Abstract
Free full text

The Yeast AMPK Homolog SNF1 Regulates Acetyl Coenzyme A Homeostasis and Histone Acetylation
Abstract
Acetyl coenzyme A (acetyl-CoA) is a key metabolite at the crossroads of metabolism, signaling, chromatin structure, and transcription. Concentration of acetyl-CoA affects histone acetylation and links intermediary metabolism and transcriptional regulation. Here we show that SNF1, the budding yeast ortholog of the mammalian AMP-activated protein kinase (AMPK), plays a role in the regulation of acetyl-CoA homeostasis and global histone acetylation. SNF1 phosphorylates and inhibits acetyl-CoA carboxylase, which catalyzes the carboxylation of acetyl-CoA to malonyl-CoA, the first and rate-limiting reaction in the de novo synthesis of fatty acids. Inactivation of SNF1 results in a reduced pool of cellular acetyl-CoA, globally decreased histone acetylation, and reduced fitness and stress resistance. The histone acetylation and transcriptional defects can be partially suppressed and the overall fitness improved in snf1Δ mutant cells by increasing the cellular concentration of acetyl-CoA, indicating that the regulation of acetyl-CoA homeostasis represents another mechanism in the SNF1 regulatory repertoire.
INTRODUCTION
SNF1 is the budding yeast ortholog of mammalian AMP-activated protein kinase (AMPK). SNF1 and AMPK are highly conserved in eukaryotes and serve as cellular energy sensors and master regulators of metabolism (1–3). The yeast SNF1 complex consists of the catalytic α subunit Snf1p; one of three different regulatory β subunits, Sip1p, Sip2p, or Gal83p; and the stimulatory γ subunit Snf4p (4). The kinase activity of Snf1p is activated by upstream kinases Sak1p, Tos3p, and Elm1p, which phosphorylate activation loop residue Thr210 (5, 6). The activity of these upstream kinases is opposed by the activity of protein phosphatases Reg1p-Glc7p and Sit4, which dephosphorylate Thr210 of Snf1p (7–9). SNF1 is regulated at the dephosphorylation step, and ADP activates SNF1 by protecting it against dephosphorylation (10).
SNF1 regulates transcription by several mechanisms, including phosphorylation of transcriptional activators and repressors; modification of chromatin; and RNA polymerase II, Srb-mediator, and preinitiation complex assembly. One of the best-studied targets of SNF1 is the repressor Mig1p. Upon glucose depletion, Mig1p is phosphorylated by SNF1, which results in the translocation of Mig1p from the nucleus to the cytoplasm (11–13) or relieves the transcriptional repression imposed by Mig1p by altering its interaction with the corepressor Tup1p/Ssn6p (14). In addition to phosphorylating and regulating the transcriptional factors, SNF1 also regulates directly the RNA polymerase II holoenzyme (15, 16).
SNF1 regulates transcription also by a chromatin-based mechanism. SNF1 phosphorylates histone H3 on serine 10 (H3S10), and this modification promotes SAGA and TATA binding protein recruitment at the INO1 gene and acetylation of histone H3 at lysine 14 (H3K14) (17–19). However, since H3S10 phosphorylation is not required for activation of SUC2, GAL1, and HIS3 genes, it is not a general requirement for transcriptional induction of SNF1-dependent genes (19–21). SNF1 phosphorylates Gcn5p in vitro and affects Gcn5p-mediated histone acetylation at the HIS3 and ADY2 promoters and expression of the corresponding genes (21, 22). The notion of the possible involvement of SNF1 in histone acetylation is also supported by the finding that the activation of many SNF1-dependent genes requires histone-acetylation-dependent binding of Adr1p and Cat8p (16, 22).
SNF1 also phosphorylates and regulates metabolic enzymes, including acetyl coenzyme A (acetyl-CoA) carboxylase (ACC) (23–26). ACC catalyzes the carboxylation of acetyl-CoA to malonyl-CoA, the first and rate-limiting reaction in the de novo synthesis of fatty acids (27), and thus affects the concentration of cytosolic acetyl-CoA. Histone acetyltransferases (HATs) use acetyl-CoA as a substrate, and the nucleocytosolic pool of acetyl-CoA regulates global histone acetylation (28–31). We have shown that attenuated expression of the yeast ACC, encoded by the ACC1 gene, results in the globally increased acetylation of chromatin histones and altered transcriptional regulation (32). Since SNF1 phosphorylates and inhibits Acc1p (25, 26), we hypothesized that SNF1 also regulates acetyl-CoA homeostasis and global histone acetylation. In this study, we show that inactivation of SNF1 results in a reduced pool of cellular acetyl-CoA and globally decreased histone acetylation. The transcriptional defects can be partially suppressed and the overall fitness improved in snf1Δ mutant cells by increasing the cellular concentration of acetyl-CoA, indicating that the cellular role of SNF1 includes regulation of acetyl-CoA homeostasis and maintenance of global histone acetylation.
MATERIALS AND METHODS
Yeast strains and media.
All of the yeast strains used in this study are listed in Table 1. Standard genetic techniques were used to manipulate yeast strains and to introduce mutations from non-W303 strains into the W303 background (40). Strains LG329 and MZ031 were derived from JHY200 by replacing the pJH33 plasmid in JHY200 with the pQQ18 or pMK439S10A plasmid, respectively, using 5-fluoroorotic (5-FOA) shuffle (21, 33). Cells were grown in YPD medium (1% yeast extract, 2% Bacto peptone, 2% glucose), in YEP medium (1% yeast extract, 2% Bacto peptone) containing 0.05% glucose, or under selection in synthetic complete (SC) medium containing 2% glucose and, when appropriate, lacking specific nutrients in order to select for a strain with a particular genotype. Repression of the tetO7 promoter is typically accomplished by the addition of doxycycline (41). However, the cells expressing the tetO7-ACC1 allele display only about 50% of the Acc1p enzymatic activity even in the absence of doxycycline (32). Hence, to avoid possible secondary effects of doxycycline on snf1Δ mutant cells, the experiments in this study were performed in the absence of the drug.
