Abstract
Purpose
Dominant optic atrophy (DOA) is the most common form of autosomal inherited optic neuropathy, mainly caused by mutations in the optic atrophy 1 (OPA1) gene. The purpose of this study was to detect OPA1 gene mutations and associated phenotypes in Chinese patients with suspected hereditary optic neuropathy.Methods
A cohort of 193 Chinese families with suspected hereditary optic neuropathy was collected, which had been excluded from the three common primary mitochondrial DNA mutations associated with Leber hereditary optic neuropathy in our prior screening. Sanger sequencing was used to analyze variants in the coding and adjacent regions of OPA1.Results
In this study, 11 heterozygous OPA1 mutations, among which eight were novel and three were known, were identified in 12 of the 193 families (6.2%) but in none of the 192 control individuals. These novel mutations consisted of two nonsense mutations (p.E707* and p.K797*), two missense mutations (p.T330S and p.V377I), two deletions (p.S64fs and p.L331fs), one small insertion (p.L17fs), and one splice site mutation (c.2614-2A>G). Of the 12 families, three had a family history of optic neuropathy while nine were sporadic cases. Analysis of the family members in the two sporadic cases demonstrated that one parent in each of the two families had the OPA1 mutation and mild phenotype of optic atrophy. A 4-year-old boy with severe ocular phenotype was found to be compound heterozygous for two OPA1 mutations, a p.S64fs frameshift deletion and a p.V377I missense mutation, possibly implying an additive effect.Conclusions
This study implies that the frequency of DOA is much lower than that of Leber hereditary optic neuropathy in Chinese compared with other ethnic groups. Lack of awareness of the mild phenotype of DOA may contribute to the low frequency of OPA1-related DOA in Chinese. The phenotype associated with compound heterozygous OPA1 mutations may suggest a possible addictive effect.Free full text

Mutation survey of the optic atrophy 1 gene in 193 Chinese families with suspected hereditary optic neuropathy
Abstract
Purpose
Dominant optic atrophy (DOA) is the most common form of autosomal inherited optic neuropathy, mainly caused by mutations in the optic atrophy 1 (OPA1) gene. The purpose of this study was to detect OPA1 gene mutations and associated phenotypes in Chinese patients with suspected hereditary optic neuropathy.
Methods
A cohort of 193 Chinese families with suspected hereditary optic neuropathy was collected, which had been excluded from the three common primary mitochondrial DNA mutations associated with Leber hereditary optic neuropathy in our prior screening. Sanger sequencing was used to analyze variants in the coding and adjacent regions of OPA1.
Results
In this study, 11 heterozygous OPA1 mutations, among which eight were novel and three were known, were identified in 12 of the 193 families (6.2%) but in none of the 192 control individuals. These novel mutations consisted of two nonsense mutations (p.E707* and p.K797*), two missense mutations (p.T330S and p.V377I), two deletions (p.S64fs and p.L331fs), one small insertion (p.L17fs), and one splice site mutation (c.2614–2A>G). Of the 12 families, three had a family history of optic neuropathy while nine were sporadic cases. Analysis of the family members in the two sporadic cases demonstrated that one parent in each of the two families had the OPA1 mutation and mild phenotype of optic atrophy. A 4-year-old boy with severe ocular phenotype was found to be compound heterozygous for two OPA1 mutations, a p.S64fs frameshift deletion and a p.V377I missense mutation, possibly implying an additive effect.
Conclusions
This study implies that the frequency of DOA is much lower than that of Leber hereditary optic neuropathy in Chinese compared with other ethnic groups. Lack of awareness of the mild phenotype of DOA may contribute to the low frequency of OPA1-related DOA in Chinese. The phenotype associated with compound heterozygous OPA1 mutations may suggest a possible addictive effect.
Introduction
Optic neuropathy, a common visual impairment, can be caused by genetic defects, toxic factors, and ocular or brain diseases [1-3]. Genetic defects may occur in nuclear genome or mitochondrial DNA (mtDNA) [4-6]. Mutations in mtDNA are known to cause Leber hereditary optic neuropathy (LHON, MIM 535000) [7]. At least eight loci (OPA1 to OPA8) have been mapped to the nuclear genome in association with optic atrophy, in which three causative genes have been identified, including the optic atrophy 1 (OPA1, OMIM 605290) gene, the optic atrophy 3 (OPA3, OMIM 606580) gene, and the transmembrane protein 126A (TMEM126A, OMIM 612988) gene [1].
Dominant optic atrophy (DOA, MIM 165500) is the most common form of hereditary optic neuropathy resulting from mutations in nuclear genome, with a prevalence of 1 in 50,000 overall [8] and as high as 1 in 35,000 in the north of England [9], similar to the frequency of LHON (1 in 25,000 to 1 in 50,000) as reported in Caucasians [1,10]. DOA typically presents as painless and slowly progressive visual loss, occurring insidiously with a mean age of onset at 6–10 years of age [1,11]. DOA exhibits marked inter- and intrafamilial variable phenotypes [9,12,13]; for instance, about 25% of patients were found to have optic atrophy with fundus examination but without a complaint of visual deterioration [11,14].
