Abstract
Purpose
The clinical relevancy of the 7-subtype classification of triple-negative breast cancer (TNBC) reported by Lehmann and colleagues is unknown. We investigated the clinical relevancy of TNBC heterogeneity by determining pathologic complete response (pCR) rates after neoadjuvant chemotherapy, based on TNBC subtypes.Experimental design
We revalidated the Lehmann and colleagues experiments using Affymetrix CEL files from public datasets. We applied these methods to 146 patients with TNBC with gene expression microarrays obtained from June 2000 to March 2010 at our institution. Of those, 130 had received standard neoadjuvant chemotherapy and had evaluable pathologic response data. We classified the TNBC samples by subtype and then correlated subtype and pCR status using Fisher exact test and a logistic regression model. We also assessed survival and compared the subtypes with PAM50 intrinsic subtypes and residual cancer burden (RCB) index.Results
TNBC subtype and pCR status were significantly associated (P = 0.04379). The basal-like 1 (BL1) subtype had the highest pCR rate (52%); basal-like 2 (BL2) and luminal androgen receptor had the lowest (0% and 10%, respectively). TNBC subtype was an independent predictor of pCR status (P = 0.022) by a likelihood ratio test. The subtypes better predicted pCR status than did the PAM50 intrinsic subtypes (basal-like vs. non basal-like).Conclusions
Classifying TNBC by 7 subtypes predicts high versus low pCR rate. We confirm the clinical relevancy of the 7 subtypes of TNBC. We need to prospectively validate whether the pCR rate differences translate into long-term outcome differences. The 7-subtype classification may spur innovative personalized medicine strategies for patients with TNBC.Free full text

Differential response to neoadjuvant chemotherapy among 7 triple-negative breast cancer molecular subtypes
Abstract
Purpose
The clinical relevancy of the 7-subtype classification of triple-negative breast cancer (TNBC) reported by Lehmann and Bauer et al is unknown. We investigated the clinical relevancy of TNBC heterogeneity by determining pathological complete response (pCR) rates after neoadjuvant chemotherapy, based on TNBC subtypes.
Experimental Design
We revalidated the Lehmann and Bauer et al. experiments using Affymetrix CEL files from public datasets. We applied these methods to 146 TNBC patients with gene expression microarrays obtained from June 2000 to March 2010 at our institution. Of those, 130 had received standard neoadjuvant chemotherapy and had evaluable pathological response data. We classified the TNBC samples by subtype, then correlated subtype and pCR status using Fisher’s exact test and a logistic regression model. We also assessed survival and compared the subtypes to PAM50 intrinsic subtypes and residual cancer burden (RCB) index.
Results
TNBC subtype and pCR status were significantly associated (P=0.04379). The basal-like 1 (BL1) subtype had the highest pCR rate (52%); basal-like 2 (BL2) and luminal androgen receptor (LAR) had the lowest (0% and 10%, respectively). TNBC subtype was an independent predictor of pCR status (P=0.022) by a likelihood ratio test. The subtypes better predicted pCR status than did the PAM50 intrinsic subtypes (basal-like vs non basal-like).
Conclusions
Classifying TNBC by 7 subtypes predicts high vs. low pCR rate. We confirm the clinical relevancy of the 7 subtypes of TNBC. We need to prospectively validate whether the pCR rate differences translate into long-term outcome differences. The 7-subtype classification may spur innovative personalized medicine strategies for TNBC patients.
Introduction
Triple-negative breast cancer (TNBC) is defined by lack of expression of estrogen receptor (ER) and of progesterone receptor (PR) and lack of amplification or overexpression of human epidermal growth factor receptor 2 (HER2). TNBC represents approximately 15–20% of all patients with breast cancer and, compared with other breast cancer types, is associated with a high recurrence rate and short survival duration (1, 2). Reasons for this unfavorable prognosis include the heterogeneity and aggressive nature of the disease and the absence of well-defined molecular targets that could form the basis for targeted therapy (3).
In previous studies, 20–30% of patients with TNBC achieved pathological complete response (pCR) neoadjuvant chemotherapy, and pCR was strongly associated with prolonged overall survival (OS) and event-free survival (4–7). In fact, patients with TNBC who achieved pCR had the same prognosis as did patients with non-TNBC (4). In contrast, among patients who did not achieve pCR, patients with TNBC had a significantly poorer outcome (a shorter survival duration driven by higher relapse rates) than did patients with non-TNBC (4). TNBC’s heterogeneous response to chemotherapy in the neoadjuvant setting suggests that different subtypes of primary TNBC may be associated with high or low pCR rates. Subtype-specific individualization by molecular profiling would help us predict benefit from standard chemotherapy and develop personalized targeted drugs for TNBC.
