Abstract
Free full text

Improving the Accuracy and Efficiency of Identity-by-Descent Detection in Population Data
Associated Data
Abstract
Segments of indentity-by-descent (IBD) detected from high-density genetic data are useful for many applications, including long-range phase determination, phasing family data, imputation, IBD mapping, and heritability analysis in founder populations. We present Refined IBD, a new method for IBD segment detection. Refined IBD achieves both computational efficiency and highly accurate IBD segment reporting by searching for IBD in two steps. The first step (identification) uses the GERMLINE algorithm to find shared haplotypes exceeding a length threshold. The second step (refinement) evaluates candidate segments with a probabilistic approach to assess the evidence for IBD. Like GERMLINE, Refined IBD allows for IBD reporting on a haplotype level, which facilitates determination of multi-individual IBD and allows for haplotype-based downstream analyses. To investigate the properties of Refined IBD, we simulate SNP data from a model with recent superexponential population growth that is designed to match United Kingdom data. The simulation results show that Refined IBD achieves a better power/accuracy profile than fastIBD or GERMLINE. We find that a single run of Refined IBD achieves greater power than 10 runs of fastIBD. We also apply Refined IBD to SNP data for samples from the United Kingdom and from Northern Finland and describe the IBD sharing in these data sets. Refined IBD is powerful, highly accurate, and easy to use and is implemented in Beagle version 4.
SEGMENTS of indentity-by-descent (IBD) may be detected in population samples, using high-density genetic data. Such segments delineate haplotypes that are shared by inheritance from a recent common ancestor. By definition, an IBD segment must be inherited from a single ancestor. Consequently, when detecting an IBD segment in population data, the IBD segment must have sufficient length to provide confidence that the segment is not a fusion of multiple short IBD segments from different ancient common ancestors, while allowing for some error in precisely identifying the segment endpoints. This length constraint implies that for detected IBD segments, the shared common ancestor will be a recent ancestor.
Detectable IBD segments are ubiquitous in genome-wide SNP data from population samples (B. L. Browning and S. R. Browning 2011). Because IBD is fundamental in genetics, detected IBD segments have a wide variety of applications (Browning and Browning 2012), including long-range phase determination (Kong et al. 2008), phasing family data (S. R. Browning and B. L. Browning 2011), imputation (Jonsson et al. 2012), detecting signals of natural selection (Albrechtsen et al. 2009; Cai et al. 2011; Han and Abney 2013), inferring past demographic history (Campbell et al. 2012; Gusev et al. 2012; Palamara et al. 2012; Ralph and Coop 2012), IBD mapping (Purcell et al. 2007; Gusev et al. 2011; Browning and Thompson 2012), and heritability analysis in founder populations (Price et al. 2011; Zuk et al. 2012; Browning and Browning 2013).
A variety of methods exist for IBD segment detection. Probabilistic methods including Beagle IBD (Browning and Browning 2010), IBD_Haplo (Brown et al. 2012), RELATE (Albrechtsen et al. 2009), IBDLD (Han and Abney 2011), and PLINK (Purcell et al. 2007) fit a hidden Markov model (HMM) for IBD status and determine posterior probabilities of IBD. Computation times for these methods scale quadratically with increasing sample size, and all except PLINK are too computationally intensive for very large data sets (Browning and Browning 2012). PLINK requires prior thinning of genetic markers to reduce linkage disequilibrium (LD), which discards information (Browning and Browning 2010).
Several nonprobabilistic IBD-detection methods have been developed for use on large data sets. GERMLINE (Gusev et al. 2009) introduced an efficient dictionary approach to IBD detection that scales much better with increasing sample size. This dictionary approach was adopted by fastIBD (B. L. Browning and S. R. Browning 2011) and is used here to detect candidate IBD tracts for evaluation by the Refined IBD algorithm. The nonprobabilistic methods differ in their criteria for recognizing IBD: GERMLINE uses genetic length of a segment, fastIBD uses haplotype frequency, and Refined IBD uses genetic length and a likelihood ratio for an IBD vs. a non-IBD model.
Because Refined IBD incorporates modeling of LD, it is able to make powerful use of the data. In particular, Refined IBD does not require thinning of markers to reduce LD, and it does not incur an increase in false positive rates due to unmodeled LD. Refined IBD achieves higher accuracy than GERMLINE because it includes a refinement step that applies a probabilistic approach rather than using only lengths of haplotype sharing. Refined IBD’s probabilistic approach gives higher accuracy than fastIBD’s haplotype frequency approach because it better accounts for haplotype phase uncertainty. Refined IBD is computationally efficient and can be used on large data sets.
We compare the results of Refined IBD with those of GERMLINE, fastIBD, and Beagle IBD on simulated data. We also apply Refined IBD to two large data sets: the Wellcome Trust Case Control Consortium phase 2 control data (5000 individuals from the United Kingdom genotyped on 1 million SNPs) (Barrett et al. 2009) and the Northern Finland Birth Cohort (5000 individuals from Northern Finland genotyped on 300,000 SNPs) (Sabatti et al. 2009).
Methods
Overview of the Refined IBD algorithm
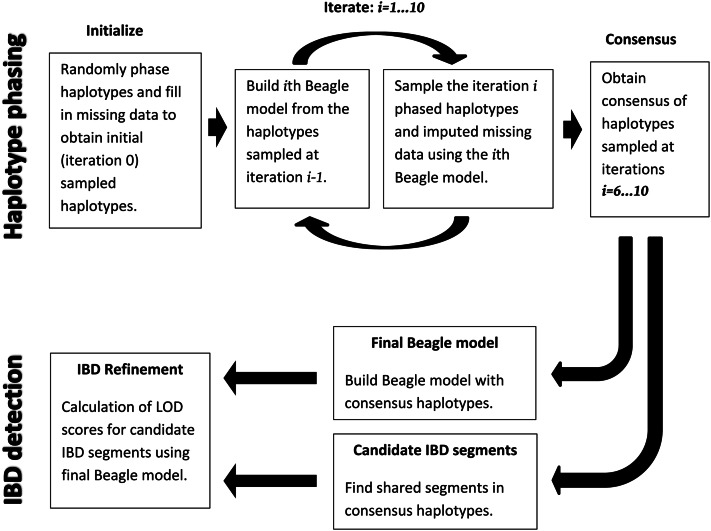
Figure 1 gives an overview of the Refined IBD algorithm. The first part of the algorithm (top row in Figure 1) estimates haplotype phase. Subsequent to haplotype phase determination, there are two steps in the Refined IBD algorithm (bottom row in Figure 1). The first is identification of candidate IBD segments. The candidate segments are regions in which two individuals share an identical statistically phased haplotype segment that is longer than a specified threshold. In the second step, we use the phased haplotypes to build a haplotype frequency model, and for each candidate IBD segment we calculate the likelihood of an IBD model (one haplotype shared IBD) and of a non-IBD model (no haplotypes shared IBD). We compute the LOD score, which is the base 10 log of the likelihood ratio. Candidate segments having LOD score greater than a specified threshold (the default threshold is 3.0) are reported as IBD segments.
It is possible to run Refined IBD several times with different random-number seeds and to merge the resulting IBD segments. Except as otherwise noted, all results presented here are from a single run. IBD segments from multiple runs for a sample pair are combined by taking the union of the IBD segments from the multiple runs and merging overlapping IBD segments in the union. Merging is performed sequentially on pairs of overlapping segments. Whenever a pair of overlapping segments is found, the pair of IBD segments is replaced with the merged IBD segment. The merged IBD segment’s chromosome interval is the union of the overlapping intervals, and the merged segment’s LOD score is the maximum LOD score of the overlapping intervals. Merging IBD segments from multiple runs results in greater power to detect long IBD segments at the cost of increased run time and loss of haplotype information.
Refined IBD reports the index (1 or 2) of the IBD haplotype in each individual. Each index identifies one of the two ordered consensus haplotypes of an individual that are reported by Beagle. However, when IBD segments from multiple runs are merged, the haplotype identification is lost as the estimated haplotype phase typically differs slightly between runs.
Identification of candidate IBD segments
When applying the GERMLINE algorithm to detect candidate IBD segments, we do not permit any mismatching alleles in the shared haplotype. Each candidate IBD segment is defined by its starting and ending genome coordinates, the pair of sample identifiers, and the haplotype index (1 or 2) of the shared haplotype for each sample.
The ibdwindow parameter in Beagle version 4 determines the number of markers included in each window when using the GERMLINE algorithm to find candidate IBD segments. The ibdwindow parameter is equivalent to the GERMLINE bits parameter. Too large a value may result in missing short segments of IBD, while too small a value will increase computation time. The default value of 64 is suitable for SNP arrays with 1 million SNPs across the genome, as at this marker density, 64 markers correspond to ~0.2 cM, which is significantly shorter than the default threshold on IBD segment length. For SNP array data, we recommend setting this parameter to approximately the average number of markers per 0.2 cM.