Of the three nuclear genes known to cause optic neuropathy when mutated, mutations in OPA3 and TMEM126A are extremely rare. OPA3 mutations result in optic atrophy associated with cataract [15,16], while TMEM126A mutations are responsible for an autosomal recessive form of optic atrophy [17,18]. Mutations in OPA1 account for the majority (60%–70%) of patients with DOA [19]. Thus far, at least 230 pathogenic mutations have been identified in OPA1 [20]. However, extremely variable phenotypes of DOA hindered the diagnosis and, therefore, the clinical test of OPA1 mutational screening. Until now, the genotype-phenotype correlations of OPA1 mutations with DOA were elusive [21,22]. Recently, analysis of OPA1 in some large case series of hereditary optic neuropathy in Caucasians suggested that scanning the most frequently mutated exons might be a good strategy for identifying OPA1 mutations [21,23]. Although exon 27, exon 8, and exon 15 may be mutational hot spots in Caucasians [21], the frequency of OPA1 mutation in Chinese patients has not been evaluated. Only a few Chinese families with DOA have been reported to be associated with OPA1 mutations [24-27], not to mention the mutational hot spots. In this study, Sanger sequencing was used to detect the mutation of OPA1 in 193 Chinese families with suspected hereditary optic neuropathy, in which the three common LHON-associated primary mutations of mtDNA had been excluded with prior screening.
Methods
Patients
Probands with suspected hereditary optic neuropathy from 193 unrelated families were examined at the Pediatric and Genetic Clinic in Zhongshan Ophthalmic Center. The criteria for participating in this study were as follows: (1) reduced visual acuity not related to refractive error or ocular media; (2) optic atrophy revealed with fundus examination; (3) exclusion of optic atrophy with known causes; and (4) absence of the three common primary LHON-associated mutations in mtDNA, which was confirmed based on methods described in our previous study [28]. All probands were assessed by experienced ophthalmologists or neuro-ophthalmologists. Informed consent conforming to the tenets of the Declaration of Helsinki and the Guidance of Sample Collection of Human Genetic Diseases (863-Plan) by the Ministry of Public Health of China was obtained from the participants or their guardians before the study. For the probands from the 193 families, 155 were sporadic cases, and 38 had a family history of hereditary optic neuropathy; 132 were male, and 61 were female.
Mutation screening
Genomic DNA from the proband of each family was prepared from peripheral blood leukocytes as previously described [23]. All the coding exons and exon-intron junctions of OPA1 (references from NCBI: NC_000003.11 for gDNA, NM_015560.1 for mRNA, and NP_056375.2 for protein) were amplified with PCR using 30 primer sets (Table 1). The primer pairs were designed to cover at least 80 bp of each exon-intron junction, aiming to detect most of the known intronic mutations. PCR reaction was performed in 20 μl volumes containing 80 ng genomic DNA. Touchdown PCR was performed, which consisted of a denaturizing step at 95 °C for 5 min and annealing temperature decreased by 2 °C after the first 5 cycles and the second 5 cycles, and then down to the optimal annealing temperature (listed in Table 1), and a final extension at 72 °C for 5 min. Sequences of the amplicons were determined with BigDye Terminator cycle sequencing kit v3.1 and an ABI3130 Genetic Analyzer (Applied Biosystems, Foster City, CA). Sequences from patients and the OPA1 reference sequence (NM_015560.1) were compared using SeqMan II software (Lasergene 8.0, DNASTAR, Madison, WI). All variations were confirmed with bidirectional sequencing, and novel mutations were then evaluated in 384 chromosomes of 192 normal individuals. The effect of a novel missense mutation on the encoded protein was predicted with the Blosum62 Table [29] as well as PolyPhen-2 [30] and the SIFT online tool [31]. Splice site mutations were predicted with the Splice Site Prediction by Neural Network [32]. In addition, the degree of evolutionary conservation at amino acid positions altered by novel OPA1 mutations was analyzed using the MegAlign program of the Lasergene package. Segregation analyses of mutations were performed on patients with available family members.
