in the 37 divergent genes and the inverse correlation of Ka/Ks ratios with protein sequence divergence could have arisen by either increased positive selection or neutral evolution. To test which of the 37 divergent genes were under positive diversifying selection, we used the McDonald-Kreitmann test (MKT) to compare sequences from the East Asian population with sequences from the non-Asian population. The MKT analyzes the neutral theory prediction that the ratio of synonymous-to-nonsynonymous polymorphisms (Ps/Pn) within groups should be the same as the ratio of synonymous-to-nonsynonymous divergence (Ds/Dn) between groups. Among the 37 divergent genes, eight were determined to be under diversifying selection (Table 4), and this number increased to 12 once outliers were excluded (Table S2). One of the 7 housekeeping genes, ppa, was also determined to be under diversifying selection, but this gene exhibited a relatively low level of amino acid divergence. Excess nonsynonymous fixation, one signature of adaptive protein evolution, causes the Neutrality Index (NI) in the MKT to be less than 1. For all statistically significant MKT comparisons, the NI was <0.702 (Table 4).
Table 4
| Annotation | Gene ID (26695) | Dn | Ds | Pn | Ps | P value | NIb | α-Valuec |
| HopZ (omp1) | HP0009 | 12.07 | 16.44 | 285 | 346 | 0.765 | 1.122 | −0.122 |
| HopD (omp2) | HP0025 | 22.21 | 25.98 | 95 | 141 | 0.561 | 0.788 | 0.211 |
| BabA (omp28) | HP1243 | 28.33 | 28.08 | 127 | 244 | 0.020 | 0.515* | 0.484 |
| HomC/HomD | HP0373 | 38.63 | 37 | 177 | 212 | 0.373 | 0.799 | 0.200 |
| HomB | NAd | 5.01 | 9.13 | 198 | 291 | 0.702 | 1.239 | −0.239 |
| SabA/HopP/(omp17) | HP0725 | 21.21 | 15.39 | 177 | 215 | 0.137 | 0.597 | 0.402 |
| HopK (omp12) | HP0923 | 13.13 | 14.62 | 58 | 77 | 0.672 | 0.838 | 0.161 |
| HopA (omp6) | HP0229 | 4.01 | 9.18 | 126 | 113 | 0.114 | 2.552 | −1.552 |
| HopL (omp26) | HP1157 | 29.20 | 24.52 | 201 | 307 | 0.036 | 0.549* | 0.450 |
| VacA-like protein | HP0609/0610 | 36.11 | 38.47 | 743 | 1321 | 0.028 | 0.599* | 0.400 |
| VacA-like protein | HP0922 | 58.41 | 57.43 | 325 | 488 | 0.032 | 0.654* | 0.345 |
| HpaA-like protein | HP0492 | 79.15 | 57.12 | 45 | 41 | 0.399 | 0.792 | 0.207 |
| alpha-(1,3)-fucosyltransferase | HP0651 | 27.51 | 24.45 | 145 | 220 | 0.070 | 0.585 | 0.414 |
| lipopolysaccharide 1,2-glucosyltransferase (rfaJ) | HP0159 | 12.11 | 6.10 | 76 | 71 | 0.232 | 0.539 | 0.460 |
| cysteine-rich protein D/beta-lactamase HcpD | HP0160 | 8.06 | 3.03 | 45 | 63 | 0.047 | 0.268* | 0.731 |
| cytotoxin associated protein A (cagA) | HP0547 | 144.15 | 79.93 | 209 | 165 | 0.041 | 0.702* | 0.297 |
| vacuolating cytotoxin A (vacA) | HP0887 | 64.97 | 51.03 | 183 | 271 | 0.002 | 0.530* | 0.469 |
| flagellar hook-length control protein | HP0906 | 13.09 | 9.18 | 106 | 103 | 0.469 | 0.721 | 0.278 |
| recombination protein RecB/helicase | HP1553 | 40.50 | 28.97 | 132 | 167 | 0.033 | 0.565* | 0.434 |
| ribonuclease H (rnhA) | HP0661 | 4.04 | 2.04 | 9 | 17 | 0.150 | 0.267 | 0.732 |
| ribonuclease HII (rnhB) | HP1323 | 6.05 | 7.29 | 34 | 50 | 0.736 | 0.819 | 0.180 |