The ibdcm parameter in Beagle version 4 controls the minimum genetic length of a candidate IBD segment. A value that is too small will result in increased computing time while not contributing much to IBD detection as small candidate segments are unlikely to pass the LOD score threshold. The default value of 1.0 cM was chosen based on the relatively low power to detect smaller segments in SNP array data (see Results).
Haplotype frequency models for IBD and non-IBD
We start with a model for haplotype frequencies. We use the Beagle HMM (S. R. Browning and B. L. Browning 2007), but our approach is general and it could be readily adapted to other HMMs for haplotype frequencies. The HMM for haplotype frequencies determines a HMM for unrelated individuals and a HMM for parent–offspring pairs (Browning and Browning 2009). We calculate the probability of the observed genotype data for a pair of individuals under a non-IBD model (the likelihood of non-IBD), using the HMM for unrelated individuals, and we calculate the probability of the observed genotype data for a pair of individuals under an IBD model (the likelihood of IBD), using the HMM for a parent–offspring pair since a parent–offspring pair also shares one haplotype identical by descent.
A HMM is defined by its state space, initial probabilities, transition probabilities, set of emitted symbols, and emission probabilities (Rabiner 1989). Since the Beagle HMMs for unrelated individuals and parent–offspring pairs have been fully described previously (S. R. Browning and B. L. Browning 2007, 2009), we give only a brief description of these HMMs here. In the Beagle HMM, there is a set of haploid hidden states Sm corresponding to each marker m. Each haploid hidden state corresponds to a cluster of haplotypes that are locally similar around marker m. The hidden state for a diploid individual at marker m is an element
In the Beagle HMM, each haploid hidden state at marker m is labeled with one of the marker’s alleles (more than one hidden state at marker m can be labeled with the same allele). The emission probability for the labeled allele is 1, while the emission probability for any other allele is 0. For parent–offspring pairs, this means that the probability of the observed genotypes given the hidden state
Each haplotype used to build the Beagle HMM has a unique path through the model. Thus each haploid state has an associated count of how many haplotypes pass through the state, and similarly each possible transition has an associated count. Consider a haploid transition in the Beagle HMM. Call the state that the transition starts from at marker m the “source” state and the state that the transition goes to at marker m + 1 the “destination” state. The transition probability is the count associated with that transition divided by the count associated with the source state for the transition. In other words, of those haplotypes passing through the source state, the transition probability is the proportion that transitions into the destination state. Given a starting marker m, the initial probability for a haploid state at marker m is equal to the proportion of haplotypes that pass through that state, which is the count associated with that state divided by the total number of haplotypes used to build the model. Transition and initial probabilities for unrelated diploid individuals (pairs of hidden states) or parent–offspring pairs (triples of hidden states) are obtained by multiplying the corresponding haploid probabilities.
Since IBD segments are typically much shorter than their corresponding chromosome, we reduce computation time by calculating likelihoods using only genotype data in the interior of the candidate IBD segment. In this way, we can also avoid modeling the recent recombination events that demarcate the boundaries of the IBD segment. It is difficult to identify the IBD segment endpoints with high accuracy. Incorrectly including some non-IBD markers at the ends of a real IBD segment can result in a severely reduced LOD score for the segment, while incorrectly removing small parts of the ends of a real IBD segment tends to result in only a small drop in LOD score. We thus trim a small fixed number of markers from each end of the candidate IBD segment and compute likelihoods of the non-IBD and IBD models in the trimmed genomic interval. The trimmed markers are restored to the IBD segment after calculation of the likelihoods. The number of markers to trim when calculating the LOD score is controlled by the ibdtrim parameter in Beagle version 4. Increasing the trim number will reduce power to detect short segments. When a short candidate segment is trimmed there may not be enough markers left to provide sufficient information to be confident of IBD. The default trim value (40) was chosen based on analyses of the Wellcome Trust Case Control Consortium 2 data, by looking for a value that would maximize the amount of IBD detected (data not shown). For SNP array data we recommend setting the trim value to approximately the average number of markers per 0.15 cM.
The likelihoods for the IBD and non-IBD models are calculated using Baum’s forward algorithm (Baum 1972). For the non-IBD model, the probability of the observed data for a pair of individuals is the product of the probabilities for each individual.
Other improvements to Beagle in version 4
As well as implementing Refined IBD, Beagle version 4 has improvements to haplotype phasing and to general usability. Beagle version 4 reports the consensus haplotype for each individual. Other haplotype phasing programs also use consensus haplotypes (Scheet and Stephens 2006; Li et al. 2010). Previous versions of Beagle have used the Viterbi algorithm (Viterbi 1967) to generate the reported phased haplotypes. However, we have found the consensus haplotypes to be more accurate than the haplotypes obtained from the Viterbi algorithm.
For consensus haplotypes, haplotypes are estimated at multiple iterations (or from multiple runs) of the phasing algorithm, and the results are merged. The top row of Figure 1 illustrates the procedure for obtaining consensus haplotypes in Beagle version 4. The haplotype phasing module involves multiple iterations of estimating (sampling) haplotypes based on a provisional model and then updating the model based on the new estimated haplotypes. By default, four pairs of haplotypes are sampled per individual per iteration. Haplotypes estimated in the first few iterations are not likely to be very accurate, because the provisional model is still in the initial stages of converging toward a good solution. Thus, in Beagle version 4, the consensus haplotypes are obtained from all sampled haplotypes after a specified number of burn-in iterations (five burn-in iterations and five additional iterations by default). Under default settings there are 20 pairs of sampled haplotypes per individual that are used for obtaining the consensus haplotypes (4 pairs per iteration × 5 iterations after burn-in).
The first step in obtaining the consensus haplotypes is to obtain consensus genotypes for those genotypes that were missing. Consensus genotypes are obtained by taking the most frequently sampled genotype, breaking ties randomly. After consensus genotypes are obtained, the consensus phasing for an individual is obtained by working along the chromosome, one pair of successive heterozygous genotypes at a time. A pair of successive heterozygous genotypes has no intervening heterozygous genotypes. Only sampled haplotype pairs having the consensus heterozygous genotype at both markers are used to determine the consensus phasing. The consensus phasing of two successive heterozygous genotypes is determined by majority vote, breaking ties randomly. For example, in phasing the genotypes AC and TG, if 16 sampled haplotype pairs have AT/CG phase, while 4 sampled haplotype pairs have AG/CT phase, the consensus haplotypes will have the AT/CG phase.
Beagle version 4 uses Variant Call Format for input and output data files (Danecek et al. 2011). Variant Call Format is a standard, widely used format for genotype data (1000 Genomes Consortium 2010), and use of this format will reduce the need for tedious data file format conversion.
Beagle version 4 also uses a sliding marker window that makes the memory usage independent of the number of markers in the data set. Decreased accuracy near the edge of the marker windows is avoided by using overlapping windows and trimming half of the overlap from each window prior to merging data in adjacent windows. Haplotypes in adjacent marker windows are aligned using a heterozygote near the middle of the overlap.
Scale factors
The Beagle HMM is represented by a directed acyclic graph. When the model is constructed, a process of node merging occurs. Two nodes of the graph, x and y, are merged if the maximum difference in downstream frequencies is less than a threshold (B. L. Browning and S. R. Browning 2007). The threshold is
where m is the scale factor, b is the shift parameter, and nx and ny are the numbers of haplotypes whose path through the graph includes nodes x and y, respectively. The scale and shift parameters were originally introduced to control the degree of parsimony of the fitted Beagle model when performing association testing (B. L. Browning and S. R. Browning 2007). Larger values of these parameters result in more merging of nodes and hence a more parsimonious model. Without the added parsimony, each haplotype cluster may contain very few observations, reducing power to detect an association.
For Beagle’s other applications (phasing, imputation, and IBD detection), we have not found the shift parameter to be useful, so we assume a shift parameter of b = 0 for the remainder of this section. For phasing and imputation, we have found that a scale factor of m = 1 performs well. For IBD detection, a different scale factor (the IBD scale factor) can be used for the final model while continuing to use a scale factor of 1 for the haplotype phasing step. We have previously used IBD scale factors of 1 (Browning and Browning 2010) and 2 (B. L. Browning and S. R. Browning 2011). Recently we have realized that for a given sample size (number of genotyped individuals), the choice of IBD scale parameter for values ≥2 is somewhat arbitrary, provided that appropriate compensatory adjustment is made to the threshold for significance of the fastIBD score or the Refined IBD LOD score (data not shown). However, as the sample size changes, the optimal choice of IBD scale factor for a fixed score threshold changes. For a given choice of IBD scale parameter, as more individuals are added to the data, the fitted model becomes larger (less parsimonious). A larger model allows for higher precision in haplotype frequency estimation, resulting in fewer false positives. On the other hand, the requirement that shared haplotypes must traverse the same path through the model to be declared IBD becomes more onerous as the model size increases, and detection rates can drop.
