Abstract
Free full text

The Congenital Heart Disease Genetic Network Study
Abstract
Congenital heart defects (CHD) are the leading cause of infant mortality among birth defects, and later morbidities and premature mortality remain problematic. Although genetic factors contribute significantly to cause CHD, specific genetic lesions are unknown for most patients. The National Heart, Lung, and Blood Institute-funded Pediatric Cardiac Genomics Consortium established the Congenital Heart Disease Genetic Network Study to investigate relationships between genetic factors, clinical features, and outcomes in CHD. The Pediatric Cardiac Genomics Consortium comprises 6 main and 4 satellite sites at which subjects are recruited, and medical data and biospecimens (blood, saliva, cardiovascular tissue) are collected. Core infrastructure includes an administrative/data-coordinating center, biorepository, data hub, and core laboratories (genotyping, whole-exome sequencing, candidate gene evaluation, and variant confirmation). Eligibility includes all forms of CHD. Annual follow-up is obtained for probands <1-year-old. Parents are enrolled whenever available. Enrollment from December 2010 to June 2012 comprised 3772 probands. One or both parents were enrolled for 72% of probands. Proband median age is 5.5 years. The one third enrolled at age <1 year are contacted annually for follow-up information. The distribution of CHD favors more complex lesions. Approximately, 11% of probands have a genetic diagnosis. Adequate DNA is available from 97% and 91% of blood and saliva samples, respectively. Genomic analyses of probands with heterotaxy, atrial septal defects, conotruncal, and left ventricular outflow tract obstructive lesions are underway. The scientific community’s use of Pediatric Cardiac Genomics Consortium resources is welcome.
Congenital heart disease (CHD) is the most common class of birth defects with an estimated incidence of 2% to 3%, resulting in the birth of 80 000 to 120 000 affected newborns in the United States each year.1,2 CHD comprises a wide range of cardiovascular malformations from critical forms presenting in the immediate newborn period to mild defects, such as bicuspid aortic valve, which may not be detected until adulthood. Despite remarkable progress in clinical care for affected individuals, CHD remains the leading cause of infant mortality among birth defects.3
For those that survive infancy, there is a high rate of comorbidities, both cardiac and extracardiac, and expected lifespan is still diminished.4 These issues have become increasingly important, as the number of adults with CHD now exceeds the number of children with CHD.5 For some time, the pathogenesis of CHD has been attributed to genetic and environmental factors, including possible interactions between them.6 Beginning with the groundbreaking work of Ruth Whittemore, who was able to study CHD recurrence rates among offspring of affected mothers,7 the recognition of the relative importance of genetic causes for CHD has been in ascendency. More recent population-based studies in Denmark have substantiated that view, revealing that the relative risk among first-degree relatives for all forms of CHD is 3.2; risks vary by lesion and lesion class, with several important ones having even higher relative risks.8 The heritability of bicuspid aortic valve and of left ventricular outflow tract obstructive lesions are estimated to be in excess of 70%.9–11 Finally, consanguinity rates among parents of children with CHD in countries where such unions are customary are 2 to 3 times higher than in their general population.12,13 Taken as a whole, the epidemiological evidence strongly points to genetic factors playing a crucial role in the pathogenesis of CHD.
The causes of CHD remain largely unknown. Aneuploidies such as trisomy 21 (causing Down syndrome) are strongly associated with CHD. Genomic defects that cannot be readily detected with karyotyping are increasingly understood to have a role in CHD. This first became apparent in the 1990s with the discovery of 22q11.2 deletions, which were found to underlie roughly 2% of CHD (excluding BAV) and more than 50% of specific conotruncal lesions.14 The use of technologies that detect copy number variants (CNVs) has uncovered numerous other segmental aneuploidies that cause CHD.15–21 Finally, studies of rare families segregating CHD in a Mendelian fashion, either isolated defects or syndromes, as well as screening of candidate genes critical for cardiovascular development in CHD cohorts have identified numerous gene mutations underlying CHD.22 In aggregate, however, these genetic and genomic abnormalities account for less than one quarter of CHD cases.23
Rationale
The Pediatric Cardiac Genomics Consortium (PCGC) was formed as part of the strategic plan of the National Heart, Lung, and Blood Institute (NHLBI) to accelerate the translation of basic research findings into clinical studies and trials. Specifically, it was recognized that there was an exciting opportunity to harness several emerging genomic technologies and resources to move toward a comprehensive understanding of the genetics of CHD in humans.24 In addition, the existence of an NHLBI-sponsored clinical trials network, the Pediatric Heart Network, and the planned formation of a research consortium devoted to a thorough elucidation of cardiac development, the Cardiovascular Development Consortium (CvDC), provided a unique opportunity to form a broad and synergistic enterprise. These 3 consortia are now termed the Bench to Bassinet Program.
As noted above, genetic factors underlying CHD largely remain to be elucidated. This is attributable, in large measure, to the challenges associated with that task. Specifically, CHD comprises a large number of cardiovascular malformations, for which the relevant genetic and genomic defects vary. It is likely that there are a large number of causative genes overall, and the expected frequency of causative variants for any one gene is low. Taken together, there are major challenges to assembling cohorts of sufficient size to power studies of most forms of CHD.
