Abstract
Free full text

Cyclic GMP-AMP Containing Mixed Phosphodiester Linkages Is An Endogenous High Affinity Ligand for STING
Abstract
The presence of microbial or self DNA in the cytoplasm of mammalian cells is a danger signal detected by the DNA sensor cyclic-GMP-AMP (cGAMP) synthase (cGAS), which catalyzes the production of cGAMP that in turn serves as a second messenger to activate innate immune responses. Here we show that endogenous cGAMP in mammalian cells contains two distinct phosphodiester linkages, one between 2′-OH of GMP and 5′-phosphate of AMP, and the other between 3′-OH of AMP and 5′-phosphate of GMP. This molecule, termed 2′3′-cGAMP, is unique in that it binds to the adaptor protein STING with a much greater affinity than cGAMP molecules containing other combinations of phosphodiester linkages. The crystal structure of STING bound to 2′3′-cGAMP revealed the structural basis of this high-affinity binding and a ligand-induced conformational change in STING that may underlie its activation.
INTRODUCTION
Innate immune sensing of microbial infections is mediated by germline-encoded pattern recognition receptors that include membrane proteins such as Toll-like receptors (TLRs) and cytosolic proteins such as NOD-like receptors (NLRs) and RIG-I like receptors (RLRs)(Iwasaki and Medzhitov, 2010; Ronald and Beutler, 2010; Takeuchi and Akira, 2010). As virtually all infectious microorganisms contain and need nucleic acids in their life cycles, the innate immune system has evolved to recognize the microbial DNA and RNA as a central strategy of host defense. Specifically, several TLRs are localized on the endosomal membrane to detect RNA or DNA in the lumen of the endosomes, whereas RLRs are responsible for detecting viral and bacterial RNA in the cytoplasm.
DNA is known to be an immune stimulatory molecule for more than a century, but how DNA activates the host immune system has not been extensively investigated until recently (O’Neill, 2013). DNA in the endosome is detected by TLR9, which then triggers the production of type-I interferons and inflammatory cytokines. When microbial or host DNA is delivered to the cytoplasm, it can also induce type-I interferons through the endoplasmic reticulum membrane protein STING (also known as MITA, ERIS or MPYS)(Barber, 2011). STING functions as an adaptor protein that recruits and activates the protein kinases IKK and TBK1, which in turn activate the transcription factors NF-κB and IRF3 to induce interferons and other cytokines.
We have recently identified cyclic GMP-AMP Synthase (cGAS) as a DNA sensor that activates STING(Sun et al., 2013; Wu et al., 2013). Specifically, we found that cGAS catalyzes the synthesis of cyclic GMP-AMP (cGAMP) from ATP and GTP in the presence of DNA. cGAMP then functions as a second messenger that binds to and activates STING. While these studies clearly demonstrate that cGAMP is an endogenous second messenger produced by cGAS in mammalian cells, the exact nature of the internal phosphodiester linkages between GMP and AMP in cGAMP was not determined in part because mass spectrometry alone could not unambiguously distinguish these linkages without the availability of all cGAMP isomers as the standard reference. Although chemically synthesized cGAMP that contains homogenous 3′−5′ linkages is capable of inducing IFNβ, it remains possible that cGAMP containing other phosphodiester linkages might also activate the STING pathway.
In this study, we further investigated the structure of cGAMP through a combination of chemical and biophysical techniques. We found that cGAMP produced by cGAS contains a phosphodiester linkage between 2′-OH of GMP and 5′-phosphate of AMP and another between 3′-OH of AMP and 5′-phosphate of GMP. We further showed that this molecule, herein referred to as 2′3′-cGAMP, was produced in mammalian cells in response to DNA in the cytoplasm. Moreover, we demonstrated that 2′3′-cGAMP binds to STING with a high affinity and is a potent inducer of interferon-β (IFNβ). We also solved the crystal structure of STING bound to the cGAS product and observed extensive interactions between 2′3′-cGAMP and STING, which provide the structural basis for their specific and high affinity binding. Importantly, the structure of the STING – cGAMP complex revealed that this natural ligand induces conformational rearrangements in STING that may underlie its activation.
RESULTS
The product of cGAS is cyclic GMP-AMP containing mixed phosphodiester bonds
Both 2′−5′ and 3′−5′ phosphodiester linkages between nucleotides are known to exist in nature while the 2′−5′ linkage is less common. The internal phosphodiester linkages of the natural cGAMP produced by cGAS remain to be determined. We therefore chemically synthesized cGAMP molecules containing all four possible phosphodiester linkages (Figure 1A; Supplemental Figure S1). The chemical synthesis of cGAMP isoforms was performed using procedures modified from published methods (see Supplemental Experimental Procedures) (Gaffney et al., 2010; Zhang et al., 2006). For simplicity, we name these cGAMP molecules according to the OH position of GMP followed by the OH position of AMP that form the phosphodiester bonds; for example, 2′3′-cGAMP contains a phosphodiester linkage between 2′−OH of GMP and 5′-phosphate of AMP and another between 3′−OH of AMP and 5′-phosphate of GMP. We also used purified cGAS protein to enzymatically synthesize the natural cGAMP from ATP and GTP in the presence of DNA(Sun et al., 2013). The purified cGAS product and synthetic cGAMP isomers were analyzed by nuclear magnetic resonance (NMR) spectroscopy (Figure 1B). Strikingly, the 1H NMR spectrum of the cGAS product was identical to that of synthetic 2′3′-cGAMP, but distinct from those of other cGAMP isomers. In particular, the anomeric proton (H1′) was a singlet with a 3′-phosphate and a doublet with 2′-phosphate. Consistently, only the phosphates of 2′,3′-cGAMP had the same 31P NMR chemical shifts as those of natural cGAMP (Supplemental Figure S2A). We also performed mass spectrometry analysis of the natural and synthetic cGAMP using Q-Exactive, an instrument with high resolution and mass accuracy. The total mass of each of these singly charged molecules ([M+H]+) was 675.107, exactly matching the theoretical mass of cGAMP (Supplemental Figure S2B). The tandem mass (MS/MS) spectra of the cGAS product, which was fragmented using higher energy collision dissociation (HCD), were identical to those of synthetic 2′3′-cGAMP, and similar but not identical to those of 2′2′-cGAMP and 3′3′-cGAMP (Figure 1C). The MS/MS spectra of 3′2′-cGAMP appeared to be most distinct from those of 2′3′-cGAMP and the cGAS product. Reverse phase HPLC analysis showed that natural cGAMP co-eluted with 2′3′-cGAMP, but not other cGAMP molecules (Supplemental Figure S2C). We also determined the configuration of the cGAS product by circular dichroism (CD), confirming that it is derived from d-ribose (Supplemental Figure S2D). The CD spectrum of the natural cGAMP overlapped well with that of 2′3′-cGAMP. The near-UV CD spectra indicate that the four cGAMPs adopt significantly different conformations, with 2′3′ and 2′2′-cGAMPs forming a CD band pattern distinct from those of 3′2′- and 3′3′-cGAMPs. Collectively, these results provide definitive proof that cGAS synthesizes 2′3′-cGAMP in vitro.