Therefore, to have a single LOD score threshold regardless of sample size, the IBD scale factor must increase as the sample size increases and the size (complexity) of the fitted Beagle model must stay approximately constant, to maintain power to detect IBD as sample size increases. Since model complexity is controlled by the merging threshold given in Equation 1, when the sample size increases by a factor of k, the IBD scale factor can be increased by a factor of
In summary, with reference to Equation 1 and Figure 1, a scale factor of 1 is used for model building during haplotype phasing (top row of Figure 1). However, an IBD scale factor >1 is used for building the final Beagle model for IBD refinement (bottom row of Figure 1), as described above. A shift of 0 is used in all cases.
Simulated SNP data
To assess false positive and true positive IBD detection rates, we simulated data. In our simulation data, we attempted to match both current and historical effective population sizes, to obtain a good match with real data. Ancient historical population sizes affect the number of common variants and the extent of LD between them. The extent of LD affects power to detect IBD and can affect false positive IBD detection rates. Current and very recent population sizes affect the number of rare variants and the amount of detectable IBD. The amount of detectable IBD in the simulation is critical. Our method is designed for large outbred populations in which the amount of detectable IBD is low, so we want the simulated data to reflect this.
Recent analyses of the allele frequency spectrum, with particular attention paid to the rare end of the spectrum, have shown that explosive population growth has occurred in the past few hundred generations (Keinan and Clark 2012). In our view, previous models fitted to sequence data do not go far enough in modeling this growth, as they allow only for a single rate of recent growth, resulting in growth rate estimates of 2% (Nelson et al. 2012) or 9% (Coventry et al. 2010) per generation. In contrast, census data show that the rate of population growth has accelerated in the past few hundred generations and is currently ~30% per generation globally (Keinan and Clark 2012).
We used Fastsimcoal (Excoffier and Foll 2011) to simulate sequence data that we then thinned to obtain simulated SNP array data. We simulated 10 regions, each with 30 Mb of sequence on 2000 diploid individuals. We used a mutation rate of 2.5 × 10−8 (Nachman and Crowell 2000) and a recombination rate of 10−8 (i.e., 1 Mb = 1 cM). Our simulation scheme was designed with European populations in mind, as our available real data are from European populations. The effective population size was initially (prior to expansion beginning 300 generations ago) 3000 diploid individuals. This reflects the European effective population size estimated using LD between common variants (Tenesa et al. 2007). In our simulations, the effective population size began to grow 300 generations ago (timing reflects the advent of large-scale organized agriculture) at a rate of 1.8% per generation (reflecting, e.g., the 1.7% growth rate estimate in Nelson et al. 2012), reaching 270,000 by 50 generations ago. We modeled population growth rate increases in the past 50 generations based on English census data, as shown in Supporting Information, Figure S1. At 50 generations ago, we increased the growth rate to 5%, giving effective population size 2 million at 10 generations ago. We increased the growth rate further, to 25% per generation, for the final 10 generations, yielding an effective population size of 24 million (2 × 106 × exp(0.25 × 10)) at the current generation.
After generating the sequence data, we created simulated SNP array data from it by removing all variants with more than two alleles and all variants with frequency <2%, and by selecting variants from those remaining to obtain ~1000 variants per 30-Mb region (corresponding to a SNP density of 1 million SNPs genome-wide) with minor allele frequencies uniformly distributed between 2% and 50%. We then added genotype error at a rate of 0.05%, reflecting the very high accuracy seen in current genotyping arrays after applying standard quality control filters (Steemers et al. 2006). Genotype error was introduced by converting homozygote genotypes to heterozygote and by converting heterozygote genotypes to a randomly chosen homozygote genotype. We also removed haplotype phase information.
We performed analyses on all 2000 simulated individuals and on a subset of 500 individuals. We used Refined IBD with minimum segment length 0.5 cM and LOD score thresholds of 3 and 4, with the remaining parameters at their default settings. We ran fastIBD (Beagle version 3.3.1) (B. L. Browning and S. R. Browning 2011) with IBD scale at the default value of 2 for n = 500 (this is very close to our new recommendation of
We ran GERMLINE version 1.5.1 (Gusev et al. 2009) with parameters used in Gusev et al. (2011). Specifically, we used options “-haploid -min_m 1 -bits 32 -err_hom 1 -err_het 1”. The “-min_m 1” option means that GERMLINE reports only IBD segments with estimated length ≥1 cM. The “-haploid” option ensures that GERMLINE makes use of the haplotype phase information. Our previous published analyses with GERMLINE used GERMLINE’s default setting that does not use haplotype phase information (B. L. Browning and S. R. Browning 2010, 2011). As seen by comparing the results presented here with the results in our earlier work, utilization of haplotype phase information greatly improves GERMLINE’s performance on SNP array data. We used the phased haplotypes output from the Beagle Refined IBD analysis as input to GERMLINE. These haplotypes are based on a consensus of haplotypes sampled at different iterations of the Beagle phasing algorithm, as described above, and have accuracy higher than that from previous versions of Beagle run with default settings. The high SNP density (equivalent to a SNP array with 1 million SNPs) also contributes to high phasing accuracy. The high accuracy of these haplotypes facilitates the strong performance of GERMLINE in these data.
We used the full simulated phase-known sequence data to determine the true IBD status, so that we could assess the accuracy of the IBD estimated from the thinned, phase-agnostic SNP data. When determining the true IBD, we ignored variants with ≤10 copies in the 2000 individuals, as very recent mutations disrupt sequence identity. For two IBD haplotypes separated by m meioses, looking along the chromosome, recombination ends the IBD at rate m times the recombination rate, while new mutations disrupt the sequence identity at rate m times the mutation rate. In our simulation, the mutation rate is 2.5 times the recombination rate, so in any IBD segment we expect an average of 2.5 identity-disrupting new mutations. Ignoring variants with ≤10 copies, we declared pairs of haplotypes with identical sequence for at least 0.1 cM to be segments of true IBD for the purpose of assessing IBD detection accuracy.
Simulated sequence data
Using the simulated phased sequence data described above, we also created simulated filtered, unphased sequence data with genotype errors. We removed variants with more than two alleles or with minor allele frequency <0.5%, added genotype error at a rate of 0.1%, and removed information about genotype phase. The minor allele frequency filter of 0.5% was chosen to be higher than the threshold used to determine true IBD (0.25%). This allows the variants with frequency in the 0.25–0.5% range to be used to assess accuracy of the detected IBD segments. Error rates in sequence data vary considerably, depending on the depth of sequence coverage. We chose to use a per-variant error rate twice that of the simulated SNP data. The number of genotype errors occurring in an IBD segment is also increased in sequence data since there are more variants and thus more opportunities for error. We analyzed 500 individuals of the simulated unphased filtered sequence data, using Refined IBD with a minimum segment length of 0.2 cM and LOD scores of 4 and 5.
Wellcome Trust Case Control Consortium 2 data
The Wellcome Trust Case Control Consortium phase 2 controls consist of 5200 individuals from the United Kingdom genotyped on a custom Illumina array (Barrett et al. 2009). After quality control, including removal of SNPs not in Hardy–Weinberg equilibrium (P < 10−5) and SNPs with minor allele frequency <1%, 885,127 autosomal SNPs remained for analysis. We ran Refined IBD with a minimum genetic length threshold of 0.5 cM. We used windows of 10,000 markers (window = 10,000) with 1000-marker overlap between adjacent windows (overlap = 1000). Other parameters were left at their default values. Genetic lengths of detected IBD segments in these data and in the Finnish data (described below) were determined using the estimated genetic distances provided by the International Haplotype Map Consortium (Frazer et al. 2007).
Northern Finland Birth Cohort data
The Northern Finland Birth Cohort data consist of 5402 individuals from Northern Finland, born in 1966, with genotypes on 320,981 autosomal SNPs from an Illumina Infinium SNP array (Sabatti et al. 2009). We excluded 503 individuals with close relatives (relatedness equivalent to first cousins or closer) in the data, as described previously (Browning and Browning 2013), leaving 4899 individuals for analysis. Due to the relatively low density of SNPs in these data, we used a smaller number of SNPs for the IBD detection windows in the GERMLINE algorithm (ibdwindow = 32) and a smaller than usual trim for the likelihood-ratio score (ibdtrim = 30). To reduce memory requirements, we took advantage of the windowing built into Beagle 4 and used 2000 marker windows (~20 Mb) with a 400-marker overlap between adjacent windows (~4 Mb). Other parameters for Refined IBD were left at their default values.
