Abstract
Free full text

FGFR2 signaling underlies p63 oncogenic function in squamous cell carcinoma
Abstract
Oncogenic transcription factors drive many human cancers, yet identifying and therapeutically targeting the resulting deregulated pathways has proven difficult. Squamous cell carcinoma (SCC) is a common and lethal human cancer, and relatively little progress has been made in improving outcomes for SCC due to a poor understanding of its underlying molecular pathogenesis. While SCCs typically lack somatic oncogene-activating mutations, they exhibit frequent overexpression of the p53-related transcription factor p63. We developed an in vivo murine tumor model to investigate the function and key transcriptional programs of p63 in SCC. Here, we show that established SCCs are exquisitely dependent on p63, as acute genetic ablation of p63 in advanced, invasive SCC induced rapid and dramatic apoptosis and tumor regression. In vivo genome-wide gene expression analysis identified a tumor-survival program involving p63-regulated FGFR2 signaling that was activated by ligand emanating from abundant tumor-associated stroma. Correspondingly, we demonstrate the therapeutic efficacy of extinguishing this signaling axis in endogenous SCCs using the clinical FGFR2 inhibitor AZD4547. Collectively, these results reveal an unanticipated role for p63-driven paracrine FGFR2 signaling as an addicting pathway in human cancer and suggest a new approach for the treatment of SCC.
Introduction
Squamous cell carcinoma (SCC) is a treatment-refractory malignancy arising within the epithelium of the skin, lung, esophagus, and upper aerodigestive tract (so-called head and neck SCC, HNSCC). SCC is largely a carcinogen-induced tumor, as major risk factors include tobacco, alcohol, and sun exposure. Little progress has been made in improving overall survival from SCC during the past 30 years, in part because of a limited understanding of the molecular pathobiology of this disease (1). SCCs do not commonly harbor somatic oncogene-activating mutations, but instead are associated with frequent mutational inactivation of tumor suppressor pathways including p53 and NOTCH, the therapeutic implications of which remain to be established (2, 3). In addition, although the EGFR is an attractive therapeutic target that is overexpressed or amplified in a subset of cases, only limited success with EGFR-directed therapy has been achieved in SCC despite intensive efforts over many years (4, 5). Recent systematic sequencing studies of HNSCC have confirmed the remarkable rarity of tumor-associated somatic mutations in well-established oncogenic drivers, which include HRAS (4% of cases) and the PI3K catalytic subunit PIK3CA (7% of cases) (2, 3). Thus, new approaches are needed to identify the key biological drivers in SCC in order to develop more effective and selective therapies for this disease.
One underexplored area for potential therapeutic intervention in SCC and other cancers involves deregulation of various lineage-specific transcription factors (6). The p53-related transcription factor p63 (TP63), a master regulator of embryonic epithelial development, is subject to overexpression and/or genomic amplification in up to 80% of primary HNSCC tumors as well as in SCCs of the skin, lung, and esophagus (7, 8). Numerous p63-regulated transcriptional target genes and programs have been identified through developmental and cell-culture studies, including differentiation, adhesion signaling, and senescence suppression (9–11). While several of these genes and pathways could in theory contribute to tumorigenesis, it is unknown which if any are relevant in the context of p63-expressing tumors in vivo. Indeed, the precise role of p63 in cancer may be context dependent, as loss rather than overexpression of p63 is associated with disease progression in some tumors, including bladder cancer (12). These seemingly paradoxical results could be in part attributable to distinct functions of different p63 protein variants. The p63 locus is expressed as multiple isoforms, most notably through 2 promoters that produce N-terminal variants either containing or lacking the p53-like transactivation domain (TAp63 or ΔNp63, respectively) (13). Data from mouse models indeed support isoform-specific functions of p63 in both development and cancer. While germline inactivation of p63 (14) or ΔNp63 (15) results in mice lacking skin and limbs and is perinatal lethal, selective TAp63 deficiency produces viable animals that exhibit an increase in metastatic tumors (16). Notably, the major p63 isoform expressed in stratified squamous epithelium and SCC is ΔNp63α (17, 18).
The interactions between tumor cells and their microenvironment likely play an essential role in SCC pathogenesis, providing another fertile area for investigation and potential therapeutic intervention. Substantial data point to essential and possibly distinct roles for immune cells and stromal cells in SCC initiation and progression (19, 20). Of particular interest in recent years has been the contribution of tumor-associated fibroblasts. In many cancers, these cells have been shown to contribute to tumor proliferation, differentiation, and invasion as well as creation of a permissive environment for tumor formation and maintenance (20). Encasement of tumor cells within a dense fibrous stroma is a hallmark of HNSCC and other squamous tumors, and high levels of stromal infiltrate are associated with a poor prognosis in these malignancies (21, 22). Despite the clinical and biological importance of tumor stroma in SCC, however, previous work has provided few therapeutically actionable insights and the precise mechanisms of this stromal contribution are poorly understood.
Given the potentially complex role of p63 in SCC, understanding its precise contribution to cancer pathogenesis in the autochthonous tumor context could provide major new insights into this disease. We therefore established an endogenous SCC model to test the effect of deleting all p63 isoforms within established, invasive tumors. This experiment demonstrated exquisite dependence of SCC on high levels of p63. Extensive in vivo analysis revealed a deregulated paracrine FGFR2 signaling program that is controlled directly by p63, activated by stroma-derived ligand, and required for tumor survival. We then validated these findings and their therapeutic relevance using a clinical FGFR2 inhibitor to induce apoptosis of advanced endogenous tumors.
Results
Development and validation of a robust murine SCC model.
SCC is a cancer caused by prolonged, repeated carcinogen exposure. Therefore, to generate a murine SCC model that would recapitulate the features of the human disease we employed a well-established protocol of weekly carcinogen (7,12-dimethyl-benz[a]anthracene [DMBA]) treatment in the oral cavity (23) or on the back (24–26). These SCCs were readily distinguishable from nonmalignant papillomas in both appearance and size (Figure (Figure1,1, A and B). Tumors were locally invasive and exhibited metastases to lymph nodes and lung in a subset of cases (~13%), (Figure (Figure1B).1B). These tumors shared the histologic features of human SCC, including variable degrees of ongoing squamous epithelial differentiation and high levels of nuclear p63 expression (Figure (Figure1B).1B). Similar to human SCC, these tumors also expressed robust levels of EGFR and cyclin D1 proteins (Supplemental Figure 1A; supplemental material available online with this article; 10.1172/JCI68899DS1), and resemble the recently described “classical SCC” that develops in carcinogen-treated mice (27). Human SCCs frequently harbor inactivating mutations in p53 (2, 3, 28), and p53 inactivation is important for SCC progression in genetic models of oral SCC, cooperating potently with events such as AKT activation (29). Consistent with these data, we found that SCC tumors arising in DMBA-treated p53+/– mice appeared at reduced latency compared with those occurring in WT mice (Figure (Figure1C)1C) (30, 31). These tumors showed evidence of p53 loss or mutation as do human SCC primary tumors and cell lines (Supplemental Figure 1). Like human SCCs, our murine tumors express higher levels of p63 mRNA and protein than normal skin, corresponding to the ΔNp63α isoform (Figure (Figure1,1, D and E, and refs. 10, 18, 32). Using FACS of disaggregated tumors to isolate cells expressing the epithelial-specific α6-integrin (CD49f) (33), we observed levels of ΔNp63 mRNA between 50- and 200-fold greater than TAp63 (Figure (Figure1F),1F), consistent with similar findings observed in human HNSCC cell lines (18). Taken together, these data demonstrate that this model shares key molecular and histologic features of human SCC.