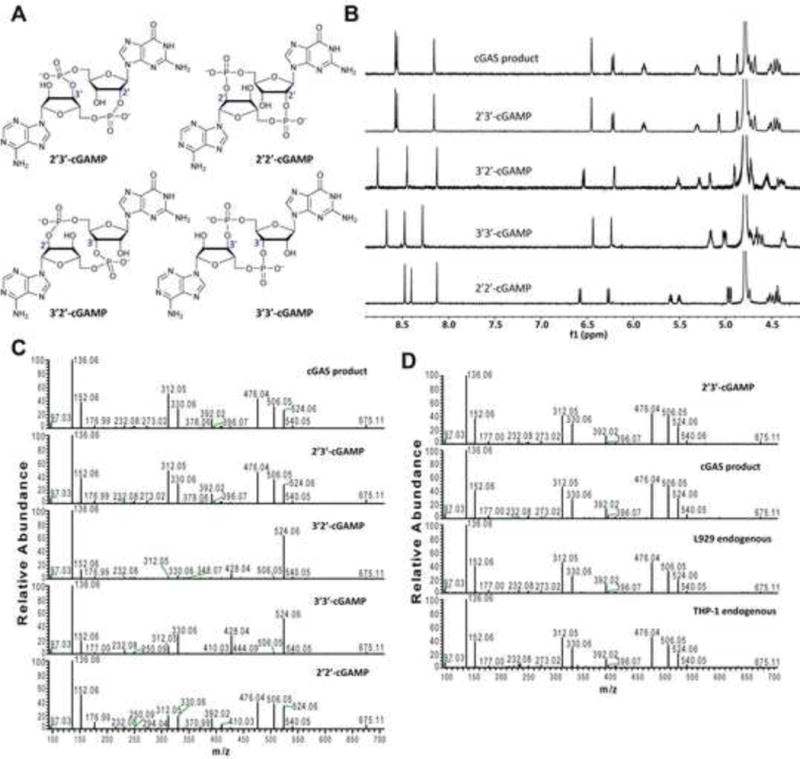
(A) Depiction of cGAMPs containing different combinations of phosphodiester bonds. (B) 1H NMR spectra of the cGAS product and synthetic cGAMPs at 50°C in 30 mM NaH2PO4/Na2HPO4/D2O buffer (pH 7.4). Substrate concentrations: natural cGAMP, 1.05 mM; 2′3′-cGAMP, 1.05 mM; 2′2′-cGAMP, 1.50 mM; 3′2′-cGAMP, 1.50 mM; 3′3′-cGAMP, 1.50 mM. (C) Tandem mass spectra of natural and synthetic cGAMPs resulting from higher energy collision dissociation (HCD) of the precursor ion ([M+H]+ = 675.107). (D) L929 and THP1 cells were transfected with HT-DNA and the cell extracts containing endogenous cGAMP were analyzed by tandem mass spectrometry along with the cGAS product and synthetic 2′3′-cGAMP. See also Figures S1 and S2.
Endogenous cGAMP produced in DNA-transfected cells contains mixed phosphodiester bonds
To test whether mammalian cells could produce endogenous cGAMP that contains the mixed phosphodiester linkages, we transfected the mouse cell line L929 and human monocytes THP1 with herring testis DNA (HT-DNA), then cell lysates were heated at 95°C to denature proteins and the supernatants were prepared for analysis of endogenous cGAMP by mass spectrometry(Wu et al., 2013). The MS/MS spectra of the endogenous molecule from both cell lines were identical to those of cGAS product and 2′3′-cGAMP, indicating that the endogenous second messenger is 2′3′-cGAMP (Figure 1D).
2′3′-cGAMP is a high affinity ligand of STING
We performed isothermal titration calorimetry experiments to measure the affinity (Kd) of STING binding to natural and synthetic cGAMP. A C-terminal domain (CTD) encompassing residues 139–379 of human STING, which was previously shown to mediate binding to the bacterial second messenger cyclic di-GMP (Burdette et al., 2011; Huang et al., 2012; Ouyang et al., 2012; Shang et al., 2012; Shu et al., 2012; Yin et al., 2012), was expressed in E. coli and purified to apparent homogeneity for the ITC experiment. Consistent with previous reports, we found that c-di-GMP bound to STING with a Kd of 1.21 μM (Figure 2A and and2D).2D). Interestingly, both natural cGAMP and synthetic 2′3′-cGAMP bound to STING with such a high affinity that curve fitting was difficult (Supplemental Figure S3A). In addition, unlike the binding of c-di-GMP, which is an exothermic process, the binding of natural and 2′3′-cGAMP to STING was endothermic, suggesting that the energy may be used for STING conformational change (see below). To obtain the Kd of natural and synthetic 2′3′-cGAMP for STING, we titrated different amounts of these compounds as competitors into the STING – c-di-GMP complex (Figure 2B and and2C).2C). These measurements yielded a Kd of 4.59 nM for the cGAS product and 3.79 nM for 2′3′-cGAMP (Figure 2D). The competition experiment was also performed for 3′2′-cGAMP, because its binding to STING generated little heat change (Supplemental Figure S3B). This compound binds to STING with a Kd of 1.61 μM. 2′2′- and 3′3′-cGAMP were titrated directly to STING and the Kd values were calculated to be 287 nM and 1.04 μM, respectively (Figure 2D; Supplemental Figure S3C and S3D). Thus, the Kd of 2′3′-cGAMP was ~300 fold lower than those of c-di-GMP, 3′2′-cGAMP and 3′3′-cGAMP, and ~75 fold lower than that of 2′2′-cGAMP.