Table 1
Yeast strains used in this study
| Strain | Genotype | Source or reference |
|---|---|---|
| W303-1a | MATa ade2-1 his3-11,15 leu2-3,112 trp1-1 ura3-1 ssd1-d2 can1-100 | R. Rothstein |
| W303-1α | MATα ade2-1 his3-11,15 leu2-3,112 trp1-1 ura3-1 ssd1-d2 can1-100 | R. Rothstein |
| W303 | MATa/MATα ade2-1/ade2-1 his3-11,15/his3-11,15 leu2-3,112/leu2-3,112 trp1-1/trp1-1ura3-1/ura3-1 can1-100/can1-100 | R. Rothstein |
| LG362 | W303-1a tetO7-ACC1 | 32 |
| YL558 | MATa trp1 leu2-3,112 ura3-52 snf1Δ-2::TRP1 | 21 |
| MZ008 | W303-1a snf1::TRP1 | This study |
| MZ011 | W303-1a tetO7-ACC1 snf1::TRP1 | This study |
| JHY200 | W303-1a hta1-htb1::Nat hta2-htb2::HPH hht1-hhf1::KAN hht2-hhf2::KAN pJH33 [URA3 CEN ARS HTA1-HTB1 HHT2-HHF2] | 33 |
| LG329 | W303-1a hta1-htb1::Nat hta2-htb2::HPH hht1-hhf1::KAN hht2-hhf2::KAN pQQ18 [CEN LEU2 HTA1-HTB1 HHT2-HHF2] | This study |
| MZ031 | W303-1a hta1-htb1::Nat hta2-htb2::HPH hht1-hhf1::KAN hht2-hhf2::KAN pMK439S10A [CEN LEU2 HTA1-HTB1 hht2-S10A-HHF2] | This study |
| FY1292 | MATa gcn5::HIS3 leu2Δ1 lys2-173 trp1Δ63 ura3-52 his3Δ200 arg4-12 | 34 |
| LG258 | W303-1α gcn5::HIS3 | This study |
| YTT2256 | W303-1a yng2::NatMX | 35 |
| DY4548 | W303-1α rpd3::LEU2 | 36 |
| DY5068 | W303-1α hda1::URA3 | 36 |
| LG167 | W303-1α hda1::URA3 | This study |
| LG170 | W303-1a rpd3::LEU2 | This study |
| MAP12 | W303-1a mig1:: kanMX | 37 |
| MZ017 | W303-1a snf1::TRP1 hda1::URA3 | This study |
| MZ020 | W303-1a snf1::TRP1 rpd3::LEU2 | This study |
| MZ048 | W303-1a snf1::TRP1 gcn5::HIS3 | This study |
| MZ059 | W303-1a snf1::TRP1 yng2::NatMX | This study |
| MZ293 | W303-1a gcn5::HIS3 tetO7-ACC1 | This study |
| MZ287 | W303-1a gcn5::HIS3 snf1::TRP1 tetO7-ACC1 | This study |
| MZ311 | W303-1a yng2::NatMX tetO7-ACC1 | This study |
| MZ316 | W303-1a yng2::NatMX snf1::TRP1 tetO7-ACC1 | This study |
| MZ184 | W303-1a mig1:: kanMX tetO7-ACC1 | This study |
| MZ245 | W303-1a mig1:: kanMX snf1::TRP1 | This study |
| MZ182 | W303-1a mig1:: kanMX snf1::TRP1 tetO7-ACC1 | This study |
| MZ297 | W303-1a mig1:: kanMX gcn5::HIS3 | This study |
| MZ298 | W303-1a mig1:: kanMX gcn5::HIS3 snf1::TRP1 | This study |
| MZ303 | W303-1a mig1:: kanMX gcn5::HIS3 tetO7-ACC1 | This study |
| MZ289 | W303-1a mig1:: kanMX gcn5::HIS3 snf1::TRP1 tetO7-ACC1 | This study |
| MZ313 | W303-1a mig1:: kanMX yng2::NatMX | This study |
| MZ306 | W303-1a mig1:: kanMX yng2::NatMX snf1::TRP1 | This study |
| MZ314 | W303-1a mig1:: kanMX yng2::NatMX tetO7-ACC1 | This study |
| MZ308 | W303-1a mig1:: kanMX yng2::NatMX snf1::TRP1 tetO7-ACC1 | This study |
| MZ411 | W303-1a mig1:: kanMX snf1::TRP1 hda1::URA3 | This study |
| MZ408 | W303-1a mig1:: kanMX snf1::TRP1 rpd3::LEU2 | This study |
| YHT751 | BY4741 acs1::HIS3 acs2::HygMX [pHT215 acs2-Ts1-CEN-URA3] pda1::kanMX pot1::LEU2 | 28 |
| MZ484 | W303-1a acs1::HIS3 | This study |
| MZ488 | W303-1a mig1:: kanMX acs1::HIS3 | This study |
| MZ490 | W303-1a snf1::TRP1 acs1::HIS3 | This study |
| MZ494 | W303-1a mig1:: kanMX snf1::TRP1 acs1::HIS3 | This study |
| BY4843 | BY4741 MATa SWI4-QQ-MYC::KAN (K1016Q:K1066Q) | 38 |
| BY4844 | BY4741 MATa SWI4-RR-MYC::KAN (K1016R:K1066R) | 38 |
| MZ438 | W303-1a SWI4-QQ-MYC::KAN (K1016Q:K1066Q) | This study |
| MZ429 | W303-1a SWI4-RR-MYC::KAN (K1016R:K1066R | This study |
| MZ439 | W303-1a snf1::TRP1 SWI4-QQ-MYC::KAN (K1016Q:K1066Q) | This study |
| MZ430 | W303-1a snf1::TRP1 SWI4-RR-MYC::KAN (K1016R:K1066R) | This study |
| LG465 | W303-1a SIP2-TAP::HisMx | This study |
| MZ373 | W303-1a snf1::TRP1 SIP2-TAP::HisMx | This study |
| YER111C | BY4741 SWI4-TAP::HisMx | Open Biosystems |
| MZ497 | W303-1a SWI4-TAP::HisMx | This study |
| MZ500 | W303-1a snf1::TRP1 SWI4-TAP::HisMx | This study |
| YSC6272 | BY4741 snf4::KanMX | Open Biosystems |
| YPDahl55 | W303-1a sak1::KanMX elm1::KanMX tos3::TRP1 | 39 |
| MZ524 | W303-1a snf1::TRP1[pSNF1-K84R] | This study, 7 |
| MZ525 | W303-1a snf1::TRP1[pSNF1-T210A] | This study, 7 |
Western blotting.
Yeast cells were inoculated to an A600 of 0.1 and grown in YPD medium to an A600 of 1.0. Four A600 units were harvested and immediately boiled in SDS sample buffer. Denatured proteins were separated on a 15% denaturing polyacrylamide gel and Western blotting with anti-histone H3 polyclonal antibody (ab1791; Abcam) at a dilution of 1:1000, anti-acetyl histone H3 (Lys14) polyclonal antibody (acH3K14; 07-353, Upstate Biotechnology) at a dilution of 1:500, and anti-hyperacetylated histone H4 polyclonal antibody (acH4K5,8,12,16; 06-946; Upstate Biotechnology) at a dilution of 1:1,000 was carried out as described previously (32). Western blot assays were quantified with NIH ImageJ software (W. S. Rasband, ImageJ, U.S. National Institutes of Health, http://imagej.nih.gov/ij/).
Real-time RT-PCR.
