Abstract
Motivation
Chromatin immunoprecipitation coupled to next-generation sequencing (ChIP-seq) is widely used to study the in vivo binding sites of transcription factors (TFs) and their regulatory targets. Recent improvements to ChIP-seq, such as increased resolution, promise deeper insights into transcriptional regulation, yet require novel computational tools to fully leverage their advantages.Results
To this aim, we have developed peakzilla, which can identify closely spaced TF binding sites at high resolution (i.e. resolves individual binding sites even if spaced closely), as we demonstrate using semisynthetic datasets, performing ChIP-seq for the TF Twist in Drosophila embryos with different experimental fragment sizes, and analyzing ChIP-exo datasets. We show that the increased resolution reached by peakzilla is highly relevant, as closely spaced Twist binding sites are strongly enriched in transcriptional enhancers, suggesting a signature to discriminate functional from abundant non-functional or neutral TF binding. Peakzilla is easy to use, as it estimates all the necessary parameters from the data and is freely available.Availability and implementation
The peakzilla program is available from https://github.com/steinmann/peakzilla or http://www.starklab.org/data/peakzilla/.Contact
[email protected].Supplementary information
Supplementary data are available at Bioinformatics online.Free full text

Identification of transcription factor binding sites from ChIP-seq data at high resolution
Abstract
Motivation: Chromatin immunoprecipitation coupled to next-generation sequencing (ChIP-seq) is widely used to study the in vivo binding sites of transcription factors (TFs) and their regulatory targets. Recent improvements to ChIP-seq, such as increased resolution, promise deeper insights into transcriptional regulation, yet require novel computational tools to fully leverage their advantages.
Chromatin immunoprecipitation coupled to next-generation sequencing (ChIP-seq) is widely used to study the in vivo binding sites of transcription factors (TFs) and their regulatory targets. Recent improvements to ChIP-seq, such as increased resolution, promise deeper insights into transcriptional regulation, yet require novel computational tools to fully leverage their advantages.
Results: To this aim, we have developed peakzilla, which can identify closely spaced TF binding sites at high resolution (i.e. resolves individual binding sites even if spaced closely), as we demonstrate using semisynthetic datasets, performing ChIP-seq for the TF Twist in Drosophila embryos with different experimental fragment sizes, and analyzing ChIP-exo datasets. We show that the increased resolution reached by peakzilla is highly relevant, as closely spaced Twist binding sites are strongly enriched in transcriptional enhancers, suggesting a signature to discriminate functional from abundant non-functional or neutral TF binding. Peakzilla is easy to use, as it estimates all the necessary parameters from the data and is freely available.
To this aim, we have developed peakzilla, which can identify closely spaced TF binding sites at high resolution (i.e. resolves individual binding sites even if spaced closely), as we demonstrate using semisynthetic datasets, performing ChIP-seq for the TF Twist in Drosophila embryos with different experimental fragment sizes, and analyzing ChIP-exo datasets. We show that the increased resolution reached by peakzilla is highly relevant, as closely spaced Twist binding sites are strongly enriched in transcriptional enhancers, suggesting a signature to discriminate functional from abundant non-functional or neutral TF binding. Peakzilla is easy to use, as it estimates all the necessary parameters from the data and is freely available.
Availability and implementation: The peakzilla program is available from https://github.com/steinmann/peakzilla or http://www.starklab.org/data/peakzilla/.
The peakzilla program is available from https://github.com/steinmann/peakzilla or http://www.starklab.org/data/peakzilla/.
Contact: gro.balkrats@krats
gro.balkrats@krats
Supplementary information: Supplementary data are available at Bioinformatics online.
1 INTRODUCTION
Gene expression is mainly regulated at the transcriptional level and achieved through the binding of transcription factors (TFs) to genomic regulatory regions called promoters and enhancers. Chromatin immunoprecipitation followed by sequencing (ChIP-seq) is extensively used to determine transcription factor binding sites (TFBSs) genome-wide (Johnson et al., 2007; Robertson et al., 2007). Compared with ChIP-chip (Iyer et al., 2001; Ren et al., 2000), ChIP-seq has dramatically improved the resolution of the identified TFBSs (from hundreds to only tens of nucleotides). A recent refinement of ChIP-seq, the ChIP-exo method (Rhee and Pugh, 2011), further increases the resolution of ChIP-seq experiments, theoretically to single nucleotides.
The specific features and strategies of TF ChIP-seq data analysis have been well described by several reviews (e.g. Pepke et al., 2009) and protocols (e.g. Bardet et al., 2012). Briefly, as typically only one of both ends of the immunoprecipitated DNA fragments is sequenced, the sequencing reads (or tags) result in a bimodal distribution that is characteristic for true TFBSs (Fig. 1a). This distribution is typically used to estimate the average size of the fragments, which subsequently allows the prediction of TFBSs across the genome.