(A) The original titration traces (top) and integrated data (bottom) of ITC experiments, in which c-di-GMP was titrated into a solution of STING dimer. (B and C) cGAS product (B) or synthetic 2′3′-cGAMP (C) was titrated into a solution of c-di-GMP bound STING, because the tight binding of 2′3′-cGAMP to STING made it difficult to do curve fitting if it were titrated into apo-STING dimer directly. (D) Summary of ligand binding affinities of STING for different cyclic di-nucleotides. For c-di-GMP, synthetic 2′2′- and 3′3′-cGAMP, the ligands were titrated into apo-STING dimer. For others, the ligands were titrated into c-di-GMP bound STING (3.55:1 in molar ratio). (E) Different concentrations of the cGAS product, synthetic cGAMPs and c-di-GMP were delivered into digitonin-permeabilized L929 cells. Four hours later, IFN-β RNA was measured by qRT-PCR. Dose response curves and the half maximal effective concentration (EC50) for each compound were generated using GraphPad Prism 5.0 software. The error bars indicate standard deviations of triplicate experiments. See also Figure S3.
cGAMPs are potent inducers of type-I interferons
We delivered different amounts of the cGAMP isomers as well as c-di-GMP into L929 cells and measured IFNβ induction by q-RT-PCR (Figure 2E). The cGAMP molecules induced IFNβ with an EC50 that ranged from 15 nM to 42 nM, whereas c-di-GMP had an EC50 of greater than 500 nM. Thus, it appeared that the binding affinity of different cyclic di-nucleotides did not correlate well with their EC50 in the cell-based assays. The reason for this is not clear, but it is possible that different compounds have different stability or distribution in the cells. Nevertheless, these experiments provide direct evidence that the cGAS product, 2′3′-cGAMP, is a high affinity ligand for STING (Kd: ~ 4 nM) and a potent inducer of IFNβ in cells (EC50: ~ 20 nM).
The crystal structure of STING-cGAMP complex reveals ligand-induced conformational rearrangements of STING
We co-crystallized the STING C-terminal domain (CTD) (residues 139–379) with the purified cGAS product in the C2 space group. The structure of the complex was solved by molecular replacement using an apo-STING structure (PDB code: 4F9E) as the search model and was refined to 1.88 Å resolution (Table 1). There is one STING protomer in the crystallographic asymmetric unit, which forms a butterfly-shaped dimer with another protomer that is related by the crystallographic two-fold symmetry. The bound cGAMP molecule sits at the two-fold axis (see details below) (Figure 3A). The ordered region of STING (from Asn152 to Glu336) adopts an overall structure similar to the apo-STING, characterized by a central twisted β sheet surrounded by four α helices (Figure 3A and Supplemental Figure S4A). However, STING in complex with cGAMP displays several striking differences from apo-STING in both the structure of the monomer and the arrangement of the dimer (Figure 3B). Compared with the apo-dimer, the two protomers in the dimer of the complex structure undergo substantial inward rotations in relation to the cGAMP binding site. This more closed arrangement creates a deeper pocket between the two protomers to embrace cGAMP. In addition, the cGAMP binding site is covered by a lid of four-stranded anti-parallel β-sheet and the connecting loops formed by residues 219–249 from each of the two protomers (Figure 3C). In contrast, this segment in the apo-structure is largely disordered (Ouyang et al., 2012; Yin et al., 2012). The formation of the β sheet is not due to crystallographic packing (Supplemental Figure S4B). The interdomain interactions within the lid involve several pairs of polar contacts, between the side group of Tyr245 and the main-chain carbonyl oxygen atom of Gly234, the side group of Ser243 and the main-chain amide nitrogen atom of Lys236, as well as the side groups of Asp237 and Lys224 (Figure 3D).

(A) Overall structure of STING CTD bound to the cGAS product. Two perpendicular views are shown and the 2′3′-cGAMP molecule is indicated. STING forms a dimer and is colored in yellow and cyan for each molecule, respectively, in the right panel. The same color scheme is applied to other figures unless indicated otherwise. (B) Structural comparison of the apo- and cGAMP-bound forms of STING. The apo-STING dimer (PDB code: 4F9E, gray) is superimposed against one protomer of the cGAMP bound STING dimer. The arrow indicates the orientation of the conformational change in STING upon 2′3′-cGAMP binding. (C) Two new β sheets in each STING protomer are induced upon cGAMP binding. 2Fo-Fc electron density map, shown in blue mesh, is contoured at 1 σ on one chain. (D) The β sheets are stabilized through interdomain interactions. The residues that mediate interdomain interactions are shown in sticks. The distances between interacting atoms are in Å. All the structure figures are prepared in PyMol(DeLano, 2002). See also Figure S4.
Table 1
Statistics of data collection and refinement of cGAMP bound STING
| Data | cGAMP bound STING |
|---|---|
| Space Group | C2 |
| Unit Cell (Å,°) | 89.525 77.927 35.974 90 96.98 90 |
| Number of molecules in ASU | 1 |
| Wavelength (Å) | 0.97918 |
| Resolution (Å) | 50–1.88 (1.91–1.88) |
| Rmerge (%) | 7.8 (65.0) |
| I/σ | 17.82 (2.20) |
| Completeness (%) | 99.4 (98.6) |
| Number of measured reflections | 99,635 |
| Number of unique reflections | 19,800 |
| Redundancy | 5.0 (4.8) |
| Wilson B factor (Å2) | 30.80 |
| R-factor (%) | 16.07 (23.09) |
| Rfree (%) | 18.15 (30.83) |
| Number of atoms | |
 Macromolecules Macromolecules | 1483 |
 Ligand Ligand | 45 |
 Water Water | 72 |
 All atoms All atoms | 1600 |
| Average B value (Å2) | |
 Macromolecules Macromolecules | 46.20 |
 Ligand Ligand | 23.10 |
 solvent solvent | 50.00 |
 All atoms All atoms | 45.70 |
| Rms deviations from ideal values | |
 Bonds (Å) Bonds (Å) | 0.007 |
 Angle (°) Angle (°) | 1.213 |
| Ramachandran plot statistics (%) | |
 Favored Favored | 97.22 |
 Allowed Allowed | 2.78 |
 Outliers Outliers | 0 |
Values in parentheses are for the highest resolution shell. R=Σ|Fobs−Fcalc|/ΣFobs, where Fcalc is the calculated protein structure factor from the atomic model (Rfree was calculated with 10% of the reflections selected).