Table 1
| Fragment | Forward primer (5′-3′) | Reverse primer (5′-3′) | Product length (bp) | Annealing temperature |
|---|---|---|---|---|
| Exon 1 | CCTCGGCCGCGGCTCTGTGC | GGGCTCCTGTCATTCTGGGTCCTCAAG | 327 | 65 °C |
| Exon 2 | TTTGCACCACATTTTCCTCATCT | GGCATCTTCCTATTAGCATCATTA | 743 | 60 °C |
| Exon 3 | GGGCAAAATTATGAAACCT | TAAAATTATGGGCTACTGG | 523 | 57 °C |
| Exon 4 | ACTGCCTGGGCTGGAAC | GGAACTGTCATTTTACTGGGC | 507 | 65 °C |
| Exon 4b | GCCCTATCGTAATATGAAATCTGAG | GCATAAGCAGCATTATAAATTTGG | 257 | 60 °C |
| Exon 5 | AGGCGATTTGATTCTTTGA | ACTTGGATGTTTTTGTATTTG | 320 | 55 °C |
| Exon 5b | AACCATCCCTCCCTAGCTTACATCT | GGCTTTACCTATACTACCCACTCCAG | 396 | 65 °C |
| Exon 6 | CTTTCATAAGAAATGACTAGAATAGCAACA | TGGGCATAAGATTCACTCAAAATAGG | 560 | 57 °C |
| Exon 7 | ATGTGAGTAGCAAGGAATTTTCCAAGTG | CCTCCAAGCACATTAGGTTAGAAAGAAA | 484 | 65 °C |
| Exon 8 | CTAAATAAACTGAATGAGAAATGGAC | ACATTACTTGGAACATGTAAGATTAC | 446 | 60 °C |
| Exon 9 | GTTTTGCTGTTCCTATTTTCAATGTGCA | GCCTCCTGGCTGTGCCTTCTACTGATTT | 464 | 65 °C |
| Exon 10–11 | CCCAGCAGTAGTGTGAAGGG | AAAACAATGCTAAAGTTTGGGG | 716 | 60 °C |
| Exon 12–13 | TGTGAGCGTCTTATCTGAATGGA | AATGAATACGAAGAGAAGGCAAAAA | 506 | 60 °C |
| Exon 14 | TTGCTATAATGTAGACACAGGGG | CCATGTACCATTTCCTTTTGTG | 395 | 60 °C |
| Exon 15–16 | TCATTCCGGGTTTTCGATAC | AAACTGCTCTCAATTCTGCC | 662 | 65 °C |
| Exon 17 | GCTACCGTATTGGAATGTTTTCCTCCTC | AAATGTTCTCATCTGTTTGAACTCTGCA | 525 | 65 °C |
| Exon 18 | GGGTCATAGGCGCACTCTC | TCTCAGAAAACACTTTCAACACTTG | 425 | 57 °C |
| Exon 19 | CCCAAATTCAGCCTAGTCAAAAA | GAGCCAAGGCAACAATAAATCAC | 293 | 65 °C |
| Exon 20 | GCTGGAGTGGAAGAACAAAGACAAA | CCCAAAACAGAGATGAGGAATAAAGAA | 616 | 65 °C |
| Exon 21 | CCATATCTGTCCCCAGCAAC | GCAACAGGTGATTTTAGAAGGG | 541 | 65 °C |
| Exon 22 | TAACAAATAAGCAGGCAAGTAAAAGAAG | CATTGGTTTTAGTAGTTACAAAGCAGTT | 464 | 60 °C |
| Exon 23 | ATGTGGGTTTTTTCCTTTA | GCATGTTTCATCTCTTGTC | 448 | 57 °C |
| Exon 24 | TGATTAAGCTTGTGTTATCTTTTATGC | CGTGACAAAAGTCAAATTAAGCAC | 372 | 60 °C |
| Exon 25 | CCTACCCTGTCTACTCCACAAG | TTTCCCCAGATGATCAAAGG | 523 | 60 °C |
| Exon 26 | TTAAGCTTAGGACATATCTACTGGTTC | TGGGAAGTATTTTGGCATCC | 291 | 60 °C |
| Exon 27 | TTTTGGGAAATCTGCACTTC | TCTGACCTTGTTTTCCACCC | 533 | 60 °C |
| Exon 28 | TTGGGTAAAAGGTGGTATGGTGAG | CAAGCAGGATGTAAATGAAGCAGAA | 309 | 65 °C |
Results
Optic atrophy 1 mutations
Upon complete sequencing analysis of OPA1 coding exons and adjacent intronic regions, 11 heterozygous mutations in OPA1 (Table 2, Figure 1) were detected in 12 of 193 (6.2%) families, among which eight were novel and three were known. These mutations consisted of three nonsense mutations, two splice site mutations, three deletions, two missense mutations, and one small insertion. The c.2708_2711del mutation in exon 27 was present in three of the 12 probands (25%). In addition, three mutations were found in a fragment less than 10 bp around the 5′ end of exon 10. Of the 12 cases, nine were sporadic cases (9/155, 5.8%), and three had a family history (3/38, 8.0%). The eight novel mutations were predicted to be pathogenic and not detected in 384 control chromosomes. Polymorphisms detected in OPA1 are listed in Table 3.