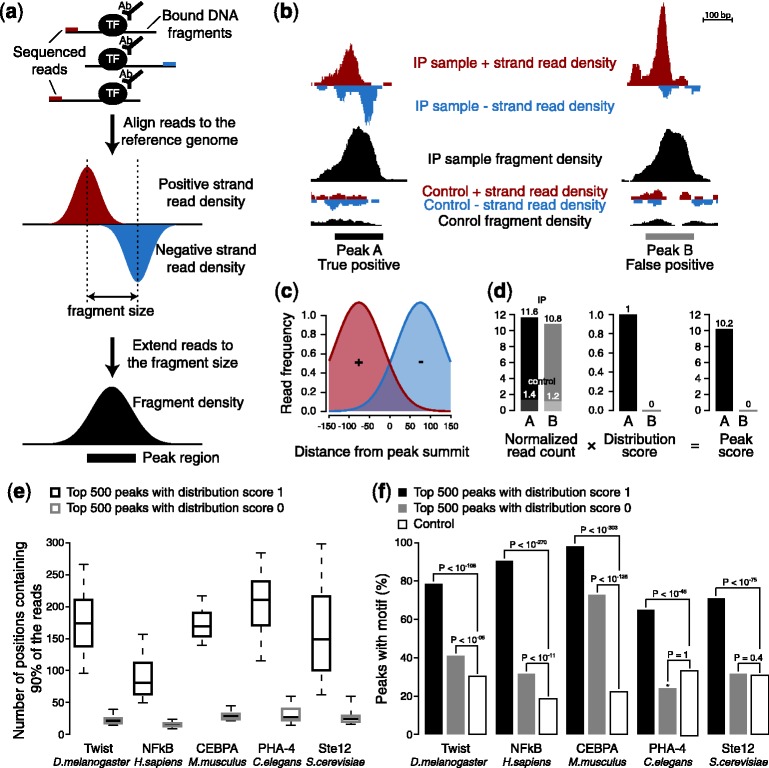
Peakzilla algorithm. (a) Overview of the ChIP-seq pipeline. TFBSs display a characteristic bimodal distribution of the positive and negative strand reads. (b) Example of a true-positive (Peak A) and false-positive (Peak B) peak in the Twist dataset in D.melanogaster (genomic coordinates chr2L:12420984-12423043 and chrX: 9899747-9905926, respectively). Peak B, unlike peak A, does not exhibit the characteristic double distribution of reads on the positive and negative strands. (c) Read distribution model using two Gaussian distributions. (d) Peak score. While both peaks A and B from (b) show the same enrichment of read count over control, the score for peak B is penalized by the distribution score, a multiplicative factor [0 … 1], as it does not fit to the specific double distribution of the model in (c). (e) Fragment diversity or data non-redundancy. Y-axis denotes the number of genomic positions that contain 90% of the reads that contribute to a peak. Peaks with a distribution score of 0 are more redundant, whereas peaks with a distribution score of 1 are more diverse. The same plot for all peaks in shown in Supplementary Figure S2 (f) Fraction of peaks with a distribution score of 0 or 1 that contain the corresponding TF motif
Many computational tools have been developed and are successfully used to predict such binding events (Wilbanks and Facciotti, 2010). However, in our experience, these tools are not optimized to take advantage of recent methodological improvements, which comprise paired-end sequencing, high sequencing depth (e.g. on Illumina HiSeq systems) and—most importantly—an increase in experimental resolution, both of conventional ChIP-seq and of ChIP-exo. Available tools typically merge closely spaced read-density peaks into large regions, which is preferable when analyzing certain chromatin features that mark extended regions (e.g. histone modifications), but means that the ability to distinguish (i.e. resolve) individual closely spaced TFBSs is lost. High resolution and precision [i.e. the correct prediction of the TFBSs’ exact locations as measured for example as the distance from the inferred TFBS (the reported peak summit) to the TF’s sequence motif] are crucial when determining individual TFBSs (e.g. Guo et al., 2012), as promoter and enhancer regions often consist of multiple TFBSs for the same TF (homotypic TFBSs clusters) (Gotea et al., 2010; He et al., 2011; Lifanov et al., 2003) or different TFs (Berman et al., 2002; Schroeder et al., 2004). Thus, to fully leverage current ChIP methodologies toward understanding the structure and function of enhancers, the ability to determine multiple closely spaced TFBSs is critical. To meet this need, we developed a new computational tool, peakzilla, which fully exploits the bimodal distribution of sequence reads characteristic of true TF binding events, to identify closely adjacent TFBSs with high resolution and precision. Peakzilla is not meant for the identification of broad enriched regions (e.g. histone marks) for which we recommend using MACS or similar programs.
We evaluate peakzilla by comparing it with three widely used peak-finders of the first generation such as MACS (Zhang et al., 2008), QuEST (Valouev et al., 2008) and cisGenome (Ji et al., 2008), as well as four methods developed more recently for the detection of high-resolution peaks such as spp (Kharchenko et al., 2008), SISSRs (Jothi et al., 2008), GPS (Guo et al., 2010) and PeakRanger (Feng et al., 2011). Peakzilla shows superior resolution and precision on conventional ChIP-seq datasets from Saccharomyces cerevisiae (Zheng et al., 2010), Caenorhabditis elegans (Zhong et al., 2010), Drosophila melanogaster (He et al., 2011), mouse (Schmidt et al., 2010) and human (Kasowski et al., 2010; Cuddapah et al., 2009) and on recent ChIP-exo datasets (Rhee and Pugh, 2011). We also specifically test the resolution limits of each method using semisynthetic ChIP-seq datasets and show experimentally that peakzilla fully reflects increased experimental resolution by performing ChIP-seq experiments for Twist in D.melanogaster at normal, medium and high resolution. These results suggest that peakzilla is best suited for the identification of TFBSs with recent ChIP methods.
2 PEAKZILLA ALGORITHM
Peakzilla uses the bimodal distribution of the reads (Fig. 1a) not only to estimate the fragment length but also to weight the read counts during peak calling and to score the candidate TFBSs. This has two important advantages: first, it enables peakzilla to more clearly discriminate between reads from adjacent TFBSs, leading to a substantial increase in resolution compared with treating reads irrespective of their directionality. Second, it avoids false positives that originate from artifacts during library preparation or sequencing, without the need to collapse or down-weight reads that map to identical genomic positions (Fig. 1b and Supplementary Fig. S1). This is especially important when working with high sequence coverage, as obtained for small genomes (e.g. yeast, flies or nematodes) and with modern next generation sequencers (Chen et al., 2012). Finally, when using peakzilla on paired-end ChIP-seq data, the estimated fragment size is directly averaged from the mapped reads.
Peakzilla first scans the genome for candidate TFBSs that show high coverage in sequencing reads of the immunoprecipitated (IP) sample compared with the control sample (note that the control sample is optional, allowing peakzilla to be used with ChIP-exo). It then scores the candidates by the normalized read count of IP sample minus the control sample if available. To discriminate between artifacts and true binding events in enriched regions, each candidate TFBS score is further weighted by a distribution score that estimates how well the observed distribution of the reads in the peak region fits to a model for the bimodal read distribution (Fig. 1c and d). Indeed, further analyses suggest that candidates who are penalized using this distribution score are likely false because they contain substantially less diverse sequence reads (Fig. 1e and Supplementary Fig. S2). Their high read densities stem from only a few highly duplicated sequences, which are likely amplification artifacts, and are significantly less enriched in the corresponding TF motif (Fig. 1f). The method is illustrated in the method section and in Supplementary Figure S3.
3 RESULTS
3.1 Overall performance
To evaluate peakzilla, we compared it with other methods using diverse ChIP-seq datasets from S.cerevisiae, C.elegans, D.melanogaster, mouse and human. Although the number of peaks called by the different methods is highly dependent on the thresholds and parameters used, the respective genomic regions overlap well (Supplementary Fig. S4), demonstrating the maturity of available tools for ‘peak calling’. For example, all known Twist enhancers are identified by all methods, except for four and three enhancers that are not found by QuEST and GPS, respectively (Supplementary Fig. S5). For most TFs tested, of all peaks identified by only one of the methods, those found exclusively by peakzilla are significantly more highly enriched in TF motif occurrences (Fig. 2a and Supplementary Fig. S6). They also have higher fold enrichments of ChIP over input than peaks found exclusively by any of the other methods (Fig. 2b and Supplementary Fig. S6).