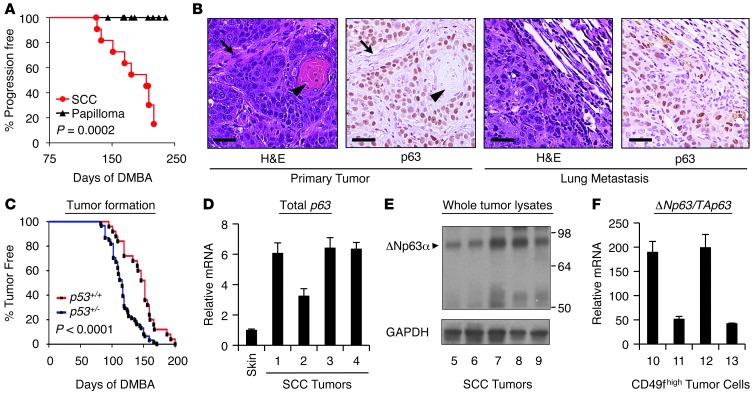
(A) Kaplan-Meier curve of progression of histologically defined papillomas compared with SCC tumors. Tumors were considered progressive when they passed 100 mm3 size threshold. (B) Primary and metastatic SCC tumors from DMBA-treated mice share the histologic features (H&E) — note stromal elements (arrows) and differentiated keratin “pearls” (arrowheads) — and nuclear p63 expression (IHC) of human SCC. Scale bars: 25 μm. (C) Haploinsufficiency for p53 reduces tumor latency following weekly DMBA treatment. p53+/+, n = 25, p53+/–, n = 61. P value assessed by log-rank test. (D) Increased expression of p63 mRNA in DMBA-induced SCCs versus normal skin. mRNA levels were assessed by QRT-PCR and normalized to β-actin. Error bars represent SEM. (E) Predominant expression of ΔNp63α protein (arrowhead) in murine SCC tumors. Whole-tumor lysates were assessed for p63 expression using an antibody that detects all isoforms. GAPDH serves as a loading control. (F) ΔNp63 is overexpressed relative to TAp63 in murine SCC tumors. Whole tumors were disaggregated and sorted for CD49fhi (α-6 integrin) epithelial cells, and mRNA levels were assessed as in D.
Exquisite sensitivity of autochthonous SCC to loss of p63.
To determine whether tumors were dependent on p63, we employed a conditional p63 allele (p63L/L) (Supplemental Figure 2A). A previous study showed that Cre-mediated recombination in homozygous p63L/L mice during embryogenesis recapitulates both the genomic structure and the phenotype of the p63-null mouse (34). In order to control p63 excision in a tissue- and temporal-specific manner, we crossed these mice to a transgenic strain expressing the Cre recombinase/estrogen receptor (CreER) fusion protein from the keratin 14 (K14) promoter (35). Notably, our murine tumors demonstrate high overlap between p63 and K14 expression (Figure (Figure2A),2A), as was previously shown in human SCC (36). Given the importance of p53 in human SCC, we then performed additional crosses to generate p63L/L K14-CreER mice on a p53+/– background for DMBA treatment. Mice were administered DMBA on the back skin or in the oral cavity until tumors reached approximately 100 mm3, a size not reached by papillomas (Figure (Figure1A),1A), then treated for 5 consecutive days with 100 mg/kg tamoxifen. Allele-specific PCR (Supplemental Figure 2A) demonstrated that p63 was excised only in tissues that normally express K14 (Figure (Figure2B2B and ref. 37). Immunohistochemical (IHC) staining 72 hours after tamoxifen treatment confirmed that p63 is rapidly and efficiently ablated from established SCC tumors in a temporally controlled manner (Figure (Figure22C).
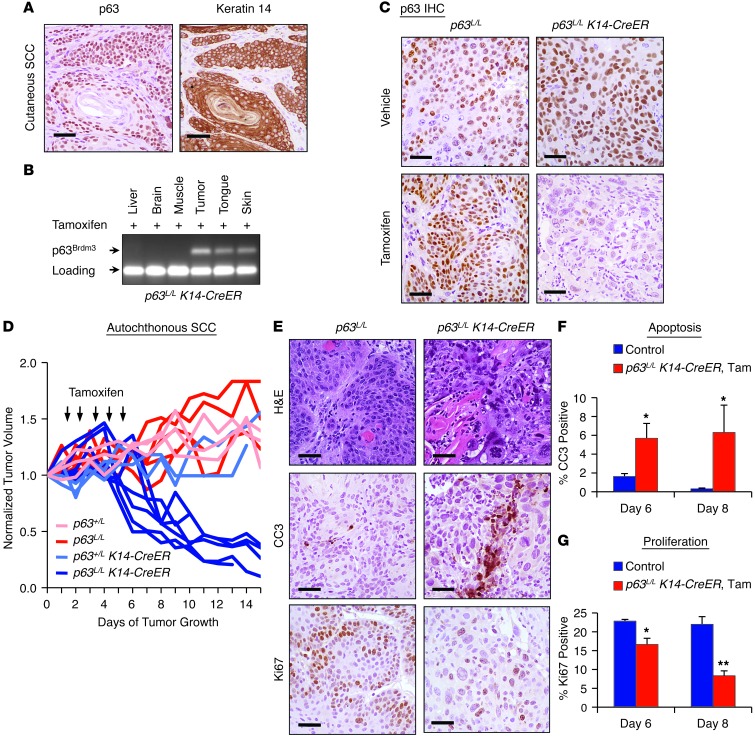
(A) IHC of K14 and p63 in SCC tumors. Scale bars: 25 μm. (B) PCR analysis of DNA from tissues of a p63L/L K14-CreER mouse treated with daily i.p. injection of 100 mg/kg tamoxifen for 5 consecutive days. The p63Brdm3 allele generated after excision and primer locations are shown in Supplemental Figure 2A. (C) IHC of p63 protein expression in tumors of indicated genotypes 72 hours after treatment with vehicle or tamoxifen as above. Scale bars: 25 μm. (D) Volume of cutaneous tumors in p63+/L (n = 4), p63L/L (n = 6), p63+/L K14-CreER (n = 3), or p63L/L K14-CreER (n = 6) mice following tamoxifen treatment (arrows). Tumor volume was normalized to size 1 day prior to treatment. P < 0.0001 for p63L/L K14-CreER compared with each control group as assessed by multiple measures ANOVA. (E) Representative histology (H&E) and IHC staining for Ki67 and cleaved caspase 3 (CC3) of tumors from tamoxifen-treated p63L/L or p63L/L K14-CreER mice. Scale bar: 25 μm. (F) Quantification of cleaved caspase 3 staining in control versus tamoxifen-treated p63L/L K14-CreER tumors, shown as percentage of field staining positive. *P < 0.05 as determined by Mann-Whitney test. (G) Quantification of Ki67-positive cells in control versus tamoxifen-treated K14-CreER tumors. *P < 0.05; **P < 0.01 as determined by Student’s unpaired t test. Error bars represent SEM. See Methods for details of controls in F and G.
A tumor-bearing cohort of p63L/L K14-CreER mice served as the experimental arm, while tumor-bearing cohorts of p63L/L, p63+/L, and p63+/L K14-CreER mice served as controls. All tumors of control mice showed progressive growth following tamoxifen treatment (Figure (Figure2D).2D). In contrast, all p63L/L K14-CreER tumors showed rapid and dramatic regression within a few days of p63 excision, which was observed in both cutaneous (Figure (Figure2D2D and Supplemental Figure 2B) and oral SCCs (Supplemental Figure 2C). Importantly, we saw no evidence of effects on normal epithelial tissues, consistent with the reported effects of p63 loss in the normal epidermis that appear only after a period of months (34). Histological analysis after the last dose of tamoxifen demonstrated apoptotic features in p63L/L K14-CreER tumors compared with control p63L/L tumors (Figure (Figure2E).2E). Staining for cleaved caspase 3 by IHC confirmed the marked induction of apoptosis in these tumors (Figure (Figure2,2, E and F). In addition, Ki67 staining demonstrated a significant decrease in proliferation in the tamoxifen-treated p63L/L K14-CreER tumors compared with controls, which became more pronounced at later time points after excision (Figure (Figure2,2, E and G). Taken together, these data demonstrate exquisite dependence of endogenous SCC on p63 for survival and proliferation.
A central role for p63-regulated developmental signaling in endogenous SCC.
It is unknown whether any of the established transcriptional targets or functions of p63 are critical for maintenance of SCC in vivo. In order to systematically identify such key programs, we performed global gene expression profiling following p63 genomic excision using an orthotopic primary tumor transplant system we developed (Figure (Figure3A3A and ref. 25). Importantly these tumors can be serially transplanted (Supplemental Figure 3A) and grow with rapid kinetics (Supplemental Figure 3, B and C). This system therefore allows us to generate multiple tumors with the same genetic alterations and endogenous tumor stroma, all growing in vivo at the same time and at the same rate in multiple mice. We can thus conduct carefully controlled replicate in vivo experiments. Notably, these tumors share the same histological features and p63 expression as the primary tumor (Figure (Figure3A).3A). Similar to autochthonous tumors (Figure (Figure2D),2D), only p63L/L K14-CreER tumors and not p63L/L tumors had reduced growth following tamoxifen treatment (Supplemental Figure 3, B and C). We then collected whole tumors at multiple defined intervals following treatment, prepared RNA, and performed global gene expression profiling using Illumina MouseWG-6 BeadChip arrays. Results were analyzed for significant differences between p63L/L K14-CreER tumors treated with vehicle (hereafter p63-intact) and tamoxifen (hereafter p63-ablated), revealing 834 transcripts that had a greater than 1.5-fold change and a P value of less than 0.05 following p63 excision.