Table 2
| OPA1 | Patient ID | Nucleotide change | Amino acid change | Status | Conser vation | Computational prediction | Allele frequency in | Reported | |||
|---|---|---|---|---|---|---|---|---|---|---|---|
| Blosum62 | PolyPhen or Splice site | SIFT | cases | controls | |||||||
| exon 2 | le1608 | c.49_50insGG | p.L17fs | Hetero | - | - | - | - | 1/386 | 0/384 | This study |
| exon 2 | le2028 | c.190_194del | p.S64fs | Hetero | - | - | - | - | 1/386 | 0/384 | This study |
| intron 9 | le2146 | c.985–1G>A | Splicing defect | Hetero | - | - | - | - | 1/386 | NA | Delettre et al. [40] |
| exon 10 | le1524 | c.989C>G | p.T330S | Hetero | Yes | 5>1 | PrD | D | 1/386 | 0/384 | This study |
| exon 10 | le1656 | c.991_992del | p.L331fs | Hetero | - | - | - | - | 1/386 | 0/384 | This study |
| exon 11 | le2028 | c.1129G>A | p.V377I | Hetero | Yes | 4>3 | PrD | T | 1/386 | 0/384 | This study |
| exon 21 | le1599 | c.2119G>T | p.E707* | Hetero | - | - | - | - | 1/386 | 0/384 | This study |
| exon 24 | le1432 | c.2389A>T | p.K797* | Hetero | - | - | - | - | 1/386 | 0/384 | This study |
| exon 24 | le1601 | c.2470C>T | p.R824* | Hetero | - | - | - | - | 1/386 | NA | Ferre et al. [21] |
| intron 25 | le1574 | c.2614–2A>G | Splicing defect | Hetero | - | - | SSA | - | 1/386 | 0/384 | This study |
| exon 27 | le1411, le1434, le2062 | c.2708_2711del | p.V903fs | Hetero | - | - | - | - | 3/386 | NA | Ferre et al. [21] |
The proband le2028 is compound heterozygous for two OPA1 mutations, a frameshift deletion and a missense mutation (Figure 2). Abbreviations: Hetero, Heterozygous; PrD, probably damaging; SSA, splicing site abolished; D, damaging; T, tolerated; NA, not analyzed.
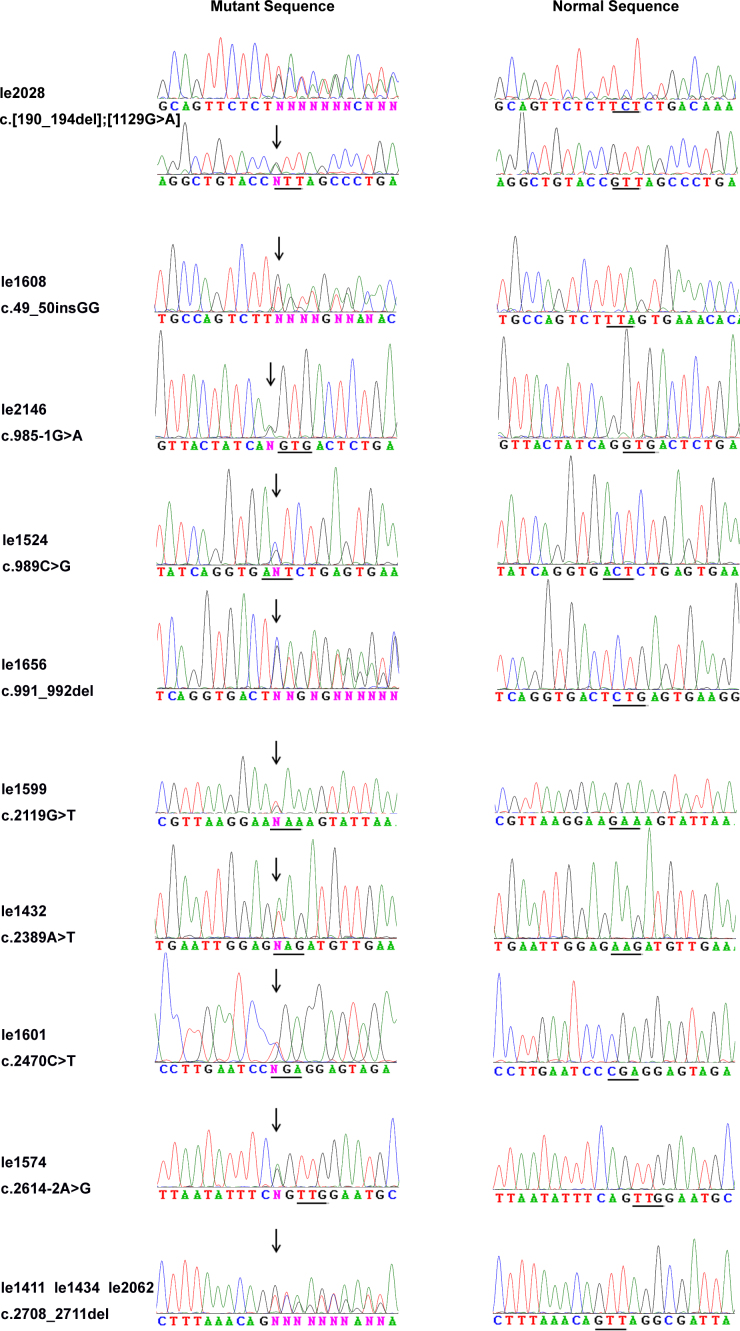
Sequence chromatograms. The 11 sequence changes detected in the probands with dominant optic atrophy are shown (left column) compared with corresponding normal sequences (right column). The mutational sites are indicated with an arrow, and the amino acid codes are depicted with a line.