Real-time reverse transcription (RT)-PCR with the following primers was performed as described previously (32): ACT1, 5′-TATGTGTAAAGCCGGTTTTGC-3′ and 5′-GACAATACCGTGTTCAATTGGG-3′; SUC2, 5′-TCAATGACAAACGAAACTAGCG-3′ and 5′-GGCATCTTTTTCATCGTACCAC-3′; GAL4, 5′-ACTGTCTTCTATCGAACAAGCATG-3′ and 5′-TCCCAGTTGTTCTTCAGACACTT-3′; REG2, 5′-ATGTGAGAAGCTCGAACAACACT-3′ and 5′-TCTCCAAAAAAGGTTCTCCAATC-3′; MAL33, 5′-CAAGTATGCATGCGACTATTGTC-3′ and 5′-GTTGCTGATATGTGCAATCGAA-3′; ACS1, 5′-CTCTGCCGTACAATCATCAAAAC-3′ and 5′-CCGAAGTCAAATGTTCATACTCAT-3′; CAT8, 5′-TCATTAGAACTCTGGGTTCACAAG-3′ and 5′-GTCTTGATCATTGCTTCCTTCAC-3′; RPS11B, 5′-TGTCCACTGAATTAACTGTTCAATC-3′ and 5′-CATCTCTTGGTTCTCTTGGAAGTC-3′; RPS22B, 5′-AGCTGATGCTTTGAATGCCA-3′ and 5′-TTCGCCAATGTAACCATGCT-3′; HHT1/2, 5′-GAAGCCTCACAGATATAAGCCAG-3′ and 5′-ATCTTGAGCGATTTCTCTGACC-3′; HHF1/2, 5′-CCAAGCGTCACAGAAAGATTCTA-3′ and 5′-ACCAGAAATACGCTTGACACCA-3′; HTA1/2, 5′-CGGTGGTAAAGGTGGTAAAGC-3′ and 5′-TGGAGCACCAGAACCAATTC-3′; HTB1/2, 5′-CAAAGTTTTGAAGCAAACTCACCC-3′ and 5′-GCCAATTTAGAAGCTTCAGTAGC-3′). Primer pairs for the canonical histones were designed so that they measure the expression of both genes for that particular histone (HTA1 and HTA2, HTB1 and HTB2, HHT1 and HHT2, and HHF1 and HHF2).
ChIP assays.
In vivo chromatin cross-linking and immunoprecipitation were performed essentially as described previously (32). Immunoprecipitation was performed with the following antibodies: for anti-histone H3 (Abcam ab1791), anti-acetyl-histone H3 (Lys14) (Upstate Cell Signaling Solutions 07-353), and anti-hyperacetylated histone H4 (Penta) (Upstate Cell Signaling Solutions 06-946). The primers used for real-time PCR are as follows: POL1, 5′-TCCTGACAAAGAAGGCAATAGAAG-3′ and 5′-TAAAACACCCTGATCCACCTCTG-3′; INO1, 5′-ACGAGCTGCTCACCAAGTACAG-3′ and 5′-TGAACACGTAGTCTTGAACAGTGG-3′; UBC8, 5′-TTGGAGGTTACATGTAGAGTTGCC-3′ and 5′-CCCGATGCGATATCAATGTTAG-3′; INT6, 5′-GGAAGCGAAATTCAAGGGTTG-3′ and 5′-TTGCATGATTGTGCATACGC-3′; SUC2, 5′-CGACTTTTTTTTTTTGGATTTCG-3′ and 5′-GCACAAGAACAAGAGAATGTTTTG-3′; REG2, 5′-ATGTGAGAAGCTCGAACAACACT-3′ and 5′-TCTCCAAAAAAGGTTCTCCAATC-3′; HXK1, 5′-CATTTAGGTCCAAAGAAACCACA-3′ and 5′-GGTCTCGCTGTCAACTGTAAACA-3′; GAL4, 5′-ACTGTCTTCTATCGAACAAGCATG-3′ and 5′-TCCCAGTTGTTCTTCAGACACTT-3′; MAL31, 5′-AAAAAAGACAGGAACGACTCACAC-3′ and 5′-ACTTTTCTTACCTTGCTCCTCCA-3′; MAL33, 5′-CAAGTATGCATGCGACTATTGTC-3′ and 5′-GTTGCTGATATGTGCAATCGAA-3′; HHT2, 5′-CTTCTTGTGACCGCAGTTGTATAT-3′ and 5′-AAAGGGGAAGAACAGTTGGAAG-3′; HHF2, 5′-GGCTGTTGTTATTCGGCTAGAT-3′ and 5′-CTTTCACCGCCTCATCCTAATAT-3′; ACT1, 5′-CTCTTGTATTCTTCCTTCCCCT TTC-3′ and 5′-ATGGTGCAAGCGCTAGAACATAC-3′; ADH1, 5′-AATCCCACGGTAAGTTGGAATAC-3′ and 5′-AAGCGTGCAAGTCAGTGTGAC-3′; RPS11B, 5′-CTTTACGTTTCCCTTCTCTGCTG-3′ and 5′-TTTCCCTGGCTTGATACGTTTC-3′), RPS22B, 5′-GCCCATGTGTTGGAGGGAAGG-3′ and 5′-ATCAGCTAAAACGGAAGAGCGAG-3′; MDN1, 5′-GTACCTCTGAGTTAATTTGACATCG-3′ and 5′-GGACATTCTCAGCAGATTATAACGG-3′; RNY1, 5′-CGGGAATCATGAGTCCTTATG-3′ and 5′-AGCACCCCACTCAGTATAGCA-3′).
Acetyl-CoA and malonyl-CoA assays.
Cells were grown in YPD medium to an A600 of 1.0. Sodium azide was added to a final concentration of 10 mM, and 3 × 108 cells were harvested by centrifugation and lysed in 200 μl of 10% perchloric acid with prechilled glass beads. The lysate was neutralized with 10 M KOH to pH 7.5. Acetyl-CoA and malonyl-CoA were assayed by enzyme-linked immunosorbent assay (ELISA) (Antibodies-online GmbH; ABIN457115 and ABIN366452).
Immunoprecipitation.
Immunoprecipitation of acetylated proteins was performed as described previously (32).
Statistical analysis.
The statistical analyses were performed by chi-square (χ2) test or analysis of variance with Microsoft Excel 2007 software, with the level of significance set at P < 0.05.
RESULTS
Inactivation of SNF1 affects acetyl-CoA homeostasis and leads to hypoacetylation of histones H3 and H4.
Histone acetylation depends on intermediary metabolism for supplying acetyl-CoA as a substrate for HATs in the nucleocytosolic compartment (28). Cytosolic acetyl-CoA is also used by acetyl-CoA carboxylase to yield malonyl-CoA, a precursor for de novo synthesis of fatty acids (27). We showed that the activity of acetyl-CoA carboxylase Acc1p regulates homeostasis of nucleocytosolic acetyl-CoA and acetylation of histones and nonhistone proteins (32). Since SNF1 in yeast and AMPK in mammalian cells directly phosphorylate and inactivate Acc1p (23–26), we speculated that inactivation of SNF1, which results in increased Acc1p activity, would promote the conversion of acetyl-CoA to malonyl-CoA. This would result in a decreased level of acetyl-CoA that is available to HATs and would lead to hypoacetylation of chromatin histones. We also hypothesized that it would be possible to partially suppress the effect of snf1Δ on the homeostasis of acetyl-CoA by reducing the activity of Acc1p by using the tetO7-ACC1 allele (26). Expression of genes under the regulatable tetO7 promoter can be repressed by the addition of doxycycline (41); however, we found that cells expressing the tetO7-ACC1 allele display only about 50% of the normal Acc1p enzymatic activity, even in the absence of doxycycline (32).