| type I restriction enzyme M protein (hsdM) | HP0463 | 11.06 | 11.25 | 149 | 142 | 0.881 | 1.067 | −0.067 |
| type I restriction enzyme M protein (hsdM) | HP0850 | 17.16 | 14.43 | 93 | 122 | 0.242 | 0.641 | 0.358 |
| type IIG restriction-modification enzyme | HP1354 | 8.02 | 6.04 | 404 | 365 | 0.738 | 0.834 | 0.165 |
| type III restriction enzyme R protein | HP1371 | 10.03 | 13.25 | 228 | 188 | 0.269 | 1.602 | −0.602 |
| metalloprotease | HP0806 | 8.09 | 4.08 | 41 | 52 | 0.141 | 0.398 | 0.601 |
| preprotein translocase subunit SecG | HP1255 | 5.03 | 4.09 | 23 | 38 | 0.314 | 0.491 | 0.508 |
| tRNA delta(2)-isopentenylpyrophosphate transferase (miaA) | HP1415 | 9.09 | 9.34 | 55 | 63 | 0.828 | 0.897 | 0.102 |
| selenocysteine synthase (SelA)/L-seryl-tRNA(Sec) selenium transferase | HP1513 | 13.13 | 12.44 | 60 | 94 | 0.237 | 0.604 | 0.395 |
| purine nucleoside phosphorylase (punB) | HP1530 | 7.07 | 4.10 | 30 | 40 | 0.202 | 0.434 | 0.565 |
| poly(A) polymerase (papS) | HP0640 | 7.03 | 10.29 | 52 | 88 | 0.778 | 0.864 | 0.135 |
| poly E-rich protein | HP0322 | 14.15 | 6.12 | 64 | 62 | 0.111 | 0.446 | 0.553 |
| tRNA(Ile)-lysidine synthase | HP0728 | 8.05 | 7.16 | 51 | 55 | 0.725 | 0.825 | 0.174 |
| probable ATP/GTP binding protein | HP0729 | 10.08 | 21.39 | 90 | 90 | 0.062 | 2.121 | −1.121 |
| bacterial SH3 domain protein | HP1250 | 17.52 | 8.44 | 39 | 26 | 0.505 | 0.722 | 0.277 |
| excinuclease ATPase subunit | HP0852 | 6.03 | 6.14 | 95 | 82 | 0.779 | 1.180 | −0.180 |
| NADH-ubiquinone oxidoreductase chain F | HP1265 | 7.05 | 3.03 | 54 | 58 | 0.186 | 0.400 | 0.599 |
| Control group | ||||||||
| ATP synthase F0F1 subunit alpha (atpA) | HP1134 | 0 | 2 | 5 | 79 | 0.721 | Null | Null |
| elongation factor P (efp) | HP0177 | 0 | 1 | 3 | 47 | 0.800 | Null | Null |
| A/G-specific adenine glycosylase (mutY) | HP0142 | 1 | 3.03 | 32 | 81 | 0.023 | 1.197 | −0.197 |
| inorganic pyrophosphatase (ppa) | HP0620 | 5.04 | 5.16 | 1 | 22 | 0.001 | 0.046* | 0.953 |
| anthranilate isomerase (trpC) | HP1279 | 5.01 | 7.11 | 62 | 109 | 0.722 | 0.807 | 0.192 |
| urease accessory protein (ureI) | HP0071 | 0 | 2.01 | 8 | 43 | 0.541 | Null | Null |
| GTP-binding protein (yphC) | HP0834 | 2 | 4.06 | 17 | 50 | 0.680 | 0.689 | 0.310 |
 =
= (Pn/Ps)/(Dn/Ds), where P is polymorphic within the population, D is divergence or fixed difference between populations, n is nonsynonymous, and s is synonymous.
(Pn/Ps)/(Dn/Ds), where P is polymorphic within the population, D is divergence or fixed difference between populations, n is nonsynonymous, and s is synonymous.Genes subject to positive, diversifying selection will exhibit a reduction in effective population size, as measured by the mean nucleotide diversity of synonymous sites (πs). The basic reason is that as positive selection sweeps alleles through a population, the silent site variation hitches alongside the selected region, thereby reducing πs in comparison to alleles under neutral evolution. Ka/Ks ratios would then negatively correlate with πs. Alternatively, elevated Ka/Ks ratios may reflect an increase in neutral evolution, and in this case, the elevated Ka/Ks ratios should be associated with elevated πs due to increased drift.