Table 3
| Nucleotide change | Amino acid change | Status | Conser- vation | Computational prediction | Allele frequency in cases | Reported or SNP ID | ||
|---|---|---|---|---|---|---|---|---|
| Blosum62 | PolyPhen or Splice site | SIFT | ||||||
| c.43C>A | p.Q15K | Hetero | No | 5>1 | B | T | 1/386 | rs75414918 |
| c.321G>A | p.(S107=) | Hetero | NA | - | NC | - | 1/386 | rs117888848 |
| c.473G>A | p.S158N | Hetero/homo | No | 4>1 | B | T | 125/386 | rs7624750 |
| c.557–19T>C | - | Hetero/homo | NA | - | NC | - | 109/386 | rs3772393 |
| c.575C>T | p.A192V | Hetero | Yes | 4>0 | B | T | 9/386 | rs34307082 |
| c.624+13G>C | - | Hetero | NA | - | NC | - | 3/386 | this study |
| c.625–4T>A | - | Hetero | NA | - | NC | - | 1/386 | this study |
| c.870+4T>C | - | Hetero/homo | NA | - | NC | - | 12/386 | Liu Y et al. [41,42] |
| c.870+32T>C | - | Hetero/homo | NA | - | NC | - | 105/386 | Liu Y et al. [41,42] |
| c.871–9T>A | - | Hetero | NA | - | NC | - | 1/386 | this study |
| c.1177A>C | p.(R393=) | Hetero | NA | - | NC | - | 1/386 | rs149752576 |
| c.1608A>C | p.(A536=) | Hetero | NA | - | NC | - | 11/386 | rs78767626 |
| c.1770+16T>G | - | Hetero/homo | NA | - | NC | - | 27/386 | rs9831772 |
| c.1884A>G | p.(V628=) | Hetero | NA | - | NC | - | 9/386 | rs73069703 |
| c.1923G>A | p.(A641=) | Hetero | NA | - | NC | - | 3/386 | rs138114609 |
| c.2109T>C | p.(A703=) | Hetero/homo | NA | - | NC | - | 121/386 | rs9851685 |
| c.2276–128_2276–127insT | - | Hetero | NA | - | NC | - | 1/386 | this study |
| c.2808G>A | p.(A936=) | Hetero/homo | NA | - | NC | - | 35/386 | rs117475774 |
| c.2796C>T | p.(R932=) | Hetero | NA | - | NC | - | 8/386 | rs35540805 |
| c.2819–4A>G | - | Hetero | NA | - | NC | - | 2/386 | rs184273607 |
| c.2883+2T>C | - | Hetero | NA | - | NC | - | 1/386 | this study |
Abbreviations: Hetero, Heterozygous; Homo, Homozygous; B, benign; NC, not changed; T, tolerated; NA, not analyzed.
Optic atrophy 1 compound heterozygous mutations
A 4-year-old boy harbored two OPA1 mutations, c.190_194del (p.S64fs) and c.1129G>A (p.V377I). Segregation analysis demonstrated that the deletion was inherited from his father and the missense mutation from his mother (Figure 2), indicating compound heterozygosity. The boy was observed suffering from poor vision as early as 2 years of age. Fundus examination revealed temporal disc pallor in both eyes when he was 4 years old. Retinal nerve fiber layer (RNFL) thinning was confirmed with optical coherence tomography (OCT). Pattern visual-evoked potential (PVEP) showed prolonged latencies and diminished amplitudes in both eyes. In contrast, his father presented a much milder phenotype: His best-corrected visual acuity was 20/30 (Snellen) for both eyes, and his refraction was −1.50 sph for the right eye and −3.50 sph for the left eye. Fundus examination of the father showed bilateral disc pallor similar to his son while OCT and PVEP tests revealed milder abnormalities compared with the son. The mother had normal visual acuity of 20/20 (Snellen) for both eyes without any detectable abnormalities on fundus observation, OCT scan, and PVEP recordings. No extraocular neurologic sign presented in the proband and his parents (Table 4).
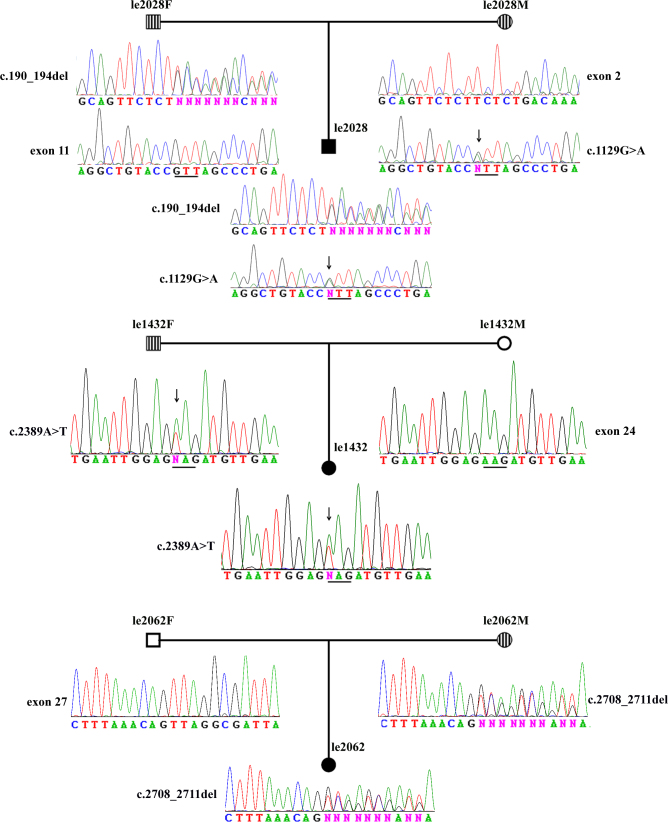
Segregation analysis of Optic Atrophy 1 mutations in three families with dominant optic atrophy. Circles represent women, and squares represent men. Two circles and a square filled in black indicate probands with suspected hereditary optic atrophy. Circles and squares filled with lines show carriers with Optic Atrophy 1 (OPA1) variants. Proband le2028 is compound heterozygous for OPA1 mutations. The father has a c.190_194del mutation in exon 2, and the mother has a c.1129G>A in exon 11. Both mutations transmitted to their son. Proband le1432 inherited a c.2389A>G mutation from her father. Proband le2062 inherited a c.2708_2711del mutation from her mother. The mutational sites are indicated with an asterisk, and amino acid codes are depicted with a line.