Extensive interactions between 2′3′-cGAMP and STING underlie their specific and high affinity binding
Since the crystallographic two-fold axis passes through the asymmetric 2′3′-cGAMP molecule, cGAMP must adopt two orientations related by the two-fold symmetry. This is consistent with the fact that the two protomers in the STING dimer are expected to have equal probabilities to interact with either the guanidine or the adenosine moiety. We therefore assigned two alternative conformations with the occupancy of 0.5 for cGAMP and several surrounding amino acid residues. Simulated annealing omit map of the refined structure shows decent density for cGAMP (Figure 4A, upper panel). 2′3′-cGAMP, but not other isoforms, fits the electron density map well (Figure 4A, lower panel; Supplemental Figure S5A and S5B). Compared to c-di-GMP bound to STING, cGAMP sits ~2.5 Å deeper in the crevice between the STING dimeric interface (Figure 4B). In addition, the two wings of the butterfly are ~20 Å closer to each other in the STING:cGAMP structure due to the more closed arrangement of the two STING protomers. Further analyses of the cGAMP binding pocket show that cGAMP is well coordinated by extensive polar and hydrophobic interactions (Supplemental Figure S5C). The rings of cGAMP purine base groups stack against four around aromatic residues, Tyr 240 and Tyr167 from each of the two protomers (Figure 4C). Notably, the two α-phosphate groups of cGAMP contact Arg238 from both of the two protomers and Arg232 from one protomer (Figure 4D, left). The free 3′-OH of GMP points to two Ser162 residues from the lower part of the pocket. The guanine base directly interacts with the side groups of Glu260 and Thr263, as well as the main-chain carbonyl oxygen of Val239 (Figure 4D, right). These unique polar contacts explain why 2′3′-cGAMP is a specific and high affinity ligand for STING. Besides, residues from the β sheet (Arg232, Arg238, Val239), which are involved in the cGAMP binding, are likely to control the formation of the lid and further activation of STING.
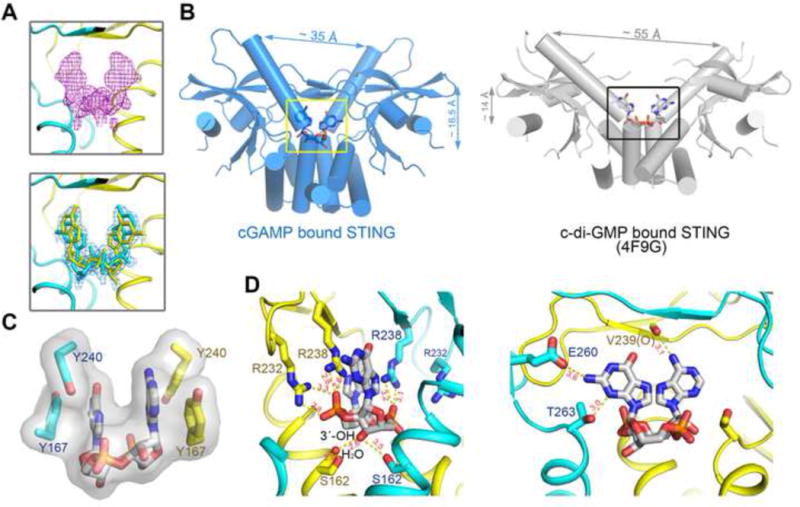
(A) Upper: the simulated annealing omit electron density map for cGAMP contoured at 3.0 σ. Lower: The 2Fo-Fc electron density map for cGAMP after refinement contoured at 1.0 σ. Two alternative conformations of cGAMP were built at 0.5 occupancy, respectively. (B) cGAMP, binding at a deeper pocket, drags the STING dimer (blue) closer to each other, compared to c-di-GMP bound STING (PDB code: 4F9G, gray). (C) The base groups of cGAMP are roughly parallel to each other and to four aromatic rings of the Tyr residues. (D) The bottom ribose ring (left panel) and the upper purine base groups (right panel) of cGAMP are coordinated by extensive polar contacts. Ser162 of STING interacts with 3′−OH of GMP. Only one conformation of 2′3′-cGAMP is shown for clarity. See also Figure S5.
Arginine 232 of STING is important for the cytosolic DNA signaling pathway
Three previous reports of the crystal structures of STING bound to cyclic-di-GMP used a rare human variant that substitutes Arg232 with a histidine(Ouyang et al., 2012; Shu et al., 2012; Yin et al., 2012). Extensive sequencing of DNA from human populations has shown that the Arg232 allele is prevalent and thus should be considered wild-type STING(Jin et al., 2011). The use of the H232 variant of STING may explain why c-di-GMP did not induce a significant conformational change of STING in these studies(Ouyang et al., 2012; Shu et al., 2012; Yin et al., 2012). A previous report showed that a mutation of Arg231 of mouse STING (equivalent to Arg232 in human STING) to alanine abolished IFNβ induction by cyclic-di-GMP, but not DNA(Burdette et al., 2011). However, based on our crystal structure of the STING-cGAMP complex, a mutation of Arg232 to histidine is expected to significantly weaken cGAMP binding and downstream signaling by STING, and a mutation of Arg232 to alanine should be even more detrimental. We therefore investigated the function of Arg232 of STING in two sets of experiments. First, we knocked down endogenous STING by RNAi in L929 cells and replaced it with WT, R232A or R232H of human STING. These stable cell lines were transfected with HT-DNA or treated with 2′3′-cGAMP, followed by measurement of IFNβ by q-RT-PCR (Figure 5A and and5B).5B). The cells expressing WT STING were able to induce IFNβ in response to DNA or cGAMP stimulation, whereas those expressing either R232A or R232H were defective. As a control, the double stranded RNA analogue poly[I:C] stimulated IFNβ expression in all of these cell lines. Second, we stably expressed WT or mutant STING in HEK293T cells, which have undetectable expression of endogenous STING and cGAS(Sun et al., 2013). The cells were then transfected with the human cGAS expression plasmid followed by measurement of IFNβ RNA (Figure 5C and and5D).5D). WT STING, but not the R232A mutant, was able to support IFNβ induction by cGAS. The R232H mutant was partially defective, possibly because the positively charged histidine may weakly substitute for some of the functions of Arg232. MAVS, an essential adaptor protein of the RIG-I pathway(Seth et al., 2005), was able to induce IFNβ in all of these cell lines. Taken together, our structural and functional data strongly support an important role of Arg232 in the functions of STING and further underscore the role of cGAS as an indispensable cytosolic DNA sensor.