Gene expression analyses have identified molecular subtypes of TNBC that are refining our understanding of breast cancer biology and enabling development of targeted therapy. Recently, Lehmann and Bauer et al (3) reported that TNBC can be classified into 7 subtypes (6 defined subtypes and an unstable group) by gene expression microarray. The 7 TNBC subtypes were characterized on the basis of gene ontologies and differential gene expression and labeled as basal-like 1 (BL1), basal-like 2 (BL2), immunomodulatory (IM), mesenchymal (M), mesenchymal stem-like (MSL), luminal androgen receptor (LAR), and unstable (UNS).
In this study, we confirmed Lehmann and Bauer’s findings and applied their methodology to a population of TNBC patients. For each molecular subtype, we then evaluated pCR rates after standard neoadjuvant chemotherapy regimens, as well as clinical outcomes. Finally, we compared the predictive performance of the 7 subtypes with that of previously defined “intrinsic subtypes” whose gene expression profiles are established through the 50-gene Prediction Analysis of Microarray (PAM50) assay (8), and also evaluated the residual breast cancer burden (RCB) index (9), a post-chemotherapy pathologic measure that considers the size and invasive proportion of the primary tumor and the number and size of nodal metastases, and has potential to predict the clinical outcome after neoadjuvant chemotherapy more accurately than the current pCR vs. non-pCR category.
Materials and Methods
Validation of Lehmann and Bauer’s Gene Expression Analysis
Datasets and normalization of data
Reproducing the method of Lehmann and Bauer et al (3), we obtained 12 public datasets (323 patients’ DNA microarrays) from their TNBC training set and all 7 public datasets (201 patients’ DNA microarrays) from their TNBC validation set. The raw data for 63 of the 386 samples from their training set were not included in our analysis. Datasets GSE5364 and GSE22513 (56 of the 63 samples) were not available to collect. The remaining 7 of the 63 samples were not used because we could not confirm them as TNBC based on Lehmann and Bauer et al’s data. In their study, triple-negative status was identified by using mRNA expression of ER, PR, and HER2; they reported these data in supplemental tables. We identified TNBC status from these tables according to their criteria.
All of the arrays used were a type of Affymetrix U133 array: U133A, U133 Plus 2.0, or U133AAofAv2. Lehmann and Bauer et al used the robust multiarray average (RMA) algorithm to normalize and quantify the data (10). The data pre-processing and quantification were performed using R statistical software. We used this approach with two modifications. First, we used the frozen robust multiarray analysis (fRMA) algorithm, which allows arrays to be analyzed individually or in small batches and then combines the data for analysis, to normalize and quantify all of the datasets. Second, considering the potential effect of platforms on the gene expression profiles, we converted all of the other platforms to HGU133A by using the “fRMA Tools” R package. We later used linear mixed models to correct for any remaining platform-specific differences.
k-means clustering
We followed the methods used by Lehmann and Bauer et al. to identify TNBC subtypes and relative gene signatures. A total of 14,644 probes were selected to represent unique genes. We performed principal component analysis to check for the existence of batch effects. If batch effects were present, feature-by-feature linear models were applied to remove them. Principal component analysis results revealed an obvious batch effect, which was reduced after the application of linear models. We applied consensus clustering using 1000 iterations of k-means clustering to assess the classification robustness and to determine the optimal number of clusters. We performed k-means clustering on the genes (n = 1192) with standard deviation>0.8 and used silhouette width (s[i]) to measure the relative closeness of individual samples to their cluster centers. K-means clustering resulted in 6 stable clusters (s[i]>0) with a total of 261 samples and 1 unstable cluster (s[i]<0) containing 62 samples. Our consensus clustering results confirmed that it is reasonable to set k to 7. We labeled these 7 clusters as subtypes 1 to 7. We computed centroids for each cluster from the consensus clustering and then determined the Pearson correlation of each centroid with each sample in the validation set from Lehmann and Bauer et al. The highest correlation (and lowest P value) was used as the criterion with which to determine the subtype that a specific sample belonged to.(detail in supplementary)
Comparison of Lehmann and Bauer’s and our subtype classification
We compared our clustering of the training and validation samples with the clustering by Lehmann and Bauer et al by using 7×7 contingency tables and X2 tests. We also used Cohen’s kappa to assess the association between our classifications and those reported by Lehmann and Bauer et al.