Table 4
| Patient ID | OPA1 mutations | Sex | Age (year) at | Inheri- tance | BCVA | Optic | VEP | RNFL thinning | Scotoma | |||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| exam | onset | OD | OS | atrophy | prolonged latencies | diminished amplitudes | OD | OS | OD | OS | ||||
| le1608 | c.49_50insGG | M | 12 | 9 | Spo | 0.3 | 0.2 | Yes | Yes | No | NA | NA | NA | NA |
| le2028 | c.[190_194del]; [1129G>A] | M | 4 | 2 | Fam | 0.05 | 0.05 | Yes | Yes | Yes | Yes | Yes | NA | NA |
| le2028F | c.190_194del | M | 39 | NA | Fam | 0.6 | 0.7 | Yes | Yes | No | Yes | Yes | No | No |
| le2028M | c.1129G>A | F | 33 | NA | Fam | 1.5 | 1.5 | No | No | No | No | No | No | No |
| le2146 | c.985–1G>A | M | 16 | 6 | Fam | 0.2 | 0.4 | Yes | Yes | Yes | NA | NA | Yes | Yes |
| le1524 | c.989C>G | M | 17 | 17 | Fam | 0.3 | 0.15 | Yes | NA | NA | NA | NA | Yes | Yes |
| le1656 | c.991_992del | M | 8 | 7 | Spo | 0.1 | 0.1 | Yes | Yes | Yes | NA | NA | Yes | Yes |
| le1599 | c.2119G>T | F | 24 | 14 | Spo | 0.3 | 0.3 | Yes | Yes | Yes | NA | NA | Yes | Yes |
| le1432 | c.2389A>T | F | 13 | 10 | Spo | 0.4 | 0.2 | Yes | NA | NA | Yes | Yes | Yes | Yes |
| le1432F | c.2389A>T | M | 41 | NA | Spo | 0.7 | 0.7 | Yes | NA | NA | Yes | Yes | NA | NA |
| le1601 | c.2470C>T | F | 35 | 35* | Spo | 0.3 | 0.2 | Yes | NA | NA | NA | NA | NA | NA |
| le1574 | c.2614–2A>G | M | 24 | 24* | Spo | 0.2 | 0.3 | Yes | NA | NA | NA | NA | NA | NA |
| le1434 | c.2708_2711del | M | 6 | 3 | Spo | 0.2 | 0.4 | Yes | Yes | Yes | Yes | Yes | NA | NA |
| le1411 | c.2708_2711del | M | 9 | 6 | Spo | 0.2 | 0.2 | Yes | NA | NA | NA | NA | NA | NA |
| le2062 | c.2708_2711del | F | 17 | 12 | Spo | 0.4 | 0.4 | Yes | Yes | Yes | NA | NA | No | No |
| le2062M | c.2708_2711del | F | 49 | NA | Spo | 0.7 | 0.7 | Yes | NA | NA | Yes | Yes | NA | NA |
Analysis of the family members in two sporadic cases (le1432 and le2062) demonstrated that one parent in each of the two families had the OPA1 mutation and mild phenotype of optic atrophy. The asterisk indicates the time when obvious visual deterioration occurred after taking antituberculosis medicine. Abbreviations: Fam, family; Spo, sporadic; BCVA, best-corrected visual acuity; RNFL, retinal nerve fiber layer; M, male; F, female; OD, right eye; OS, left eye; NA, not available.
Clinical characteristics of optic atrophy 1-positive patients
All probands with OPA1 mutations had clinical symptoms and signs of DOA (Table 4). The disease occurred insidiously in most probands, with reduced visual acuity noticed at about 12 years old (12±9.5 years; ranging between 2 and 35 years). The mean age for the probands’ first visit to the eye clinic was about 15 years (15±8.8 years; ranging from 4 to 35 years). Interestingly, two probands (le1574 and le1601) experienced significant visual deterioration after taking antituberculosis medicine. Additional segregation analysis of the mutations was performed in two sporadic cases, i.e., le1432 and le2062, through contact tracing of the OPA1-positive probands (Figure 2). The two patients had no siblings, but their parents were available for further analysis. All four parents were farmers and were visually asymptomatic by self-report. However, the father of le1432 and the mother of le2062 harbored an OPA1 mutation. Ophthalmic examination of the two “healthy carriers” demonstrated mild phenotype of optic atrophy: mild reduced visual acuity, attenuated retinal vessels, temporal disc pallor, and thinning RNFL on the OCT scan (Table 4).