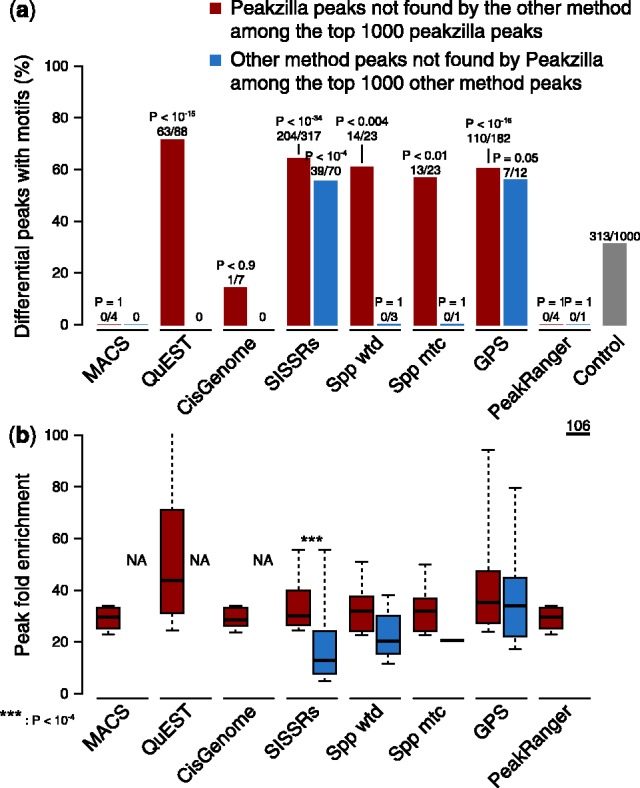
High precision of peakzilla peaks. Analyses performed on the Twist dataset in D.melanogaster. (a) Enrichment of motifs in differential peaks between peakzilla and other methods. Bionomial P-values of enrichment over control and number of differential peaks with a motif are shown on top of the bars. See Supplementary Figure S6 for other datasets and species. (b) Fold enrichment values of differential peaks and associated Wilcoxon P-values (NA: no peak available)
3.2 High precision of peakzilla peaks
When identifying TFBSs at high resolution, the correct prediction of the precise TFBSs’ location is important and critical for subsequent analyses of sequence features of TF binding. We found that most methods, including peakzilla, place the summits of the peak regions closely to the nearest motif occurrence (Supplementary Fig. S7, which also shows that the fraction of peaks that contain a motif is comparable across methods), arguing that the high resolution of peakzilla’s peak (see below) does not come at the expense of precision.
3.3 Multiple peak regions function as transcriptional enhancers
The main strength of peakzilla is its ability to find peaks at high resolution. As the locations of sequencing reads that originate from a single TFBS are limited by the fragment size used for library preparation, we adjust our search window for counting and scoring sequence reads accordingly, and report peak regions as the average fragment size centered on the summit position. This is different from the large peak regions reported by MACS and to a lesser extent QuEST, CisGenome and PeakRanger (and from reporting only the summit positions as do SISSRs and spp; Fig. 3a), and is one of the key features that allow peakzilla to resolve closely spaced peaks up to distances that correspond to half the fragment size. This is the highest resolution that can easily be obtained without losing the ability to uniquely assign reads to individual TFBSs. A further gain in resolution would require the deconvolution of overlapping read distributions by model fitting, a computationally intensive approach used, for example, by GPS (Guo et al., 2010).
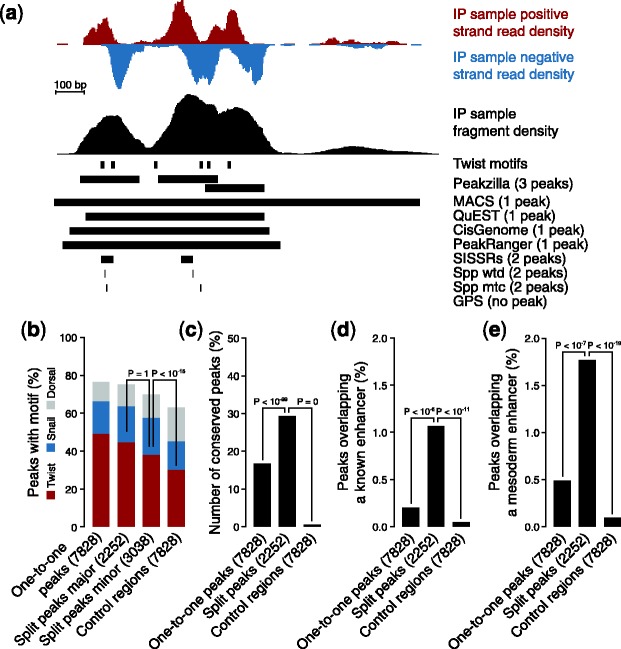
Functionality of multiple peak regions. Analyses performed on the Twist dataset in D.melanogaster. (a) Example of peak split. Peakzilla detects three adjacent peaks, while MACS, QuEST, CisGenome and PeakRanger report a single large peak region, and SISSRs and spp report two peak regions (GPS did not call any peak in that region; we considered all peaks called with standard parameters for each method). (b) Split peaks match motif occurrences. All peakzilla peaks corresponding to a single MACS peak (major: same summit; minor: additional summit) are more highly enriched in Twist motifs than control regions, suggesting that they constitute true independent TFBSs. The same is true for motifs of Snail and Dorsal, which are TFs known to cooperate with Twist. (c) Split peaks are highly conserved. (d) Split peaks are enriched for known enhancers. (e) Split peaks are enriched for mesodermal enhancers
Peakzilla splits a substantial number of MACS peaks (e.g. 22% for Twist) into several peaks, each constituting a putative TFBS (Supplementary Fig. S8). Indeed, as expected for independent TFBSs, both the split peaks that correspond to the MACS summits (major peaks) and the minor peaks were significantly enriched for the Twist motif (Fig. 3b). Thus, many split MACS peaks may represent homotypic clusters of Twist binding sites. In addition, minor peaks frequently contained motifs for the TFs Snail and Dorsal, which are known to cooperate with Twist and might have been co-precipitated after cross-linking (Fig. 3b).
Most importantly, MACS peaks split by peakzilla appear to be more often functional than MACS peaks that are not split (one-to-one peaks) and control regions (Fig. 3c): TF binding to split peaks is more highly conserved in other Drosophila species (He et al., 2011) and they are significantly more enriched for known Twist or mesodermal enhancers than one-to-one peaks one or controls (Fig.3d and e). This is highly relevant as the large fraction of TFBS that are identified by ChIP approaches yet do not appear to be functioning as transcriptional enhancers (‘neutral binding’ (Li et al., 2008; Kvon et al., 2012)) have been a major obstacle to studying the direct regulatory targets of TFs and to understanding the true number and density of enhancers in animal genomes. All together, these results suggest that peakzilla is ideal for identifying regions with multiple binding sites and that such information is important for detecting functional enhancers.