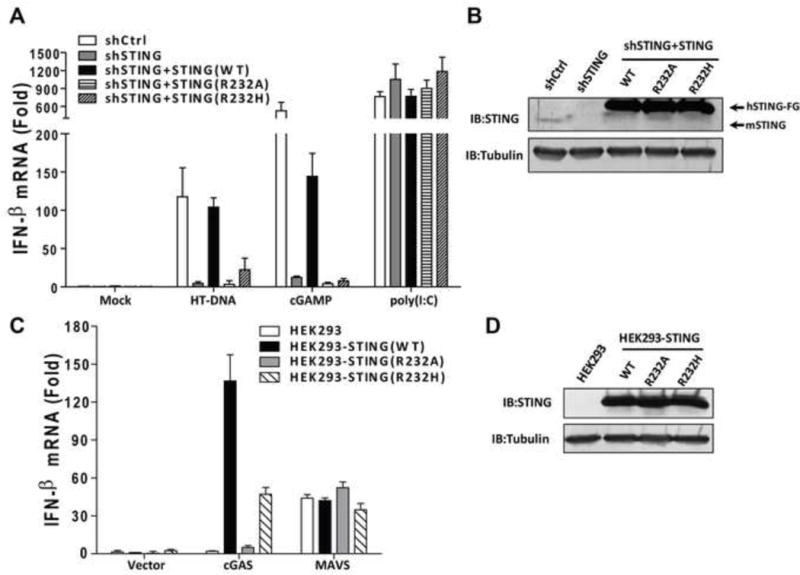
(A) L929 cells in which endogenous STING was stably knocked down by shRNA and replaced or poly(I-C) for 8 hr. IFN-β RNA levels were measured by qRT-PCR. (B) Extracts from the cell lines shown in (A) were immunoblotted with antibodies against STING or β-tubulin. (C) HEK293T cells stably expressing WT or mutant human STING-Flag were transfected with expression vectors for human cGAS or MAVS. 24 hr after transfection, IFN-β RNA levels were measured by qRT-PCR. (D) Extracts from the cell lines shown in (C) were immunoblotted with antibodies against STING or β-tubulin. The error bars indicate standard deviations of triplicate experiments.
DISCUSSION
Our previous studies identified cGAS as a cytosolic DNA sensor and a cyclase that synthesizes cGAMP using ATP and GTP as the substrates(Sun et al., 2013; Wu et al., 2013). cGAMP then functions as a second messenger that binds to and activates STING. Here we employed chemical synthesis and several biophysical approaches to further characterize the internal phosphodiester linkages of the cGAS product and determined that it is 2′3′-cGAMP. While this manuscript was in preparation, Gao et al reported the structures of cGAS in its apo- and DNA-bound forms, which confirmed that cGAS is indeed a DNA-activated cyclic-GMP-AMP synthase that catalyzes the synthesis of cGAMP from ATP and GTP (Gao et al., 2013). This elegant study also elucidated the structural mechanism by which DNA binding leads to the activation of cGAS. Using a different approach, Gao et al also found that the truncated cGAS protein synthesizes 2′3′-cGAMP in vitro. However, they did not test whether 2′3′-cGAMP has any biological or biochemical activity, nor did they show whether endogenous 2′3-cGAMP is produced in mammalian cells. In this report, we show that stimulation of mouse and human cells with DNA leads to the production of endogenous 2′3′-cGAMP. Moreover, we demonstrate that 2′3′-cGAMP binds to STING with a much greater affinity than other cGAMP isomers and c-di-GMP. We further show that 2′3′-cGAMP and other cGAMP isomers are much more potent than c-di-GMP in inducing IFNβ in cells.
Further insights into the structure and function of 2′3′-cGAMP are gained from the crystal structure of the STING CTD bound to this endogenous ligand. This crystal structure has a resolution of 1.88Å, allowing for a detailed view of the ligand structure, including both 2′−5′ and 3′−5′phosphodiester linkages. The structure reveals specific residues on STING that mediate the binding of 2′3′-cGAMP. Furthermore, a comparison of this structure to the previously published STING CTD structures in its apo form reveals extensive conformational rearrangements induced by the natural ligand. Specifically, the two arms of the V shaped STING dimer move closer by about 20 Å and a new four β-stranded sheet forms a lid above the cGAMP binding site in the ligand-bound STING structure. These features are absent in the previously determined STING:c-di-GMP structures, which used a STING variant containing the R232H mutation. In these structures, c-di-GMP binding does not induce any obvious conformational rearrangement in STING(Ouyang et al., 2012; Shu et al., 2012; Yin et al., 2012). However, in two other structures containing the WT STING (Arg232) and c-di-GMP, one exhibits similar conformational changes as observed in the STING-cGAMP complex(Huang et al., 2012), and the other shows a distinct conformational change in that Arg232 is oriented differently(Shang et al., 2012). The “closed” conformation observed by Huang et al may have captured the active state of STING induced by c-di-GMP, which is capable of activating STING, albeit more weakly than cGAMP.
The extensive interactions between STING and 2′3′-cGAMP provide the structural basis for their high affinity binding. In particular, Glu260, Thr263 and Val239 interact with the guanine base of GMP and Ser162 interacts with the free 3′−OH group of GMP, explaining why cGAMP containing a phosphodiester bond between 2′−OH of GMP and 5′-phosphate of AMP is a high affinity ligand. In addition, the two α-phosphate groups interact with Arg232 from one protomer and Arg238 from both protomers. This structural analysis explains that the R232A or R232H mutations strongly impair the function of STING in response to DNA or cGAMP. Our data highlight the importance of using the wild-type (Arg232) STING in structural and functional studies.