Results
Simulation study
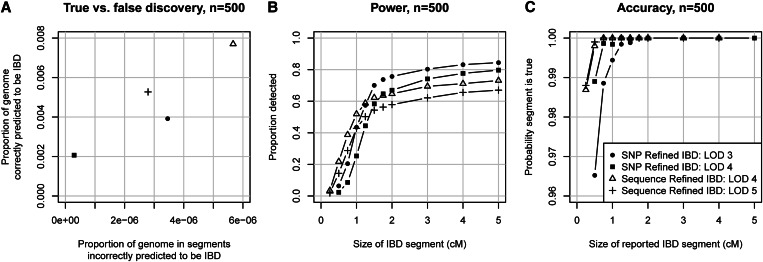
To compare the proposed Refined IBD method with existing methods, we generated simulated SNP data (see Methods) on ten 30-Mb regions, with 500 and with 2000 simulated individuals. Figure 2 summarizes the accuracy of the methods. Figure 2, A and D, shows that Refined IBD with LOD score threshold 3 has higher accuracy and higher power than GERMLINE. Figure 2, B, C, E, and F, shows that Refined IBD has higher power than fastIBD to detect short (<1–2 cM) IBD segments for comparable levels of accuracy. In contrast, fastIBD has better ability to detect close to 100% of the larger segments (>2 cM) whereas Refined IBD typically misses 10–20% of these larger segments. FastIBD does a good job of not missing parts of large segments because the algorithm is run 10 times, so that phasing errors in one run may be avoided in a different run. When we ran Refined IBD 3 times with different random-number seeds and merged the results, much of the missed IBD was recovered (Figure 2). Similar results can be obtained with less computation by joining high-confidence IBD segments with nearby low-confidence segments from a single run of Refined IBD (data not shown). Overall accuracy for all the methods considered here is very high, with at least 94% of reported segments reflecting true underlying IBD. For several of the methods, depending on parameter settings, power is high to detect segments of size ≥1 cM, particularly when the larger sample size is used.
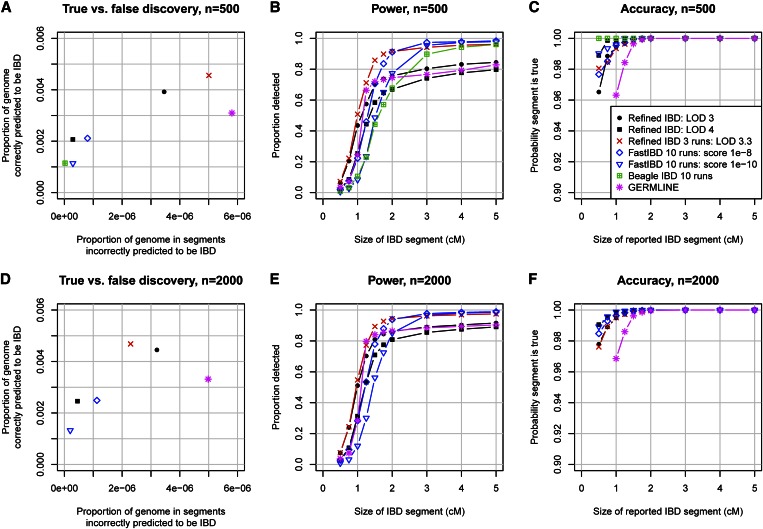
Identity-by-descent detection accuracy. (A–C) Sample size of 500 individuals; (D–F) sample size of 2000 individuals. A and D show true vs. false discovery. False discovery (x-axis) is measured by the average proportion of the genome that, for a pair of individuals, is in detected IBD segments that are determined to be false. Here falsely detected IBD segments are segments for which at most 25% of the detected segment is true IBD as determined from the simulated phase-known sequence data. True discovery (y-axis) is measured by the average proportion of the region that, for a pair of individuals, is in detected IBD that is also true IBD. Any part of a detected IBD segment that is not part of a true IBD segment is not included in this measure. B and E show power to detect IBD as a function of the underlying size of the true IBD segment. The average proportion of the segment that is detected is shown on the y-axis. Undetected segments (proportion 0) are included in this measure. C and F measure the accuracy of detected segments of a given reported size. The y-axis gives the probability that a reported segment is true, which is defined here as the probability that at least 50% of the segment is true IBD.
The most challenging part of IBD detection is determination of the IBD endpoints. Inferred haplotype allele identity may extend beyond the true IBD region, leading to overestimation of IBD endpoints. Furthermore, determination of haplotype phase is difficult at the IBD endpoints because the recent recombination demarcating the IBD endpoints disrupts the haplotypes, and consequent phase errors lead to incorrect determination of IBD endpoints. In Figure 2, we classified a reported IBD segment as true or false by whether it at least partly reflected some underlying IBD. This approach was designed to avoid conflating accuracy at the boundaries of the reported segment with accuracy of the segment itself. In Figure 3, we consider under- and overestimation of the IBD segment. Both types of error are primarily due to incorrect determination of segment endpoints. Overestimation is always on the ends of an IBD segment, while underestimation can occur in the middle of an IBD segment (if two or more short segments are reported, with intervening gaps, instead of one long segment) as well as at the ends. It can be seen that fastIBD with the recommended 10 iterations tends to significantly overestimate endpoints and misses very little of the true underlying segments when the segments are at least partly found. In contrast, GERMLINE and Refined IBD have much less overestimation of endpoints, but tend to miss large parts of the segment. Refined IBD with three runs is almost as good as fastIBD with respect to underestimation and is better than fastIBD with respect to overestimation. It also has a better true vs. false discovery profile than fastIBD due to better ability to detect short segments (Figure 2). Refined IBD with a single run misses much less IBD from longer segments with a sample size of 2000 individuals than with a sample size of 500 individuals. This is probably because haplotype phase estimation accuracy increases with sample size (S. R. Browning and B. L. Browning 2007), and increased haplotype phase accuracy increases the ability to find most or all of a larger IBD segment.
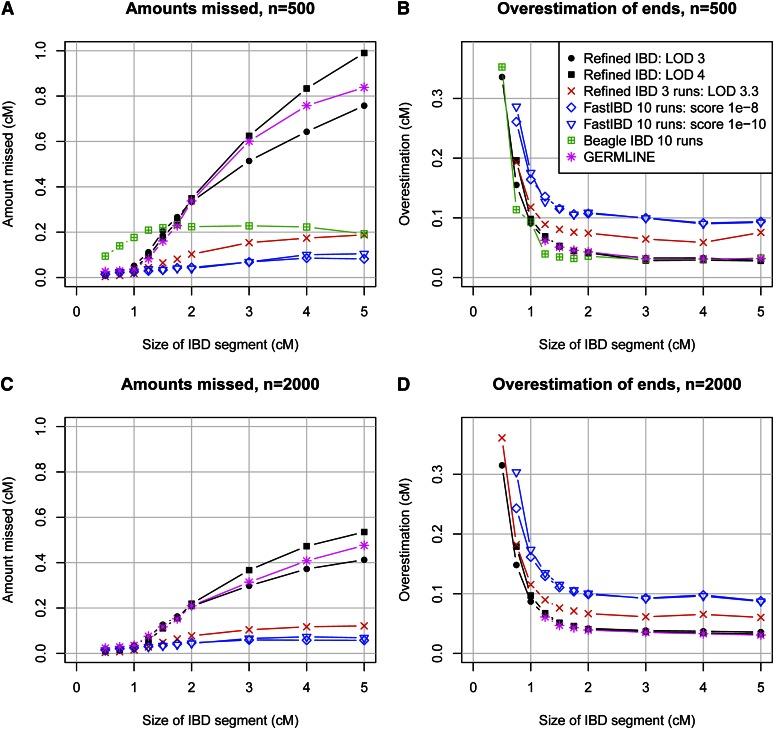
Under- and overestimation of IBD segment lengths. (A and B) Sample size of 500 individuals; (C and D) sample size of 2000 individuals. A and C show the average amount of IBD segment missed, for segments of a given size, conditional on at least part of the segment being found. The missed amount includes gaps in the middle of a segment and underestimation of endpoints of a segment. B and D show the average amount of overestimation of a segment, for segments of a given size, conditional on at least part of the segment being found. Overestimation of ends includes the bridging of two segments: in such a case the true IBD in one segment contributes to the end overestimation of the other segment.
Depending on the application, one may wish to be conservative in reporting the IBD segment endpoints. One approach is to trim a fixed number of markers from each end of the detected segments. Figure 4 shows how the accuracy (including overestimation and falsely detected segments) and the true IBD detection rate vary by LOD score threshold and amount trimmed. The optimal combination depends on the level of accuracy required. For higher detection at the cost of lower accuracy, a less stringent LOD score threshold such as 2 combined with a light to moderate level of trimming (up to 100 markers) is best. For high accuracy, such as a false discovery rate <1%, one needs a stringent LOD score threshold such as 4 combined with a high level of trimming (≥100 markers). A high level of trimming both removes potential overestimation from the detected segments and removes short segments that are slightly less likely to reflect true underlying IBD (Figure 2, C and F).