3.4 High resolution of peakzilla
The evaluation of TFBS predicted from ChIP-seq data (i.e. peaks) is complicated by the fact that a ground truth typically does not exist for experimental ChIP-seq datasets. This is especially true for the evaluation of peak calls at different resolutions that predict a single TFBS versus several closely spaced TFBS. Although the occurrence of TF motifs is a good proxy for independent binding events, we sought to demonstrate the validity of peak splitting more directly by generating semisynthetic datasets with defined peak-to-peak distances. We found that peakzilla is still able to separate all peak pairs at 150 bp without calling any false positives, which is the best resolution compared with all others methods (Fig. 4).
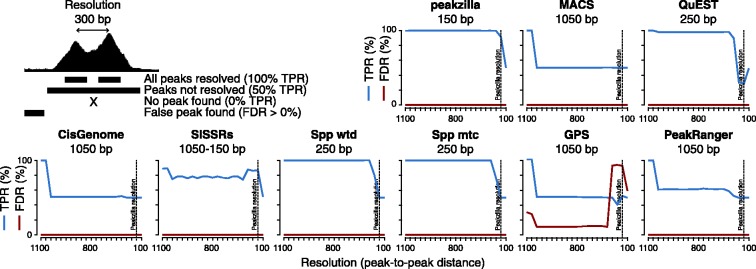
High resolution of peakzilla. We evaluated the different methods on semisynthetic datasets that contained peak pairs at decreasing peak-to-peak distances (i.e. resolution). For each method, we determined a true-positive rate (TPR; number of correct peak calls divided by the total number of true peaks) and FDR (number of false peak calls divided by the number of total peak calls) and indicate the best resolution reached (in base pairs below each method’s name)
3.5 Peakzilla leverages increased resolution of ChIP experiments
To directly demonstrate peakzilla’s ability to make use of increased experimental ChIP resolution, we performed conventional ChIP-seq for Twist from Drosophila embryos with increasingly smaller fragment sizes. For this, the chromatin was sonicated into relatively small DNA fragments and then further trimmed by DNase I digestion before ChIP (see ‘Methods’ section). This yielded three ChIP datasets with estimated fragment sizes of 102, 72 and 49 bp, from which we called peaks with the different peak finders (Fig. 5a). We expected that with decreasing fragment sizes, the width of the identified peaks should decrease and the resolution, i.e. the ability to resolve closely spaced binding sites, should increase. Indeed, the peak regions reported by peakzilla, but not those reported by most other methods, showed decreased width with decreasing fragment sizes (Fig. 5a). More importantly, the resolution, as measured by the minimal peak-to-peak distance (after removing 1% outliers), increased with decreasing fragment sizes for peakzilla, but not for other methods (Fig. 5b). SISSRs, GPS and peakzilla performed well on the small-fragment sample, with peakzilla reaching the highest resolution of all methods. These results demonstrate that the maximum benefit of experimental methods with higher resolution can only be obtained when used together with high-resolution computational methods such as peakzilla.
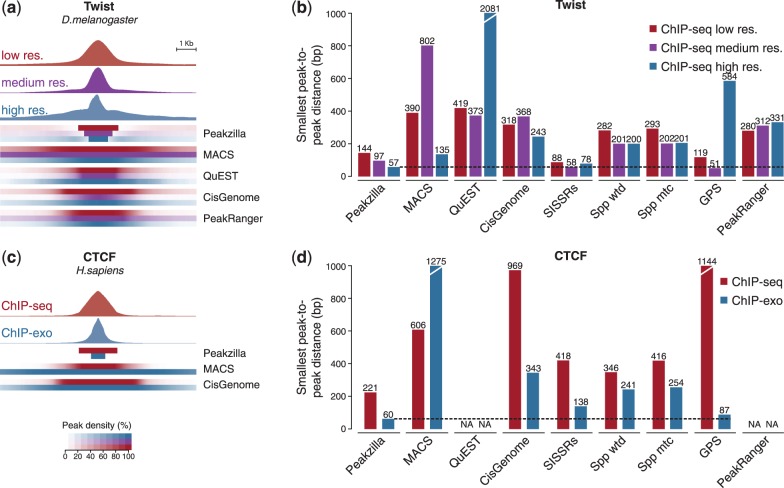
Application to high-resolution data. (a) Average fragment densities and peak regions from low- (red), medium- (purple) and high-resolution (blue) peaks for Twist (best 1000 peaks of each method). SISSRs, spp and GPS are not shown, as they do not report peak regions but only summit positions. (b) Resolution achieved by the different methods at low- (red), medium- (purple) and high-resolution (blue) as calculated as the minimal peak-to-peak distance (after removing 1% outliers for each method). (c) Average fragment densities and peak regions from ChIP-seq (red) and ChIP-exo (blue) peaks for CTCF (best 1000 peaks of each method; QuEST and PeakRanger cannot be used without a control sample). (d) Resolution of the methods calculated as in (b)
3.6 Peakzilla as a peak caller for ChIP-exo data
The recently developed ChIP-exo method adds a lamda exonuclease digestion step after ChIP, which trims the 5′ DNA strand until the cross-linked TFBS (Rhee and Pugh, 2011). This digestion end point can be mapped to the genome using the remaining single-stranded overhang. Because each TFBS can be mapped from both sides, the resulting distribution of mapped breakpoints is also bimodal and resembles that of conventional ChIP-seq, with the ‘fragment sizes’ corresponding directly to the actual sizes of the TF footprints. To our knowledge, no computational method has been specifically developed for the analysis of ChIP-exo data.
We therefore assessed how well peakzilla and other methods perform on ChIP-exo datasets. We called human CTCF binding sites from published ChIP-seq data (Cuddapah et al., 2009) and ChIP-exo data (Rhee and Pugh, 2011). Peakzilla reported estimated fragment sizes of 98 bp for ChIP-seq and 36 bp for ChIP-exo (Fig. 5c) and had the highest resolution (smallest peak-to-peak distance) among all methods tested (Fig. 5d). This suggests that peakzilla is well suited for high-resolution ChIP-seq data, including ChIP-exo.