Although 2′3-cGAMP binds to STING with a much higher affinity than cGAMP isomers containing other phosphodiester linkages, all four cGAMP isomers induced IFNβ with similar EC50 values, which were much lower than that of c-di-GMP. Thus, all cGAMP isoforms are potent inducers of IFNβ, raising the possibility that cGAMP containing distinct phosphodiester linkages might exist in nature, perhaps in some lower organisms. Indeed, Vibrio cholera contains a cyclase that synthesize 3′3′-cGAMP involved in bacterial chemotaxis and colonization (Davieset al., 2012). At present, it is not clear why mammals have evolved to produce 2′3′-cGAMP as the endogenous second messenger to trigger innate immune responses.
In summary, our results demonstrate that 1) the endogenous second messenger produced in mammalian cells in response to cytosolic DNA stimulation is 2′3′-cGAMP; 2) 2′3′-cGAMP is a high affinity ligand for STING; 3) 2′3′-cGAMP is a potent inducer of IFNβ in mammalian cells; 4) 2′3′-cGAMP induces conformational rearrangements in STING that might underlie its activation; and 5) extensive interactions between 2′3′-cGAMP and STING observed in the crystal structure of the complex explains their specific and high affinity binding.
EXPERIMENTAL PROCEDURES
Enzymatic Synthesis and Purification of cGAMP
To generate natural cGAMP using the enzyme cGAS, a reaction containing 20mM Tris-Cl, pH7.5, 5mM MgCl2, 10mM CoCl2, 0.01mg/ml herring testis DNA, 1mM ATP, 1mM GTP, and 0.1μM recombinant SUMO-tagged human cGAS (aa147–522) was incubated at 37°C for 1hr. The mixture was fractionated on a Hitrap Q column using a linear 0–0.5M NaCl gradient; a UV peak corresponding to cGAMP was collected and loaded onto a C18 column (201TP510, 1cmX25cm, Phenomenex, Hesperia, CA), and eluted with a linear 0–100% methanol gradient.
Preparation of Endogenous cGAMP
Endogenous cGAMP was prepared from DNA transfected L929 and THP-1 cells, respectively. After HT-DNA transfection for 4 hours, about 3× 107 cells were lysed in hypotonic buffer [10mM Tris-HCl, pH7.4, 10mM KCl, 1.5mM MgCl2]. The lysates were heated at 95°C for 5 min and centrifuged again at 17,000g for 10 min to remove denatured proteins. The heat-resistant supernatant was fractionated on a C-18 column (Eclipse Plus 4.6×30 mm, 3.5μm, Agilent Technologies) equilibrated with 0.1% formic acid and eluted with a linear gradient of 0–100% methanol. The presence of cGAMP in each fraction was monitored by activity assay (Wu et al., 2013), and the fraction with peak activity was used for further MS and MS/MS analysis.
MS and MS/MS Analysis
High resolution mass measurement was performed on Q-Exactive mass spectrometer (ThermoFisher Scientific) as described before (Wu et al, 2013). Tandem MS/MS analysis was conducted using the same LC-MS system. Full scan mass spectra were acquired from m/z 300–700 with a resolution of 70,000 at m/z = 200 in the Orbitrap. MS/MS spectra (resolution: 35,000 at m/z = 200) were acquired in a data-dependent mode whereby the top 5 most abundant parent ions were subjected to further fragmentation by higher energy collision dissociation (HCD). The normalized collision energy was set at 30.
Protein Expression and Purification
The complementary DNA of wild type human STING C-terminal domain (STING CTD, 139–379) was cloned into pET-SUMO vector (InvitrogenTM). Overexpression of STING CTD was induced in E. coli BL21 (DE3) pLysS with 0.8 mM isopropyl-β-D-thiogalactoside when the cell density reached an OD600nm of 1.2. After growth at 20°C for 12 h, the cells were collected, resuspended in a buffer containing 25mM Hepes, pH 7.8, and 150mM NaCl, and disrupted by French Press with 2 passes at 15,000 p.s.i. Cell debris was removed by centrifugation at 27,000g for 50 min. The supernatant was loaded onto a Ni2+-nitrilotriacetate affinity resin (Qiagen). Subsequently, the resin was rinsed three times with 20 ml buffer containing 25 mM Hepes, pH 7.8, 500 mM NaCl, 20 mM imidazole-HCl, pH 8.0. The SUMO tag was then removed by digesting the proteins using the SUMO protease at 4°C overnight. The elute was concentrated to about 15 mg/ml before applying to gel-filtration chromatography (Superdex-200 10/30, GE Healthcare), which was equilibrated in the buffer containing 25 mM Hepes, pH 7.8, 150 mM NaCl. The peak fractions of the protein were collected and concentrated to 10 mg/ml for crystallization trials, or to 0.42–1.4 mg/ml (15–50 μM) for ITC experiment.
Isothermal titration calorimetry (ITC)
Isothermal titration calorimetry (ITC) was employed to measure the binding affinities between STING and cGAMP isomers or c-di-GAMP using a VP-ITC microcalorimeter (GE Healthcare). The protein and the ligand concentrations are shown in Figure 2D. The titrations were performed at 20°C in the buffer containing 25 mM Hepes, pH 7.8, 150 mM NaCl. 32 injections were performed with 4 minutes spacing time. The titration traces were integrated by NITPIC(Keller et al., 2012) and then the curves were fitted by SEDFIT(Houtman et al., 2007). The figures were prepared using GUSSI (http://biophysics.swmed.edu/MBR/software.html).
Crystallization
Natural cGAMP synthesized by cGAS was added into STING CTD in 3:1 molar ratio (cGAMP: STING dimer) followed by incubation at room temperature for 30 minutes. Crystals were grown at 20°C by the hanging-drop vapor diffusion method. The complex yielded rod-cluster crystals in the buffer containing 27% PEG 3,350, 0.1 M Bis-Tris, pH 6.8, 0.2 M NaCl. The crystals appeared overnight and grew to 20 μm×20 μm×60 μm after 3 days. The crystals were flash frozen in liquid nitrogen after being transferred to the crystallization buffer plus 20% ethylene glycol.