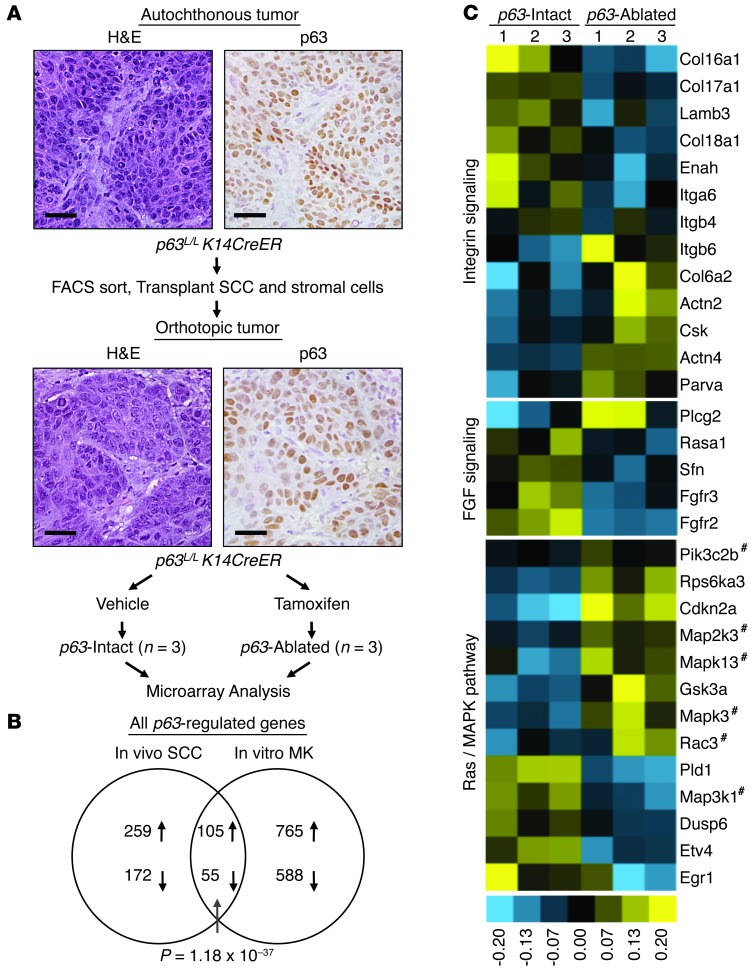
(A) Schematic of orthotopic tumor generation for microarray analysis. Representative histology (H&E) and p63 IHC staining of a primary tumor and its derivative orthotopic tumor are shown. Scale bars: 25 μm. (B) Overlap between p63-regulated genes identified in murine SCC tumors and previously identified p63-regulated genes in cultured MK (81). P value for overlap assessed by hypergeometric test. (C) Heat map of integrin signaling, FGF signaling, and RAS/MAPK signaling genes identified by DAVID analysis (39) as differentially expressed between p63-intact and p63-ablated tumors. The p63L/L K14-CreER consists of vehicle-treated p63-ablated (n = 3), and p63-ablated group consists of tamoxifen-treated p63L/L K14-CreER (n = 3). Hatch marks indicate genes shared by all 3 pathways.
The physiologic relevance of this p63-ablated versus p63-intact gene expression data set is supported by the significant overlap we observed between genes changed following p63 excision in our SCC tumors (Figure (Figure3B)3B) and those regulated in normal murine keratinocytes (MK) following p63-directed siRNA (38). We then used the Database for Annotation, Visualization, and Integrated Discovery (DAVID) (39) tool to carry out pathway analysis on these genes. Indeed, Gene Ontogeny (GO) analysis revealed “epithelium development” and “epithelial cell differentiation” as 2 of the most significant GO terms for both SCC and MK gene sets and for the set of overlapping genes (Table (Table1).1). This finding suggests that deregulation of its normal developmental programs may contribute to p63-dependent tumor maintenance. Consistent with this possibility, pathway analysis identified genes involved in integrin signaling (e.g., Col6α2, Col17α1, Lamβ3), an important p63-regulated developmental program (9, 15, 40), as enriched following p63 excision in SCC (Table (Table11 and Figure Figure3C).3C). This analysis also uncovered a functional signature associated with Ras/MAPK signaling, involving genes including Dusp6, Egr1, and Etv4, and Pld1 (Table (Table11 and Figure Figure3C).3C). Notably, this transcriptional program has not been previously ascribed to p63, but may contribute to p63 function in SCC, given that each of these genes has functional links to RAS/MAPK pathway activation in cancer (41, 42).
Table 1
p63-regulated pathways in murine SCC assessed by DAVID
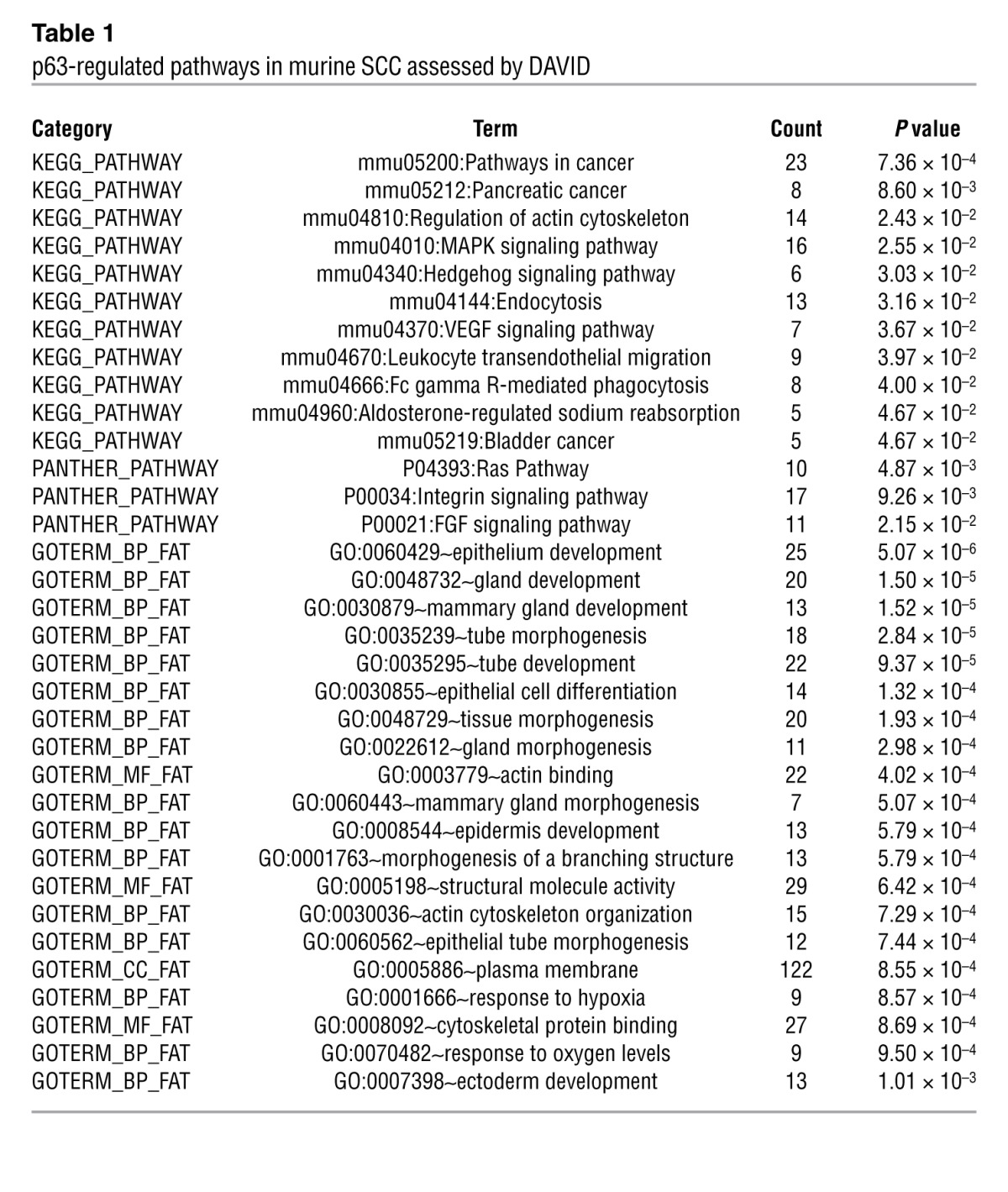
A third p63-dependent transcriptional program emerging from this analysis involved genes associated with FGF signaling, including Fgfr2 and Fgfr3 (Table (Table11 and Figure Figure3C).3C). FGF signaling seemed to be a particularly noteworthy and potentially important pathway controlled by p63 in SCC for several reasons. First, this signaling pathway and these factors in particular are likely to be relevant in p63-expressing cells, as Fgfr2 IIIb–/– mice bear striking similarities to p63–/– mice, including dramatic failure of limb and skin development (14, 43). Indeed, both FGFR2 and FGFR3 expression have been linked to p63 in a developmental context (40, 44). Second, we discovered through analysis of public data sets that FGFR2 is highly significantly overexpressed together with p63 in human SCC tumors versus normal epithelia (Supplemental Figure 3, D and E). Third, although neither FGFR2 nor FGFR3 has previously been associated with p63 in human cancer, both genes encode bona fide oncogenes and are subject to activating somatic alterations in human tumors (45, 46). Finally, there is significant overlap among all 3 of the p63-regulated gene sets we identified (integrin signaling, RAS/MAPK activation, FGF signaling), suggesting that these programs may converge on a common program that is essential for tumor maintenance (Figure (Figure33C).