To test whether the snf1Δ mutation affects acetyl-CoA homeostasis, we determined the cellular level of acetyl-CoA and malonyl-CoA in wild-type and snf1Δ, tetO7-ACC1, and snf1Δ tetO7-ACC1 mutant cells (Fig. 1A and andB).B). As expected, the cellular level of acetyl-CoA in snf1Δ mutant cells was reduced to 30% of the wild-type level. On the other hand, the malonyl-CoA level in snf1Δ mutant cells was 8-fold higher than the level in wild-type cells. Cells expressing the tetO7-ACC1 allele displayed an about 2-fold higher level of acetyl-CoA, while the malonyl-CoA level was reduced to about 30% of the wild-type level. These changes are consistent with a reduced enzymatic activity of Acc1p in tetO7-ACC1 cells (32). Introduction of the tetO7-ACC1 allele into snf1Δ mutant cells mitigated the effect of snf1Δ mutation, elevated the acetyl-CoA level to 137% of the wild-type level, and reduced the malonyl-CoA to about 42% of the wild-type level. These results suggest that SNF1 plays a role in the regulation of acetyl-CoA homeostasis even when cells are grown in the presence of glucose and SNF1 is not fully active. Our results reflect the total cellular levels of acetyl-CoA and malonyl-CoA, since current methods do not allow the measurement of acetyl-CoA separately in the nucleocytosolic compartment and in mitochondria. However, glucose represses the tricarboxylic acid cycle and respiration in S. cerevisiae and only a very small fraction of glycolytically produced pyruvate is translocated into mitochondria and converted to acetyl-CoA by the pyruvate dehydrogenase complex. Thus, the mitochondrial pool of acetyl-CoA in yeast cells is very small and does not contribute significantly to the cellular level of acetyl-CoA (42). In addition, malonyl-CoA is only cytosolic and its increased level indicates increased conversion of acetyl-CoA to malonyl-CoA in snf1Δ mutant cells (Fig. 1B).
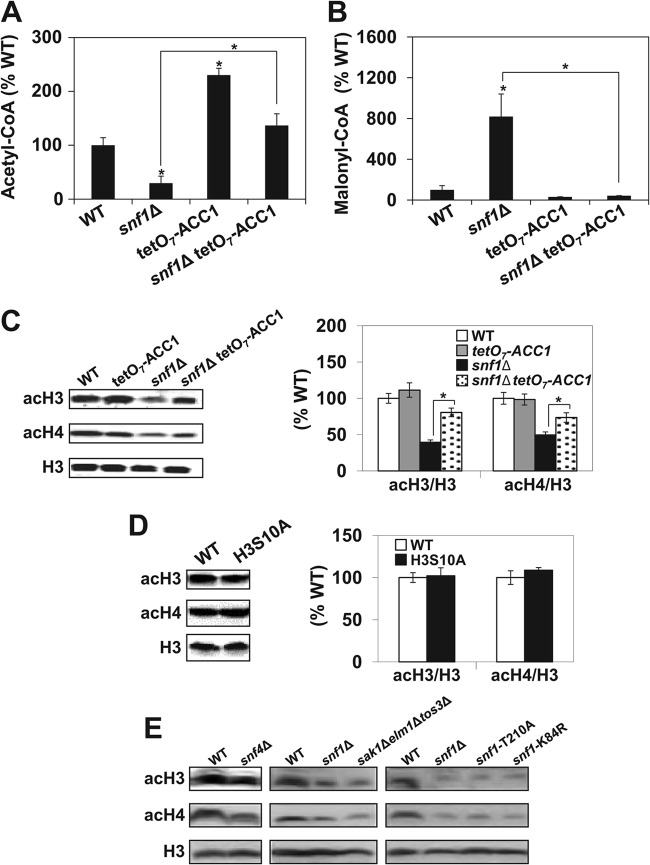
Inactivation of SNF1 affects acetyl-CoA homeostasis and results in hypoacetylation of histones H3 and H4. Acetyl-CoA (A) and malonyl-CoA (B) were assayed in cell lysates of the indicated strains by ELISA. The concentrations of acetyl-CoA and malonyl-CoA in wild-type (WT) cells were 1.8 nmol/107 cells and 21.3 pmol/107 cells, respectively. (C) snf1Δ mutant cells display lower levels of acH3 and acH4. The indicated strains were grown in YPD medium to an A600 of 1.0 at 30°C and analyzed by Western blotting with antibodies against histone H3 acetylated at lysine 14 (acH3), hyperacetylated histone H4 (acH4), and total histone H3. The experiment was performed three times, and representative results are shown. The intensity of each acH3 and acH4 band was quantified by densitometry and normalized with H3 as a loading control of each lane. (D) Phosphorylation of serine 10 of histone H3 does not affect the acetylation of bulk histones. Wild-type and H3S10A mutant cells were grown and analyzed as described above. (A to D) The experiments were repeated three times, and the results are shown as means ± standard deviations. Values that are statistically significantly different (P < 0.05) from the wild-type values are indicated by an asterisk. Values that are statistically significantly different (P < 0.05) from each other are indicated by a bracket and an asterisk. (E) Mutations that affect the activity of the SNF1 complex result in the hypoacetylation of histones H3 and H4. The indicated strains were grown in YPD medium to an A600 of 1.0 at 30°C and analyzed by Western blotting with antibodies against histone H3 acetylated at lysine 14 (acH3), hyperacetylated histone H4 (acH4), and total histone H3. The experiment was performed three times, and representative results are shown.
To determine whether the lower cellular level of acetyl-CoA in snf1Δ mutant cells affects histone acetylation, we determined the acetylation of bulk histones (Fig. 1C). Western blot analysis showed that the acetylation of both histones H3 and H4 is reduced in snf1Δ mutant cells and this defect is partially suppressed by the tetO7-ACC1 allele (Fig. 1C). However, SNF1 phosphorylates histone H3 on serine 10 (H3S10) and this modification promotes SAGA recruitment and acetylation of histone H3 at lysine 14 (H3K14) at some, but not all, promoters (17–19). To address whether the changes in the acetylation of bulk histones are due to the Snf1p-mediated H3S10 phosphorylation, we tested the histone acetylation level in a strain that expresses a nonphosphorylatable mutant form (serine 10 to alanine) of histone H3 (H3S10A) (21). Comparable histone H3 and H4 acetylation levels were found in both wild-type and mutant cells (Fig. 1D). This result shows that H3S10 phosphorylation is not responsible for the decreased acetylation of bulk histones H3 and H4 in snf1Δ mutant cells.
To test whether other mutations that affect SNF1 activity also result in global histone hypoacetylation, we measured the acetylation of bulk histones in snf4Δ, snf1-T210A, snf1-K84R, and sak1Δelm1Δtos3Δ mutants. SNF4 encodes the stimulatory subunit of the SNF1 complex (4). Phosphorylation of the activation loop Thr210 of Snf1p by the upstream kinases Sak1p, Elm1p, and Tos3p activates SNF1 kinase activity (5, 6). The snf1-K84R allele has a mutation in the conserved lysine residue that eliminates the catalytic activity of Snf1p by disrupting the ATP binding domain (43). All of the mutants tested displayed hypoacetylation of bulk histones comparable to that of the snf1Δ mutant (Fig. 1E).
snf1Δ mutant cells display globally decreased untargeted acetylation of chromatin histones.