Application of the Method to Our Population
Patients and samples
We next applied the Lehmann and Bauer et al method to a separate population of patients with TNBC. This group consisted of 146 patients treated for TNBC at MD Anderson. This dataset derived from a prospective study by Hatzis et al (11), which was conducted from June 2000 to March 2010 to develop genomic predictors for neoadjuvant chemotherapy; 97 patients overlapped the group reported by Hatzis et al. We also added new samples that were included in the cohort since the 2011 data were released (n=49). Among the 146 patients, 132 patients had received neoadjuvant chemotherapy containing sequential taxane and anthracycline-based regimens and had evaluable neoadjuvant pathological response. We excluded 2 of these patients because they had received neoadjuvant radiotherapy. Thus, 130 patients were included in our study. Patients had provided written informed consent to participate in an institutional review board–approved research protocol (LAB99-402, USO-02-103, 2003-0321, I-SPY-1) that allowed obtaining a tumor biopsy sample by fine-needle aspiration or core biopsy prior to any systemic therapy, for genomic studies to develop and test predictors of treatment outcome (9).
We identified TNBC by immunohistochemical (IHC)or fluorescence in situ hybridization (FISH) testing. ER status and PR status were considered negative if fewer than 10% of cells stained positively on IHC. HER2 status was considered negative if (a) IHC results were 0 to +1, (b) IHC results were +2 and FISH results were negative. To exclude samples falsely identified as negative by IHC, we implemented a secondary filter that removed samples in which ER expression (205225_at) was greater than the 75th percentile at the transcriptome level, as it was previously demonstrated that >96% of TNBC samples from the original 386-patient TNBC cohort had ER expression below the 75th percentile of all genes (12).
We collected the patient age, clinical stage, treatment regimen, and nuclear grade. We defined pCR as no evidence of invasive carcinoma in the breast and the axillary lymph nodes at the time of surgery. The median follow-up from diagnosis to death was 68.1 months (range 5.1–147.5).
All gene expression microarrays were profiled in the Department of Pathology at MD Anderson. Gene expression levels were derived by exposing the microarray to fluorescent probe sets (multiple oligonucleotide probes that hybridize to specific sequences of gene transcripts) (13, 14).
Gene expression analysis
We classified 146 TNBC samples as defined by Lehmann and Bauer’s gene signatures. As for the public datasets, we performed data pre-processing and quantification in R and applied the fRMA algorithm. Before classification, we applied gene-by-gene mean-centering to our data.
Association of subtype with pCR status and clinical outcome
We constructed a contingency table and performed the Fisher exact test to assess the association between TNBC subtype and pCR status. To assess the independent utility of TNBC subtype for predicting pCR status, we fit a logistic regression model to our data and used age, clinical stage, treatment regimens, and nuclear grade as potential explanatory factors. In this context, using a likelihood ratio test, we examined whether adding TNBC subtype provided a significant improvement in predictive value over a model already containing the other 4 explanatory factors.
We used Cox proportional hazards models to estimate the association between TNBC subtype and OS and distant metastasis-free survival (DMFS). We identified OS and DMFS from the diagnosis date to death or to diagnosis of distant metastasis (15).
Relationship between the 7 subtypes and PAM50 subtypes
Hatzis et al (11) identified intrinsic subtypes in their study; thus, for 92 of our patients, we had data on PAM50 subtypes (luminal A, luminal B, HER2-enriched, basal-like, and normal-like) (8). We investigated the relationship between the PAM50 intrinsic subtypes (grouped as basal-like vs. non-basal-like) and the 7 subtypes identified by Lehmann and Bauer et al.
Relationship between the 7 subtypes and RCB index
We also assessed the association between TNBC subtype and the RCB index reported by Symmans et al (9). Primary tumor and nodal metastasis samples were evaluated for residual disease as previously described, and tumors were classified as pCR, RCB-I (minimal residual disease), RCB-II (moderate residual disease), or RCB-III (extensive residual disease).
Results
Validation of Lehmann and Bauer’s TNBC classification
In our analysis, 7×7 contingency tables (Table 1) indicated high correlation (P < 2.2×10−16) between our clustering and the original clustering. We also used Cohen’s kappa coefficient to assess the association between our classifications and those reported by Lehmann and Bauer et al. For the training set, the κ value was 0.662 (n = 277); for the validation set, 0.462 (n = 200). Using large-sample normal approximations, the kappa2 function in the R package gave approximate z-values of 26.1 and 16.2, respectively; the P values were essentially 0.