The PCGC was formed to enable the robust study of the genetics of CHD. By recruiting from several main research centers as well as additional satellite sites, the PCGC has the capacity to enroll large number of probands with CHD, as well as pertinent relatives, from whom extensive phenotypic data and biosamples can be assembled. In addition, coordinated efforts in genetically analyzing those biosamples through shared core resources greatly enhance efficiency. Genetic studies of CHD can be advanced in some instances by collaborating with other research consortia with similar goals, as well as by serving as a community resource for ancillary studies. In this manner, the rationale of forming the PCGC was to accelerate the elucidation of genetic factors underlying CHD.
In this article, the design and implementation of the PCGC’s Congenital Heart Disease GEnetic NEtwork Study (CHD GENES) protocol is described. This is intended to be a reference work that will enable collaborations with investigators in the United States and internationally, as well as provide a possible template for other groups considering similar undertakings for CHD or other birth defects.
Study Overview
Network Organization and Study Management
The PCGC is funded through a U01 cooperative agreement with NHLBI, whose program officers provide leadership in overall program management. The consortium comprises 5 main clinical sites (Harvard Medical School [Boston Children’s Hospital and Brigham and Women’s Hospital], Yale School of Medicine, Columbia University Medical Center, Icahn School of Medicine at Mount Sinai, and the Children’s Hospital of Philadelphia) and 4 satellite clinical sites at the University of Rochester Medical Center, Cohen Children’s Medical Center, Children’s Hospital of Los Angeles, and University College London. Clinical sites are responsible for subject enrollment, as well as the collection of all data and biospecimens. The New England Research Institutes (NERI) is the administrative and data-coordinating center, responsible for training and certifying the clinical sites, designing case report forms, monitoring the study, managing the clinical data, contracting with and monitoring of the core facilities, and supporting meetings and communications.
CHD GENES is supported by several core facilities. Biospecimens are processed and stored at the Coriell Institute for Medical Research. Genomic and clinical data are stored in the DataHub at Children’s Hospital of Philadelphia. Cores for data generation include the Candidate Gene Evaluation and RNAseq Cores at Harvard that performs candidate gene re-sequencing and sequencing of cDNAs derived from cardiac tissues, the Genotyping Array Core at Children’s Hospital of Philadelphia that performs single nucleotide polymorphism (SNP) genotyping, the Whole Exome Sequencing Core at Yale, and the Confirmation Core at Columbia that confirms single basepair variants and small insertions/deletions with dideoxy sequencing and confirms CNVs using quantitative PCR. Data are initially processed at the Cores, and primary analysis data are deposited in the DataHub and made available publicly through Database of Genotypes and Phenotypes.
CHD GENES is managed with a set of committees and working groups. The Steering Committee is the decision-making body of the PCGC, with a chair and cochair serving 1-year terms. The Steering Committee is composed of the NHLBI program officer, the NERI principal investigator, and the site principal investigators. Decisions of the Steering Committee, including review of study designs, ancillary studies, and protocol amendments, are made by majority vote. The Protocol Committee developed and oversees the protocol for clinical recruitment, biosample collection, and phenotypic characterization of participants. The Cardiac Diagnosis Committee developed the categorization of cardiac lesions. The Study Coordinator Committee addresses practical issues around patient recruitment, clinical data collection, and biospecimen collection. The Ancillary Studies Committee reviews ancillary study proposals and makes recommendations to the Steering Committee about approval.
A series of working groups organized by anatomic lesion (heterotaxy, atrial septal defect, conotruncal, and left ventricular outflow tract obstruction) are responsible for analyzing genomic data within and across cardiac lesions. As the repository for genomic and phenotypic data, the PCGC DataHub has an integral role in supporting and facilitating data analysis.
Objectives
The overall objective of this multicenter, prospective cohort study is to investigate the relationships between genetic factors, clinical features, and outcomes in individuals with CHD. These aims are summarized in Table 1.
Table 1
Aims of CHD GENES
General aims
|
Specific aims
|
CHD indicates congenital heart disease; and CHD GENES, Congenital Heart Disease Genetic Network Study.
Patient Population and Eligibility
Patients with any form of CHD regardless of sex, age, race, or ethnicity are invited to participate in CHD GENES. Individuals with an isolated patent foramen ovale, an isolated and prematurity-related patent ductus arteriosus, pulmonary stenosis related to a twin–twin transfusion, and cardiomyopathy (without CHD) are excluded from the study. Recruitment strategies are center-specific, but most often include case ascertainment at the time of hospital admission or during an out-patient visit. Additional strategies utilize the Bench to Bassinet website (http://www.benchtobassinet.org/) or individual center websites, brochures, and other printed materials, as well as formal and informal presentations by study investigators to their patients and colleagues. A biological sample and medical data about the proband are required for enrollment. Whenever possible, the parents are also enrolled. Other family members with CHD are also eligible for enrollment.
Data Collection
Proband demographics, cardiac diagnosis, and other clinical findings; medical and developmental histories; and pregnancy and birth history are collected with structured electronic case report forms. Considerable emphasis is placed on collecting detailed cardiac diagnoses from echocardiographic, other imaging, and operative reports. The nomenclature for cardiac diagnoses is based on the International Paediatric and Congenital Cardiac Codes (http://www.ipccc.net/). Details about extracardiac anomalies and results from available genetic tests and from available genetic physical exams are also recorded. Three-generation pedigrees are constructed for each proband. Additional details about pregnancy, birth, and cardiac history are collected for subjects enrolled at age <1 year. Most data are collected through subject and family interviews in conjunction with additional medical record abstraction.