4 DISCUSSION
Understanding how combinations of TFs bind to DNA to regulate gene expression is one of the most pressing questions of today’s biology. Its importance is witnessed by recent community efforts that aim to determine all functional elements in the genomes of model organisms and the human [e.g. ENCODE (ENCODE Project Consortium, 2004), modENCODE (Celniker et al., 2009), Mouse ENCODE (Mouse ENCODE Consortium et al., 2012)]. The availability of high-throughput sequencing at low cost widely promoted the use of the ChIP-seq methodology and an enormous number of datasets for different TFs from various species, developmental stages or tissues are becoming available. This enables the identification of in vivo binding sites and thus enhancers that contain multiple binding sites for a single TF or multiple different TFs. While it is widely accepted that enhancers are characterized by clusters of TF binding motifs (Berman et al., 2002; Schroeder et al., 2004), it has remained less clear to what extent each of several clustered TF binding motifs is bound in vivo (Yáñez-Cuna et al., 2012). Similarly, potential constraints on the relative distance or orientation of co-bound TFs have remained unclear, yet might be crucial to understand the molecular mechanisms and to decipher the sequence basis of gene regulation (Yáñez-Cuna et al., 2013).
To experimentally address this question, it is important to resolve closely spaced binding sites and precisely predict their location from ChIP-seq data, challenges that current improvements to the ChIP methodology have started to address [e.g. ChIP-exo (Rhee and Pugh, 2011)]. However, while many computational tools exist to identify enriched regions (peaks) from ChIP data (‘peak calling’), many of them are not designed to fully leverage these improvements, e.g. the increased resolution or the vastly increased number of deep sequencing reads of modern deep sequencing (Chen et al., 2012). To meet these challenges, especially the need for discovering TFBSs at high resolution, we have developed a new computational method, peakzilla.
The importance of high resolution and precision is also supported by alternative efforts to correctly position predicted TFBSs, for example, by taking the location of enriched sequence motifs into account (Boeva et al., 2010; Guo et al., 2012; Wu et al., 2010). Although TFBSs predicted by peakzilla coincide well with the known sequence motifs of the respective TFs, it is important to note that we chose to predict the TFBS locations from the ChIP-seq data alone, without taking sequence motifs into account: it is well established that within TFBSs, motifs for different TFs can be more highly enriched in TFBSs than motifs for the precipitated TF itself. For example, the binding sites of most TFs in the early Drosophila embryo are highly enriched in motifs for the TF Zelda (Bradley et al., 2010; Li et al., 2008; Kvon et al., 2012; modENCODE Consortium et al., 2010; Satija and Bradley, 2012; Yáñez-Cuna et al., 2012), such that the Zelda motif–which is sometimes more highly enriched than the motif of the TF of interest–might bias the correct prediction of the TFBSs and possibly hinder the study of relative positioning and orientation of TFBSs.
The combination of maximum experimental resolution and a peak caller like peakzilla thus makes full use of recent ChIP-seq approaches and will be invaluable for testing hypotheses on how combinatorial TF binding realizes the developmental blueprint encoded in the regulatory regions of our genomes. Indeed, closely spaced Twist binding sites resolved by peakzilla coincided and were strongly enriched in known enhancers, corroborating the prevalent model that functional enhancers are characterized by clusters of TFBSs. The increasing number of ChIP studies that determine the in vivo binding sites of TFs at high resolution will prove invaluable for our understanding of enhancer function and transcriptional regulation.
5 METHODS
5.1 Peakzilla algorithm
Initially, peakzilla reads the coordinate files of the mapped reads of the IP and–optionally–control sample. These can stem from either single-end (BED format) or paired-end (BEDPE format) deep sequencing data.
Peakzilla then first determines the average fragment size of the sequencing library to determine the peak size that should result from a true TFBS. For paired-end data, this corresponds directly to the average fragment size (i.e. the average distance of the two mapped ends of each fragment). For single-end data, the average fragment size is estimated from the shift size of positive and negative reads in the top 200 enriched regions in the ChIP sample as described before. Peakzilla then defines peak size as two times the fragment size, as all reads from the ends of fragments immunoprecipitated due to a single TFBS will on average lie in this region.
In a second step, the distribution of positive and negative strand reads that are to be expected is modeled. By default, two normal distributions are used with standard deviations stdev = peak size/5 and locations of their means at one-fourth and three-fourth of the peak size, respectively. Alternatively, the user can choose to estimate the model empirically from the average distribution of reads within the top 200 candidate peaks in the ChIP.
To call TFBSs, peakzilla first scans the genome counting reads within a ‘double-window’: each putative (candidate) TFBS receives the counts of positive strand reads within a window of the fragment size downstream of the candidate TFBS and the negative strand reads within an equivalent window upstream of the candidate TFBS. This scores all candidates with a raw score defined as the normalized read count in the IP sample (normalized to a library size of 1 million reads) minus the normalized read count in the control (i.e. input) sample (note that the correction with a control sample is optional). Final peaks are the candidates with summits that are local maxima at least one fragment length (half peak size) apart from each other. This scanning mode allows for both fast and comprehensive investigation of large genomes at single base resolution.
To obtain a final peak score, each raw score is corrected with a multiplicative distribution score [0 … 1] that assesses the fit of the observed read count distribution to the distribution expected from the model (see above). This fit is assessed by a chi-square test and the chi-square P-value becomes the distribution score, which provides a measure of how likely the candidate peak is a true TFBS (distribution score: 1) or the result of a sequencing artifact (distribution score: 0). Note that to robustly estimate the average fragment size we only count distinct reads, i.e. remove duplicates (before model building). For peak calling, however, we did not remove duplicates but use the model to penalize polymerase chain reaction duplicates. This strategy is more sensitive for datasets on small genomes or with high coverage (see main text).
If a control sample is provided, an empirical false discovery rate (FDR) is calculated for each peak by repeating the peak-calling step (after fragment size estimation) with swapped IP and control sample and scoring the resulting control peaks by the raw and distribution score. This provides for each final peak score the number of true and control peaks that achieve this score or better and thus an FDR estimate.
Peakzilla reports the TFBSs in a BED-like format including the genomic positions, raw, distribution and final score, FDR and a peak number according to each peak’s rank. In addition, the control peaks and a log are reported.
The method is illustrated in a flowchart in Supplementary Figure S3.