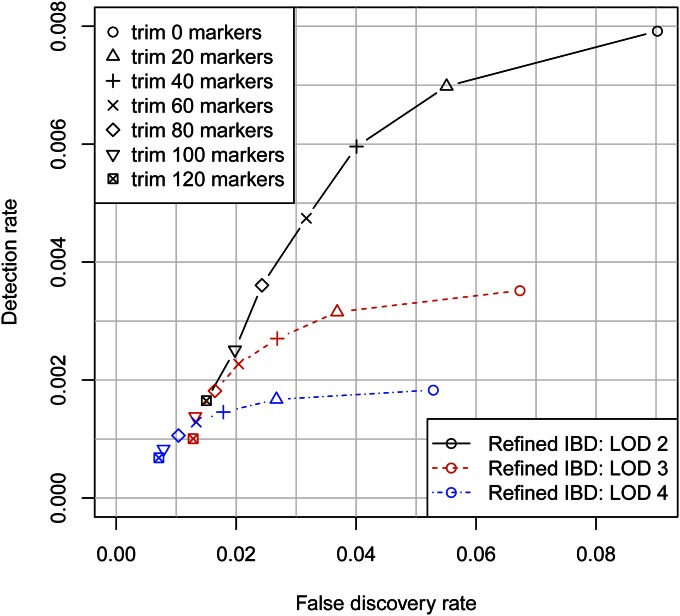
Identity-by-descent detection accuracy, including the effects of overestimation. Whereas in Figure 1 overestimation is not factored into accuracy metrics, here the false discovery rate is the proportion of the total detected IBD that does not cover a true underlying IBD segment as determined from the underlying phased sequence data. Thus, here the false discovery rate on the x-axis includes both falsely detected segments and overestimation of the endpoints of true detected segments. The detection rate on the y-axis is the average length of true IBD found per pair of individuals, divided by the length of the region.
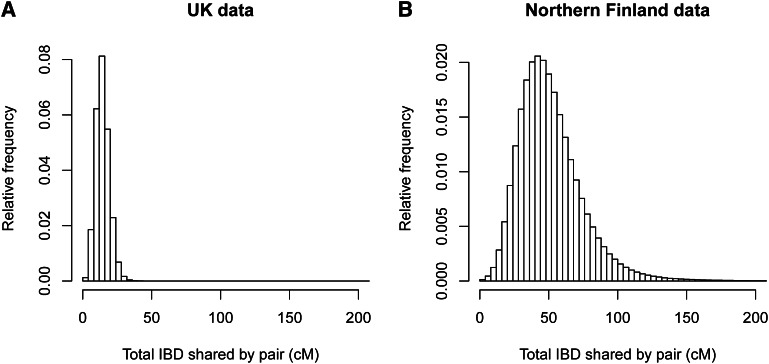
When using a LOD score threshold of 3 (to match the threshold used in the United Kingdom and Finnish data analyses), we found IBD at a rate of 0.0043. (The IBD detection rate is the probability that a randomly chosen pair of individuals has detectable IBD at a randomly chosen position.) The distribution of IBD segment lengths is shown in Figure 5A. Overall, the simulated data have a slightly higher rate of IBD detection and a somewhat lower average IBD segment size compared to the United Kingdom data (see United Kingdom results below).
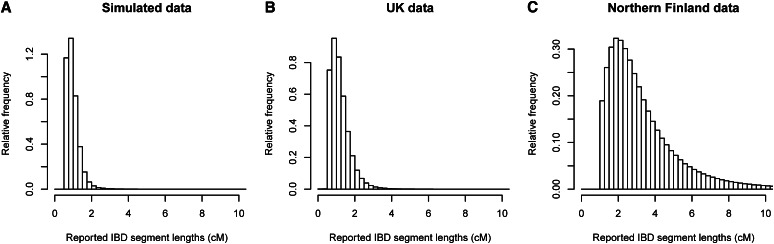
Lengths of detected IBD segments. (A) In the simulated SNP data, with a sample size of 2000. (B) In the Wellcome Trust Case Control Consortium 2 United Kingdom data. (C) In the Northern Finland Birth Cohort data. A LOD score threshold of 3 was used in all three cases.
In the results described above, the rate of genotype error was very low (0.05%), which reflects the high level of accuracy that can be achieved with quality-control–filtered SNP array data. In Figure S2, we investigate the effects of a higher rate of error (0.5%) on IBD segment detection with Refined IBD. We find that increasing the genotype error rate does not adversely decrease accuracy, but does decrease power to detect IBD.
Simulated sequence data
We generated simulated sequence data on 500 individuals. For the Refined IBD analysis, we excluded all variants with minor allele frequency ≤0.5% and added genotype error at a rate of 0.1%, which is twice that of the SNP data. Figure 6 compares IBD detection results for the simulated sequence data with those for the simulated SNP data. In the sequence data we found we needed to use a higher LOD score threshold than in SNP data to control false IBD segment discovery to a similar level. However, it should be noted that the overall level of reported segments is much higher in the sequence data, so the ratio of false to true discoveries is still well controlled with a LOD score of 3. For the same false positive IBD detection level, we detect ~50% more true IBD in the sequence data (Figure 6A). Figure 6B shows that the increase in power is due to increased ability to detect segments <1 cM. On the other hand, some of the IBD in segments >2 cM is being missed. One reason for the missed parts of long segments is the higher level of genotype error, due to both a higher error rate per variant and a higher density of variants. The phasing of the sequence data may also have a higher number of switch errors per centimorgan because low-frequency variants are generally more difficult to phase than high-frequency variants. Figure 6C shows that the accuracy of the IBD segments detected from the simulated sequence data is very high, even for segments as short as 0.25 cM.
Wellcome Trust Case Control Consortium 2 data
We analyzed ~900,000 autosomal SNPs genotyped on 5200 individuals from the United Kingdom. The average amount of IBD detected on the autosomes was 14.4 cM per pair of individuals. This equates to an IBD detection rate of 0.0041. Only 224 pairs (0.0017% of pairs) had no detected IBD. Figure 5B shows the distribution of detected IBD lengths, while Figure 7A shows the distribution of the amount of detected IBD per pair of individuals.
Northern Finland Birth Cohort data
We analyzed ~300,000 autosomal SNPs genotyped on 4899 individuals from Northern Finland after excluding close relatives. The average amount of IBD detected on the autosomes was 51.5 cM per pair of individuals. This equates to an IBD detection rate of 0.015, which is 3.6 times as high as that in the United Kingdom data, even though the United Kingdom data have much higher SNP density and thus much better power to detect small IBD segments. Only 675 pairs (0.0056% of pairs) had no detected IBD. Figure 5C shows the distribution of detected IBD lengths, while Figure 7B shows the distribution of the amount of detected IBD per pair of individuals.
Computation time
Computing requirements for IBD detection with Refined IBD are the same order of magnitude as the Beagle phasing time for the data sets we analyzed, although the actual IBD detection time depends on the amount of IBD found in the data, which in turn depends on the minimum IBD length parameter, the SNP density, the effective size of the population from which the sample was drawn, and the sample size. For example, in the United Kingdom data on chromosome 1, phasing took 78 hr while IBD detection took 94 hr with a minimum IBD length parameter of 0.5 cM. In the Northern Finland data on chromosome 1, phasing took 28 hr while IBD detection took 55 hr with a minimum IBD length parameter of 1.0 cM. In the simulated SNP data with 2000 individuals on 30 Mb, phasing took 113 min while IBD detection took 158 min with a minimum IBD length parameter of 0.5 cM. In simulated SNP data with 500 individuals on 30 Mb, phasing took 10 min while IBD detection took 7 min. In simulated sequence data with 500 individuals on 30 Mb, phasing took 68 min while IBD detection took 70 min with a minimum IBD length parameter of 0.2 cM. All computation times are from runs on a 2.4-GHz computer.
In general, computation time scales linearly with the chromosome length and quadratically in the number of individuals. Candidate IBD segments are efficiently identified using the GERMLINE hashing algorithm (Gusev et al. 2009), while calculation of LOD scores is linear in the number of candidate segments and hence quadratic in the number of individuals.
Discussion
In our simulated SNP data, Refined IBD has significantly higher power than existing computationally efficient IBD detection methods while maintaining the same high level of accuracy. The gain in power is seen primarily in the smaller segment sizes, such as 0.5–1 cM. This makes Refined IBD useful for analyses in outbred populations, in which there are few long IBD segments but many short IBD segments. The additional detected short IBD segments will improve the power of IBD mapping (Browning and Thompson 2012), facilitate haplotype phasing in population samples (Kong et al. 2008), and permit higher resolution when estimating population structure (Gusev et al. 2012; Palamara et al. 2012).
When accurate detection of long (e.g., >3 cM) segments of IBD is required, we recommend merging results from multiple runs of Refined IBD and filling any short (e.g., <2 cM) gaps between IBD segments. This approach greatly increases power to find the complete long segment of IBD, but can result in a small amount of overestimation of the length of the segment (see Figure 3).