Study coordinators, who are trained and certified for data collection and entry, enter data via a customized secure web application to NERI. NERI uses robust strategies to ensure data completion and accuracy. These include various data checks at each site and centrally with automated flagging of inconsistent or questionable values.
Patient Follow-Up
Probands who were enrolled into the study at age 12 months or less and consented to be contacted in the future are followed-up annually for at least 5 years using questionnaires completed by the parents. The questionnaire seeks to detail cardiac and noncardiac medical and surgical events, medication usage, as well as neurodevelopmental status. For deceased subjects, the relevant records are obtained, if possible, to document the cause of death.
Ethical Issues
Disclosure of Planned Whole-Genome Analyses
It was felt to be important to disclose to all potential subjects that the planned genetic studies include whole-genome analyses, and thus identifying information will theoretically be obtained. The consent form explains not only that genetic testing will be completed on DNA samples, but also that different laboratory techniques will be used, possibly including whole-genome sequencing, for the purpose of finding the causes of disorders of heart development and related conditions.
Confidentiality
The results of research genetic testing are not placed in the medical record. Evidence suggesting nonpaternity is kept confidential and not divulged to subjects or their families. All data are stripped of identifiers and labeled with a study number. Databases are strictly password-protected. Biospecimens are identified by a different number and stored securely at the biorepository. The DataHub records utilize a separate unique identifier that is independent of study site. NERI holds the link between the study numbers and the identifiers used for DataHub records.
Return of Results
After much discussion, the PCGC decided that results of research testing on biological specimens would not be shared with study participants, unless required by the local Institutional Review Board. This decision was based, in part, on a strict interpretation of Clinical Laboratory Improvement Amendments regulations.25 Indeed, several local Institutional Review Boards had policies preventing such reporting when genetic analyses are performed in a non-Clinical Laboratory Improvement Amendments–approved fashion. For the site that does return results of clinical significance, all results are independently confirmed in a Clinical Laboratory Improvement Amendments-approved clinical laboratory using a new blood sample.
Interactions With the CvDC
One of the unique features of the Bench to Bassinet program is the integration of human genetic studies in the PCGC with studies of the fundamental mechanisms underlying cardiac development in the CvDC (outlined in Figure). The high degree of evolutionary conservation observed in cardiac development allows such an approach. Several lines of investigation in the CvDC are directly relevant for the PCGC. Heart-specific transcriptome datasets are being created from embryonic mouse and chick tissues, as well as from chamber-specific human heart tissues obtained from subjects enrolled in CHD GENES. These data will permit a focused review of new genes identified by the PCGC from human genomic analyses to determine which of the CHD candidate genes are expressed in the heart at appropriate time points during development to potentially result in the relevant forms of CHD.
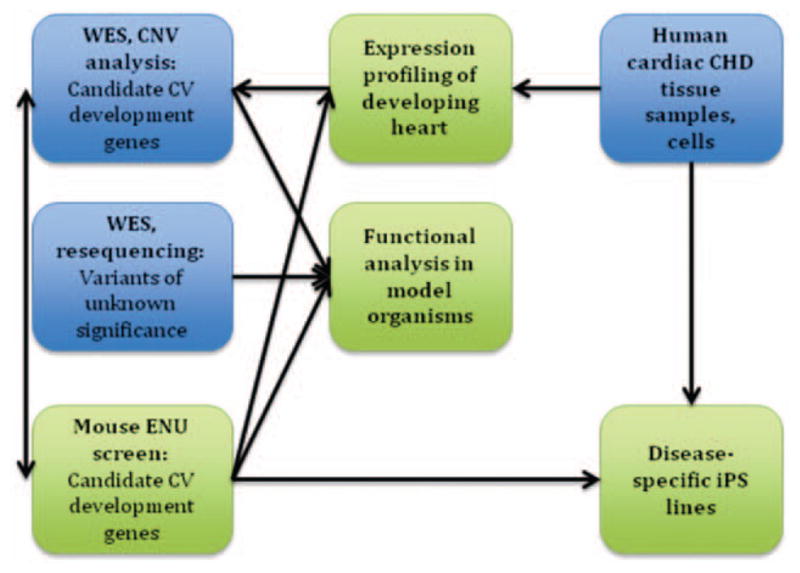
Outline of ongoing interactions between Cardiovascular Development Consortium (CvDC) centers (green) and Pediatric Cardiac Genomics Consortium (PCGC) programs (blue).