Table 1
The 7 ×7 contingency tables showing the clustering of samples in the training set and validation set using 2 sets of gene signatures as determined by Lehmann and Bauer and by us.
| Lehmann, Bauer et al | |||||||||
|---|---|---|---|---|---|---|---|---|---|
| MD Anderson | Training set | BL1 | BL2 | M | IM | MSL | LAR | UNS | P value |
| BL1 | 40 | 10 | 1 | 8 | 0 | 0 | 9 | P<2.2×10−16 | |
| BL2 | 0 | 3 | 0 | 9 | 0 | 4 | 0 | ||
| M | 0 | 3 | 49 | 0 | 0 | 0 | 3 | ||
| IM | 4 | 0 | 0 | 36 | 0 | 0 | 0 | ||
| MSL | 0 | 3 | 3 | 0 | 20 | 0 | 0 | ||
| LAR | 0 | 4 | 0 | 0 | 0 | 26 | 0 | ||
| UNS | 5 | 9 | 4 | 0 | 0 | 4 | 24 | ||
| Lehmann, Bauer et al | |||||||||
|---|---|---|---|---|---|---|---|---|---|
| MD Anderson | Validation set | BL1 | BL2 | M | IM | MSL | LAR | UNS | P value |
| BL1 | 40 | 10 | 1 | 8 | 0 | 0 | 9 | P<2.2×10−16 | |
| BL2 | 0 | 3 | 0 | 9 | 0 | 4 | 0 | ||
| M | 0 | 3 | 49 | 0 | 0 | 0 | 3 | ||
| IM | 4 | 0 | 0 | 36 | 0 | 0 | 0 | ||
| MSL | 0 | 3 | 3 | 0 | 20 | 0 | 0 | ||
| LAR | 0 | 4 | 0 | 0 | 0 | 26 | 0 | ||
| UNS | 5 | 9 | 4 | 0 | 0 | 4 | 24 | ||
The contingency table for the training set (Table 1) indicated that the BL1, M, IM, MSL, and LAR subtypes are more stable than the BL2 and UNS subtypes. The contingency table for the validation set (Table 1) indicated that subtypes IM, MSL, and LAR are more stable than the other subtypes.
In summary, there was high correlation between the results of the Lehmann and Bauer et al study and the results we obtained by using approximately the same methods.
TNBC Subtypes Predict pCR Status of Patients Treated with Neoadjuvant Chemotherapy
Of the 130 patients with evaluable pathological response after neoadjuvant chemotherapy, 16 patients received an anthracycline regimen alone, 3 patients received a taxane regimen alone, and 111 patients received both anthracycline and taxane regimens. We classified patients into subtypes as follows: BL1, 21 patients; BL2, 8 patients; M, 26 patients; IM, 27 patients; MSL, 13 patients; LAR, 20 patients; and UNS, 15 patients. There was no statistically significant difference in treatment regimens between subtypes (P=0.651).
The pCR rate for all patients was 28% (37/130). BL1 had the highest pCR rate (52%), and BL2 and LAR had the lowest pCR rates (0 and 10%, respectively). The Fisher exact test indicated a significant association between TNBC subtype and pCR status (P=0.04379) (Table 3). TNBC subtype was an independent predictor of pCR status (P=0.022) by a likelihood ratio test (based on logistic regression models with and without TNBC subtype as the variant).
Table 3
Distribution of pCR/non-pCR status by TNBC subtype
| pCR | Non-pCR | pCR rate | 95%CI | P value | |
|---|---|---|---|---|---|
| BL1 | 11 | 10 | 0.52 | 0.31–0.73 | P=0.043 |
| BL2 | 0 | 8 | 0.00 | 0.00–0.00 | |
| M | 8 | 18 | 0.31 | 0.13–0.48 | |
| IM | 8 | 19 | 0.30 | 0.12–0.46 | |
| MSL | 3 | 10 | 0.23 | 0.0001–0.45 | |
| LAR | 2 | 18 | 0.10 | 0.03–0.23 | |
| UNS | 5 | 10 | 0.33 | 0.09–0.57 |
Likelihood ratio test:
Adjusting clinical features: age, clinical stage, nuclear grade, and treatment type.