Our analysis of simulated sequence data shows that Refined IBD can be used for IBD detection in sequence data. However, further development of IBD detection methodology for sequence data is needed. Such methods should be designed to take full advantage of the information contained in rare variants while accounting for both the higher genotype error rate in sequence data and the possibility of mutations occurring in an IBD segment since the common ancestor.
Our analysis of data from United Kingdom individuals found significantly more IBD than a previous study. Here we found IBD at a rate of 0.0041 (probability that a randomly chosen pair of individuals has detectable IBD at a randomly chosen position), whereas the previous rate was 0.00035 (B. L. Browning and S. R. Browning 2011). This difference is due in part to the improved sensitivity of Refined IBD over that of fastIBD (see Figure 2). An even more significant contributing factor is the relative SNP densities in the two analyses. The earlier analysis was of ~500,000 SNPs genome-wide, while the analysis presented here uses almost twice as many SNPs. At higher SNP densities, power to detect small segments increases. While small segments individually contribute little IBD, there are many more small segments than large, since the number of ancestors of an individual can increase exponentially with the number of generations to the ancestors. Thus SNP density can have a large effect on the rate of IBD detected.
Compared to the United Kingdom data, the Northern Finland data show a much higher rate of detected IBD (0.015 vs. 0.0041). This high level of IBD is to be expected in an isolated population. A high level of detected IBD enables the application of IBD-based heritability estimation. In previous IBD-based heritability analysis of these data, we found significant heritability for cholesterol and fasting glucose levels (Browning and Browning 2013).
The haplotype-based output of Refined IBD is useful for downstream analyses. In individual-based IBD detection, one does not know whether three individuals who are all IBD with each other are IBD for the same haplotype or not, as illustrated in Figure 8. A previous approach to the multiindividual IBD problem was joint analysis of multiple individuals (Moltke et al. 2011); however, this is computationally demanding. With haplotype-based IBD, there is some uncertainty in determining the multi-individual haplotype IBD because some IBD is not detected, while false positive IBD can occur. Gusev et al. (2011) apply clustering to deal with this issue. With determination of multi-individual IBD, it becomes possible to extend IBD mapping from the existing pairwise approaches (Purcell et al. 2007; Browning and Thompson 2012) to a multi-individual approach (Gusev et al. 2011), potentially making better use of the information in the data. Another advantage of haplotype-based output is that one can directly match the haplotypes to the IBD, so that if one finds an interesting pattern of IBD sharing, one can identify the underlying shared haplotypes.
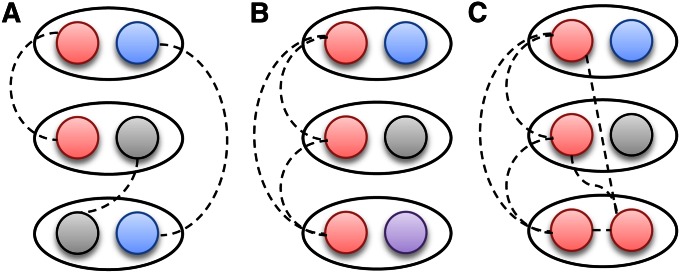
Patterns of IBD sharing between three individuals. Individuals are shown as ovals, while their haplotypes are shown as circles. IBD at a haplotype level is shown by dashed lines connecting the IBD haplotypes and by the use of the same color for IBD haplotypes. In all cases, there is IBD between all three pairs of individuals. (A) Each pair of individuals shares a different haplotype. (B) The three individuals share a single haplotype. (C) As in B, but the third individual is homozygous by descent. These three scenarios cannot be distinguished without further data when IBD is reported only at the individual level, but are clearly different with IBD at the haplotype level.
Web resources
The Beagle webpage is http://faculty.washington.edu/browning/beagle/beagle.html Variant call format specification: http://vcftools.sourceforge.net/specs.html.
Acknowledgments
The Northern Finland Birth Cohort (NFBC1966) Study is conducted and supported by the National Heart, Lung, and Blood Institute (NHLBI) in collaboration with the Broad Institute, University of California at Los Angeles (UCLA), University of Oulu, and the National Institute for Health and Welfare in Finland. This article does not necessarily reflect the opinions or views of the NFBC1966 Study Investigators, Broad Institute, UCLA, University of Oulu, the National Institute for Health and Welfare in Finland, and the NHLBI. This study makes use of data generated by the Wellcome Trust Case Control Consortium. A full list of the investigators who contributed to the generation of the data is available from www.wtccc.org.uk. Funding for the project was provided by the Wellcome Trust under awards 076113 and 085475. This study was supported by research grants HG004960, HG005701, GM099568, and GM075091 from the National Institutes of Health.
Footnotes
Communicating editor: N. A. Rosenberg
Literature Cited
- 1000 Genomes Consortium , 2010.
 A map of human genome variation from population-scale sequencing.
Nature
467: 1061–1073. [Europe PMC free article] [Abstract] [Google Scholar]
A map of human genome variation from population-scale sequencing.
Nature
467: 1061–1073. [Europe PMC free article] [Abstract] [Google Scholar] - Albrechtsen A., Korneliussen T. S., Moltle I., Hansen T. V., Nielsen F. C., et al. , 2009.
 Relatedness mapping and tracts of relatedness for genome-wide data in the presence of linkage disequilibrium.
Genet. Epidemiol.
33: 266–274. [Abstract] [Google Scholar]
Relatedness mapping and tracts of relatedness for genome-wide data in the presence of linkage disequilibrium.
Genet. Epidemiol.
33: 266–274. [Abstract] [Google Scholar] - Barrett J. C., Lee J. C., Lees C. W., Prescott N. J., Anderson C. A., et al. , 2009.
 Genome-wide association study of ulcerative colitis identifies three new susceptibility loci, including the HNF4A region.
Nat. Genet.
41: 1330–1334. [Europe PMC free article] [Abstract] [Google Scholar]
Genome-wide association study of ulcerative colitis identifies three new susceptibility loci, including the HNF4A region.
Nat. Genet.
41: 1330–1334. [Europe PMC free article] [Abstract] [Google Scholar] - Baum, L. E., 1972
 An inequality and associated maximization technique in statistical estimation for probabilistic functions of Markov processes, pp. 1–8 in Inequalities III: Proceedings of the Third Symposium on Inequalities held at the University of California, Los Angeles, September 1–9, 1969, edited by O. Shisha. Academic Press, San Diego. [Google Scholar]
An inequality and associated maximization technique in statistical estimation for probabilistic functions of Markov processes, pp. 1–8 in Inequalities III: Proceedings of the Third Symposium on Inequalities held at the University of California, Los Angeles, September 1–9, 1969, edited by O. Shisha. Academic Press, San Diego. [Google Scholar] - Brown M. D., Glazner C. G., Zheng C., Thompson E. A., 2012.
 Inferring coancestry in population samples in the presence of linkage disequilibrium.
Genetics
190: 1447–1460. [Europe PMC free article] [Abstract] [Google Scholar]
Inferring coancestry in population samples in the presence of linkage disequilibrium.
Genetics
190: 1447–1460. [Europe PMC free article] [Abstract] [Google Scholar] - Browning B. L., Browning S. R., 2007.
 Efficient multilocus association testing for whole genome association studies using localized haplotype clustering.
Genet. Epidemiol.
31: 365–375. [Abstract] [Google Scholar]
Efficient multilocus association testing for whole genome association studies using localized haplotype clustering.
Genet. Epidemiol.
31: 365–375. [Abstract] [Google Scholar] - Browning B. L., Browning S. R., 2009.
 A unified approach to genotype imputation and haplotype-phase inference for large data sets of trios and unrelated individuals.
Am. J. Hum. Genet.
84: 210–223. [Europe PMC free article] [Abstract] [Google Scholar]
A unified approach to genotype imputation and haplotype-phase inference for large data sets of trios and unrelated individuals.
Am. J. Hum. Genet.
84: 210–223. [Europe PMC free article] [Abstract] [Google Scholar] - Browning B. L., Browning S. R., 2011.
 A fast, powerful method for detecting identity by descent.
Am. J. Hum. Genet.
88: 173–182. [Europe PMC free article] [Abstract] [Google Scholar]
A fast, powerful method for detecting identity by descent.
Am. J. Hum. Genet.
88: 173–182. [Europe PMC free article] [Abstract] [Google Scholar] - Browning S. R., Browning B. L., 2007.
 Rapid and accurate haplotype phasing and missing-data inference for whole-genome association studies by use of localized haplotype clustering.
Am. J. Hum. Genet.
81: 1084–1097. [Europe PMC free article] [Abstract] [Google Scholar]
Rapid and accurate haplotype phasing and missing-data inference for whole-genome association studies by use of localized haplotype clustering.
Am. J. Hum. Genet.