Direct p63-mediated regulation and requirement for FGFR2 signaling in SCC.
We next validated the microarray results, using quantitative RT-PCR (QRT-PCR) to confirm reduced mRNA levels of Fgfr2/3, the adhesion- and RAS/MAPK-associated genes in p63-ablated tumors compared with p63-intact tumors (Figure (Figure4A).4A). In contrast, we saw no significant changes in control genes, including Fgfr1, Fgfr4, or Egfr, following inactivation of p63 (Figure (Figure4A).4A). Reduction in Col17α1, Pld1, Fgfr2, and Fgfr3 mRNA was further confirmed in primary autochthonous murine SCC tumors (Supplemental Figure 4A), and reduction of the corresponding proteins was confirmed by Western analysis (Figure (Figure4B).4B). We then wished to determine whether the p63-regulated genes were subject to direct transcriptional control by p63 in SCC, since neither Col17α1 nor Pld1 had previously been identified as p63-regulated genes. We performed ChIP using α-p63 antibodies in murine B9 SCC cells (Supplemental Figure 4B) and observed significant p63 binding upstream of the Col17α1 and Fgfr2 transcription start sites and in the first intron of Pld1 (Figure (Figure4C).4C). To confirm the specificity of these findings, we expressed FLAG-epitope–tagged WT ΔNp63α (WT) or a DNA-binding deficient point mutant ΔNp63α (R304W) (47) in B9 cells and performed ChIP using antibodies against the FLAG epitope (Supplemental Figure 4B). Only cells expressing WT ΔNp63α demonstrated significant binding of ΔNp63α to the Pld1 and Fgfr2 loci (Figure (Figure4D).4D). Collectively, these data identify Col17α1, Pld1, and Fgfr2 as direct, p63-activated target genes in SCC.
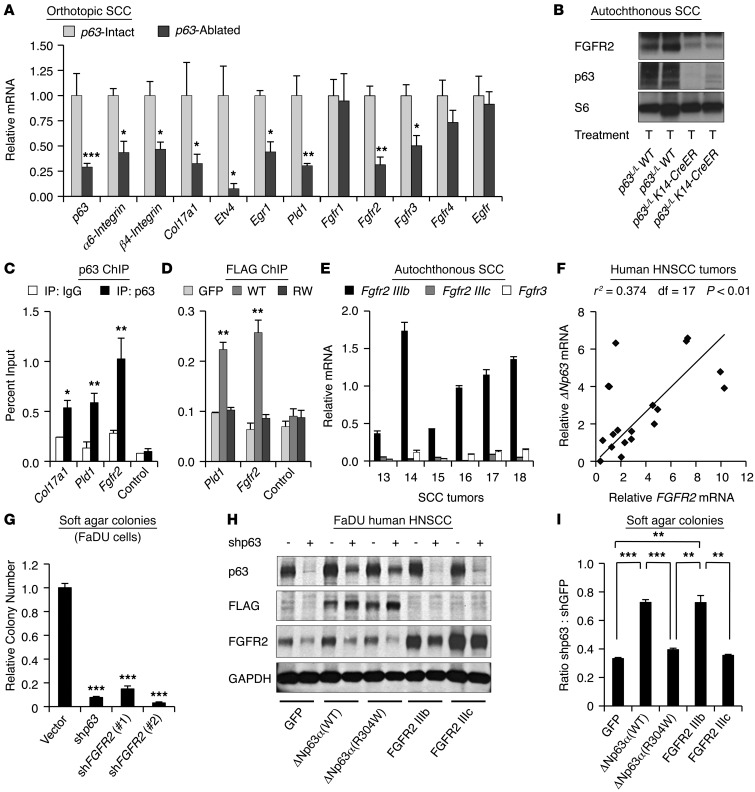
(A) QRT-PCR assessment of mRNA levels of indicated genes in p63-intact (n = 3–5) or p63-excised (n = 3–4) orthotopic tumor samples normalized to β-actin. (B) Western blot of indicated proteins in SCC tumors of indicated genotype. Ribosomal S6 serves as a loading control. T, tamoxifen. (C) ChIP of endogenous p63 at the Col17α1, Pld1, and Fgfr2 loci in murine B9 SCC cells using indicated antibodies. Control designates highly conserved region –2.4 kb upstream of Fgfr2 promoter. (D) ChIP of exogenously expressed, FLAG-epitope tagged WT ΔNp63α or DNA-binding–deficient ΔNp63α (R304W) in B9 cells. GFP-expressing cells serve as a negative control. (E) Relative Fgfr mRNA levels in autochthonous murine tumors, normalized to β-actin. (F) Correlation between ΔNp63 and FGFR2 mRNA levels in human primary HNSCC tumors as assessed by TaqMan QRT-PCR. P value was calculated using Pearson product-moment correlation coefficient. (G) Human FaDU HNSCC cells plated in soft agar 72 hours after infection with p63 or FGFR2-directed shRNAs. (H) Western blot analysis of FaDU HNSCC cells stably expressing indicated proteins 72 hours following infection with p63-directed shRNA or control shRNA. Exogenous ΔNp63α (WT) and ΔNp63α (R304W) contain a FLAG epitope tag and are resistant to p63 shRNA (47). GAPDH serves as a loading control. (I) Colony forming assay of samples in H 10 days after infection with p63-directed or control shRNA. *P < 0.05; **P < 0.01; ***P < 0.001 as assessed by Student’s unpaired t test for all experiments except where indicated. Error bars represent SEM.
The Fgfr2 transcript undergoes alternative splicing, resulting in an Fgfr2 IIIb isoform, preferentially expressed on epithelial cells, and an Fgfr2 IIIc isoform expressed on mesenchymal cells (48). Examination of mRNA levels in autochthonous SCC tumors (Figure (Figure4E)4E) showed that the epithelial-specific Fgfr2 IIIb isoform was expressed at substantially higher levels than Fgfr2 IIIc or Fgfr3, suggesting that Fgfr2 IIIb is the most relevant p63-regulated isoform in SCC. In order to establish the relevance of FGFR2 in human tumors, we first examined the regulation of FGFR2 by p63 in HNSCC cell lines. As was seen in murine tumors, lentiviral-mediated knockdown of p63 resulted in substantially reduced FGFR2 levels in human HNSCC cells (Supplemental Figure 4C). Next, we obtained primary human HNSCC tumor samples and examined the relationship between ΔNp63 and FGFR2 expression. Using QRT-PCR we found that FGFR2 and ΔNp63 mRNA levels were directly and highly statistically significantly correlated in these tumors (Figure (Figure4F),4F), consistent with a direct regulatory relationship.
We then wished to test the functional contribution of the p63-FGFR2 axis in SCC. Significantly, we found that lentiviral knockdown of either p63 or FGFR2 in human HNSCC cells was associated with similar dramatic suppression of anchorage-independent growth (Figure (Figure4G)4G) and colony forming ability (Supplemental Figure 4D). To determine whether p63 is sufficient to induce FGFR2 expression in this setting, we expressed either a WT ΔNp63α isoform, the DNA-binding–deficient mutant ΔNp63α (R304W), or TAp63α (WT) in HaCAT-immortalized human keratinocytes. Only WT ΔNp63α induced significant expression of FGFR2 (Supplemental Figure 4E), suggesting that ΔNp63α alone can activate FGFR2. To directly test the requirement for FGFR2 signaling in p63-dependent tumor maintenance, we performed a genetic rescue experiment in human FaDU HNSCC cells. We stably expressed either a control GFP protein or an shRNA-resistant FLAG-epitope tagged ΔNp63α (WT) or ΔNp63α (R304W) (47), followed by lentiviral-mediated knockdown of endogenous p63 (Figure (Figure4H).4H). Expression of ΔNp63α (WT) was able to significantly rescue the decreased soft agar colony formation following p63 knockdown, while the DNA-binding–deficient ΔNp63α (R304W) was not (Figure (Figure4I).4I). Most remarkably, overexpression of the IIIb isoform of FGFR2, but not the IIIc isoform, was able to rescue colony formation to the same extent as ΔNp63α itself (Figure (Figure4I).4I). Collectively, these data suggest that FGFR2 IIIb is a key activated target of p63 in SCC, and they demonstrate the essential role of p63-dependent FGFR2 IIIb signaling in both human and murine SCC.