To test whether SNF1 regulates histone acetylation globally or only at specific loci, we used chromatin immunoprecipitation (ChIP) to evaluate the occupancy of histone H3 acetylated at lysine 14 (acH3K14), as well as hyperacetylated histone H4 (acH4K5,8,12,16), in the promoter regions of SNF1-dependent genes (Fig. 2A) and SNF1-independent loci (Fig. 2B). The SNF1-dependent genes included INO1, UBC8, SUC2, REG2, HXK1, GAL4, MAL31, MAL33, HHT2, and HHF2. The SNF1-independent loci included promoters of ACT1, ADH1, RPS11B, MDN1, and RNY1 and the intergenic region in chromosome VI (INT6; coordinates 261412 to 261538). Expression of INO1 and UBC8 is upregulated in the rpd3Δ mutant and downregulated in the snf1Δ mutant (44, 45). SUC2, REG2, HXK1, GAL4, MAL31, and MAL33 are repressed by glucose in a Mig1p/Mig2p-dependent manner (46). The intergenic region was selected since it is 11 kb distant from the nearest coding region. We used anti-H3 antibody, which recognizes the C-terminal region of H3, which is not posttranslationally modified. The ChIP signal obtained with this antibody thus represents the total H3 occupancy and can be used to calculate the histone acetylation levels per nucleosome content (47). To account for differences in nucleosome density at different genomic loci, we corrected the acH3 and acH4 occupancies for histone H3 content and generated values that represent acetylation per nucleosome (Fig. 2). The acetylation of histones H3 and H4 per nucleosome in promoters of SNF1-dependent genes was 1.2 to 1.9 and 1.3 to 2.1 times lower in snf1Δ mutant cells than in wild-type cells, respectively. The histone H3 and H4 acetylation in snf1Δ mutant cells was increased 1.1- to 1.9- and 1.1- to 2.1-fold by the introduction of the tetO7-ACC1 allele, respectively (Fig. 2A). The acetylation of histones H3 and H4 per nucleosome in promoters of SNF1-independent loci was 1.2 to 1.9 and 1.4 to 1.8 times lower in snf1Δ mutant cells than in wild-type cells, respectively. The histone H3 and H4 acetylation in promoters of SNF1-independent genes of snf1Δ mutant cells was increased 1.1- to 2.8- and 1.2- to 1.8-fold by introducing the tetO7-ACC1 allele, respectively (Fig. 2B).

snf1Δ mutant cells display decreased untargeted acetylation of chromatin histones. The indicated strains were grown at 30°C in YPD medium to an A600 of 1.0. ChIP experiments were performed with antibodies against total histone H3 (H3), histone H3 acetylated at lysine 14 (acH3), and hyperacetylated histone H4 (acH4). Occupancies of H3, acH3, and acH4 were determined in the promoter regions of SNF1-dependent genes (A) and SNF1-independent loci (B). Acetylation per nucleosome was calculated as ratios of acH3 to total H3 and acH4 to total H3. The experiments were repeated three times, and the results are shown as means ± standard deviations. Values that are statistically significantly different (P < 0.05) from the wild-type (WT) values are indicated by an asterisk. Values that are statistically significantly different (P < 0.05) from each other are indicated by a bracket and an asterisk.
To test whether the decreased acetylation of chromatin histones found in promoters and intergenic regions of snf1Δ mutant cells is due to altered H3S10 phosphorylation, we used ChIP to evaluate the occupancy of acetylated histones also in the H3S10A mutant. The acetylation of histones H3 and H4 per nucleosome in promoters and intergenic regions differed less than 3% and 6% between H3S10A mutant and wild-type cells, respectively (Fig. 3A and andB).B). This indicates that phosphorylation of histone H3 at serine 10 is not responsible for the decreased acetylation of histones H3 and H4 at the tested loci in snf1Δ mutant cells. The decreased acetylation of both histones H3 and H4 in snf1Δ mutant cells and suppression by the tetO7-ACC1 allele at all SNF1-dependent loci, as well as SNF1-independent loci, suggest that inactivation of SNF1 decreases the acetylation of chromatin histones in a global untargeted manner and are in agreement with the decreased acetylation of bulk histones measured by Western blot analysis (Fig. 1C).
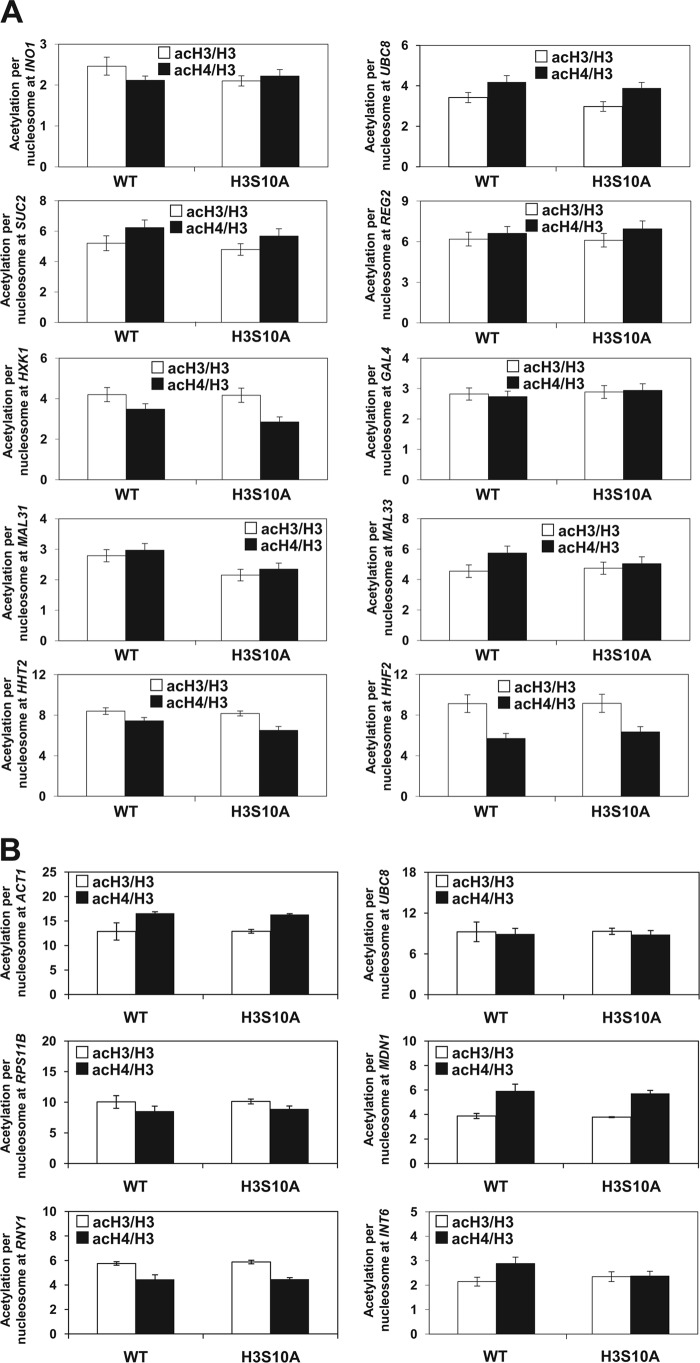
Phosphorylation of histone H3 at serine 10 does not affect the acetylation of chromatin histones. The indicated strains were grown at 30°C in YPD medium to an A600 of 1.0. ChIP experiments were performed with antibodies against total histone H3 (H3), histone H3 acetylated at lysine 14 (acH3), and hyperacetylated histone H4 (acH4). Occupancies of H3, acH3, and acH4 were determined in the promoter regions of SNF1-dependent genes (A) and SNF1-independent loci (B). Acetylation per nucleosome was calculated as ratios of AcH3 to total H3 and acH4 to total H3. The experiments were repeated three times, and the results are shown as means ± standard deviations. The acetylation per nucleosome in wild-type (WT) and H3S10A cells was not statistically significantly different at any of the loci tested.