One of the most vexing problems in any large-scale human genetic study is how to identify which of the many variants identified in probands are actually diseasecausing. By re-sequencing putative new CHD genes, we expect to identify additional deleterious variants in other cases. Functional validation in model organisms provides a complementary avenue to genetic data and is especially important in the setting of a genetically heterogeneous disease, such as CHD. To perform functional validation on the large number of new CHD candidate genes that will be discovered by the PCGC, the CvDC zebrafish core (University of Utah) will provide high-throughput analysis in zebrafish using morpholino knockdown followed by evaluation of the cardiac phenotype. The CvDC center at the University of Pittsburgh is performing an N-ethyl-N-nitrosourea mouse mutagenesis screen using fetal echocardiography. Over 100 mouse lines with cardiac defects have been recovered, and the underlying mutations are being identified. The list of genes identified in CHD GENES’ subjects will be cross-referenced with those mouse mutations on an ongoing basis. Data from the CvDC cores are deposited in the CvDC data hub, which is available to the PCGC, facilitating seamless integration of model organism and human data.
Ancillary Studies
An important facet of the mission of the PCGC is to facilitate ancillary studies that can be undertaken by individual investigators within or outside of the PCGC centers, as well as with larger research consortia. The URLs for the PCGC’s website for ancillary studies, including one that provides a PDF with the protocol and another that provides the application form, are provided in Table 2. Applications are reviewed by the PCGC’s Ancillary Studies Committee within a month of submission.
Table 2
PCGC Internet Resources
| Resource Title | URL |
|---|---|
| Bench-to-Bassinet program | http://www.benchtobassinet.net |
| PCGC research | http://www.benchtobassinet.net/PCGCresearch.asp |
| PCGC consent documents | Download forms from http://www.benchtobassinet.net/CHDGenesInfCons.asp |
| PCGC ancillary studies | http://www.benchtobassinet.net/PCGCAncillaryStudies.asp |
| Policies | http://www.benchtobassinet.net/PDF/CHAPTER%205_110110.pdf |
| Application | Download form from the Ancillary Studies page |
PCGC indicates Pediatric Cardiac Genomics Consortium.
Interested investigators are encouraged to contact a PCGC Principal Investigator or NHLBI Project Scientist (listed at the PCGC website) to obtain additional information about the existing data and assistance with the application process. For instance, investigators may need to know how many subjects have been recruited with a specific form of CHD, or details about whether specific phenotypic or genotypic data are available. Applications to assay genomic DNAs for variants in one or a very limited number of genes will be entertained, but it is hoped that exome or genome-sequencing data generated within the PCGC will obviate the need for resequencing over time. Until then, the PCGC will balance requests for aliquots of the limited and nonrenewable DNA resources in its biobank against its own ongoing studies and other ancillary studies. The CHD GENES consent document enables provision of PCGC data and DNA to outside investigators in a deidentified manner for cardiovascular research. At some PCGC centers, subjects or the consenting parents, where relevant, have the right to opt out of the use of their data and samples for such external collaboration, but the percentage who are choosing this option is very low.
In general, ancillary studies requiring recall of subjects or extensive additional extraction of data from medical records will be challenging and would require funding provided by the applicant. As identifying information is not transmitted to NERI, ancillary studies of this type will require collaboration with the original study sites. Of note, the consent document for CHD GENES includes the option to opt out of recontact, so some subjects cannot be recalled under any circumstance.
Results to Date
Enrollment
CHD GENES enrollment commenced in December 2010. The planned accrual duration is 4 years and 10 months, to end on September 30, 2015. Enrollment targets were designed to increase over the first 18 months of the study, up to a target of 570 per quarter by July 2012. As of June 8, 2012, we have enrolled 3772 probands with CHD, ranging from those identified as fetuses to older adults. CHD GENES has enrolled consistently above target, ranging from 547 to 703 per quarter, whereas targets ranged from 320 to 570 per quarter. The ancillary sites have contributed approximately 30% of the study subjects. Specimen collection for CHD GENES, primarily peripheral blood samples, is continuous, although does not always occur immediately after consent. Of 3772 probands enrolled, specimens have been submitted for 95% (3586 cases).
We attempt to obtain samples for DNA from both parents of the proband. Thus far, parent–child trios and one parent–child duos constitute 46% and 26% of enrollment, respectively (Table 3). The vast majority of enrolled relatives are parents (98%). Among the parent enrollments, 58% are the mothers.
Table 3
Demographic Characteristics of CHD GENES Probands
| Biosample Obtained
| |||||
|---|---|---|---|---|---|
| Enrolled | Total With Sample | Proband Only | Duo | Trio | |
| Number | 3772 | 3586 | 1005 | 916 | 1665 |
| Percentage male | 55.2 | 55.1 | 51.4 | 56.1 | 56.8 |
| Age at consent (in years) | |||||
 Mean Mean | 11.1 | 11.3 | 18.7 | 12.1 | 6.4 |
 (standard deviation) (standard deviation) | (13.4) | (13.5) | (18.1) | (11.0) | (8.5) |
 Median Median | 5.5 | 5.6 | 14.2 | 11.7 | 2.6 |
 (interquartile range) (interquartile range) | (0.5, 17.1) | (0.5, 17.4) | (2.3, 31.4) | (1.7, 18.5) | (0.3, 11.0) |
 Percentage <1 year Percentage <1 year | 30.9 | 30.6 | 20.6 | 21.7 | 41.4 |
| Ethnicity (%)* | |||||
 White, non-Hispanic White, non-Hispanic | 61.6 | 61.5 | 50.3 | 58.2 | 69.8 |
 White, Hispanic White, Hispanic | 17.8 | 18.0 | 25.1 | 18.1 | 13.7 |
 Black, non-Hispanic Black, non-Hispanic | 6.2 | 6.1 | 5.6 | 9.9 | 4.2 |
 Asian, non-Hispanic Asian, non-Hispanic | 4.8 | 4.9 | 7.4 | 3.2 | 4.4 |
 Mixed ethnicity or other Mixed ethnicity or other | 6.8 | 6.8 | 8.0 | 7.3 | 5.9 |
 Incomplete information Incomplete information | 2.8 | 2.7 | 3.5 | 3.2 | 2.0 |
CHD GENES indicates Congenital Heart Disease Genetic Network Study.