A paracrine FGF7-FGFR2 signaling axis is upregulated in autochthonous tumors.
We noted substantially increased expression of both p63 and its target FGFR2 in murine (Figure (Figure5A)5A) and human (Supplemental Figure 5A) autochthonous tumors relative to multiple SCC cell lines, leading us to hypothesize that the p63-dependent FGFR2 signaling axis might be more highly activated in tumors as compared with cultured cells. Indeed, immunofluorescent (IF) staining of primary tumors, using K14 as an epithelial cell marker and vimentin as a stromal marker, demonstrated dramatic membrane FGFR2 staining and nuclear p63 staining selectively within malignant epithelial cells (Figure (Figure5B).5B). The ligands for the epithelial-expressed FGFR2 IIIb include FGF1 and FGF7 and are typically produced by mesenchymal cells during normal development, resulting in paracrine signaling (49, 50). Consistent with the hypothesis that stromal-derived ligands signal in a paracrine fashion to FGFR2 IIIb in autochthonous SCC, IF showed that tumor cells were surrounded by a dense vimentin-positive stroma (Figure (Figure5B).5B). The tumor stroma appeared to comprise activated fibroblasts, as evidenced by SMA staining (Supplemental Figure 5B). Furthermore, we documented highly elevated expression of multiple FGFR2 ligands, including FGF1, FGF7, and FGF10 in whole tumors compared with cultured cell lines, with FGF1 and FGF7 being the most highly expressed (Figure (Figure5C5C and Supplemental Figure 5C). We then examined expression of Fgf7, the most specific of the ligands for FGFR2 IIIb, and p63 in FACS-isolated cell populations from autochthonous tumors, comparing CD49f (α6-integrin) positive (tumor) versus negative (stromal) cells following exclusion of hematopoietic and endothelial cells. We observed robust expression of Fgf7 in stromal cells and virtually no expression in tumor cells, while the reverse was true for p63 (Figure (Figure5D).5D). Thus, FGFR2 and its ligand FGF7 are overexpressed in different cell populations in endogenous SCC, suggesting activated paracrine signaling in vivo.
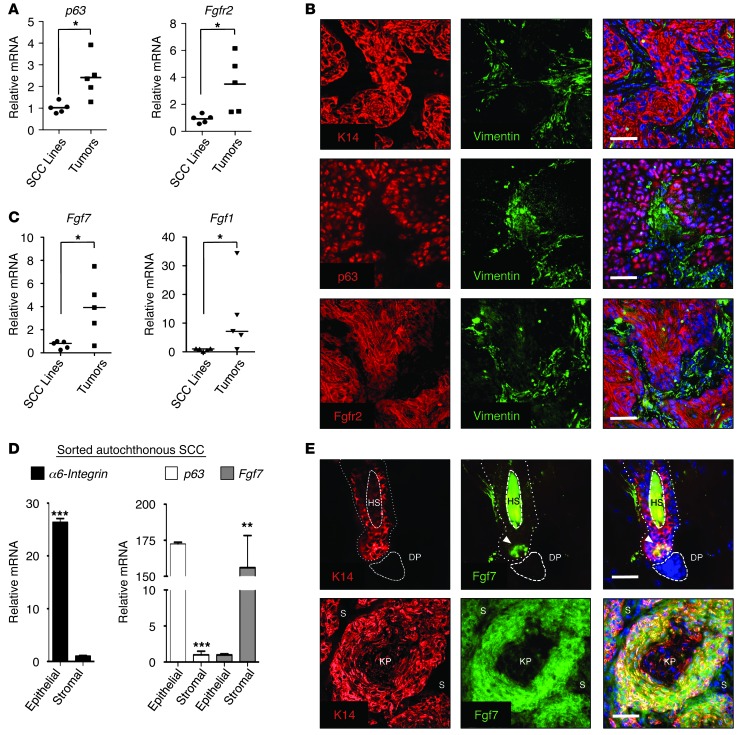
(A) Upregulation of p63 (left) and FGFR2 (right) in SCC tumors versus cell lines. Relative mRNA expression was determined by QRT-PCR in murine SCC cell lines (n = 5) and tumors (n = 5). Bar indicates mean value. *P < 0.05 as assessed by Student’s unpaired t test. (B) IF staining of autochthonous murine SCC tumors, showing colocalization of K14, nuclear p63, and membrane-associated FGFR2. Vimentin (green) identifies stromal cells. Hoechst dye (blue) identifies nuclei. Scale bars: 25 μm. (C) Upregulation of FGFR2 ligands in SCC tumors. Relative mRNA expression of Fgf7 (left) and Fgf1 (right) in murine SCC cell lines (n = 5) and tumors (n = 5). Bars indicate mean value. *P < 0.05 as assessed by Student’s unpaired t test. (D) Stroma-specific expression of FGF7 in autochthonous SCC. Primary tumors were disaggregated, then separated by FACS into epithelial CD49fhi (α-6 integrin) and stromal (CD49flo) populations following elimination of hematopoietic (CD45+) and endothelial (CD31+) cells prior to RNA analysis. Note epithelial-specific expression of p63. Error bars indicate SEM. **P < 0.01, ***P < 0.001. (E) Spatially restricted FGF7 engagement in normal epithelia is abolished in SCC. IF staining of murine hair follicle in telogen (top) and autochthonous murine SCC tumors (bottom) for FGF7. K14 staining identifies epithelial cells. Note highly restricted expression in hair follicle (arrow) compared with tumor. HS, hair shaft; DP, dermal papilla; KP, keratin pearl; S, stromal cells. Scale bars: 25 μm.
In the normal adult epithelium (e.g., the regenerating hair follicle), FGF7 is produced by a select group of mesenchymal cells in the dermal papilla, resulting in restricted receptor engagement and limited signaling within the hair germ at the base of the follicle (51). We confirmed this finding by IF staining of murine skin, demonstrating that only a small fraction of epithelial cells at the hair follicle base are exposed to and uptake FGF7, whereas the majority of follicle epithelial cells (marked by K14) entirely lack FGF7 staining (Figure (Figure5E5E and Supplemental Figure 5D). In dramatic contrast, we observed that the ligand-producing stroma surrounding the SCC cells in autochthonous tumors resulted in FGF7 engagement at high levels in essentially all tumor cells (Figure (Figure5E).5E). Of note, membrane FGFR2 staining appeared less intense in tumor cells adjacent to FGF7-releasing stroma, in keeping with receptor internalization following ligand engagement (Supplemental Figure 5E). Together, these findings uncover a prominent role for disorganized and deregulated paracrine signaling via p63-induced FGFR2 in the pathogenesis of endogenous SCC.
Therapeutic inhibition of FGFR signaling induces apoptosis in autochthonous SCC.
In order to directly validate our model of hyperactivated ligand-induced FGFR2 signaling in SCC, we tested the effect of pharmacologic FGFR2 inhibition in these tumors. Notably, as an established oncogenic tumor driver, FGFR2 represents a potentially attractive therapeutic target in human cancer (52). We obtained an orally bioavailable FGFR inhibitor, AZD4547, which exhibits high specificity for FGFR1, -2, and -3 and low nanomolar potency for FGFR2 inhibition (53). We first examined FGFR2 signaling in FaDU HNSCC cells, which exhibit FGFR2 levels approaching those we observed in vivo (Supplemental Figure 6A). Cells were serum starved, then stimulated with FGF7 in the presence of increasing doses of AZD4547. Like many growth factor receptor tyrosine kinases, FGFR2 is reported to signal through multiple downstream pathways, including MAPK, PI3K, and PLCγ (52). Low nanomolar doses of AZD4547 potently abolished FGF7-stimulated signaling to AKT and ERK, but were less effective at inhibiting signaling through other reported downstream pathways including p38 and STAT3 (Figure (Figure6A6A and Supplemental Figure 6B). Inhibition by AZD4547 was specific for FGFR2-dependent signaling, as this compound did not block EGF-dependent signaling (Supplemental Figure 6B) and had similar effects on both human and murine SCC cell lines (Supplemental Figure 6C). These data demonstrate that AZD4547 potently and specifically blocks ligand-mediated FGFR2 signaling in SCC cells.