TNBC subtype was an independent predictor of pCR status (P=0.022)
Clinical Outcomes by Subtype
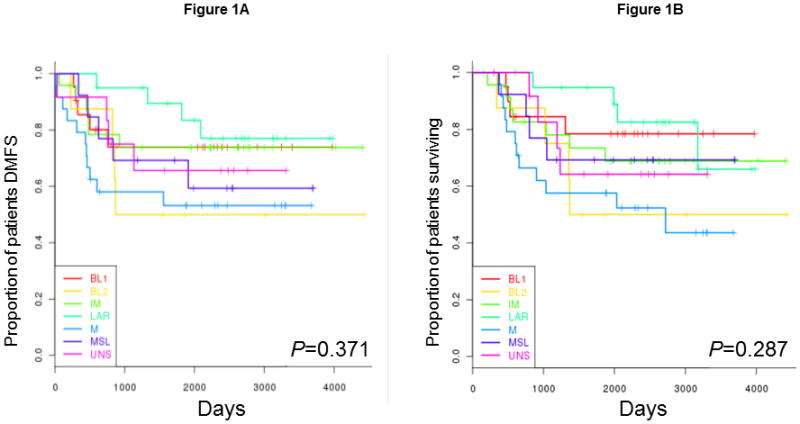
We excluded 8 patients from the survival analysis because of their history of other types of cancer. The Cox models showed no significant association between TNBC subtype and OS (P= 0.287) or DMFS (P= 0.371). The Kaplan-Meier plots for each subtype with respect to OS and DMFS are shown in Figures 1A and 1B, respectively. Owing to the limited sample size, the median survival rates were not available for all TNBC subtypes. However, despite its lower pCR rate, LAR had the best OS rate; M had the worst.
Relationship between the 7 Subtypes and PAM50 Subtype
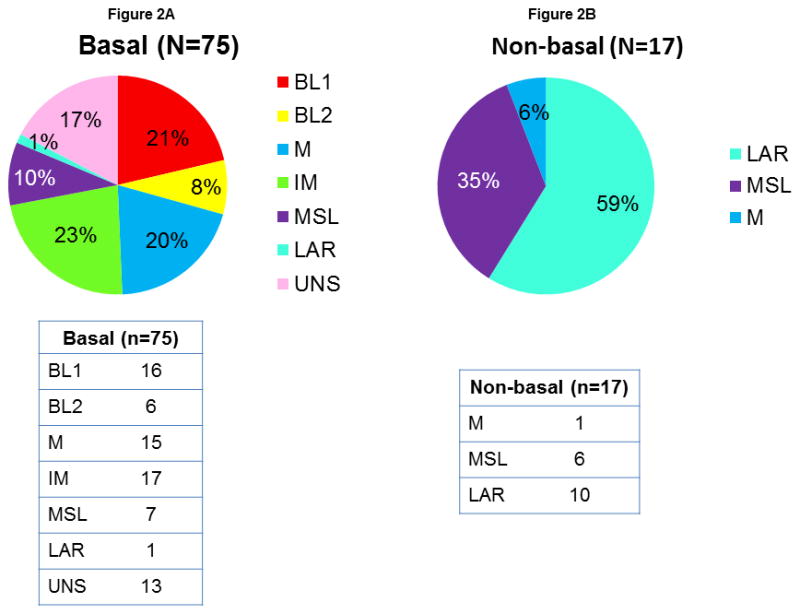
Since PAM50 gene expression analysis has been incorporated into the clinic and shown to be strongly related to the clinical outcome of breast cancer, we determined the relationship between these classification systems. Figure 2 shows the distribution of the 7 Lehmann and Bauer TNBC subtypes between the PAM50 basal-like subtype and non-basal-like subtypes (grouped). All tumors in the BL1 and BL2 subtypes belonged to the basal-like PAM50 subtype, and most tumors in the LAR subtype belonged to the non-basal-like PAM50 group. In the non-basal-like group, there were only 3 TNBC subtypes, LAR, MSL, and M; the majority (59%) of these tumors were the LAR subtype.