81: 1084–1097. [Europe PMC free article] [Abstract] [Google Scholar] - Browning S. R., Browning B. L., 2010.
 High-resolution detection of identity by descent in unrelated individuals.
Am. J. Hum. Genet.
86: 526–539. [Europe PMC free article] [Abstract] [Google Scholar]
High-resolution detection of identity by descent in unrelated individuals.
Am. J. Hum. Genet.
86: 526–539. [Europe PMC free article] [Abstract] [Google Scholar] - Browning S. R., Browning B. L., 2011.
 Haplotype phasing: existing methods and new developments.
Nat. Rev. Genet.
12: 703–714. [Europe PMC free article] [Abstract] [Google Scholar]
Haplotype phasing: existing methods and new developments.
Nat. Rev. Genet.
12: 703–714. [Europe PMC free article] [Abstract] [Google Scholar] - Browning S. R., Browning B. L., 2012.
 Identity by descent between distant relatives: detection and applications.
Annu. Rev. Genet.
46: 617–633. [Abstract] [Google Scholar]
Identity by descent between distant relatives: detection and applications.
Annu. Rev. Genet.
46: 617–633. [Abstract] [Google Scholar] - Browning S. R., Browning B. L., 2013.
 Identity-by-descent-based heritability analysis in the Northern Finland Birth Cohort.
Hum. Genet.
132: 129–138. [Europe PMC free article] [Abstract] [Google Scholar]
Identity-by-descent-based heritability analysis in the Northern Finland Birth Cohort.
Hum. Genet.
132: 129–138. [Europe PMC free article] [Abstract] [Google Scholar] - Browning S. R., Thompson E. A., 2012.
 Detecting rare variant associations by identity-by-descent mapping in case-control studies.
Genetics
190: 1521–1531. [Europe PMC free article] [Abstract] [Google Scholar]
Detecting rare variant associations by identity-by-descent mapping in case-control studies.
Genetics
190: 1521–1531. [Europe PMC free article] [Abstract] [Google Scholar] - Cai Z., Camp N. J., Cannon-Albright L., Thomas A., 2011.
 Identification of regions of positive selection using Shared Genomic Segment analysis.
Eur. J. Hum. Genet.
19: 667–671. [Europe PMC free article] [Abstract] [Google Scholar]
Identification of regions of positive selection using Shared Genomic Segment analysis.
Eur. J. Hum. Genet.
19: 667–671. [Europe PMC free article] [Abstract] [Google Scholar] - Campbell C. L., Palamara P. F., Dubrovsky M., Botigue L. R., Fellous M., et al. , 2012.
 North African Jewish and non-Jewish populations form distinctive, orthogonal clusters.
Proc. Natl. Acad. Sci. USA
109: 13865–13870. [Europe PMC free article] [Abstract] [Google Scholar]
North African Jewish and non-Jewish populations form distinctive, orthogonal clusters.
Proc. Natl. Acad. Sci. USA
109: 13865–13870. [Europe PMC free article] [Abstract] [Google Scholar] - Coventry A., Bull-Otterson L. M., Liu X., Clark A. G., Maxwell T. J., et al. , 2010.
 Deep resequencing reveals excess rare recent variants consistent with explosive population growth. Nat. Commun. 1: 131. [Europe PMC free article] [Abstract] [Google Scholar]
Deep resequencing reveals excess rare recent variants consistent with explosive population growth. Nat. Commun. 1: 131. [Europe PMC free article] [Abstract] [Google Scholar] - Danecek P., Auton A., Abecasis G., Albers C. A., Banks E., et al. , 2011.
 The variant call format and VCFtools.
Bioinformatics
27: 2156–2158. [Europe PMC free article] [Abstract] [Google Scholar]
The variant call format and VCFtools.
Bioinformatics
27: 2156–2158. [Europe PMC free article] [Abstract] [Google Scholar] - Excoffier L., Foll M., 2011.
 fastsimcoal: a continuous-time coalescent simulator of genomic diversity under arbitrarily complex evolutionary scenarios.
Bioinformatics
27: 1332–1334. [Abstract] [Google Scholar]
fastsimcoal: a continuous-time coalescent simulator of genomic diversity under arbitrarily complex evolutionary scenarios.
Bioinformatics
27: 1332–1334. [Abstract] [Google Scholar] - Frazer K. A., Ballinger D. G., Cox D. R., Hinds D. A., Stuve L. L., et al. , 2007.
 A second generation human haplotype map of over 3.1 million SNPs.
Nature
449: 851–861. [Europe PMC free article] [Abstract] [Google Scholar]
A second generation human haplotype map of over 3.1 million SNPs.
Nature
449: 851–861. [Europe PMC free article] [Abstract] [Google Scholar] - Gusev A., Lowe J. K., Stoffel M., Daly M. J., Altshuler D., et al. , 2009.
 Whole population, genome-wide mapping of hidden relatedness.
Genome Res.
19: 318–326. [Europe PMC free article] [Abstract] [Google Scholar]
Whole population, genome-wide mapping of hidden relatedness.
Genome Res.
19: 318–326. [Europe PMC free article] [Abstract] [Google Scholar] - Gusev A., Kenny E. E., Lowe J. K., Salit J., Saxena R., et al. , 2011.
 DASH: a method for identical-by-descent haplotype mapping uncovers association with recent variation.
Am. J. Hum. Genet.
88: 706–717. [Europe PMC free article] [Abstract] [Google Scholar]
DASH: a method for identical-by-descent haplotype mapping uncovers association with recent variation.
Am. J. Hum. Genet.
88: 706–717. [Europe PMC free article] [Abstract] [Google Scholar] - Gusev A., Palamara P. F., Aponte G., Zhuang Z., Darvasi A., et al. , 2012.
 The architecture of long-range haplotypes shared within and across populations.
Mol. Biol. Evol.
29: 473–486. [Europe PMC free article] [Abstract] [Google Scholar]
The architecture of long-range haplotypes shared within and across populations.
Mol. Biol. Evol.
29: 473–486. [Europe PMC free article] [Abstract] [Google Scholar] - Han L., Abney M., 2011.
 Identity by descent estimation with dense genome-wide genotype data.
Genet. Epidemiol.
35: 557–567. [Europe PMC free article] [Abstract] [Google Scholar]
Identity by descent estimation with dense genome-wide genotype data.
Genet. Epidemiol.
35: 557–567. [Europe PMC free article] [Abstract] [Google Scholar] - Han L., Abney M., 2013.
 Using identity by descent estimation with dense genotype data to detect positive selection.
Eur. J. Hum. Genet.
21: 205–211. [Europe PMC free article] [Abstract] [Google Scholar]
Using identity by descent estimation with dense genotype data to detect positive selection.
Eur. J. Hum. Genet.
21: 205–211. [Europe PMC free article] [Abstract] [Google Scholar] - Jonsson T., Atwal J. K., Steinberg S., Snaedal J., Jonsson P. V., et al. , 2012.
 A mutation in APP protects against Alzheimer’s disease and age-related cognitive decline.
Nature
488: 96–99. [Abstract] [Google Scholar]
A mutation in APP protects against Alzheimer’s disease and age-related cognitive decline.
Nature
488: 96–99. [Abstract] [Google Scholar] - Keinan A., Clark A. G., 2012.
 Recent explosive human population growth has resulted in an excess of rare genetic variants.
Science
336: 740–743. [Europe PMC free article] [Abstract] [Google Scholar]
Recent explosive human population growth has resulted in an excess of rare genetic variants.
Science
336: 740–743. [Europe PMC free article] [Abstract] [Google Scholar] - Kong A., Masson G., Frigge M. L., Gylfason A., Zusmanovich P., et al. , 2008.
 Detection of sharing by descent, long-range phasing and haplotype imputation.
Nat. Genet.
40: 1068–1075. [Abstract] [Google Scholar]
Detection of sharing by descent, long-range phasing and haplotype imputation.
Nat. Genet.
40: 1068–1075. [Abstract] [Google Scholar] - Li Y., Willer C. J., Ding J., Scheet P., Abecasis G. R., 2010.
 MaCH: using sequence and genotype data to estimate haplotypes and unobserved genotypes.
Genet. Epidemiol.
34: 816–834. [Europe PMC free article] [Abstract] [Google Scholar]
MaCH: using sequence and genotype data to estimate haplotypes and unobserved genotypes.
Genet. Epidemiol.
34: 816–834. [Europe PMC free article] [Abstract] [Google Scholar] - Moltke I., Albrechtsen A., Hansen T. V., Nielsen F. C., Nielsen R., 2011.
 A method for detecting IBD regions simultaneously in multiple individuals–with applications to disease genetics.
Genome Res.
21: 1168–1180. [Europe PMC free article] [Abstract] [Google Scholar]
A method for detecting IBD regions simultaneously in multiple individuals–with applications to disease genetics.
Genome Res.