Among the divergent genes that were shown to be subject to positive diversifying selection based on the MKT, there was an inverse correlation when comparing πs values to Ka/Ks values, and this was observed for both non-Asian and East Asian strains (Fig. 4). Thus, the genes with elevated Ka/Ks values have reduced silent site diversity due to positive selection. When comparing πs values of the divergent genes isolated from non-Asian strains with the πs values from the corresponding East Asian sequences, there was a significant positive correlation (p<0.001, Fig. 4), which indicates that the same genes experience reduced silent site diversity and increased positive selection in both East Asian and non-Asian populations. These results provide evidence that diversifying selection has spurred the rapid evolution of amino acid sequence changes in these two populations.

(A,B) A linear regression analysis showed a significant correlation between Ka/Ks values and πs values, when analyzing sequences from non-Asian strains (p<0.05). There was a non-significant correlation when analyzing these sequences from East Asian strains (p =
= 0.07). (C) There was a strong positive correlation when comparing πs values of these 12 genes from either East Asian strains with the corresponding πs values from non-Asian strains (p<0.001).
0.07). (C) There was a strong positive correlation when comparing πs values of these 12 genes from either East Asian strains with the corresponding πs values from non-Asian strains (p<0.001).
We performed similar analyses for the set of genes that were not found to be under diversifying selection, based on MKT analysis (Fig. 5). In contrast to what was observed with the group of genes under diversifying selection (Fig. 4a and 4b), we did not detect any significant correlation between Ka/Ks values and πs values when analyzing this group of genes. Thus, within this group of genes, as Ka/Ks ratios increase, the average πs values across the gene do not change, which indicates that the genes are subject to more neutral evolution, as expected for the gene set that is not experiencing positive diversifying selection.
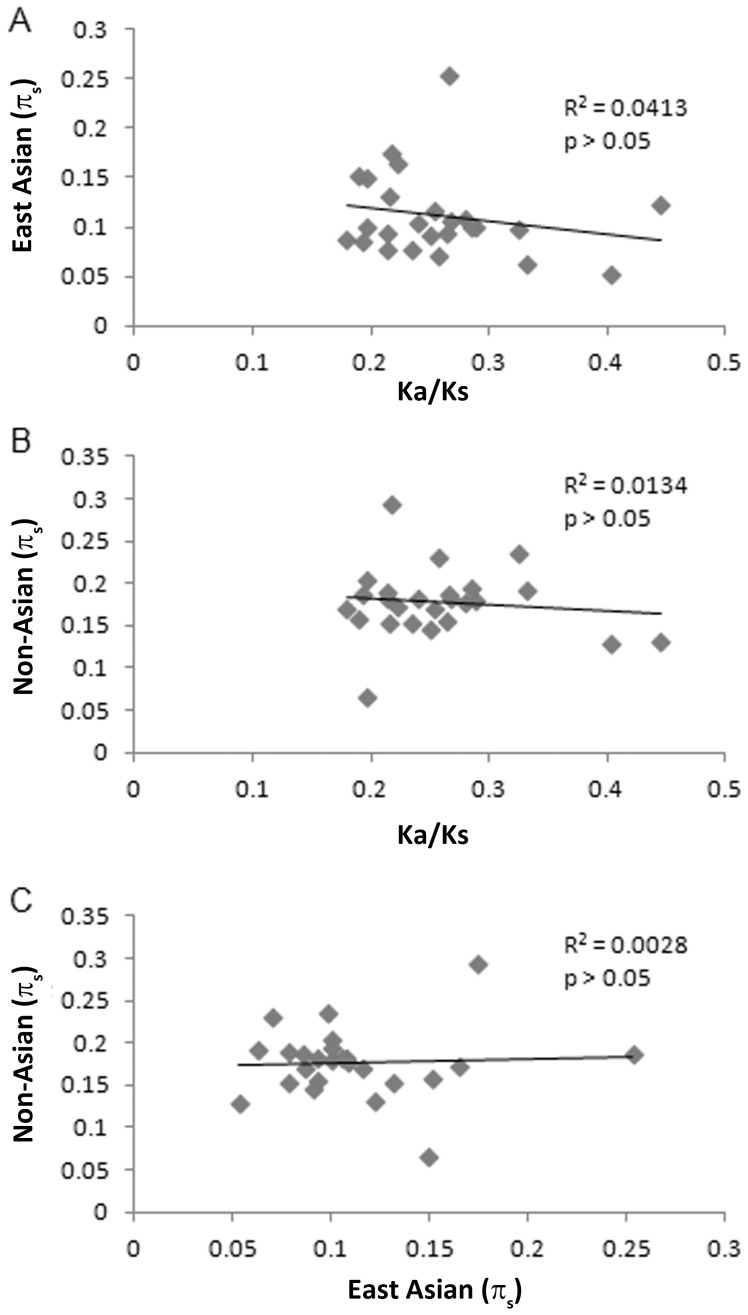
(A,B) Linear regression analyses showed non-significant trends when comparing Ka/Ks values to πs values (p>0.05). (C) There was no significant correlation between the πs values of these sequences from East Asian strains with the corresponding πs values from non-Asian strains (p>0.05).