Characteristics of the Subjects
Table 3 shows selected demographic characteristics of the probands. Among the enrolled probands, 55.2% are male. The median age is 5.5 years, with 31% <1 year old and 23% >18 years at enrollment. One third of probands were enrolled at age <1 year. The proportions of non-Hispanic white and Hispanic white probands are 62% and 18%, respectively. Blacks, Asians, and individuals of mixed or other ethnic characteristics each constitute 5% to 7%. The characteristics of probands from whom biospecimens have been obtained are similar demographically to the larger cohort of all enrollees. Probands for whom DNA was procured from at least one parent are younger, with 41% of probands among completed parent–child trios being <1 year of age.
The percentages of probands with specific isolated forms of CHD or more complex cardiovascular lesions belonging to CHD classes are shown in Table 4. For this, we relied on the classification system used by the National Birth Defects Prevention Study.26 The distribution of CHD types diverges significantly from the distribution in the general population, with some lesions being underrepresented (atrial septal defect and ventricular septal defect population-based frequencies of 16% and 28%, respectively) and others overrepresented (population-based frequencies of conotruncal defects 15%; left ventricular outflow tract obstructive lesions 9%; heterotaxy 2%).27
Table 4
Clinical Characteristics of CHD GENES Probands
| % | |
|---|---|
| Primary cardiac defect | |
 Conotruncal defect Conotruncal defect | 30.9 |
 Left ventricular outflow tract obstructive lesion Left ventricular outflow tract obstructive lesion | 27.1 |
 Heterotaxy Heterotaxy | 6.8 |
 Atrial septal defect, isolated (secundum or sinus venosus) Atrial septal defect, isolated (secundum or sinus venosus) | 5.3 |
 Ventricular septal defect, isolated Ventricular septal defect, isolated | 4.7 |
| Extracardiac defects | 41.0 |
| Abnormal clinical genetic testing | |
 Autosomal or sex chromosome aneuploidy Autosomal or sex chromosome aneuploidy | 3.9 |
 Deletions excluding 22q11.2 and duplications Deletions excluding 22q11.2 and duplications | 2.5 |
 22q11.2 deletion 22q11.2 deletion | 2.1 |
 Single gene mutations Single gene mutations | 2.1 |
 Unbalanced rearrangement Unbalanced rearrangement | 0.5 |
CHD GENES indicates Congenital Heart Disease Genetic Network Study.
About 41% of probands, including those with genetic diagnoses, have at least one extracardiac anomaly. Extracardiac anomalies included major and minor malformations, dysmorphic features, and endocrinologic and hematologic anomalies. This rate is approximately double than that observed in a population-based study of CHD.27
Approximately, 11% of probands were known to have a genetic abnormality based on previous testing, usually in the context of clinical care. This proportion is likely a lower bound for the prevalence of known genetic anomalies in the cohort because the data derive from parental report and medical records at the recruiting institutions, both of which may be incomplete. Among probands with identified genetic anomalies, the most common abnormalities are aneuploidy (primarily autosomal) and 22q11.2 deletions.
Biospecimens
The preferred type of biospecimen in CHD GENES for genomic DNA extraction is whole blood; saliva is the alternative from participants aged >5 years. Samples are collected at each of the clinical sites and processed centrally at Coriell. Genomic DNA is extracted and assessed for quality control by assessing DNA quantity and quality by measuring A260, A280, assessing DNA degradation by gel electrophoresis, and performing SNP genotyping. Of the 3772 probands enrolled, whole blood has been collected on 88% of participants and saliva on 8% of participants. Forty-four percent of probands have whole-blood DNA available from the proband and both parents. Quality control metrics were met for 97% of whole-blood samples and 91% of saliva samples. We have collected a minimum of 10 μg of DNA that passed quality control on 98% of participants, and a minimum of 20 μg on 95% of participants.
Cardiac tissues, which can be used for gene-expression analyses or to identify somatic mutations, are procured at the time of surgery for some subjects. These tissues include those that are resected during routine surgical procedures for CHD (eg, ventricular muscle, diseased valve tissues, abnormal great vessels, right atrial appendage), as well as small portions of the atrial septum primum that can be biopsied during secundum atrial septal defect repair, if specific consent is obtained. All tissues are either snap frozen with liquid nitrogen or stored in RNAlater.
Current Ongoing Studies
Exome Sequencing of Trios
The PCGC is exploring the hypothesis that de novo mutations with strong effect size underlie a significant percentage of CHD. To test this, we selected nearly 400 probands with severe forms of CHD, and from whom DNA samples from both parents were available (ie, trios).