Peakzilla can be downloaded from http://github.com/steinmann/peakzilla or http://www.starklab.org/data/peakzilla/
5.2 Program implementation
Peakzilla is implemented in Python 2 and runs on both the standard CPython and the fast PyPy interpreter. The program is freely available under the terms of the General Public License at http://github.com/steinmann/peakzilla. It runs from the command line under any Linux distribution or OSX. The only required argument is the name of the file with the aligned reads from the ChIP sample and–optionally–from the control sample (both BED format). In addition, the following parameters can be used: −m to specify the number of candidate binding sites to use to estimate fragment size (default: 200); −l to limit the candidate regions to lengths above a certain minimum length (which may be necessary if the dataset contains a large number of strong polymerase chain reaction artifacts; default: off [1]); −f to set a FDR cutoff (default: off [100]); −c to set an enrichment cutoff (default: 2); −s to set a score cutoff (default: 1); −l to specify logfile (default: log.txt); −e to use an empirical estimate derived from the data for the model instead of a normal distribution (default: off); −p to specify that the data corresponds to fragments sequenced at both ends (paired-end sequencing; default: off). For a human ChIP-seq dataset with 19.7 million reads and 7.8 in the control peakzilla runs in 4 min and consumes <800 MB of memory under CPython. Using the faster PyPy interpreter reduces time needed for analysis and memory requirements by half. Peakzilla can therefore be run efficiently on any modern desktop computer.
5.3 ChIP-seq datasets
Raw sequencing reads for Twist in D.melanogaster (He et al., 2011) (ArrayExpress accession code E-MTAB-376), CEBPA in Mus musculus (Schmidt et al., 2010) (GEO accession code GSE22078) and PHA-4 in C.elegans (Zhong et al., 2010) (GEO accession code GSE14545) were aligned uniquely using bowtie allowing for three mismatches to the corresponding genomes (assemblies dm3, mm9 and ce6, respectively). For NFkB in Homo sapiens (Kasowski et al., 2010) (GEO accession code GSE19486), Ste12 in S.cerevisiae (Zheng et al., 2010) (GEO accession code GSE19636) and CTCF ChIP-seq (Cuddapah et al., 2009) (GEO accession code GSE12889) and ChIP-exo (Rhee and Pugh, 2011) (which the authors kindly shared) in H.sapiens, already mapped reads were used (assemblies hg18, sarCer2 and hg18, respectively).
5.4 Semisynthetic datasets
We generated semisynthetic control (input) and ChIP samples by subsampling input samples and ChIP peaks: we generated two 30 million bp artificial chromosomes by repeatedly randomly subsampling an arbitrarily selected region from the Twist input sample that showed no strong enrichment, one as a semisynthetic input control and the second as a ChIP background. We next selected several highly ranking peaks from the Twist ChIP sample that were found by all methods, had no other peak nearby and showed a regular fragment density distribution. We subsampled them to yield peaks with an enrichment of 5-fold or higher over background and placed such peaks as pairs with defined peak-to-peak distances into the background every 30 000 bp for 1000 peak-pairs. For each distance between the semisynthetic peak-pairs, we combined the semisynthetic chromosome with the experimental ChIP data for peak calling of the combined set (i.e. parameters are estimated primarily on the experimental dataset as it contains higher peaks).
5.5 High-resolution ChIP-seq
Embryos aged 2–4 h after egg laying were processed and immunoprecipitated with Twist antibodies based on the protocol by He et al. (2011) with slight modifications. Sonication occurs in three microfuges, each with ~80 mg chromatin extracts resuspended in 250 µl A2 buffer, in a Biorupter sonicator for 15 min on high (30 s on and off) at 4°C. After 15 min cooling, the sonication step is repeated, followed by high-speed centrifugation at 4°C for 10 min and pooling of the supernatant (the DNA fragments should be mostly between 200–500 bp). Six hundred microliters of supernatant are then incubated with 120 µl of DNAse I (RNAse-free from NEB, to 0.3 U/µl final concentration) and 80 µl of DNAse I buffer for 30 min at 37°C. To stop DNAse I activity, A2 buffer with 10% sodium dodecyl sulfate is added to a give a final concentration of 1% sodium dodecyl sulfate. The extract is then directly used for ChIP (the DNA fragments should now be mostly between 50 and 200 bp). During Illumina library preparation, the samples are run on a 2% gel at 90 V for ~2 h, and fragments corresponding to ~50, ~75 and ~100 bp inserts are cut out of the gel (slices are ~25 bp thick). The final libraries are run a BioAnalyzer to measure the actual average insert size. The high-resolution ChIP-seq data for Twist is deposited on GEO under the accession code GSE40664.
5.6 Peak calling
The format of the mapped reads was adapted to each method. Peakzilla, SISSRs (Jothi et al., 2008) version 1.4, cisGenome (Ji et al., 2008) version 2.0, Spp (Kharchenko et al., 2011) version 1.8 and GPS (Guo et al., 2010) version 0.10.1 were run with default parameters. MACS (Zhang et al., 2008) version 1.4.1 was run with an mfold parameter 3,30 and the gsize parameter was adapted for each genome. QuEST (Valouev et al., 2008) version 2.4 was run with the following interactive choices: TFBSs with recommended (or relaxed) peak calling parameters. PeakRanger (Feng et al., 2011) read extension length parameter was run using peakzilla’s estimated fragment length. Both QuEST and PeakRanger could not be used for the CTCF samples without a control dataset.
5.7 Functional analyses
We used the known motif CACATGT for Twist and the motifs from JASPAR (Sandelin et al., 2004): snail (sna MA0086.1), dorsal (dl_1 MA0022.1), NFkB (NFKB1 MA0105.1), CEBPA (CEBPA MA0102.2), pha-4 (Foxa2 MA0047.1), Ste12 (STE12 MA0393.1). We searched for motif de novo using MEME (Bailey and Gribskov, 1998) within 31 bp around peak summits and for occurrences of the known motifs using MAST (Bailey and Gribskov, 1998) (from the MEME suite programs version 4.1.1) with a P-value of 10−3 (10−2 for Twist, which corresponds to allowing for one mismatch) in an area of 151 bp (average genomic fragment length) around each peak summit. We called a peak conserved when it overlapped with a peak region in all other Drosophila species from He et al. (2011) (Drosophila simulans, Drosophila yakuba, Drosophila erecta, Drosophila ananassae and Drosophila pseudoobscura). We overlap peaks with known Twist enhancers from He et al. (2011) and the known mesodermal enhancers from Bonn et al. (2012) (only M for mesoderm at stage 5, 6 and 7). To create a set of control peaks, we shuffled peaks randomly within the same chromosomes.
ACKNOWLEDGEMENTS
The authors are grateful to Ho Sung Rhee and Franklin Pugh for kindly sharing the mapped read coordinates for ChIP-exo of human CTCF (Rhee and Pugh, 2011). They would like to thank J. Omar Yáñez-Cuna and Gerald Stampfel (IMP) for discussions, help and advice. A.F.B., J.S., J.A.K., J.Z. and A.S. conceived the project. J.S. wrote peakzilla. A.F.B. benchmarked peakzilla and performed all computational analyses. A.F.B. and A.S. analyzed the data. S.B. and J.Z. designed and performed the high-resolution ChIP-seq experiments. A.F.B., J.Z. and A.S. wrote the manuscript.