Data collection and processing
The data set was collected to 1.88 Å resolution with a wavelength of 0.97918 Å at 19-ID of Argonne National Laboratory, Structural Biology Center at the Advanced Photon Source (APS). The crystal are in the space group C2 with unit cell dimensions a=89.525 Å, b=77.927 Å, c=35.974 Å, a=90°, b=96.98°, c=90°. The data was integrated and scaled with HKL3000 package(Minor et al., 2006). Further calculations were performed using programs from PHENIX(Adams et al., 2010), unless explicitly specified. Data collection and statistics are summarized in Table 1.
Structural determination and refinement
Apo-STING monomer (PDB accession code 4F9E) was transferred into the lattice of the cGAMP bound STING by molecular replacement. One distinct solution was identified and the resulting electron density was of sufficiently good quality and most of the side chains were clearly shown. The manual building of residues 229–238 was performed in COOT(Emsley et al., 2010). The structure refinement was performed using PHENIX(Adams et al., 2010) after excluding 10% of the data for the R-free calculation.
Acknowledgments
Results shown in this report are derived from work performed at Argonne National Laboratory, Structural Biology Center at the Advanced Photon Source. Argonne is operated by University of Chicago Argonne, LLC, for the U.S. Department of Energy, Office of Biological and Environmental Research under contract DE-AC02-06CH11357. We thank Drs. Diana Tomchick, James Chen and Chad Brautigam at the Structural Biology Core at UT Southwestern Medical Center for assistance in crystallography and ITC experiments. We also thank Fenghe Du and Xiang Chen for technical assistance. This work was supported by an NIH grant (AI-093967) to Z.J.C.
The structural and biophysical studies of STING/cGAMP complex were performed by X.Z. Chemical synthesis, NMR, HPLC and CD spectroscopy of cGAMP isomers were carried out by H.S under the supervision of C.C. Mass spectrometry of cGAMPs and functional assays of cGAMPs and STING mutants were performed by J.W. L.S. performed enzymatic synthesis and purification of the cGAS product. X-W. Z. assisted in the crystal structure analyses. Z.J.C., X.Z., and C.C. wrote the manuscript and all authors agree with the contents.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errorsmaybe discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
SUPPLEMENTAL INFORMATION
Supplemental information includes five supplemental figures and experimental procedures for chemical synthesis of cGAMP isoforms.
ACCESSION NUMBER
The coordinates of 2′3′-cGAMP bound human STING CTD structure have been deposited in the RCSB protein data bank (PDB: 4KSY).
References
- GUSSI. on World Wide Web http://biophysicsswmededu/MBR/softwarehtml.
- Adams PD, Afonine PV, Bunkoczi G, Chen VB, Davis IW, Echols N, Headd JJ, Hung LW, Kapral GJ, Grosse-Kunstleve RW, et al. PHENIX: a comprehensive Python-based system for macromolecular structure solution. Acta crystallographica. 2010;66:213–221. [Europe PMC free article] [Abstract] [Google Scholar]
- Barber GN. Cytoplasmic DNA innate immune pathways. Immunological reviews. 2011;243:99–108. [Abstract] [Google Scholar]
- Burdette DL, Monroe KM, Sotelo-Troha K, Iwig JS, Eckert B, Hyodo M, Hayakawa Y, Vance RE. STING is a direct innate immune sensor of cyclic di-GMP. Nature. 2011;478:515–518. [Europe PMC free article] [Abstract] [Google Scholar]
- Davies BW, Bogard RW, Young TS, Mekalanos JJ. Coordinated regulation of accessory genetic elements produces cyclic di-nucleotides for V. cholerae virulence. Cell. 2012;149:358–370. [Europe PMC free article] [Abstract] [Google Scholar]
- DeLano WL. The PyMOL Molecular Graphics System. 2002 on World Wide Web http://wwwpymolorg.
- Emsley P, Lohkamp B, Scott WG, Cowtan K. Features and development of Coot. Acta crystallographica. 2010;66:486–501. [Europe PMC free article] [Abstract] [Google Scholar]
- Gaffney BL, Veliath E, Zhao J, Jones RA. One-flask syntheses of c-di-GMP and the [Rp,Rp] and [Rp,Sp] thiophosphate analogues. Org Lett. 2010;12:3269–3271. [Europe PMC free article] [Abstract] [Google Scholar]
- Gao P, Ascano M, Wu Y, Barchet W, Gaffney BL, Zillinger T, Serganov AA, Liu Y, Jones RA, Hartmann G, et al. Cyclic [G(2′,5′)pA(3′,5′)p] Is the Metazoan Second Messenger Produced by DNA-Activated Cyclic GMP-AMP Synthase. Cell 2013 [Europe PMC free article] [Abstract] [Google Scholar]
- Houtman JC, Brown PH, Bowden B, Yamaguchi H, Appella E, Samelson LE, Schuck P. Studying multisite binary and ternary protein interactions by global analysis of isothermal titration calorimetry data in SEDPHAT: application to adaptor protein complexes in cell signaling. Protein Sci. 2007;16:30–42. [Europe PMC free article] [Abstract] [Google Scholar]
- Huang YH, Liu XY, Du XX, Jiang ZF, Su XD. The structural basis for the sensing and binding of cyclic di-GMP by STING. Nature structural & molecular biology. 2012;19:728–730. [Abstract] [Google Scholar]
- Iwasaki A, Medzhitov R. Regulation of adaptive immunity by the innate immune system. Science. 2010;327:291–295. [Europe PMC free article] [Abstract] [Google Scholar]
- Jin L, Xu LG, Yang IV, Davidson EJ, Schwartz DA, Wurfel MM, Cambier JC. Identification and characterization of a loss-of-function human MPYS variant. Genes and immunity. 2011;12:263–269. [Europe PMC free article] [Abstract] [Google Scholar]