Inactivation of SNF1 leads to hypoacetylation of nonhistone proteins.
In addition to histones, acetylation has been shown to regulate other proteins (38, 48, 49). To test whether the decreased cellular level of acetyl-CoA in snf1Δ mutant cells affects the acetylation of nonhistone proteins, we assayed the acetylation of Sip2p and Swi4p by immunoprecipitation with anti-pan-acetyl-lysine antibody (48). Sip2p is one of the regulatory β subunits of the SNF1 complex. Sip2p is acetylated by NuA4 (49), and the elevated level of acetyl-CoA in tetO7-ACC1 cells results in increased Sip2p acetylation (32). Swi4p binds Swi6p to form transcriptional complex SBF (50). The acetylation of Swi4p regulates the interaction with Swi6p and thus the SBF-dependent gene transcription (38). Our results show that the acetylation of both Sip2p and Swi4p is decreased in snf1Δ mutant cells in comparison with wild-type cells (Fig. 4) and suggests that in addition to histones, SNF1 is important also for the normal acetylation of nonhistone proteins.

snf1Δ mutant cells display reduced acetylation of nonchromatin proteins. Wild-type (WT) and snf1Δ mutant cells were grown in YPD medium at 30°C to an A600 of 1.0. Acetylated proteins were immunoprecipitated (IP) with anti-acetyllysine antibody or control antibody (normal mouse IgG) and analyzed by Western blotting with antibodies against the tandem affinity purification (TAP) tag. Typical results from three independent experiments are shown. The intensity of each immunoprecipitated band was quantified by densitometry and expressed as a percentage of the wild-type value ± the standard deviation. The asterisks indicate that the values were statistically significantly different (P < 0.05) from the wild-type value.
SNF1 genetically interacts with HATs and HDACs.
If SNF1 promotes histone acetylation and plays a role in establishing the global level of histone acetylation, then inactivation of histone acetyltransferases (HATs) in snf1Δ mutant cells should result in a synthetic growth defect, while inactivation of histone deacetylases (HDACs) should improve the growth rate and fitness of snf1Δ mutant cells. To test this hypothesis, we introduced the yng2Δ, gcn5Δ, hda1Δ, and rpd3Δ mutations into snf1Δ mutant cells. Yng2p is a subunit of NuA4 and piccolo NuA4 complexes and regulates the HAT activity of Esa1p. Gcn5p is the catalytic subunit of the related HATs SAGA, ADA, and SLIK. Rpd3p is the catalytic subunit of the class I Sin3/Rpd3 HDAC, and Hda1p is the catalytic subunit of the class II HDA1 HDAC complex. SAGA and NuA4 are the major HATs in yeast, responsible for most of the HAT activity, while the Sin3/Rpd3 and Hda1 complexes account for most of the HDAC activity (51). Introduction of the hda1Δ or rpd3Δ mutation into snf1Δ mutant cells did not affect the growth rate at 30°C; however, it partially suppressed their temperature sensitivity (Fig. 5A). On the other hand, both the yng2Δ and gcn5Δ mutations produced a negative synthetic phenotype with the snf1Δ mutation (Fig. 5B). The genetic interactions between the snf1Δ mutation and mutations in the major yeast HDACs and HATs thus indicate that SNF1 is involved in the control of the global level of histone acetylation.

The snf1Δ mutation displays genetic interactions with mutations in HATs and HDACs. (A) Inactivation of HDA1 or RPD3 partially suppresses the temperature sensitivity of snf1Δ mutant cells. Tenfold serial dilutions of the indicated strains were spotted onto YPD plates and grown for 3 days at 30 or 35°C. (B) Strains with YNG2 or GCN5 deleted display a synthetic growth defect with the snf1Δ mutation. Tenfold serial dilutions of the indicated strains were spotted onto YPD plates and grown for 3 days at 30°C. Typical results from three independent experiments are shown. WT, wild type.
Reduced ACC1 expression suppresses the stress sensitivity of snf1Δ mutant cells.
In addition to its importance in the regulation of metabolism, SNF1 was also implicated in responses to a variety of stresses (52–55). The cellular response to many stresses requires rapid reprogramming of the transcriptional machinery (56, 57). Since active transcription generally correlates with increased acetylation of promoter histones (58, 59), we reasoned that a reduced supply of acetyl-CoA and globally decreased histone acetylation in snf1Δ mutant cells would interfere with the appropriate stress response. To test whether the histone hypoacetylation of snf1Δ mutant cells is partially responsible for stress sensitivity, we measured the growth rates of the snf1Δ and snf1Δ tetO7-ACC1 mutant strains under different stress conditions. The snf1Δ mutant grows somewhat slower than the wild type at 30°C but is not able to grow at 35°C. The introduction of the tetO7-ACC1 allele into snf1Δ mutant cells improves their growth rate at 30°C only slightly but clearly suppresses the growth defect of snf1Δ mutant cells at 35°C, at pH 7.5, or in the presence of 50 mM hydroxyurea (Fig. 6). This suppression was abolished by the introduction of the gcn5Δ or yng2Δ mutation, indicating that the activities of SAGA and NuA4 are required for suppression of the stress sensitivity of snf1Δ mutant cells by the tetO7-ACC1 allele. The tetO7-ACC1 allele in snf1Δ mutant cells increased the acetyl-CoA level above the wild-type level (Fig. 1A) but failed to completely suppress the growth defect of snf1Δ mutant cells (Fig. 6). This result was expected, since SNF1 phosphorylates and regulates a number of other proteins. Altogether, the results suggest that the sensitivity of snf1Δ mutant cells to different stresses is suppressed by the increased level of acetyl-CoA brought about by the reduced activity of Acc1p and is not caused by reduced H3S10 phosphorylation (Fig. 6).
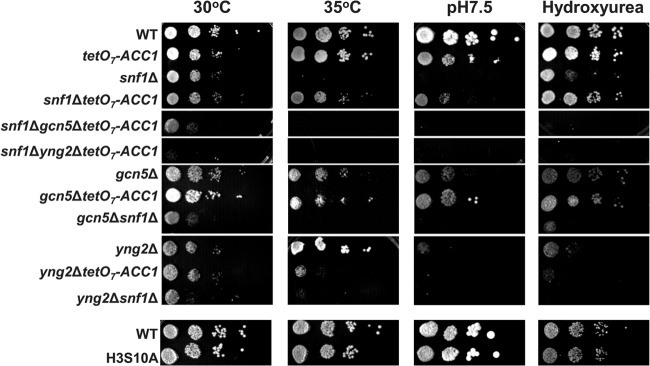
Reduced ACC1 expression partially suppresses the stress sensitivity of snf1Δ mutant cells in a HAT-dependent manner. Tenfold serial dilutions of the indicated strains were spotted onto YPD plates and grown for 2 days at 30 or 35°C, grown for 6 days on YPD plates at pH 7.5, or grown for 3 days on YPD plates containing 50 mM hydroxyurea. Typical results from three independent experiments are shown. WT, wild type.