Funding: Austrian Ministry for Science and Research through the Genome Research in Austria (GEN-AU) Bioinformatics Integration Network III (to A.S. and A.F.B.); Austrian Research Fund (FWF) (Z_153_B09 to J.A.K. and J.S.); NIH New Innovator (1DP2 OD004561-01 to J.Z., a Pew scholar); European Research Council (ERC) Starting Grant from the European Community’s Seventh Framework Programme (FP7/2007-2013;)/ERC (242922) (to A.S.). Basic research at the IMP is supported by Boehringer Ingelheim.
Conflict of Interest: none declared.
REFERENCES
- Bailey TL, Gribskov M. Combining evidence using p-values: application to sequence homology searches. Bioinformatics. 1998;14:48–54. [Abstract] [Google Scholar]
- Bardet AF, et al. A computational pipeline for comparative ChIP-seq analyses. Nat. Protoc. 2012;7:45–61. [Abstract] [Google Scholar]
- Berman BP, et al. Exploiting transcription factor binding site clustering to identify cis-regulatory modules involved in pattern formation in the Drosophila genome. Proc. Natl Acad. Sci. USA. 2002;99:757–762. [Europe PMC free article] [Abstract] [Google Scholar]
- Boeva V, et al. De novo motif identification improves the accuracy of predicting transcription factor binding sites in ChIP-Seq data analysis. Nucleic Acids Res. 2010;38:e126. [Europe PMC free article] [Abstract] [Google Scholar]
- Bonn S, et al. Tissue-specific analysis of chromatin state identifies temporal signatures of enhancer activity during embryonic development. Nat. Genet. 2012;44:148–156. [Abstract] [Google Scholar]
- Bradley RK, et al. Binding site turnover produces pervasive quantitative changes in transcription factor binding between closely related Drosophila species. PLoS Biol. 2010;8:e1000343. [Europe PMC free article] [Abstract] [Google Scholar]
- Celniker SE, et al. Unlocking the secrets of the genome. Nature. 2009;459:927–930. [Europe PMC free article] [Abstract] [Google Scholar]
- Chen Y, et al. Systematic evaluation of factors influencing ChIP-seq fidelity. Nat. Methods. 2012;9:609–614. [Europe PMC free article] [Abstract] [Google Scholar]
- Cuddapah S, et al. Global analysis of the insulator binding protein CTCF in chromatin barrier regions reveals demarcation of active and repressive domains. Genome Res. 2009;19:24–32. [Europe PMC free article] [Abstract] [Google Scholar]
- ENCODE Project Consortium. The ENCODE (ENCyclopedia Of DNA Elements) Project. Science. 2004;306:636–640. [Abstract] [Google Scholar]
- Feng X, et al. PeakRanger: a cloud-enabled peak caller for ChIP-seq data. BMC Bioinformatics. 2011;12:139. [Europe PMC free article] [Abstract] [Google Scholar]
- Gotea V, et al. Homotypic clusters of transcription factor binding sites are a key component of human promoters and enhancers. Genome Res. 2010;20:565–577. [Europe PMC free article] [Abstract] [Google Scholar]
- Guo Y, et al. Discovering homotypic binding events at high spatial resolution. Bioinformatics. 2010;26:3028–3034. [Europe PMC free article] [Abstract] [Google Scholar]
- Guo Y, et al. High resolution genome wide binding event finding and motif discovery reveals transcription factor spatial binding constraints. PLoS Comput. Biol. 2012;8:e1002638. [Europe PMC free article] [Abstract] [Google Scholar]
- He Q, et al. High conservation of transcription factor binding and evidence for combinatorial regulation across six Drosophila species. Nat. Genet. 2011;43:414–420. [Abstract] [Google Scholar]
- Iyer VR, et al. Genomic binding sites of the yeast cell-cycle transcription factors SBF and MBF. Nature. 2001;409:533–538. [Abstract] [Google Scholar]
- Ji H, et al. An integrated software system for analyzing ChIP-chip and ChIP-seq data. Nat. Biotechnol. 2008;26:1293–1300. [Europe PMC free article] [Abstract] [Google Scholar]
- Johnson DS, et al. Genome-wide mapping of in vivo protein-DNA interactions. Science. 2007;316:1497–1502. [Abstract] [Google Scholar]
- Jothi R, et al. Genome-wide identification of in vivo protein-DNA binding sites from ChIP-Seq data. Nucleic Acids Res. 2008;36:5221–5231. [Europe PMC free article] [Abstract] [Google Scholar]
- Kasowski M, et al. Variation in transcription factor binding among humans. Science. 2010;328:232–235. [Europe PMC free article] [Abstract] [Google Scholar]
- Kharchenko PV, et al. Comprehensive analysis of the chromatin landscape in Drosophila melanogaster. Nature. 2011;471:480–485. [Europe PMC free article] [Abstract] [Google Scholar]
- Kharchenko PV, et al. Design and analysis of ChIP-seq experiments for DNA-binding proteins. Nat. Biotechnol. 2008;26:1351–1359. [Europe PMC free article] [Abstract] [Google Scholar]
- Kvon EZ, et al. HOT regions function as patterned developmental enhancers and have a distinct cis-regulatory signature. Genes Dev. 2012;26:908–913. [Europe PMC free article] [Abstract] [Google Scholar]
- Li X-Y, et al. Transcription factors bind thousands of active and inactive regions in the Drosophila blastoderm. PLoS Biol. 2008;6:e27. [Europe PMC free article] [Abstract] [Google Scholar]
- Lifanov AP, et al. Homotypic regulatory clusters in Drosophila. Genome Res. 2003;13:579–588. [Europe PMC free article] [Abstract] [Google Scholar]
- modENCODE Consortium et al. Identification of functional elements and regulatory circuits by Drosophila modENCODE. Science. 2010;330:1787–1797. [Europe PMC free article] [Abstract] [Google Scholar]
- Mouse ENCODE Consortium et al. An encyclopedia of mouse DNA elements (Mouse ENCODE) Genome Biol. 2012;13:418. [Europe PMC free article] [Abstract] [Google Scholar]
- Pepke S, et al. Computation for ChIP-seq and RNA-seq studies. Nat. Methods. 2009;6:S22–S32. [Europe PMC free article] [Abstract] [Google Scholar]
- Ren B, et al. Genome-wide location and function of DNA binding proteins. Science. 2000;290:2306–2309. [Abstract] [Google Scholar]