21: 1168–1180. [Europe PMC free article] [Abstract] [Google Scholar] - Nachman M. W., Crowell S. L., 2000.
 Estimate of the mutation rate per nucleotide in humans.
Genetics
156: 297–304. [Europe PMC free article] [Abstract] [Google Scholar]
Estimate of the mutation rate per nucleotide in humans.
Genetics
156: 297–304. [Europe PMC free article] [Abstract] [Google Scholar] - Nelson M. R., Wegmann D., Ehm M. G., Kessner D., St Jean P., et al. , 2012.
 An abundance of rare functional variants in 202 drug target genes sequenced in 14,002 people.
Science
337: 100–104. [Abstract] [Google Scholar]
An abundance of rare functional variants in 202 drug target genes sequenced in 14,002 people.
Science
337: 100–104. [Abstract] [Google Scholar] - Palamara P. F., Lencz T., Darvasi A., Pe’er I., 2012.
 Length distributions of identity by descent reveal fine-scale demographic history.
Am. J. Hum. Genet.
91: 809–822. [Europe PMC free article] [Abstract] [Google Scholar]
Length distributions of identity by descent reveal fine-scale demographic history.
Am. J. Hum. Genet.
91: 809–822. [Europe PMC free article] [Abstract] [Google Scholar] - Price A. L., Helgason A., Thorleifsson G., McCarroll S. A., Kong A., et al. , 2011.
 Single-tissue and cross-tissue heritability of gene expression via identity-by-descent in related or unrelated individuals.
PLoS Genet.
7: e1001317. [Europe PMC free article] [Abstract] [Google Scholar]
Single-tissue and cross-tissue heritability of gene expression via identity-by-descent in related or unrelated individuals.
PLoS Genet.
7: e1001317. [Europe PMC free article] [Abstract] [Google Scholar] - Purcell S., Neale B., Todd-Brown K., Thomas L., Ferreira M. A. R., et al. , 2007.
 PLINK: a tool set for whole-genome association and population-based linkage analyses.
Am. J. Hum. Genet.
81: 559–575. [Europe PMC free article] [Abstract] [Google Scholar]
PLINK: a tool set for whole-genome association and population-based linkage analyses.
Am. J. Hum. Genet.
81: 559–575. [Europe PMC free article] [Abstract] [Google Scholar] - Rabiner L. R., 1989.
 A tutorial on hidden Markov-models and selected applications in speech recognition.
Proc. IEEE
77: 257–286. [Google Scholar]
A tutorial on hidden Markov-models and selected applications in speech recognition.
Proc. IEEE
77: 257–286. [Google Scholar] - Ralph, P., and G. Coop, 2012
 The geography of recent genetic ancestry across Europe. arXiv:1207.3815 [q-bio.PE]. [Europe PMC free article] [Abstract]
The geography of recent genetic ancestry across Europe. arXiv:1207.3815 [q-bio.PE]. [Europe PMC free article] [Abstract] - Sabatti C., Service S. K., Hartikainen A. L., Pouta A., Ripatti S., et al. , 2009.
 Genome-wide association analysis of metabolic traits in a birth cohort from a founder population.
Nat. Genet.
41: 35–46. [Europe PMC free article] [Abstract] [Google Scholar]
Genome-wide association analysis of metabolic traits in a birth cohort from a founder population.
Nat. Genet.
41: 35–46. [Europe PMC free article] [Abstract] [Google Scholar] - Scheet P., Stephens M., 2006.
 A fast and flexible statistical model for large-scale population genotype data: applications to inferring missing genotypes and haplotypic phase.
Am. J. Hum. Genet.
78: 629–644. [Europe PMC free article] [Abstract] [Google Scholar]
A fast and flexible statistical model for large-scale population genotype data: applications to inferring missing genotypes and haplotypic phase.
Am. J. Hum. Genet.
78: 629–644. [Europe PMC free article] [Abstract] [Google Scholar] - Steemers F. J., Chang W., Lee G., Barker D. L., Shen R., et al. , 2006.
 Whole-genome genotyping with the single-base extension assay.
Nat. Methods
3: 31–33. [Abstract] [Google Scholar]
Whole-genome genotyping with the single-base extension assay.
Nat. Methods
3: 31–33. [Abstract] [Google Scholar] - Tenesa A., Navarro P., Hayes B. J., Duffy D. L., Clarke G. M., et al. , 2007.
 Recent human effective population size estimated from linkage disequilibrium.
Genome Res.
17: 520–526. [Europe PMC free article] [Abstract] [Google Scholar]
Recent human effective population size estimated from linkage disequilibrium.
Genome Res.
17: 520–526. [Europe PMC free article] [Abstract] [Google Scholar] - Viterbi A. J., 1967.
 Error bounds for convolutional codes and an asymptotically optimum decoding algorithm.
IEEE Trans. Inf. Theory
13: 260. [Google Scholar]
Error bounds for convolutional codes and an asymptotically optimum decoding algorithm.
IEEE Trans. Inf. Theory
13: 260. [Google Scholar] - Zuk O., Hechter E., Sunyaev S. R., Lander E. S., 2012.
 The mystery of missing heritability: genetic interactions create phantom heritability.
Proc. Natl. Acad. Sci. USA
109: 1193–1198. [Europe PMC free article] [Abstract] [Google Scholar]
The mystery of missing heritability: genetic interactions create phantom heritability.
Proc. Natl. Acad. Sci. USA
109: 1193–1198. [Europe PMC free article] [Abstract] [Google Scholar]
Articles from Genetics are provided here courtesy of Oxford University Press
Full text links
Read article at publisher's site: https://doi.org/10.1534/genetics.113.150029
Read article for free, from open access legal sources, via Unpaywall:
https://www.genetics.org/content/genetics/194/2/459.full.pdf
Citations & impact
Impact metrics
Citations of article over time
Alternative metrics
Smart citations by scite.ai
Explore citation contexts and check if this article has been
supported or disputed.
https://scite.ai/reports/10.1534/genetics.113.150029
Article citations
Fine-scale genetic structure and rare variant frequencies.
PLoS One, 19(11):e0313133, 05 Nov 2024
Cited by: 0 articles | PMID: 39499706 | PMCID: PMC11537391
Analysis of Gyimes Csango population samples on a high-resolution genome-wide basis.
BMC Genomics, 25(1):942, 07 Oct 2024
Cited by: 0 articles | PMID: 39375616 | PMCID: PMC11459897
9,000 years of genetic continuity in southernmost Africa demonstrated at Oakhurst rockshelter.
Nat Ecol Evol, 8(11):2121-2134, 19 Sep 2024
Cited by: 1 article | PMID: 39300260 | PMCID: PMC11541196
Genetic history and biological adaptive landscape of the Tujia people inferred from shared haplotypes and alleles.
Hum Genomics, 18(1):104, 18 Sep 2024
Cited by: 0 articles | PMID: 39289776 | PMCID: PMC11409738
Large-scale genome sequencing of giant pandas improves the understanding of population structure and future conservation initiatives.
Proc Natl Acad Sci U S A, 121(36):e2406343121, 26 Aug 2024
Cited by: 0 articles | PMID: 39186654
Go to all (327) article citations
Data
Data behind the article
This data has been text mined from the article, or deposited into data resources.
BioStudies: supplemental material and supporting data
Similar Articles
To arrive at the top five similar articles we use a word-weighted algorithm to compare words from the Title and Abstract of each citation.
A fast and accurate method for detection of IBD shared haplotypes in genome-wide SNP data.
Eur J Hum Genet, 25(5):617-624, 08 Feb 2017
Cited by: 9 articles | PMID: 28176766 | PMCID: PMC5437913
Identity-by-descent-based phasing and imputation in founder populations using graphical models.
Genet Epidemiol, 35(8):853-860, 17 Oct 2011
Cited by: 22 articles | PMID: 22006673 | PMCID: PMC3368215
Fast and Accurate Shared Segment Detection and Relatedness Estimation in Un-phased Genetic Data via TRUFFLE.
Am J Hum Genet, 105(1):78-88, 06 Jun 2019
Cited by: 23 articles | PMID: 31178127 | PMCID: PMC6612710
Identity by descent between distant relatives: detection and applications.
Annu Rev Genet, 46:617-633, 17 Sep 2012
Cited by: 97 articles | PMID: 22994355
Review
Funding
Funders who supported this work.
NHGRI NIH HHS (4)
Grant ID: R01 HG005701
Grant ID: HG005701
Grant ID: R01 HG004960
Grant ID: HG004960
NIGMS NIH HHS (4)
Grant ID: P01 GM099568
Grant ID: GM075091
Grant ID: GM099568
Grant ID: R01 GM075091
Wellcome Trust (2)
WTCCC2 core activities
Dr Paul Flicek, European Bioinformatics Institute
Grant ID: 085475
The genetics of weight, BMI, adiposity and obesity
Dr Mark Maccarthy, University of Oxford
Grant ID: 076113