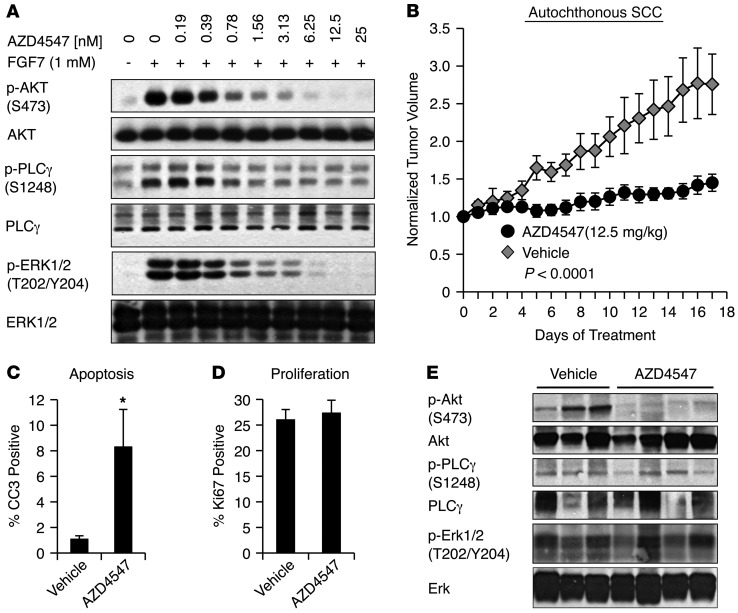
(A) Inhibition of ligand-dependent FGFR2 signaling with AZD4547 extinguishes AKT, ERK1/2, and PLCγ phosphorylation. FaDU cells were serum starved and pretreated for 1 hour with the indicated doses of AZD4547, followed by a 15-minute stimulation with FGF7 ligand. (B) AZD4547 treatment blocks progression of autochthonous SCC. Mice bearing DMBA-induced SCC tumors received daily oral AZD4547 (12.5 mg/kg, n = 7) or vehicle control (n = 6). Error bars represent SEM. P < 0.0001 as assessed by multiple-measures ANOVA. (C) AZD4547 treatment induces autochthonous tumor apoptosis in vivo. Tumors harvested after 17 days treatment as in B were assessed for cleaved caspase 3 staining by IHC. AZD4547 (n = 7), vehicle control (n = 5). *P < 0.05 as determined by Mann-Whitney test. (D) Quantification of Ki67-positive cells in tumors as in C. AZD4547 (n = 7), vehicle control (n = 5). P value not significant as determined by Student’s unpaired t test. (E) AZD4547 treatment inhibits PI3K signaling downstream of ligand-activated FGFR2 in SCC. Western blot analysis of autochthonous tumor lysates following 17 days of daily oral treatment with vehicle control or AZD4547 (12.5 mg/kg).
The rationale for therapeutic targeting of FGFR2 has focused largely on relatively uncommon tumors that exhibit activating somatic mutations resulting in ligand-independent signaling (45). Our findings suggest that FGFR2 upregulation and activated paracrine signaling in SCCs may confer pathway dependence in the absence of receptor mutation. To test this hypothesis, we established a cohort of mice bearing autochthonous DMBA-induced SCCs and we treated these mice with either 12.5 mg/kg/d AZD4547 or control vehicle. This dose was well tolerated by mice, which exhibited no overt physiologic effects compared with vehicle-treated mice. Remarkably, however, AZD4547 treatment resulted in a virtual complete arrest of tumor progression, which was apparent as early as 5 days following treatment initiation and which was associated with a dramatic increase in progress-free survival of AZD4547-treated mice (Figure (Figure6B6B and Supplemental Figure 6D). Analysis of tumors following AZD4547 treatment showed an increase in apoptosis as assessed by cleaved caspase 3 staining, while there was no discernible effect on proliferation (Figure (Figure6,6, C and D). We then determined which of the potential FGFR2-activated signaling pathways was affected in vivo by AZD4547 treatment of autochthonous tumors. We observed suppression of AKT phosphorylation following AZD4547 treatment, but little or no change in phosphorylation of ERK1/2 and PLCγ (Figure (Figure6E).6E). The dramatic suppression of Akt phosphorylation in all AZD4547-treated compared with vehicle-treated tumors is keeping with the established role of the PI3K/AKT pathway in cell survival and supports an essential role for p63-induced FGFR2 in maintaining survival of autochthonous SCCs through this pathway in vivo.
Discussion
Although it has been known for several years that p63 is overexpressed in a large proportion of primary SCCs (18, 32, 54), p63 is also a marker of basal epithelial cells whose expression might simply reflect the lineage of origin for these tumors. Here, we use a sophisticated genetic model to show definitively that autochthonous SCCs are dependent upon high levels of endogenous p63. Thus, deletion of p63 induces rapid tumor regression without any appreciable effects on normal p63-expressing epithelia (Figure (Figure2).2). This finding is in stark contrast to the long latency required for phenotypic changes seen in normal tissues following in vivo excision of p63 in the epithelium (34). These results define SCCs as functionally “p63-addicted” and furthermore suggest that therapies targeting the p63 pathway may benefit from a favorable therapeutic window. Indeed, the lack of toxicity we observe upon inhibiting the p63-FGFR2 axis using AZ4547, despite its dramatic effect on tumors, underscores this point (Figure (Figure66).
While our model involved deletion of all p63 isoforms, our findings support a predominant role for the ΔNp63α isoform in the observed effects. ΔNp63α is by far the major isoform expressed in these tumors, and it binds directly to a conserved regulatory region to induce expression of FGFR2, which we show to be a central mediator of p63-dependent tumor survival (Figure (Figure4).4). In contrast, TAp63α is unable to modulate expression of FGFR2 (Supplemental Figure 3). The particular contribution of ΔNp63α to squamous carcinogenesis is also supported by data presented here (Figure (Figure4)4) and previously (10, 47) showing that ΔNp63α expression can rescue cell death induced by endogenous p63 knockdown and that this isoform can cooperate with oncogenic Ras to promote squamous tumorigenesis (55).
Genome-wide in vivo gene-expression profiling of tumors following p63 excision revealed multiple p63-dependent pathways with a plausible role in tumor maintenance, including integrin and MAPK signaling, as well as direct p63-regulated genes functioning within these pathways, including Col17α1 and Pld1, respectively (Figure (Figure33).
The most remarkable finding in our analysis, however, is a prominent growth factor signaling program converging on FGFR2, which we show to be a key and direct transcriptional target of p63 in these tumors. The relevance of the p63-FGFR2 axis in human SCC is further supported by the strong correlation we observe between p63 and FGFR2 expression in primary human HNSCC tumors (Figure (Figure4).4). We provide direct evidence arguing that FGFR2 mediates effects of p63 in SCC, showing that ectopic FGFR2 expression is sufficient to rescue effects of p63 knockdown and that FGFR2 rescues as effectively as ectopic expression of ΔNp63α itself (Figure (Figure4).4). Additionally, we demonstrate that both FGFR2 and its ligands are produced at high levels in primary human and mouse tumors. By sorting such tumors, we demonstrated that in vivo the FGFR2 ligand FGF7 is produced exclusively from the stroma, yielding a paracrine signaling axis that is not recapitulated in cultured tumor cells (56). Indeed, a subset of SCC cell lines adapted to grow in the absence of stroma is known to activate autocrine FGFR signaling, consistent with our finding that this signaling is an important survival mechanism in these cancers (57). Most notably, whereas the normal developmental pattern of FGFR2 signaling involves limited and spatially restricted ligand exposure (51), we show that in endogenous SCC, the FGFR2-expressing tumor cells are surrounded by ligand-producing stroma and as a result exhibit high levels of receptor-associated ligand expression (Figure (Figure55).
Widespread activation of p63-induced FGFR2 by overexpressed stromal ligand, together with other stroma-associated effects (58), may well explain the poor prognosis associated with an abundant stromal component in human SCCs (21, 22). Other recent studies provide further evidence supporting our finding that stromal-derived growth factors and FGF7 in particular contribute to SCC pathogenesis. Deregulated signaling in stroma cells within the dermis induces a field cancerization effect associated with increased secretion of growth factors including FGF7 from the stroma (59), and transgenic expression of FGF7 itself results in transformation of the epithelium (60, 61). We have also found that other FGFR2 ligands, including FGF1 and FGF10, are elevated in the tumor microenvironment, suggesting that multiple ligands could potentially contribute to tumor progression. Finally, in addition to quantitative changes in FGFR2 and FGF expression, deregulated receptor spatial orientation and stromal juxtaposition within depolarized, disordered epithelia are likely to play a role in heightened signaling through this pathway (62).