- Rhee HS, Pugh BF. Comprehensive genome-wide protein-DNA interactions detected at single-nucleotide resolution. Cell. 2011;147:1408–1419. [Europe PMC free article] [Abstract] [Google Scholar]
- Robertson G, et al. Genome-wide profiles of STAT1 DNA association using chromatin immunoprecipitation and massively parallel sequencing. Nat. Methods. 2007;4:651–657. [Abstract] [Google Scholar]
- Sandelin A, et al. JASPAR: an open-access database for eukaryotic transcription factor binding profiles. Nucleic Acids Res. 2004;32:D91–4. [Europe PMC free article] [Abstract] [Google Scholar]
- Satija R, Bradley RK. The TAGteam motif facilitates binding of 21 sequence-specific transcription factors in the Drosophila embryo. Genome Res. 2012;22:656–665. [Europe PMC free article] [Abstract] [Google Scholar]
- Schmidt D, et al. Five-vertebrate ChIP-seq reveals the evolutionary dynamics of transcription factor binding. Science. 2010;328:1036–1040. [Europe PMC free article] [Abstract] [Google Scholar]
- Schroeder MD, et al. Transcriptional control in the segmentation gene network of Drosophila. PLoS Biol. 2004;2:E271. [Europe PMC free article] [Abstract] [Google Scholar]
- Valouev A, et al. Genome-wide analysis of transcription factor binding sites based on ChIP-Seq data. Nat. Methods. 2008;5:829–834. [Europe PMC free article] [Abstract] [Google Scholar]
- Wilbanks EG, Facciotti MT. Evaluation of algorithm performance in ChIP-seq peak detection. PLoS One. 2010;5:e11471. [Europe PMC free article] [Abstract] [Google Scholar]
- Wu S, et al. ChIP-PaM: an algorithm to identify protein-DNA interaction using ChIP-Seq data. Theor. Biol. Med. Model. 2010;7:18. [Europe PMC free article] [Abstract] [Google Scholar]
- Yáñez-Cuna JO, et al. Uncovering cis-regulatory sequence requirements for context-specific transcription factor binding. Genome Res. 2012;22:2018–2030. [Europe PMC free article] [Abstract] [Google Scholar]
- Yáñez-Cuna JO, et al. Deciphering the transcriptional cis-regulatory code. Trends Genet. 2013;29:11–22. [Abstract] [Google Scholar]
- Zhang Y, et al. Model-based analysis of ChIP-Seq (MACS) Genome Biol. 2008;9:R137. [Europe PMC free article] [Abstract] [Google Scholar]
- Zheng W, et al. Genetic analysis of variation in transcription factor binding in yeast. Nature. 2010;464:1187–1191. [Europe PMC free article] [Abstract] [Google Scholar]
- Zhong M, et al. Genome-wide identification of binding sites defines distinct functions for Caenorhabditis elegans PHA-4/FOXA in development and environmental response. PLoS Genet. 2010;6:e1000848. [Europe PMC free article] [Abstract] [Google Scholar]
Articles from Bioinformatics are provided here courtesy of Oxford University Press
Full text links
Read article at publisher's site: https://doi.org/10.1093/bioinformatics/btt470
Read article for free, from open access legal sources, via Unpaywall:
https://academic.oup.com/bioinformatics/article-pdf/29/21/2705/18533323/btt470.pdf
Citations & impact
Impact metrics
Citations of article over time
Alternative metrics
Article citations
MAE-seq refines regulatory elements across the genome.
Nucleic Acids Res, 52(2):e9, 01 Jan 2024
Cited by: 1 article | PMID: 38038259 | PMCID: PMC10810209
Widespread prevalence of a methylation-dependent switch to activate an essential DNA damage response in bacteria.
PLoS Biol, 22(3):e3002540, 11 Mar 2024
Cited by: 0 articles | PMID: 38466718 | PMCID: PMC10957082
Epigenetic targets in B- and T-cell lymphomas: latest developments.
Ther Adv Hematol, 14:20406207231173485, 30 May 2023
Cited by: 0 articles | PMID: 37273421 | PMCID: PMC10236259
Review Free full text in Europe PMC
An ancestral Wnt-Brachyury feedback loop in axial patterning and recruitment of mesoderm-determining target genes.
Nat Ecol Evol, 6(12):1921-1939, 17 Nov 2022
Cited by: 7 articles | PMID: 36396969
Pan-cancer predictions of transcription factors mediating aberrant DNA methylation.
Epigenetics Chromatin, 15(1):10, 24 Mar 2022
Cited by: 5 articles | PMID: 35331302 | PMCID: PMC8944071
Go to all (37) article citations
Data
Data behind the article
This data has been text mined from the article, or deposited into data resources.
BioStudies: supplemental material and supporting data
Functional Genomics Experiments
- (1 citation) ArrayExpress - E-MTAB-376
GEO - Gene Expression Omnibus (Showing 6 of 6)
- (1 citation) GEO - GSE19486
- (1 citation) GEO - GSE40664
- (1 citation) GEO - GSE22078
- (1 citation) GEO - GSE14545
- (1 citation) GEO - GSE19636
- (1 citation) GEO - GSE12889
Show less
Similar Articles
To arrive at the top five similar articles we use a word-weighted algorithm to compare words from the Title and Abstract of each citation.
BinDNase: a discriminatory approach for transcription factor binding prediction using DNase I hypersensitivity data.
Bioinformatics, 31(17):2852-2859, 07 May 2015
Cited by: 31 articles | PMID: 25957350
annoPeak: a web application to annotate and visualize peaks from ChIP-seq/ChIP-exo-seq.
Bioinformatics, 33(10):1570-1571, 01 May 2017
Cited by: 4 articles | PMID: 28169395 | PMCID: PMC5860050
Identification of C2H2-ZF binding preferences from ChIP-seq data using RCADE.
Bioinformatics, 31(17):2879-2881, 06 May 2015
Cited by: 11 articles | PMID: 25953800 | PMCID: PMC4547615
ChIP-Seq Data Analysis to Define Transcriptional Regulatory Networks.
Adv Biochem Eng Biotechnol, 160:1-14, 01 Jan 2017
Cited by: 7 articles | PMID: 28070596
Review
Funding
Funders who supported this work.
Austrian Science Fund FWF (1)
Asymmetric Cell Division
Dr Juergen Knoblich, Institute of Molecular Biotechnology
Grant ID: Z 153
European Research Council (1)
Regulatory Genomics in Drosophila (Regulatory Genomics)
Dr Alexander Stark, Research Institute of Molecular Pathology GmbH - Vienna
Grant ID: 242922
NIH HHS (1)
Grant ID: DP2 OD004561
 1,†§
1,†§ 