- Keller S, Vargas C, Zhao H, Piszczek G, Brautigam CA, Schuck P. High-precision isothermal titration calorimetry with automated peak-shape analysis. Analytical chemistry. 2012;84:5066–5073. [Europe PMC free article] [Abstract] [Google Scholar]
- Minor W, Cymborowski M, Otwinowski Z, Chruszcz M. HKL-3000: the integration of data reduction and structure solution--from diffraction images to an initial model in minutes. Acta crystallographica. 2006;62:859–866. [Abstract] [Google Scholar]
- O’Neill LA. Immunology. Sensing the dark side of DNA. Science. 2013;339:763–764. [Abstract] [Google Scholar]
- Ouyang S, Song X, Wang Y, Ru H, Shaw N, Jiang Y, Niu F, Zhu Y, Qiu W, Parvatiyar K, et al. Structural analysis of the STING adaptor protein reveals a hydrophobic dimer interface and mode of cyclic di-GMP binding. Immunity. 2012;36:1073–1086. [Europe PMC free article] [Abstract] [Google Scholar]
- Ronald PC, Beutler B. Plant and animal sensors of conserved microbial signatures. Science. 2010;330:1061–1064. [Abstract] [Google Scholar]
- Seth RB, Sun L, Ea CK, Chen ZJ. Identification and characterization of MAVS, a mitochondrial antiviral signaling protein that activates NF-kappaB and IRF 3. Cell. 2005;122:669–682. [Abstract] [Google Scholar]
- Shang G, Zhu D, Li N, Zhang J, Zhu C, Lu D, Liu C, Yu Q, Zhao Y, Xu S, et al. Crystal structures of STING protein reveal basis for recognition of cyclic di-GMP. Nature structural & molecular biology. 2012;19:725–727. [Abstract] [Google Scholar]
- Shu C, Yi G, Watts T, Kao CC, Li P. Structure of STING bound to cyclic di-GMP reveals the mechanism of cyclic dinucleotide recognition by the immune system. Nature structural & molecular biology. 2012;19:722–724. [Europe PMC free article] [Abstract] [Google Scholar]
- Sun L, Wu J, Du F, Chen X, Chen ZJ. Cyclic GMP-AMP synthase is a cytosolic DNA sensor that activates the type I interferon pathway. Science. 2013;339:786–791. [Europe PMC free article] [Abstract] [Google Scholar]
- Takeuchi O, Akira S. Pattern recognition receptors and inflammation. Cell. 2010;140:805–820. [Abstract] [Google Scholar]
- Wu J, Sun L, Chen X, Du F, Shi H, Chen C, Chen ZJ. Cyclic GMP-AMP is an endogenous second messenger in innate immune signaling by cytosolic DNA. Science. 2013;339:826–830. [Europe PMC free article] [Abstract] [Google Scholar]
- Yin Q, Tian Y, Kabaleeswaran V, Jiang X, Tu D, Eck MJ, Chen ZJ, Wu H. Cyclic di-GMP sensing via the innate immune signaling protein STING. Molecular cell. 2012;46:735–745. [Europe PMC free article] [Abstract] [Google Scholar]
- Zhang Z, Kim S, Gaffney BL, Jones RA. Polymorphism of the signaling molecule c-di-GMP. J Am Chem Soc. 2006;128:7015–7024. [Europe PMC free article] [Abstract] [Google Scholar]
Full text links
Read article at publisher's site: https://doi.org/10.1016/j.molcel.2013.05.022
Read article for free, from open access legal sources, via Unpaywall:
http://www.cell.com/article/S1097276513004097/pdf
Citations & impact
Impact metrics
Citations of article over time
Alternative metrics
Article citations
Cytoplasmic DNA and AIM2 inflammasome in RA: where they come from and where they go?
Front Immunol, 15:1343325, 10 Oct 2024
Cited by: 0 articles | PMID: 39450183 | PMCID: PMC11499118
Review Free full text in Europe PMC
2'3'-cGAMP interactome identifies 2'3'-cGAMP/Rab18/FosB signaling in cell migration control independent of innate immunity.
Sci Adv, 10(42):eado7024, 16 Oct 2024
Cited by: 0 articles | PMID: 39413198 | PMCID: PMC11482326
The cGAS-STING pathway and female reproductive system diseases.
Front Immunol, 15:1447719, 09 Oct 2024
Cited by: 0 articles | PMID: 39445027 | PMCID: PMC11496054
Review Free full text in Europe PMC
AARS1 and AARS2 sense L-lactate to regulate cGAS as global lysine lactyltransferases.
Nature, 634(8036):1229-1237, 25 Sep 2024
Cited by: 1 article | PMID: 39322678
2024 Lasker Award Recipient Zhijian Chen elucidates how DNA stimulates immunity.
J Clin Invest, 134(19):e186104, 19 Sep 2024
Cited by: 0 articles | PMID: 39295301 | PMCID: PMC11446599
Go to all (547) article citations
Other citations
Data
Data behind the article
This data has been text mined from the article, or deposited into data resources.
BioStudies: supplemental material and supporting data
Protein structures in PDBe (3)
-
(3 citations)
PDBe - 4F9EView structure
-
(1 citation)
PDBe - 4KSYView structure
-
(1 citation)
PDBe - 4F9GView structure
Similar Articles
To arrive at the top five similar articles we use a word-weighted algorithm to compare words from the Title and Abstract of each citation.
The cGAS-STING pathway for DNA sensing.
Mol Cell, 51(2):135-139, 01 Jul 2013
Cited by: 108 articles | PMID: 23870141 | PMCID: PMC3782533
Review Free full text in Europe PMC
cGAS produces a 2'-5'-linked cyclic dinucleotide second messenger that activates STING.
Nature, 498(7454):380-384, 30 May 2013
Cited by: 862 articles | PMID: 23722158 | PMCID: PMC4143541
Molecular basis for the specific recognition of the metazoan cyclic GMP-AMP by the innate immune adaptor protein STING.
Proc Natl Acad Sci U S A, 112(29):8947-8952, 06 Jul 2015
Cited by: 35 articles | PMID: 26150511 | PMCID: PMC4517257
Cyclic GMP-AMP as an Endogenous Second Messenger in Innate Immune Signaling by Cytosolic DNA.
Annu Rev Biochem, 86:541-566, 07 Apr 2017
Cited by: 128 articles | PMID: 28399655
Review
Funding
Funders who supported this work.
NIAID NIH HHS (2)
Grant ID: R01 AI093967
Grant ID: R01-AI-093967
NIGMS NIH HHS (2)
Grant ID: R01-GM-079554
Grant ID: R01 GM079554