Relationship between the 7 Subtypes and RCB Index
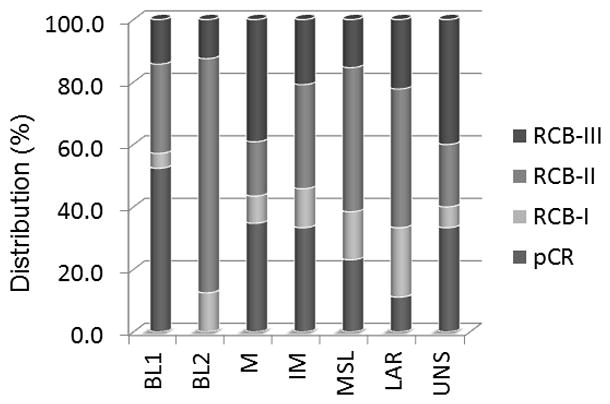
Figure 3 shows the relationship between the 7 subtypes and RCB index. Consistent with Symmans et al’s previous study, RCB index clearly predicted the clinical outcomes in the TNBC population. There was no statistical difference in OS rate between patients with pCR and those with RCB-I status, and patients with RCB-III showed the lowest OS rate. As seen in Figure 3, combining the distribution of pCR and RCB-I revealed that current neoadjuvant chemotherapy has low efficacy for the BL2 subtype in particular.
Discussion
This is the first report that the TNBC subtype can serve as an independent predictor of pCR status in patients who receive current standard chemotherapy regimens. We confirmed that TNBC is heterogeneous and can be classified with high correlation into 7 subtypes, including 1 unstable subtype, by Lehmann and Bauer’s algorithm. In our population, the subtypes better predicted pCR status than did the PAM50 intrinsic subtypes. RCB index predicted prognosis more accurately.
Although Lehmann and Bauer’s classification is not the only strategy for defining molecular subtypes of TNBC, it has had a strong impact because it classifies the entire TNBC population into homogeneous subtypes and establishes 7 subtypes, and because subtype classification had a preclinical predictive effect on the outcome of therapy selected to incorporate specific targeted treatments, such as an androgen receptor antagonist and aPI3K/mTOR inhibitor. Our study has extended these findings by showing that the 7 subtypes predicted the rate of pCR to current standard chemotherapy regimens. Gene expression analyses have identified molecular subtypes of TNBC, such as the claud in-low subtype (16, 17) and basal-like subtype. However, these subsets are still not well defined enough to enable development of targeted therapy or prediction of pathological response.
It was interesting that while the BL1 subtype had the highest chemosensitivity (pCR rate), as we hypothesized, the BL2 subtype had the lowest pCR rate, contrary to our hypothesis. These subtypes have similar biology, such as high Ki-67 mRNA expression and enrichment of proliferation genes; however, BL2 has unique gene ontologies involving growth factor signaling, such as the EGF, MET, and IGF1R pathways. The difference in gene ontologies might explain the difference in pCR rates and could provide a basis for individualized therapy. For example, BL2 tumors could be targeted with EGFR or IGF1R inhibitors.
Tumors of the LAR subtype are heavily enriched in hormonally regulated pathways. Consistent with LAR’s low pCR rate, which was in accord with our hypothesis, the luminal A and Bintrinsic subtypes, which are hormonally regulated tumors, showed less response to chemotherapy (6, 8). However, when we evaluated the RCB index, 33% of patients belonged to the pCR and RCB-I categories. This result might have affected the clinical outcomes. The LAR group had delayed recurrences compared with the other groups and did not have the lowest OS rate despite having a low pCR rate. Also, 75% of distant metastasis in the LAR subtype occurred more than 3 years after diagnosis. The LAR group showed a clearly different clinical process from that of the other subtypes. These results suggest that within TNBC, we need to distinguish the LAR subtype and design a different treatment strategy for this group.
For other groups, consistent with the current knowledge about TNBC, most recurrences and deaths happened within 3 years of diagnosis. For these groups, to achieve pCR is the most desirable result for improving prognosis; thus, we need ways of predicting chemotherapy sensitivity, resistance, or both to guide selection of a treatment regimen, and these predictors also should be associated with DMFS and OS. The 7 subtypes have the potential to solve these problems. Further studies are needed to better characterize them.
In a previous study of the PAM50 intrinsic subtypes (2), the basal-like intrinsic subtype showed high chemosensitivity and the difference between the basal-like and non-basal-like subtypes was reported to be the main biological difference seen among patients with TNBC. (18)In our study, all patients in the group with the highest pCR rate, BL1, had tumors characterized as basal-like, and all but 1 of the patients in the LAR population, which had a low pCR rate, had non-basal-like tumors. Interestingly, although all those with the BL1 and BL2 subtypes belonged to the basal-like intrinsic subtype, BL1 had the highest pCR rate and BL2 the lowest. The 7 subtypes more accurately predicted pCR status.
RCB index was significantly associated with OS rate. This index has potential to predict clinical outcomes more accurately than pCR status after neoadjuvant chemotherapy. Combining the distribution of pCR and RCB-I revealed that current neoadjuvant chemotherapy has low efficacy for the BL2 subtype in particular. We conclude that identifying the BL2 population and developing specific treatments for this group would improve the clinical outcome in TNBC patients as a whole.