Derepression of ACS1 elevates the acetyl-CoA level and histone acetylation and suppresses the snf1Δ mutation.
One of the key substrates of SNF1 is the repressor Mig1p. To explore whether the hda1Δ or rpd3Δ mutation would synergize with the mig1Δ mutation to suppress snf1Δ, we constructed the corresponding strains and measured their growth rates. Introduction of the mig1Δ mutation partially suppressed the growth defect of snf1Δ mutant cells; however, introduction of the hda1Δ or rpd3Δ mutation into a snf1Δ mig1Δ mutant strain did not provide additional suppression under any of the conditions tested (Fig. 7A). This unexpected result could be explained by assuming that mig1Δ suppresses snf1Δ by increasing the level of acetyl-CoA. We hypothesized that this could occur by derepression of ACS1. ACS1 and ACS2 encode two yeast acetyl-CoA synthetases that catalyze the ligation of acetate and CoA to yield acetyl-CoA. On glucose-containing laboratory media, ACS2 is essential and ACS1 is repressed (28, 60). Upon glucose exhaustion, the expression of ACS1 is activated by transcriptional factor Cat8p (61). In the presence of glucose, the expression of CAT8 is repressed by Mig1p/Mig2p (46). We hypothesized that the deletion of MIG1 would increase the expression of CAT8 and consequently ACS1 (Fig. 7B). In this model, the suppression of snf1Δ by mig1Δ would require ACS1. To test this possibility, we introduced the acs1Δ mutation into the snf1Δ mig1Δ mutant strain and measured its growth rate. Indeed, the full suppression of snf1Δ by mig1Δ required ACS1 (Fig. 7C). This observation is in agreement with the increased expression of the CAT8 and ACS1 genes in snf1Δ mig1Δ mutant cells in comparison with snf1Δ mutant cells (Fig. 7D). The Km of acetyl-CoA synthetase 1 (Acs1p) for acetate is about 30-fold lower than that of Acs2p (60). Therefore, even a small increase in ACS1 expression would be expected to have a significant impact on the cellular level of acetyl-CoA. Measurement of acetyl-CoA levels showed that introduction of the mig1Δ mutation into snf1Δ mutant cells results in an increase in the acetyl-CoA level, which depends on ACS1 (Fig. 7E). These results are consistent with the proposed model of the role of Mig1p in the regulation of ACS1 expression and acetyl-CoA homeostasis (Fig. 7). In wild-type cells grown in the presence of glucose, ACS1 is repressed and therefore the acs1Δ mutation does not affect the cellular level of acetyl-CoA. However, ACS1 is derepressed by the introduction of the mig1Δ mutation (Fig. 7D) and Acs1p contributes to the cellular pool of acetyl-CoA (compare wild-type and mig1Δ mutant cells in Fig. 7E). Inactivation of ACS1 thus affects the acetyl-CoA level in mig1Δ mutant cells but not in wild-type cells. These results are consistent with the acetylation of bulk histones. In snf1Δ mutant cells, histone hypoacetylation was partially suppressed by the mig1Δ mutation, and this suppression was abrogated by the acs1Δ mutation (Fig. 7F). Cumulatively, these results provide another piece of evidence that many of the snf1Δ mutant phenotypes can be suppressed by elevating the cellular level of acetyl-CoA.

Inactivation of MIG1 requires ACS1 to partially suppress the stress sensitivity and histone hypoacetylation of snf1Δ mutant cells. (A) The hda1Δ or rpd3Δ mutation does not synergize with the mig1Δ mutation in the suppression of the snf1Δ mutation. Tenfold serial dilutions of the indicated strains were spotted onto YPD plates and grown for 2 days at 30 or 35°C, grown for 6 days on YPD plates at pH 7.5, or grown for 3 days on YPD plates containing 50 mM hydroxyurea. Typical results from three independent experiments are shown. (B) Diagram indicating regulation of Acs1p by Mig1p and Cat8p. (C) Suppression of the snf1Δ mutant's slow-growth phenotype by the mig1Δ mutation requires ACS1. Tenfold serial dilutions of the indicated strains were spotted onto YPD plates and grown for 2 days at 30°C. Typical results from three independent experiments are shown. (D) Inactivation of MIG1 increases the expression of CAT8 and ACS1 in snf1Δ mutant cells. The indicated strains were grown in YPD medium to an A600 of 1.0 at 30°C. Total RNA was isolated and assayed for CAT8 and ACS1 transcripts by real-time RT-PCR. The results were normalized to ACT1 RNA and expressed relative to the value for the wild-type (WT) strain. (E) Inactivation of MIG1 increases acetyl-CoA levels in snf1Δ mutant cells in an ACS1-dependent manner. Acetyl-CoA was assayed in cell lysates of the indicated strains by ELISA. The concentration of acetyl-CoA in wild-type cells is 1.8 nmol/107 cells. (F) Inactivation of MIG1 increases the acetylation of histones in snf1Δ mutant cells in an ACS1-dependent manner. The indicated strains were grown in YPD medium to an A600 of 1.0 at 30°C and analyzed by Western blotting with antibodies against histone H3 acetylated at lysine 14 (acH3), hyperacetylated histone H4 (acH4), and total histone H3. The experiment was performed three times, and representative results are shown. (D and E) The experiments were repeated three times, and the results are shown as means ± standard deviations. Values that are statistically significantly different (P < 0.05) from the wild-type values are indicated by an asterisk. Values that are statistically significantly different (P < 0.05) from each other are indicated by a bracket and an asterisk.
Acetyl-CoA homeostasis contributes to the regulation of SNF1-dependent genes.
To test the contribution of acetyl-CoA homeostasis and global histone acetylation to the transcriptional regulation of SNF1-dependent genes, we used published data sets from DNA microarray experiments and compared set of genes upregulated upon the inactivation of SNF1 (62) with set of genes upregulated in a strain expressing histone H4 with mutations that change the acetylatable lysine residues into nonacetylatable arginine residues within the N-terminal tail (H4-K5R/K8R/K12R/K16R) (63). The first data set was generated with cells that express the snf1as allele instead of wild-type SNF1. The snf1as allele carries a mutation that renders mutant cells sensitive to the inhibitor mMe-PP1 (62). snf1as mutant cells were grown in SC medium with 3% glycerol and harvested 60 min after the addition of mMe-PP1 (62). We compared the set of genes upregulated at least 2-fold in this experiment with the set of genes upregulated at least 2-fold in histone H4 mutant H4-K5R/K8R/K12R/K16R (63) and found a significant overlap (Fig. 8A). More than 45% of the genes upregulated upon snf1as inactivation are upregulated in the H4-K5R/K8R/K12R/K16R mutant. As indicated by the χ2 test (P < 0.001), the correlation between the two data sets is much higher than that expected in an overlap of two random data sets of the same size (Fig. 8A, values in parentheses). We conclude that the mechanism responsible for upregulation of the genes upon the inactivation of snf1as involves globally decreased histone acetylation. The notion that SNF1 affects histone acetylation is supported by a recent study that showed decreased histone acetylation in the promoters of ADH2, POX1, and ACS1 upon the inhibition of snf1as (64).