Gene-wide ratios of Ka/Ks are overly conservative because they do not reveal instances of positive selection at a few, specific amino acid sites in a protein sequence. Therefore, we next used site-by-site methods of detecting positive selection to analyze the group of 25 divergent genes that were not considered to be under diversifying selection based on MKT analysis. Overall, 20 out of the 25 genes exhibited at least one site with a Ka/Ks ratio above 1.0. Figure 6 shows a subset of the sliding window analyses, and illustrates that vacA, cagA, hpaA-like gene, and hopK have specific regions under positive selection, whereas trpC and yphC (housekeeping genes), rnhB and HP1265 do not.
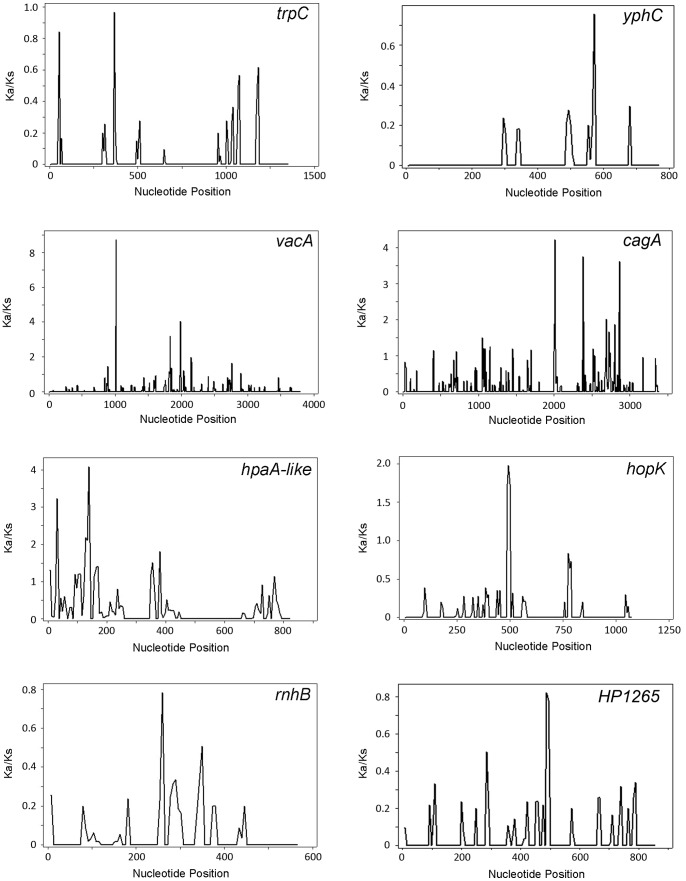
Sliding window analysis was performed to analyze the sequences of representative housekeeping genes (trpC and yphC) and several representative highly divergent genes, including vacA and cagA (under positive selection by MKT analysis) and an hpaA-like gene, hopK, rnhB, and HP1265 (not under positive selection by MKT analysis). Sequences from strain F16 (East Asian) and 26695 (non-Asian) were aligned and Ka/Ks ratios were calculated using DnaSP. In cases where sequences were not available from strains F16 or 26695, other representative East Asian or non-Asian sequences were analyzed. Parameters for the sliding window analysis were set at 50 bases (window size) and a step size of 10 bases. A Ka/Ks value of >1 indicates positive selection.