Although we reproduced Lehmann and Bauer’s 7 subtypes and could apply their algorithm to new data, the results didn’t match perfectly. Microarray-based molecular classification always involves this risk (19, 20). Even for the well-known intrinsic subtypes, several types of gene signatures have been derived in each study, and none of the classification systems tested have produced perfect agreement (8, 21–23). Although we had 130 patients, and there were a number of additional new samples that were included in the cohort since the 2011 data were released, once the patients were classified into 7 groups, we didn’t have enough power to show clinical relevance. Further, one of our most interesting groups, BL2, had the smallest sample size. It is almost impossible to collect samples from consistently treated patients for whom uniform, clear clinical information is also available. Thus, prospective future studies are needed to establish the clinical relevance of using gene profiling to divide patients by subtype. Further, prospective validation by methods such as reverse-phase protein array or IHC staining is needed to measure whether the targets noted in the 7 subtypes are truly overexpressed or are functionally activated at the protein expression level. The clinical utility of these TNBC subtypes will not reach its full potential until markers for each subtype can be validated on reliable platforms, which can define the protein expression level or functionality of determine the specific targets which drived the tumorigenicity and metastasis of each subtype (IHC or NanoString assay).
In summary, our results suggest that we especially need to distinguish the BL2 and LAR subtypes in order to apply specific treatment strategies for them. These 2 subtypes showed the lowest pCR rates; however, appropriate treatment strategies might be quite different for these 2 subtypes. For the BL2 subtype, we need to develop a novel targeted therapy in the neoadjuvant setting to achieve a higher pCR rate. In addition, we need to distinguish the LAR subtype from other TNBCs and apply a new treatment strategy similar to that for the luminal intrinsic subtype, such as targeting the androgen receptor pathway as a long-term adjuvant treatment. Prospective validation of our findings is needed. We conclude that the 7 subtypes may lead to innovative personalized medicine clinical trials for patients with TNBC.
Table 2
Patient characteristics by TNBC subtype (n = 130)
| Characteristic | Category | BL1 n=21 | BL2 n=8 | M n=26 | IM n=27 | MSL n=13 | LAR n=20 | UNS n=15 | P-value |
|---|---|---|---|---|---|---|---|---|---|
| Age (yr) | >50 | 14 | 2 | 13 | 12 | 7 | 6 | 7 | |
| ≤50 | 7 | 6 | 13 | 15 | 6 | 14 | 8 | 0.283 | |
| Clinical stage | I | 0 | 0 | 0 | 2 | 1 | 0 | 0 | |
| IIA | 3 | 2 | 6 | 9 | 4 | 7 | 2 | ||
| IIB | 8 | 4 | 13 | 5 | 1 | 6 | 7 | ||
| IIIA | 4 | 1 | 2 | 3 | 1 | 2 | 1 | ||
| IIIB | 1 | 0 | 3 | 6 | 1 | 3 | 1 | ||
| IIIC | 5 | 1 | 2 | 2 | 5 | 2 | 4 | 0.276 | |
| Nuclear grade | 2 | 2 | 1 | 4 | 0 | 2 | 7 | 0 | |
| 3 | 19 | 7 | 22 | 27 | 11 | 13 | 15 | 0.008 | |
| Treatment regimen | A | 1 | 1 | 4 | 4 | 0 | 4 | 1 | |
| T | 0 | 0 | 2 | 1 | 0 | 0 | 0 | ||
| A+T | 20 | 7 | 20 | 22 | 13 | 16 | 14 | 0.651 | |
| pCR | Yes | 11 | 0 | 8 | 8 | 3 | 2 | 5 | |
| No | 10 | 8 | 18 | 19 | 10 | 18 | 10 | 0.043 | |
| Distant recurrence | Yes | 5 | 4 | 12 | 6 | 4 | 4 | 4 | |
| No | 16 | 4 | 14 | 21 | 9 | 16 | 11 | 0.348 | |
| Local recurrence | Yes | 3 | 2 | 1 | 3 | 0 | 1 | 3 | |
| No | 18 | 6 | 25 | 24 | 13 | 19 | 12 | 0.269 | |
| Survival status | Dead | 4 | 4 | 12 | 9 | 4 | 4 | 5 | |
| Alive | 17 | 4 | 14 | 18 | 9 | 16 | 10 | 0.369 |
pCR: pathological complete response A: anthracycline T: taxane
Acknowledgments
Grant support: This research was supported by the National Institutes of Health through R01 grant CA123318 (to NT Ueno) and through Cancer Center Support Grant CA016672 (to MD Anderson Cancer Center), by the Morgan Welch Inflammatory Breast Cancer Research Program and Clinic, by a State of Texas Rare and Aggressive Breast Cancer Research Program grant, by Susan G. Komen for The Cure (to WFS) and The Breast Cancer Research Foundation (to WFS).