Discussion
DOA and LHON are the most common hereditary optic neuropathies in the general population [4,33,34], with similar prevalence in Caucasians [1,9]. Ferre et al. reported identification of genetic defects in 440 of 980 cases (45%) with suspected hereditary optic neuropathy, including OPA1 mutations in 295 (30%) patients, mtDNA mutations in 131 (13%) patients, and OPA3 mutations in 14 (1.4%) patients [21]. These results suggest that OPA1 mutations are the most common cause for patients with suspected hereditary optic atrophy. However, there is no molecular epidemiological study for DOA in Chinese populations even though there are large case series of such studies for Chinese patients with LHON [28]. It is unclear whether DOA is rare in Chinese or may not be easily recognized in clinics compared to LHON. Clinically, it may be difficult to distinguish DOA from LHON in many cases, especially in the atrophic phase [35,36]. In our previous report of a molecular epidemiological analysis of 903 families with optic neuropathy, the three primary mutations (G11778A, T14484C, and G3460A) in mtDNA were identified in 346 families (38.3%) [28]. In this study, mutations in OPA1 were detected in only 12 of the 193 families (6.2%). Since patients with one of the three primary mtDNA mutations (G11778A, T14484C, and G3460A) were not included in the cohort, the actual detection rate of OPA1 mutations in Chinese patients with hereditary optic neuropathy should be even lower, indicating a significantly lower frequency of OPA1 mutations (<6.2%) and a comparatively higher frequency of mtDNA mutations (38%) in Chinese patients with suspected hereditary optic neuropathy compared with French Caucasians (30% for OPA1 and 13% for mtDNA) [21]. Lack of awareness of the mild phenotype of DOA may contribute to the low frequency of OPA1-related DOA in Chinese and relatively high frequency of OPA1 mutations in sporadic cases, which is implied by the segregation analysis of mutations in the family of le1432 and le2062. A family history showing autosomal dominant inheritance is a key indicator leading to clinical diagnosis of DOA and genetic analysis of OPA1. For the probands from the 12 families with OPA1 mutations identified in this study, however, most (9/12, 75%) were sporadic cases (nine out of 155 sporadic cases). Similar frequency of OPA1 mutations in singleton cases was also mentioned in other studies [16,21,24,37]. Since individuals who harbor OPA1 mutations may have mild phenotypes that the individuals are unaware of, it is important to keep OPA1 in mind for patients with suspected hereditary optic neuropathy without a family history and evaluate the family members of these singleton cases carefully. Moreover, such evaluation and genetic consultation to family members may be crucial since more severe phenotypes might be induced by toxic materials, such as alcohol, smoking, and some drugs, as seen in two patients who had taken medicine for tuberculosis in this study.
Although OPA1 mutations contribute to the most (60%–70%) cases with DOA [19], the genetic diagnosis of DOA is still a challenge. OPA1 mutations are spread over 30 exons [21]. In addition, about a quarter of the mutations are located in the intronic regions adjacent to exon-intron boundaries, which usually affect splicing [20]. Under this circumstance, a cost-effective method for identifying mutations responsible for DOA is of great value. Screening the most frequently mutated exons of OPA1 initially would be a feasible strategy. Although exons 27, 8, and 15 were suggested to be regions of mutation hot spots in a previous study by Ferre et al. [21], we are unable to suggest ethnic-specific mutation hot spots based on limited number of mutations identified in our cohort of Chinese patients. The c.2708_2711del mutation in exon 27 was present in approximately 27% of families in previous studies. The same mutation was found in three probands in our study, which accounts for 25% among the OPA1-positive cases, further confirming that it is a mutational hot spot [21,24,37]. Interestingly, three novel mutations, discovered in three unrelated patients, arose from a region less than 10 bp around the 5′ end of exon 10, which may imply another mutational hot spot. Thus far, this is the largest series of patients screened for OPA1 in an Asian population. As our results provide evidence of ethnic variations in the mutation frequency of OPA1, analyzing causative genes and their exons in a systematic and population-specific fashion is necessary, especially in the context of genetic counseling for different ethnic groups.
To our knowledge, the patient with confirmed compound heterozygosity of OPA1 mutations in our study is the fourth case reported so far [38,39], which represents a rare event. All cases with compound heterozygous OPA1 mutations reported thus far had more severe phenotypes than their parents with a single heterozygous OPA1 mutation, which may suggest an additive effect [39]. Although the mother is a possible case of non-penetrance in optic atrophy, the c.1129G>A (p.V377I) mutation is still likely to be pathogenic. First, the mutation is rare as it is not found in 384 control chromosomes or the Single Nucleotide Polymorphism database. Second, the mutation is located in an evolutionarily conserved region and is predicted to be probably damaging by PolyPhen-2. Third, the severity of the phenotype of the son could, at least partly, reflect the addictive effect of the mutation from his mother. However, it is not the only interpretation of this compound heterozygous case. A c.190_194del (p.S64fs) mutation was identified in the father, presumably causing premature termination. However, the phenotype of the father was much milder than the other probands with frameshift deletions or nonsense mutations in our study. This may suggest the existence of modifier alleles at other loci. If the son did not inherit the beneficial modifier alleles from his father, this could also result in the severity of the son’s phenotype. The effect of modifier alleles may also explain the marked intrafamilial phenotypical heterozygosity between the probands and other mutation carriers, as shown in le1432 and le2062.