Discussion
H. pylori strains isolated in different parts of the world can be classified into distinct groups based on MLST analysis [13], [14]. Thus far, there has been relatively little comparative analysis of these groups at a whole-genome level. In this study, we undertook a systematic analysis designed to identify gene products that were highly divergent in East Asian H. pylori strains compared to non-Asian strains. Our analysis did not include a survey of gene deletions or gene disruptions in this panel of strains, but instead, we focused the analysis on genes that exhibited a high level of sequence divergence. We identified 57 predicted proteins, including CagA and VacA, with sequences that are highly divergent in East Asian H. pylori strains compared to non-Asian strains, and relatively conserved among the East Asian strains (Table 2). The most highly represented groups of divergent proteins were hypothetical proteins, cell envelope proteins, and proteins involved in DNA metabolism (Table 2). Among the 15 proteins classified in the cell envelope category, nine have been annotated as outer membrane proteins (OMPs).
Diversification of cell envelope proteins is of particular interest because of the important roles of the cell envelope in bacteria-host interactions. Three of the divergent outer membrane proteins identified in this study (HopZ, BabA and SabA) are reported to function as adhesins [54], [55], [56], and two other outer membrane proteins (HopA and HopD) are reported to have porin-like properties [57]. Two “VacA-like proteins” (HP0609/0610 and HP0922) were found to be highly divergent in East Asian strains compared to non-Asian strains. Similar to the vacuolating toxin VacA, these VacA-like proteins are predicted to be secreted by an autotransporter mechanism and localized to the bacteria surface [40]. The functions of these two VacA-like proteins and multiple other proteins annotated as “outer membrane proteins” have not yet been investigated. An HpaA-like protein (HP0492) was one of the most highly divergent proteins identified in this study. This protein of unknown function is considered a paralogue of HpaA, which in various studies has been reported to be either an adhesin or a flagellar-associated lipoprotein [58], [59]. Other divergent proteins within the cell envelope class [alpha-(1,3)-fucosyltransferase (FutB) and LPS 1,2-glucosyltransferase (RfaJ)] are predicted to have roles in LPS biosynthesis, and the former protein is reported to function as a molecular ruler for Lewis antigen biosynthesis [60]. Among the 15 divergent proteins in the cell envelope category, several (including RfaJ and the VacA-like proteins) are reported to be essential for H. pylori colonization of the stomach in animal models [61].
In addition to cell envelope proteins, other divergent proteins identified in this study are known or predicted to have important roles in bacteria-host interactions. For example, a protein originally annotated as “polyE-rich protein (HP0322)” and renamed “ChePep” was recently shown to be critical for H. pylori chemotaxis and is required for H. pylori colonization of deep gastric glands [62]. One of the proteins originally classified as a hypothetical protein (HP0721) is a secreted protein that has sialic acid-binding properties [63].
Numerous highly divergent proteins identified in this study are predicted to have functions related to DNA metabolism (including DNA replication, recombination and repair and restriction-modification). H. pylori is recombinogenic and is naturally competent for the uptake of DNA [64]. The ability to undergo DNA uptake and recombination promotes diversification of H. pylori and may allow the bacteria to adapt rapidly to the gastric environment of new hosts or changing conditions within a host. Several genes required for DNA uptake and recombination have important roles in promoting H. pylori colonization of the stomach in animal models [65], [66]. In addition, restriction-modification genes presumably have an important role in protecting H. pylori against phage and plasmids, and may promote the preferential uptake and chromosomal integration of H. pylori DNA rather than exogenous DNA from other sources. Interestingly, comparative genomic analyses of Neisseria isolates have also revealed marked differences among strains in restriction modification systems, and it was proposed that these systems may have a role in limiting gene flow [67].
Several previous studies have detected genes or proteins that are divergent in East Asian strains compared to non-Asian strains. For example, in a previous study, we analyzed the genome of a single East Asian strain (98-10) and detected 8 encoded proteins that were highly divergent compared to orthologues encoded by 3 non-Asian genomes that were available at the time [35]. A recent study analyzed multiple East Asian strains and reported the identification of additional divergent genes [37]. The use of different methodology in the current study allowed us to identify a set of divergent genes that partially overlaps those identified in these previous studies, and also includes multiple divergent genes that have not been previously recognized.
In previous studies, several proteins within the Sel1-like (SLR) gene family were reported to be highly divergent when comparing gene sequences from H. pylori strains classified as African, East Asian, and European, and it was reported that positive selection has driven the divergence of these proteins [36]. Most of these Sel1-like proteins were not identified in the current analysis due to the stringent criteria that we used for detecting proteins that were highly divergent in East Asian and non-Asian populations and for detecting proteins that were conserved within the East Asian population (described in Methods). For example, HP0519 is highly divergent in H. pylori strains from Japan compared to non-Asian H. pylori strains, but orthologous sequences in Korean strains resemble those of non-Asian strains [36]; therefore, this protein did not meet the criteria utilized in the current study.