Footnotes
Conflicts of interest: There is no conflict in our study.
Information of Microarray Data
Data sets for this study have been deposited into the GEO database (http://www.ncbi.nlm.nih.gov/geo/) under accession identification numbers GSE25066, GSE25055, GSE25065, GSE43502 and GSE31519.
References
Full text links
Read article at publisher's site: https://doi.org/10.1158/1078-0432.ccr-13-0799
Read article for free, from open access legal sources, via Unpaywall:
https://clincancerres.aacrjournals.org/content/clincanres/19/19/5533.full.pdf
Citations & impact
Impact metrics
Article citations
The Immune-Related 27-Gene Signature DetermaIO Predicts Response to Neoadjuvant Atezolizumab plus Chemotherapy in Triple-Negative Breast Cancer.
Clin Cancer Res, 30(21):4900-4909, 01 Nov 2024
Cited by: 0 articles | PMID: 39308141 | PMCID: PMC11528202
Synergistic effect of bazedoxifene and abemaciclib co‑treatment in triple‑negative breast cancer cells in vitro.
Oncol Lett, 28(6):554, 19 Sep 2024
Cited by: 0 articles | PMID: 39355786 | PMCID: PMC11443307
FOXM1 Transcriptionally Co-Upregulates Centrosome Amplification and Clustering Genes and Is a Biomarker for Poor Prognosis in Androgen Receptor-Low Triple-Negative Breast Cancer.
Cancers (Basel), 16(18):3191, 18 Sep 2024
Cited by: 0 articles | PMID: 39335162 | PMCID: PMC11429756
Review Free full text in Europe PMC
The Anticancer Effects and Therapeutic Potential of Kaempferol in Triple-Negative Breast Cancer.
Nutrients, 16(15):2392, 23 Jul 2024
Cited by: 1 article | PMID: 39125273 | PMCID: PMC11314279
Review Free full text in Europe PMC
Phase 2 study of neoadjuvant enzalutamide and paclitaxel for luminal androgen receptor-enriched TNBC: Trial results and insights into "ARness".
Cell Rep Med, 5(6):101595, 04 Jun 2024
Cited by: 0 articles | PMID: 38838676 | PMCID: PMC11228653
Go to all (405) article citations
Data
Data behind the article
This data has been text mined from the article, or deposited into data resources.
BioStudies: supplemental material and supporting data
GEO - Gene Expression Omnibus (5)
- (1 citation) GEO - GSE25065
- (1 citation) GEO - GSE25066
- (1 citation) GEO - GSE25055
- (1 citation) GEO - GSE43502
- (1 citation) GEO - GSE31519
Similar Articles
To arrive at the top five similar articles we use a word-weighted algorithm to compare words from the Title and Abstract of each citation.
Refinement of Triple-Negative Breast Cancer Molecular Subtypes: Implications for Neoadjuvant Chemotherapy Selection.
PLoS One, 11(6):e0157368, 16 Jun 2016
Cited by: 646 articles | PMID: 27310713 | PMCID: PMC4911051
Pathological Response in a Triple-Negative Breast Cancer Cohort Treated with Neoadjuvant Carboplatin and Docetaxel According to Lehmann's Refined Classification.
Clin Cancer Res, 24(8):1845-1852, 29 Jan 2018
Cited by: 56 articles | PMID: 29378733 | PMCID: PMC5899625
Response and survival of breast cancer intrinsic subtypes following multi-agent neoadjuvant chemotherapy.
BMC Med, 13:303, 18 Dec 2015
Cited by: 85 articles | PMID: 26684470 | PMCID: PMC4683815
Clinical implications of the intrinsic molecular subtypes of breast cancer.
Breast, 24 Suppl 2:S26-35, 05 Aug 2015
Cited by: 498 articles | PMID: 26253814
Review
Funding
Funders who supported this work.
NCI NIH HHS (4)
Grant ID: P30 CA016672
Grant ID: CA016672
Grant ID: R01 CA123318
Grant ID: CA123318