To look for de novo mutations, whole-exome sequencing (WES) is being performed for each member of the trio at the PCGC’s Whole Exome Sequencing Core. Analysis is focusing on high-probability single basepair substitutions and small insertions/deletions (indels) in the proband that were not present in the parents, and filtered for variants that are rare and likely to be deleterious (nonsense, frameshift, splicing in the highly conserved motifs, or missense changes altering conserved residues and predicted to be damaging). Several point variants and indel calls at different levels of confidence in the calling algorithm are being confirmed with Sanger sequencing at the PCGC’s Confirmation Core to establish which variants will be pursued.
Although the primary purpose of this study was to identify de novo point mutations and indels underlying CHD, the completed WES trio dataset will be potentially valuable for looking at other types of genetic defects and mechanisms. For instance, methods have been developed to use WES for the identification of CNVs.28,29 Using these approaches, we intend to look for de novo or inherited CNVs relevant for CHD. For heterotaxy, previous work has implicated defects in ciliary biology, and we will examine those candidate genes.30,31 Studies in countries where consanguineous matings are culturally acceptable have routinely shown increased rates of CHD among couples who are consanguineous.12,13 Using our WES data, we intend to look for probands with long stretches of homozygosity for rare variants, and then focus on those which are predicted to be damaging and alter genes relevant for cardiovascular development.
Copy Number Variants
The PCGC is exploring the hypotheses that rare CNVs, inherited or de novo, contribute to the pathogenesis of CHD. To test this, probands are being genotyped using Illumina SNP arrays at the PCGC’s Genotyping Array Core. Of the nearly 900 subjects being genotyped, in 46% both parents are also being genotyped. The focus of the ongoing analysis is on rare de novo CNVs. CNVs of interest will be assayed with a PCR-based method in the PCGC’s Confirmation Core Laboratory. In the future, we will assess the CNVs observed in probands for whom no parent was genotyped, either performing burden analysis using controls or secondarily determining status in the parents for CNVs of interest.
Candidate Gene Resequencing
The PCGC is exploring hypotheses related to burden of point mutations and indels in candidate genes previously related to CHD. The criteria for including genes for resequencing were as follows: known mutations in syndromic or nonsyndromic CHD, positional candidate genes from genetic linkage analysis, or genes of critical importance for cardiovascular development. To study this, resequencing panels, comprising approximately 130 genes, were developed focusing on specific forms or classes of CHD. Custom Agilent SureSelect reagents were designed to cover those genes. Newly discovered CHD genes will be added to the resequencing platform. Next generation sequencing for probands and controls (children with normal echocardiograms) are ongoing at the PCGC’s Candidate Gene Evaluation Core.
Tissue Gene Expression
The PCGC is exploring 2 hypotheses using DNAs and RNAs extracted from tissues: (1) Somatic mutations contribute to the pathogenesis of CHD; (2) skewed allelic expression, either through imprinting or alterations in promoters and enhancers, is important for the pathogenesis of CHD.
To look for possible somatic mutations, WES is being performed from genomic DNA extracted from one or more tissues of probands for whom WES has been completed using genomic DNA extracted from peripheral blood leukocytes. Comparison of the results will enable the identification of somatic mosaicism or heart-specific somatic mutations. Although prior work with fixed tissues has suggested that such mutations are important for CHD,32–36 comparable studies with fresh, frozen tissues suggested that somatic defects are not common.37 Of note, no genome-wide study of somatic variation in cardiac tissues has been performed.
To search for skewed allelic expression, next generation sequencing of cDNAs generated from RNAs extracted from the cardiovascular tissues is being performed at the PCGC’s RNAseq Core. For subjects for whom WES or SNP genotyping has been performed, we will be able to detect genes whose expression is skewed strongly from the expected 50:50 ratio using heterozygous cSNPs. For RNA sequencing data for which neither WES nor SNP genotypes are available, we can still use very rare alleles constituting 100% or nearly 100% of the sequences as a marker of skewed expression. Clustering of genes with skewed expression will identify imprinted genomic regions. Using whole-genome sequencing, we will attempt to understand why genes that are not imprinted demonstrate skewed expression. Our collaboration with the CvDC, which is identifying enhancer regions for genes important for cardiovascular development, will inform this analyis. Finally, we will look for skewed inactivation of the X-chromosome among female subjects for whom RNA sequencing is performed.
Limitations
The full compendium of genetic variants underlying CHD is expected to be very large. Although we expect CHD GENES to discover significant numbers of novel causative variants, achieving the complete set, particularly for rarer forms of CHD, will not likely be achieved. The power of this study to detect variants relevant for CHD will depend on a host of factors. For instance, genome-wide association studies searching for common variants for disease typically require thousands of subjects.38 Although CHD GENES will have sufficient numbers of probands to power a study of common variants in CHD, in general, studies seeking such variants for specific lesions or even lesion categories like conotruncal defects probably could not be adequately powered with only CHD GENES’ resources. Studies of CNVs in CHD have suggested excess frequencies of large genomic defects in 5% to 15% of affected individuals,15–19 so the existing collection of trios should enable substantial discovery efforts. Nonetheless, some CNVs relevant to CHD are likely to be rare and may not be observed many times or even more than once, limiting our ability to describe genotype–phenotype correlations. Our convenience-based recruitment will not produce a population-based cohort. One implication of that is that our cohort is inappropriate for studies of nongenetic exposures in relation to CHD. Another implication is that we will be unable to estimate attributable risks for the genetic causes of CHD that we identify.