Previous studies have shown that the divergence of two important virulence determinants of H. pylori, cagA and vacA, has been driven by positive selection [18], [23], [25], [68]. Similarly, we show, based on an analysis of Ka/Ks ratios, that most of the divergent genes with known or predicted functions exhibit evidence of positive selection. Furthermore, we show, based on MKT analysis, that 12 of the divergent genes, including cagA and vacA, are under positive, diversifying selection. Consistent with what one would expect, we show that genes subject to diversifying selection often exhibit a reduction in effective population size, as estimated by πs analysis. Among the 37 divergent genes with known or predicted functions, 32 (86%) exhibit evidence of positive selection, based on the MKT analysis combined with the sliding window analyses of Ka/Ks.
As illustrated in Figure 1, there was a higher level of intragroup relatedness among East Asian strains than among non-East Asian strains. Correspondingly, the mean silent site diversities in the divergent gene set were 0.11 and 0.18 for East Asian and non-East Asian strains, respectively, and in the control housekeeping set were 0.08 and 0.15 for East Asian and non-East Asian strains, respectively (Table 3). There was also a trend toward higher levels of silent site diversity in the divergent genes than in the control housekeeping genes. Specifically, the average silent site diversities were 0.15 and 0.11 for the divergent and control gene sets, respectively. There are at least three possible explanations for why there is an average increased rate of silent site diversity in the divergent gene set. One possibility is that there might be differences in recombination within control and divergent gene sets. To address this possibility, we tested for the presence of recombination within each nucleotide alignment using the program PHItest. An evaluation of the performance of several recombination programs using both simulated and empirical data found that PHItest effectively determines recombination under diverse conditions and performs markedly better than Max v2 and NSS at avoiding false positives of recombination under models of substitution rate heterogeneity [69]. The results of this analysis showed that all six of the divergent genes analyzed in Figure 2 and six out of seven genes in the control gene set exhibit significant recombination. Thus, recombination is widespread in H. pylori, as expected, and wholesale differences in recombination between the divergent and control gene sets do not explain the average increase in silent site diversity in the divergent gene set. An alternative explanation is that slight differences in genetic drift could explain the increase in silent site diversity in the divergent gene set. For example, the control gene set is composed of housekeeping genes under constant levels of purifying/negative selection. Neutral substitutions can accumulate under this form of selection but silent sites linked to negatively selected amino acid changes will be purified. In contrast, the divergent set experienced bouts of positive selection that were intermittent over the course of the gene’s evolution and localized to certain amino acid positions rather than the entire gene. Thus, bouts of adaptive evolution could be followed by bouts of neutral evolution across different gene regions, leading to the observed average increase in silent site diversity. Finally, a third possible explanation for why there is a slight increase in silent site diversity in the divergent genes is that mutation rates can differ between different genes within the same genome [70].
We hypothesize that in many cases, genes which are highly divergent in East Asian strains compared to non-Asian strains encode proteins that differ in functional activity. Several examples of this phenomenon have been reported previously. For example, East Asian forms of CagA (containing EPIYA-D motifs) are reported to differ in activity compared to Western forms of CagA (containing EPIYA-C motifs) [31]. Similarly, East Asian forms of SabA are reported to differ in activity compared to non-Asian forms of SabA [34]. We speculate that East Asian H. pylori strains are subject to different host or environmental conditions compared to non-Asian strains, and these conditions may have driven diversification of certain genes. Consistent with this hypothesis, diversification of OMPs has been associated with the adaptation of H. pylori to different host selective pressures [71], [72].
Finally, it is notable that the incidence of gastric cancer is markedly higher in many parts of East Asia than in non-Asian countries [5], [7], [8]. Distinctive properties of H. pylori strains circulating in various regions of East Asia may contribute to this high incidence of gastric cancer. As one example, forms of CagA containing an EPIYA-D motif (which are found in East Asian strains) exhibit increased activity in vitro compared to other forms of CagA [30], [31]. It seems possible that other proteins encoded by East Asian strains may differ in activity compared to the corresponding proteins encoded by non-East Asian strains, and such differences in activity may influence pathologic processes linked to the development of gastric cancer. In future studies, it will be important to specifically test the functional roles of the divergent proteins identified in this study, to investigate their role in H. pylori-host interactions, and to investigate their potential roles as determinants of gastric cancer risk. In addition, it will be important to investigate further the diversity of these genes in other geographic populations of H. pylori strains.