In summary, this study implies that the frequency of DOA is much lower than that of LHON in Chinese compared with other ethnic groups. Lack of awareness of the mild phenotype of DOA may contribute to the low frequency of OPA1-related DOA in Chinese. Further analysis of OPA1 in individuals with mild visual impairment and temporal disc pallor may be helpful in disclosing the real frequency of DOA in Chinese. Routine clinical test of OPA1 variations in such cases may also enhance the care of eyes through consultation and pretreatment, especially for individuals who are unaware of visual problems but harbor OPA1 mutations.
Acknowledgments
The authors are grateful to the patients for their participation. This study was supported by the Department of Education of Guangdong Province, NSFC30971588, and the Fundamental Research Funds of State Key Lab of Ophthalmology, Sun Yat-sen University.
References
Articles from Molecular Vision are provided here courtesy of Emory University and the Zhongshan Ophthalmic Center, Sun Yat-sen University, P.R. China
Citations & impact
Impact metrics
Citations of article over time
Article citations
Autosomal dominant optic atrophy caused by six novel pathogenic OPA1 variants and genotype-phenotype correlation analysis.
BMC Ophthalmol, 22(1):322, 26 Jul 2022
Cited by: 2 articles | PMID: 35883160 | PMCID: PMC9327245
Pathogenicity evaluation and the genotype-phenotype analysis of OPA1 variants.
Mol Genet Genomics, 296(4):845-862, 21 Apr 2021
Cited by: 6 articles | PMID: 33884488
Autosomal dominant optic atrophy with OPA1 gene mutations accompanied by auditory neuropathy and other systemic complications in a Japanese cohort.
Mol Vis, 25:559-573, 05 Oct 2019
Cited by: 9 articles | PMID: 31673222 | PMCID: PMC6798706
Clinical and genetic features of eight Chinese autosomal-dominant optic atrophy pedigrees with six novel OPA1 pathogenic variants.
Ophthalmic Genet, 39(5):569-576, 28 Jun 2018
Cited by: 3 articles | PMID: 29952689 | PMCID: PMC6239416
Analysis of Genetic Mutations in a Cohort of Hereditary Optic Neuropathy in Shanghai, China.
J Ophthalmol, 2017:6186052, 04 Dec 2017
Cited by: 1 article | PMID: 29348930 | PMCID: PMC5733633
Go to all (9) article citations
Data
Data behind the article
This data has been text mined from the article, or deposited into data resources.
BioStudies: supplemental material and supporting data
Diseases (5)
- (1 citation) OMIM - 535000
- (1 citation) OMIM - 606580
- (1 citation) OMIM - 165500
- (1 citation) OMIM - 605290
- (1 citation) OMIM - 612988
RefSeq - NCBI Reference Sequence Database
- (1 citation) RefSeq - NM_015560.1
SNPs (Showing 14 of 14)
- (1 citation) dbSNP - rs75414918
- (1 citation) dbSNP - rs117888848
- (1 citation) dbSNP - rs78767626
- (1 citation) dbSNP - rs3772393
- (1 citation) dbSNP - rs34307082
- (1 citation) dbSNP - rs138114609
- (1 citation) dbSNP - rs117475774
- (1 citation) dbSNP - rs35540805
- (1 citation) dbSNP - rs9831772
- (1 citation) dbSNP - rs7624750
- (1 citation) dbSNP - rs149752576
- (1 citation) dbSNP - rs73069703
- (1 citation) dbSNP - rs9851685
- (1 citation) dbSNP - rs184273607
Show less
Similar Articles
To arrive at the top five similar articles we use a word-weighted algorithm to compare words from the Title and Abstract of each citation.
Genetic and Clinical Analyses of DOA and LHON in 304 Chinese Patients with Suspected Childhood-Onset Hereditary Optic Neuropathy.
PLoS One, 12(1):e0170090, 12 Jan 2017
Cited by: 12 articles | PMID: 28081242 | PMCID: PMC5230780
Genetic screening for OPA1 and OPA3 mutations in patients with suspected inherited optic neuropathies.
Ophthalmology, 118(3):558-563, 30 Oct 2010
Cited by: 33 articles | PMID: 21036400 | PMCID: PMC3044822
Novel mutations in the OPA1 gene and associated clinical features in Japanese patients with optic atrophy.
Ophthalmology, 113(3):483-488.e1, 01 Mar 2006
Cited by: 14 articles | PMID: 16513463
Recessive optic atrophy, sensorimotor neuropathy and cataract associated with novel compound heterozygous mutations in OPA1.
Mol Med Rep, 14(1):33-40, 04 May 2016
Cited by: 10 articles | PMID: 27150940 | PMCID: PMC4918608
Review Free full text in Europe PMC