While EGFR has long been considered the most important therapeutic target in SCC, the relatively modest preclinical (63, 64) and clinical activity of EGFR inhibitors against most SCCs, including HSNCC, might argue for the presence of an additional or alternative RTK-dependent pathway driving these tumors (65–67). Much of the attention in therapeutic targeting of FGFR-dependent pathways in SCC and other cancer contexts has focused on the relatively uncommon tumors exhibiting either amplification or somatic mutation of these receptors (68, 69). Our study suggests, however, that high levels of p63 itself are sufficient to drive FGFR2 expression, signaling, and tumor cell dependence. In this regard, it is notable that the inhibition of tumor growth we observed following treatment of mice with AZD4547 is comparable to that observed using the same dose of this compound in a xenograft model that employed an FGFR-amplified cell line (53). Our data also indicate that a major impact of AZD4547 is to potently inhibit PI3K/AKT signaling. Consequently, this compound induced apoptosis when used to treat autochthonous tumors (Figure (Figure6).6). This observation may have may have direct relevance regarding which patient populations may benefit from treatment with this drug. A small but significant fraction of HNSCCs harbor either mutations in the catalytic PI3K subunit PI3KCA (2, 3) or loss of PTEN (70) and thus may exhibit resistance to this compound due to constitutive PI3K activity. Given that AZD4547 and other potent FGFR inhibitors are now entering the clinic, it is important to define the specific clinical context for its application. More broadly, our identification of a p63-driven pathway for growth factor signaling highlights the potential for therapeutic targeting of oncogenic transcriptional programs, even while targeting the transcription factors themselves remains a challenge.
Methods
Mice
To induce epidermal SCC tumors, 6-week-old mice were shaved and 25 mg DMBA (Sigma-Aldrich) dissolved in acetone was applied to the skin once per week. For oral SCC tumors, DMBA was applied to the inside of the oral cavity weekly. Tumors were measured daily, and tumor volumes were calculated using the following formula: tumor volume (mm3) = 4/3π × length/2 × width/2. In order to excise p63, mice were treated once per day with 100 mg/kg tamoxifen (Sigma-Aldrich) dissolved in sunflower seed oil (Sigma-Aldrich) by intraperitoneal injection for 5 days. Orthotopic transplants were performed as described previously (25). For in vivo AZD4547 (AstraZeneca Pharmaceuticals LP) treatment, tumor-bearing mice were treated daily by oral gavage with 12.5 mg/kg AZD4547 dissolved in 1% Tween-80. Generation of p53–/–, p63L/L mice, and K14-CreER mice has been previously described (30, 35, 37). Analysis of genomic p63 excision by PCR has been previously described (34).
AZD4547 treatment of cell lines
Inhibition of signaling by AZD4547 was performed by plating cells in 0.1% FBS for 22 hours, then serum-free medium for 1 hour. Freshly made AZD4547 was dissolved in DMSO and added to the medium for 1 additional hour. Then, 1 μM recombinant human FGF7 (Sigma-Aldrich) was added to cells for 15 minutes followed by lysis with RIPA buffer. IC50 values of SCC cells treated with AZD4547 were determined 72 hours after plating of 500 cells/well in medium containing 10% FBS using the CellTiter-Glo Luminescent Cell Viability Assay (Promega) according to the manufacturer’s instructions.
Human tumor analysis
Primary HNSCC tumors used in this study have been previously described (25). Following pathological review, total RNA was extracted from macrodissected tumor-enriched portions. Quantification of mRNA from tumor samples was performed using the TaqMan (Applied Biosystems) primer-probe sets according to the manufacturer’s protocol. β-Actin mRNA expression in tumors served as a normalization control. Primer and probe sets are described in Supplemental Table 1.
IHC and IF
Assistance in processing of murine tumor samples was provided by the Dana-Farber/Harvard Cancer Center Specialized Histopathology Core Facility. Five-micron sections were cut from formalin-fixed, paraffin-embedded tumors and stained using standard protocols. Staining for cleaved caspase 3 was performed using the SignalStain Cleaved Caspase-3 (Asp175) IHC detection kit (Cell Signaling) according to the manufacturer’s instructions. For IF staining, murine tumors were snap-frozen in HistoPrep (Fisher Scientific), and 5-μM sections were cut. Samples were fixed in 4% paraformaldehyde and permeabilized with 0.1% Triton X-100 or fixed and permeabilized with methanol (K14; SMA). Nuclei were visualized by staining with Hoechst 33342 dye (Invitrogen). Stains with the SMA antibody used the Mouse on Mouse (M.O.M.) Basic Kit (Vector Laboratories) according to the manufacturer’s instructions.
Cell lines
Murine cell lines B9 (26) and SCC/80 (71) were gifts of Nabeel Bardeesy (Massachusetts General Hospital). Murine cell lines mSCC (25), 392-1, and 1459A were generated from DMBA-induced tumors and established in R&G medium (71). Following establishment of a cell line, cells were switched to RPMI medium. JHU-029 (18) cells were a gift of David Sidranski (Johns Hopkins University, Baltimore, Maryland, USA). FaDU (72) and HaCAT (73) cells were a gift of James Rocco (Massachusetts General Hospital). HO1N1 (74), BICR-78 (75), CAL-33 (76), KYSE-30, and KYSE-150 (77) were a gift of S. Michael Rothenberg (Massachusetts General Hospital). Cells were maintained at 37°C in 5% CO2 in either RPMI or DMEM (HaCAT) supplemented with 10% FBS, penicillin, and streptomycin. All human cell lines were maintained by the MGH Center for Molecular Therapeutics cell bank and underwent high-density SNP typing to confirm their identity.
Real-time PCR
Isolated tumor cells from autochthonous tumors were generated by disaggregation of primary tumors with collagenase, and cells were FACS sorted for viable CD31neg, CD45neg, and CD49fhi cells. For all experiments, RNA was extracted from whole tumors using QIAGEN RNeasy Fibrous Tissue Mini Kit according to the manufacturer’s instructions. QRT-PCR analysis was performed using IQ SYBR Green Supermix (Bio-Rad) according to the manufacturer’s instructions. Primers used for QRT-PCR are listed in Supplemental Table 1.
Western blotting and ChIP
Protein was extracted from tumors by incubation for 1 hour at 4°C in RIPA buffer (10 mM Tris-HCl, pH 7.5, 150 mM NaCl, 1 mM EDTA, 1% [w/v] sodium deoxycholate, 0.1% [w/v] SDS, 1% [v/v] NP-40, proteinase inhibitor cocktail, phosphatase inhibitor cocktail) following physical grinding. ChIP was performed as previously described (47). Data represent 3 to 5 independently immunoprecipitated samples per antibody. Primer sets used can be found in Supplemental Table 1.
Lentiviral, retroviral production, colony forming assays, and mRNA QRT-PCR
Production of virus and mRNA analysis were performed as described previously (18). Soft agar colony-forming assays were performed by plating 3 × 104 cells per well in a 6-well plate in 0.4% agarose 72 hours following infection with lentiviral shRNA constructs. Rescue experiments in Figure Figure4,4, H and I, were supplemented with 1 μM FGF7. Data represent average of 3 random fields in 3 independently seeded wells, performed twice with similar results.
Plasmids
Retroviral pBABE (puro)-FGFR2 IIIb and pBABE (puro)-FGFR2 IIIc plasmids (45) were a gift of Heidi Greulich (Broad Institute, Cambridge, Massachusetts, USA). pLPC-ΔNp63α (WT)-FLAG, ΔNp63α (R304W)-FLAG, and pLPC-TAp63α (WT)-FLAG plasmids have been previously described (47).
Microarray
RNA was extracted from whole orthotopic tumors using QIAGEN RNeasy Fibrous Tissue Mini Kit according to the manufacturer’s instructions. Synthesis of cDNA, hybridization, and reading of signal intensity were performed by the Partners HealthCare Center for Personalized Genetic Medicine. RNA quality was assessed on a 2100 Bioanalyzer (Agilent), and total RNA was hybridized to Illumina MouseWG-6 v2 BeadChip arrays. Chips were scanned with Illumina BeadArray Reader to measure the signal intensity by the labeled target. Data were processed using the R statistical software environment, version 2.12.0 (78). Data were background corrected, normalized, and underwent variance stabilizing transformations using the Lumi package (79). Differential gene expression was determined based on a moderated t test using the Limma package (80). The entire set of unprocessed raw data is available (GEO GSE45929).