Supporting Information
Figure S1
Phylogenetic analysis of cagA, vacA, and housekeeping gene sequences. Neighbor-joining phylogenetic trees were constructed for cagA (panel A) and vacA (panel B), and a set of seven concatenated housekeeping gene fragments (panel C). The sequences of East Asian strains (boxed) are highly divergent when compared to corresponding nucleotide sequences of non-Asian strains. (Note the difference in scales used for the three trees).
(TIF)
Table S1
Analysis of nucleotide diversity (outliers removed).
(DOCX)
Table S2
Analysis of positive selection using McDonald-Kreitman test (outliers removed).
(DOCX)
Funding Statement
Supported by National Institutes of Health (NIH) AI068009, AI039657, and CA116087, and the Department of Veterans Affairs to TLC and NIH GM085163 to SRB. The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
References
Articles from PLOS ONE are provided here courtesy of PLOS
Full text links
Read article at publisher's site: https://doi.org/10.1371/journal.pone.0055120
Read article for free, from open access legal sources, via Unpaywall:
https://journals.plos.org/plosone/article/file?id=10.1371/journal.pone.0055120&type=printable
Citations & impact
Impact metrics
Citations of article over time
Alternative metrics
Smart citations by scite.ai
Explore citation contexts and check if this article has been
supported or disputed.
https://scite.ai/reports/10.1371/journal.pone.0055120
Article citations
Pathogenomics of Helicobacter pylori.
Curr Top Microbiol Immunol, 444:117-155, 01 Jan 2023
Cited by: 2 articles | PMID: 38231217
Review
Repeated out-of-Africa expansions of Helicobacter pylori driven by replacement of deleterious mutations.
Nat Commun, 13(1):6842, 11 Nov 2022
Cited by: 10 articles | PMID: 36369175 | PMCID: PMC9652371
Biomarker Characterization and Prediction of Virulence and Antibiotic Resistance from Helicobacter pylori Next Generation Sequencing Data.
Biomolecules, 12(5):691, 11 May 2022
Cited by: 12 articles | PMID: 35625618 | PMCID: PMC9138241
Review Free full text in Europe PMC
CagL polymorphisms between East Asian and Western Helicobacter pylori are associated with different abilities to induce IL-8 secretion.
J Microbiol, 59(8):763-770, 01 Jun 2021
Cited by: 3 articles | PMID: 34061339
Detection and variability analyses of CRISPR-like loci in the H. pylori genome.
PeerJ, 7:e6221, 11 Jan 2019
Cited by: 4 articles | PMID: 30648020 | PMCID: PMC6330956
Go to all (23) article citations
Data
Data behind the article
This data has been text mined from the article, or deposited into data resources.
BioStudies: supplemental material and supporting data
Lay summaries
Plain language description
Similar Articles
To arrive at the top five similar articles we use a word-weighted algorithm to compare words from the Title and Abstract of each citation.
Genome sequence analysis of Helicobacter pylori strains associated with gastric ulceration and gastric cancer.
BMC Genomics, 10:3, 05 Jan 2009
Cited by: 72 articles | PMID: 19123947 | PMCID: PMC2627912
Expansion of European vacA and cagA alleles to East-Asian Helicobacter pylori strains in Cambodia.
Infect Genet Evol, 11(8):1899-1905, 17 Aug 2011
Cited by: 5 articles | PMID: 21871583
Analysis of hopQ alleles in East Asian and Western strains of Helicobacter pylori.
FEMS Microbiol Lett, 251(1):37-43, 01 Oct 2005
Cited by: 27 articles | PMID: 16102915
Helicobacter pylori typing as a tool for tracking human migration.
Clin Microbiol Infect, 15(9):829-834, 01 Sep 2009
Cited by: 54 articles | PMID: 19702588 | PMCID: PMC3128245
Review Free full text in Europe PMC
Funding
Funders who supported this work.
NCI NIH HHS (2)
Grant ID: P01 CA116087
Grant ID: CA116087
NIAID NIH HHS (4)
Grant ID: AI039657
Grant ID: R01 AI068009
Grant ID: R01 AI039657
Grant ID: AI068009
NIGMS NIH HHS (3)
Grant ID: T32 GM007347
Grant ID: R01 GM085163
Grant ID: GM085163