The availability of CHD tissue will provide limited assessments of the cellular and molecular responses to CHD mutations and of the role that somatic mutations plays in CHD. Of note, we are accessing pathological samples removed at surgery, which means postnatally. Genes of relevance for cardiovascular development may no longer be expressed after birth, or may be expressed in cell types or tissues not removed at surgery. The underlying pathologies, which invoke responses such as cardiac hypertrophy in response to volume or pressure overload, clearly alter a wide range of genes and proteins. In addition, processes transpiring during surgery prior to tissue removal (eg, instillation of cardioplegia, cardiopulmonary bypass) could alter genes and proteins in ways that are not predictable.
Finally, CHD GENES is not systematically obtaining data about environmental factors that might contribute to CHD. To the extent that gene×environment interactions contribute to the pathogenesis of CHD, this study will not be well-positioned to find them.
In summary, the PCGC’s CHD GENES protocol is an international, multicenter, prospective cohort study of subjects with CHD, designed to identify and quantify genes and mutations responsible for CHD, investigate phenotype–genotype correlations, and create a biorepository of DNA specimens for current and future research. Because of the unique relationship with the other 2 consortia of the NHLBI’s Bench to Bassinet program, the results of the CHD GENES study can be quickly translated to basic and clinical studies of the origins and treatment of CHD. Finally, the nature of the biorepository and the structure of the PCGC allow for ancillary studies to be initiated by members of the general scientific community.
Acknowledgments
We thank the physicians who provided us with access to their patients, as well as the individuals and families who chose to participate in this research.
Sources of Funding
This work was supported by grant numbers HL098123, HL098147, HL098153, HL098162, HL098163, and HL098188 from NHLBI; Columbia University’s CTSA grant UL1 RR024156; and the Howard Hughes Medical Institute. The contents of this publication are solely the responsibility of the authors and do not necessarily represent the official views of the NHLBI.
Appendix
Writing Committee: Bruce Gelb MD (Chair), Martina Brueckner, MD, Wendy Chung, MD, PhD, Elizabeth Goldmuntz, MD, Jonathan Kaltman, MD, Juan Pablo Kaski, MD, Richard Kim, MD, Jennie Kline, PhD, Laura Mercer-Rosa, MD, George Porter, MD, PhD, Amy Roberts, MD, Ellen Rosenberg, Howard Seiden, MD, Christine Seidman, MD, Lynn Sleeper, ScD, Sharon Tennstedt, PhD
National Heart, Lung, and Blood Institute: Jonathan Kaltman, MD, Charlene Schramm, PhD, Kristin Burns, MD, Gail Pearson, MD, ScD, Ellen Rosenberg RN
Clinical Sites: Children’s Hospital Boston, Jane Newburger, MD, MPH (PI), Roger Breitbart, MD, Steven Colan, MD, Judith Geva, MSW, Angela Monafo, Amy Roberts, MD, Janice Stryker, MGC, Brigham and Women’s Hospital, Christine Seidman, MD (PI), Barbara McDonough, RN, Jonathan Seidman, PhD, Children’s Hospital Philadelphia, Elizabeth Goldmuntz, MD (PI), Sharon Edman, MS, Jennifer Garbarini, MS, CGC, Hakon Hakonarson, MD, PhD, Laura Mercer-Rosa, MD, Laura Mitchell, PhD, Jessica Tusi, MSGC, Peter White, PhD, Stacy Woyciechowski, MS, CGC, Columbia University, Wendy Chung, MD, PhD (PI), Dorothy Warburton, PhD (PI), Danielle Awad, MPH, Katrina Celia, MD, Davina Etwaru, Jaswinder Kaur Sond, MBBS, Jennie Kline, PhD, Rosalind Korsin RN, BSN, Alyssa Lanz, Emma Marquez, Ismee Williams, MD, Abigail Wilpers, Roslyn Yee, MPH, Icahn School of Medicine at Mount Sinai, Bruce Gelb, MD (PI), Denise Guevara, Ariel Julian, Meghan Mac Neal, MSGC, MPH, Cassie Mintz, MSGC, Inga Peter, PhD, Ravi Sachidanandam, PhD, Howard Seiden, MD, Cohen Children’s Medical Center, Angela Romano-Adesman, MD (PI), Dorota Gruber, MS, CGC, Nancy Stellato, RN, MSN, Yale University, Martina Brueckner, MD (PI), Richard Lifton, MD, PhD (PI), Nancy Cross, RN, University College London, John Deanfield, MD (PI), Alessandro Giardini, M.D (PI), Karen Flack, RN, University of Rochester, George Porter, MD, PhD (PI), Eileen Taillie, MGS, Children’s Hospital Los Angeles, Richard Kim, MD (PI), Nhu Tran, RN
Data Coordinating Center: New England Research Institutes, Sharon Tennstedt, PhD (PI), Roger Breitbart, MD, Kimberly Dandreo, MSc, Dianne Gallagher, MS, Minmin Lu MS, Lynn Sleeper, ScD
Core Laboratories for PCGC: Biorepository, Coriell Institutes: Dorit Berlin, PhD (PI), Christine Beiswanger, PhD