Quantification of proliferation and apoptosis following p63 excision
For quantification of cleaved caspase 3 staining in Figure Figure2F,2F, the day-6 control group consisted of vehicle-treated p633L/L K14-CreER (n = 3) and tamoxifen-treated p63L/L (n = 2) mice compared with tamoxifen-treated p63L/L K14-CreER mice (n = 5). The day 8 control group consisted of vehicle-treated p63L/L K14-CreER (n = 2) and tamoxifen-treated p63L/L mice (n = 2) compared with tamoxifen-treated p63L/L K14-CreER mice (n = 4). For quantification of Ki67 staining in Figure Figure2G,2G, the day-6 control group consisted of vehicle-treated p63L/L K14-CreER (n = 2) and tamoxifen-treated p63L/L (n = 1) compared with tamoxifen-treated p63L/L K14-CreER mice (n = 4). The day-8 control group consisted of vehicle-treated p63L/L K14-CreER (n = 2) and tamoxifen-treated p63L/L mice (n = 1) compared with tamoxifen-treated p63L/L K14-CreER mice (n = 4).
Antibodies
Western blotting.
PARP (Cell Signaling), GAPDH (FL335; Santa Cruz Biotechnology Inc.), p53 (DO-1; Santa Cruz Biotechnology Inc.), p63 (4A4; Sigma-Aldrich), BCL-2 (C2; Santa Cruz Biotechnology Inc.), Erk1/2 (K-23; Santa Cruz Biotechnology Inc.), FLAG (M2; Sigma-Aldrich), FGFR2 (Abcam), AKT (Cell Signaling), AKT-pSer473 (Cell Signaling), ERK1/2-pThr202/Tyr204 (20G11; Cell Signaling), PLCγ (D9H10; Cell Signaling), PLCγ-pSer1248 (Cell Signaling), p38-pThr180/Tyr182 (3D7; Cell Signaling), p38 (5F11; Cell Signaling), STAT3-pTyr705 (3E2; Cell Signaling), STAT3 (124H6; Cell Signaling), EGFR (1005, Santa Cruz Biotechnology Inc.), and EGFR-pTyr845 (Cell Signaling) were used.
ChIP.
p63α (H-129; Santa Cruz Biotechnology Inc.) and FLAG epitope (M2; Sigma-Aldrich) were used.
Immunohistochemistry.
Ki67 (NCL-Ki67p; Novacastra), K14 (Covance), and p63 (4A4; Sigma-Aldrich) were used.
IF.
FGF7 (H-73 or C-19; Santa Cruz Biotechnology Inc.), FGFR2 (ab52256; Abcam), p63 (H-129; Santa Cruz Biotechnology Inc.), Vimentin (S-20; Santa Cruz Biotechnology Inc.), K14 (Covance), and α-SMA (ab7817; Abcam) were used.
FACS.
CD45 (biotin-conjugated; eBioscience), CD31 (biotin-conjugated;, eBioscience), and CD49f (PE-conjugated; BD Biosciences — Pharmingen) were used.
Statistics
All P values were determined using the 2-tailed Student’s unpaired t test with the exceptions noted here. The values in Figure Figure2F2F and Figure Figure6C6C had non-Gaussian distributions and were analyzed using the Mann-Whitney test. For in vivo experiments in Figure Figure2D2D and Figure Figure6B,6B, changes in tumor volume were analyzed using multiple-measures ANOVA. For Figure Figure4F,4F, the Pearson product moment correlation coefficient (R2) was calculated, and the P value was determined from a probability table. P values for GO, KEGG, and Panther Pathways as analyzed using DAVID were determined using Fisher’s exact test. P < 0.05 was considered significant for all experiments.
Study approval
The Massachusetts General Hospital IRB approved use of tissues and clinical data for this study. All human tissue studies used exclusively discarded material collected in the course of routine clinical care, for which our IRB determined that signed informed consent was not required. Housing and treatment for all animals was approved by the Subcommittee on Research Animal Care at the Massachusetts General Hospital.
Acknowledgments
We wish to thank Nabeel Bardeesy, James Rocco, William Michaud, Mike Dennis, and Zach Nash for advice, reagents, and technical support; and Nabeel Bardeesy, Andrea McClatchey, and members of the Ellisen laboratory for comments on the manuscript. The Dana-Farber/Harvard Cancer Center Specialized Histopathology Core Facility assisted in processing and staining of murine tumors, and the Harvard Stem Cell Institute/Center for Regenerative Medicine (HSCI/CRM) Flow Cytometry Core Facility assisted with tumor sample sorting. AZD4547 used in this study was provided by AstraZeneca Pharmaceuticals LP. This work was supported by NIDCR R01 DE015945 (to L.W. Ellisen), by NCI K99 CA 157730 (to M.R. Ramsey), and by the American Cancer Society/Mass Biotech Council Cancer Research Challenge-AstraZeneca Pharmaceuticals LP Fellowship PF-09–100-01 MGO (to M.R. Ramsey).
Footnotes
Conflict of interest: The authors have declared that no conflict of interest exists.
Citation for this article: J Clin Invest. 2013;123(8):3525–3538. 10.1172/JCI68899.
Catherine Wilson’s present address is: Discovery Oncology, Genentech, Inc., South San Francisco, California, USA.
Benjamin Ory’s present address is: INSERM, UMR 957, Université de Nantes, Nantes, France.
References
Articles from The Journal of Clinical Investigation are provided here courtesy of American Society for Clinical Investigation
Full text links
Read article at publisher's site: https://doi.org/10.1172/jci68899
Read article for free, from open access legal sources, via Unpaywall:
http://www.jci.org/articles/view/68899/files/pdf
Citations & impact
Impact metrics
Citations of article over time
Alternative metrics
Article citations
Interaction between MED12 and ΔNp63 activates basal identity in pancreatic ductal adenocarcinoma.
Nat Genet, 56(7):1377-1385, 17 Jun 2024
Cited by: 0 articles | PMID: 38886586
Hypoxia-activated XBP1s recruits HDAC2-EZH2 to engage epigenetic suppression of ΔNp63α expression and promote breast cancer metastasis independent of HIF1α.
Cell Death Differ, 31(4):447-459, 27 Feb 2024
Cited by: 0 articles | PMID: 38413797
Genome-wide p63-Target Gene Analyses Reveal TAp63/NRF2-Dependent Oxidative Stress Responses.
Cancer Res Commun, 4(2):264-278, 01 Feb 2024
Cited by: 1 article | PMID: 38165157 | PMCID: PMC10832605
A novel ΔNp63-dependent immune mechanism improves prognosis of HPV-related head and neck cancer.
Front Immunol, 14:1264093, 25 Oct 2023
Cited by: 2 articles | PMID: 38022675 | PMCID: PMC10630910
Oncogenic Ras and ΔNp63α cooperate to recruit immunosuppressive polymorphonuclear myeloid-derived suppressor cells in a mouse model of squamous cancer pathogenesis.
Front Immunol, 14:1200970, 10 Aug 2023
Cited by: 0 articles | PMID: 37638000 | PMCID: PMC10449460
Go to all (67) article citations
Data
Data behind the article
This data has been text mined from the article, or deposited into data resources.
BioStudies: supplemental material and supporting data
GEO - Gene Expression Omnibus
- (1 citation) GEO - GSE45929
Similar Articles
To arrive at the top five similar articles we use a word-weighted algorithm to compare words from the Title and Abstract of each citation.
Antitumor Effects and Mechanisms of AZD4547 on FGFR2-Deregulated Endometrial Cancer Cells.
Mol Cancer Ther, 14(10):2292-2302, 20 Aug 2015
Cited by: 22 articles | PMID: 26294741
FGF18-FGFR2 signaling triggers the activation of c-Jun-YAP1 axis to promote carcinogenesis in a subgroup of gastric cancer patients and indicates translational potential.
Oncogene, 39(43):6647-6663, 15 Sep 2020
Cited by: 29 articles | PMID: 32934314 | PMCID: PMC7581496
Fibroblast growth factor receptor promotes progression of cutaneous squamous cell carcinoma.
Mol Carcinog, 58(10):1715-1725, 29 Jun 2019
Cited by: 3 articles | PMID: 31254372 | PMCID: PMC6721978
Molecular Mechanisms of p63-Mediated Squamous Cancer Pathogenesis.
Int J Mol Sci, 20(14):E3590, 23 Jul 2019
Cited by: 47 articles | PMID: 31340447 | PMCID: PMC6678256
Review Free full text in Europe PMC
Funding
Funders who supported this work.
NCI NIH HHS (4)
Grant ID: R00 CA157730
Grant ID: K99 CA157730
Grant ID: P30 CA045508
Grant ID: K99 CA 157730
NIDCR NIH HHS (1)
Grant ID: R01 DE015945