Whole Exome Sequencing, Yale University: Richard Lifton, MD, PhD (PI)
Candidate Gene Evaluation and RNASeq, Harvard Medical School: Jonathan Seidman PhD (PI)
Genotyping Array, Children’s Hospital Philadelphia: Hakon Hakonarson, MD, PhD (PI)
Data Hub, Children’s Hospital Philadelphia: Peter White, PhD (Director), Mike Italia
Confirmation, Columbia University: Wendy Chung, MD, PhD (PI)
Whole Genome Sequencing Harvard Medical School and Brigham & Women’s Hospital: Christine Seidman, MD (PI)
Observational Study Monitoring Board: Maria Brooks (Chair), PhD, University of Pittsburgh, Michelle Olive, PhD, National Heart, Lung, and Blood Institute, Jeffrey Botkin, MD, MPH, University of Utah School of Medicine, Josee Dupuis, PhD, Boston University School of Public Health, Vidu Garg, MD, Ohio State University College of Medicine, Mike Watson, PhD, American College of Medical Genetics
External Advisory Committee: James Bristow, MD, DOE Joint Genome Institute, Todd Evans, PhD, Weill Cornell Medical College, Christina Kendziorski, PhD, University of Wisconsin, Elaine Mardis, PhD, Washington University School of Medicine, Jeffrey Murray, MD, University of Iowa, Joel Saltz, MD, PhD, Emory University, Hector Wong, MD, Cincinnati Children’s Hospital
Footnotes
The complete list of individuals contributing to this research consortium are listed in the Appendix.
Disclosures
None.
References
Full text links
Read article at publisher's site: https://doi.org/10.1161/circresaha.111.300297
Read article for free, from open access legal sources, via Unpaywall:
https://www.ahajournals.org/doi/pdf/10.1161/CIRCRESAHA.111.300297
Citations & impact
Impact metrics
Citations of article over time
Alternative metrics
Smart citations by scite.ai
Explore citation contexts and check if this article has been
supported or disputed.
https://scite.ai/reports/10.1161/circresaha.111.300297
Article citations
Increased endothelial sclerostin caused by elevated DSCAM mediates multiple trisomy 21 phenotypes.
J Clin Invest, 134(11):e167811, 03 Jun 2024
Cited by: 0 articles | PMID: 38828726 | PMCID: PMC11142749
Mutations in genes related to myocyte contraction and ventricular septum development in non-syndromic tetralogy of Fallot.
Front Cardiovasc Med, 10:1249605, 28 Sep 2023
Cited by: 1 article | PMID: 37840956 | PMCID: PMC10569225
The Influence of Maternal Condition on Fetal Cardiac Function during the Second Trimester.
Diagnostics (Basel), 13(17):2755, 25 Aug 2023
Cited by: 0 articles | PMID: 37685293 | PMCID: PMC10486346
Heterozygous rare variants in NR2F2 cause a recognizable multiple congenital anomaly syndrome with developmental delays.
Eur J Hum Genet, 31(10):1117-1124, 27 Jul 2023
Cited by: 5 articles | PMID: 37500725 | PMCID: PMC10545729
VBASS enables integration of single cell gene expression data in Bayesian association analysis of rare variants.
Commun Biol, 6(1):774, 25 Jul 2023
Cited by: 2 articles | PMID: 37491581 | PMCID: PMC10368729
Go to all (100) article citations
Similar Articles
To arrive at the top five similar articles we use a word-weighted algorithm to compare words from the Title and Abstract of each citation.
Assessment of large copy number variants in patients with apparently isolated congenital left-sided cardiac lesions reveals clinically relevant genomic events.
Am J Med Genet A, 173(8):2176-2188, 27 Jun 2017
Cited by: 9 articles | PMID: 28653806 | PMCID: PMC5560080
Diagnostic yield after next-generation sequencing in pediatric cardiovascular disease.
HGG Adv, 5(3):100286, 23 Mar 2024
Cited by: 1 article | PMID: 38521975 | PMCID: PMC11024993
The Congenital Heart Disease Genetic Network Study: Cohort description.
PLoS One, 13(1):e0191319, 19 Jan 2018
Cited by: 55 articles | PMID: 29351346 | PMCID: PMC5774789
A Biobank for Long-term and Sustainable Research in the Field of Congenital Heart Disease in Germany.
Genomics Proteomics Bioinformatics, 14(4):181-190, 27 Apr 2016
Cited by: 4 articles | PMID: 27132144 | PMCID: PMC4996858
Review Free full text in Europe PMC
Funding
Funders who supported this work.
Howard Hughes Medical Institute
NCRR NIH HHS (1)
Grant ID: UL1 RR024156
NHLBI NIH HHS (15)
Grant ID: U01 HL098123
Grant ID: U01 HL098153
Grant ID: U01 HL098163
Grant ID: HL098153
Grant ID: HL098162
Grant ID: HL098163
Grant ID: U01 HL098147
Grant ID: U01 HL098162
Grant ID: HL098123,
Grant ID: HL098147
Grant ID: HL098188
Grant ID: U01 HL098188
Grant ID: UM1 HL098162
Grant ID: UM1 HL098123
Grant ID: UM1 HL098147