Acetyl-CoA homeostasis and histone acetylation contribute to the regulation of SNF1-dependent genes. (A) Inactivation of SNF1 results in the increased expression of genes that are also upregulated in a nonacetylatable histone H4 mutant (H4-K5R/K8R/K12R/K16R). The Venn diagram is based on previously published data sets (62, 63) and shows the extent of overlap between genes upregulated at least 2-fold upon the inactivation of Snf1p in the snf1as mutant strain and genes upregulated at least 2-fold in the H4-K5R/K8R/K12R/K16R strain. The values in parentheses are theoretical values calculated for the chance overlap of two random data sets of the corresponding sizes. (B to F) The indicated strains were grown in YPD medium to an A600 of 1.0 at 30°C. Total RNA was isolated and assayed for ACT1, REG2, GAL4, SUC2, MAL33, HHT1/2, HHF1/2, HTA1/2, and HTB1/2 transcripts by real-time RT-PCR. The results were normalized to ACT1 RNA and expressed relative to the value for the wild-type (WT) strain. (G) The snf1Δ mutant displays a synthetic growth defect with the SWI4-RR mutation. Tenfold serial dilutions of the indicated strains were spotted onto YPD plates and grown for 2 days at 30°C. Typical results from three independent experiments are shown. (B to F) The experiments were repeated three times, and the results are shown as means ± standard deviations. Values that are statistically significantly different (P < 0.05) from the wild-type values are indicated by an asterisk. Values that are statistically significantly different (P < 0.05) from each other are indicated by a bracket and an asterisk.
To test the contribution of acetyl-CoA homeostasis to the transcriptional regulation of genes that require SNF1 for full expression (45, 62), we measured the expression of two groups of genes, glucose-repressed and non-glucose-repressed genes, in wild-type and snf1Δ, tetO7-ACC1, and snf1Δ tetO7-ACC1 mutant cells. The genes in the first group include REG2, GAL4, SUC2, and MAL33 and are repressed by glucose in a Mig1p/Mig2p-dependent manner; REG2 and GAL4 are repressed by Mig1p, and SUC2 and MAL33 are repressed redundantly by Mig1p/Mig2p (46). The genes in the second group include the histone genes HHT1/2, HHF1/2, HTA1/2, and HTB1/2. Similarly to INO1, the expression of the histone genes is not repressed by glucose but requires SNF1 (45). The mRNA levels for the individual genes in snf1Δ mutant cells were reduced to 14 to 79% of the wild-type levels, and the defect was partially suppressed by the tetO7-ACC1 allele (Fig. 8B and andC).C). The H3S10 phosphorylation did not affect the expression of the genes tested, since their expression did not differ in wild-type cells and the H3S10A mutant (Fig. 8D and andE).E). Similar result was reported previously for SUC2; the H3S10A mutation did not significantly affect the SUC2 mRNA level and histone H3 acetylation in the SUC2 promoter (20). These results suggest that the decreased cellular level of acetyl-CoA and histone hypoacetylation contribute to the altered transcriptional regulation in snf1Δ mutant cells.
Recently, the G1/S-specific transcriptional factor Swi4p was shown to be acetylated (38). Swi4p binds Swi6p to form transcriptional complex SBF. The acetylation of Swi4p regulates the interaction with Swi6p and thus SBF-dependent gene transcription. The SBF complex is involved in the regulation of transcription of the histone genes (65). Since the acetylation of Swi4p is affected in snf1Δ mutant cells (Fig. 4), we wanted to determine whether decreased acetylation of Swi4p affects the expression of the histone genes. The expression of all four histone genes was reduced in a strain that expresses Swi4p with mutations that change two acetylatable lysine residues into nonacetylatable arginine residues (Swi4p-RR) (38) (Fig. 8F). The Swi4p mutation that changes the two acetylatable lysines into glutamine residues (Swi4p-QQ), which mimics constitutive acetylation, did not affect the expression of the histone genes. Introduction of the snf1Δ mutation into the Swi4p-RR strain resulted in additive decreases in the expression of the histone genes (Fig. 8F). Interestingly, unlike Swi4p-QQ, the Swi4p-RR mutation displayed a synthetic growth defect with the snf1Δ mutation (Fig. 8G). These results suggest that the snf1Δ mutation affects the acetylation of histones, as well as Swi4p, both of which are required for normal expression of the histone genes.
HATs are required to mediate the effect of an increased acetyl-CoA level.
Since the regulation of SUC2 transcription is relatively well understood (20), we used it to analyze in more detail the role of acetyl-CoA homeostasis. While the tetO7-ACC1 allele does not significantly change the extent or kinetics of SUC2 induction, it allows weak expression in the snf1Δ mutant (Fig. 9A and andB).B). The effect is relatively small, but only partial suppression would be expected because the Mig1p-mediated repression is not relieved in the snf1Δ mutant. Since SNF1 is required for normal acetyl-CoA homeostasis and a normal level of untargeted histone acetylation when cells are grown in YPD medium (Fig. 1 and and2),2), we analyzed SUC2 regulation under repressing conditions in the mig1Δ mutant background (Fig. 9C). The major known roles of SNF1 in the regulation of SUC2 expression are phosphorylation of Mig1p and relief of the suppression mediated by the Mig1p-Tup1p/Ssn6p complex (11, 14). The snf1Δ mutation reduced SUC2 expression similarly in both the MIG1 (Fig. 8B) and mig1Δ (Fig. 9C) mutant strains. However, while the tetO7-ACC1 allele suppressed this effect only partially in MIG1 mutant cells (Fig. 8B), the suppression was more robust in mig1Δ mutant cells (compare the expression in mig1Δ mutant cells with the expression in mig1Δ snf1Δ tetO7-ACC1 cells; Fig. 9C). This result suggests that in the absence of the Mig1p repressor, the defect in SUC2 expression due to the snf1Δ mutation can be completely suppressed by introducing the tetO7-ACC1 allele and increasing the cellular level of acetyl-CoA and histone acetylation. To test whether HATs are required to mediate the effect of an increased acetyl-CoA concentration, we determined SUC2 expression in the mig1Δ gcn5Δ and mig1Δ yng2Δ mutants. As expected, the gcn5Δ and yng2Δ mutations significantly decreased SUC2 expression in the mig1Δ mutant strain and the effect was additive with the snf1Δ mutation. Interestingly, the tetO7-ACC1 allele did not significantly increase expression in the mig1Δ gcn5Δ and mig1Δ yng2Δ mutants, suggesting that the effect of the increased cellular level of acetyl-CoA on SUC2 expression requires the activities of SAGA and NuA4 (Fig. 9C). Cumulatively, these results suggest that in addition to relieving the repression mediated by the Mig1p-Tup1p/Ssn6p complex, SNF1 also supports SUC2 expression by maintaining a normal level of histone acetylation.

Loss of SUC2 expression in snf1Δ mutant cells is partially suppressed by the tetO7-ACC1 allele. (A) The indicated strains were grown in YPD medium to an A600 of 1.0 at 30°C. The cells were harvested, washed, and resuspended and grown in YEP medium containing 0.05% glucose. Total RNA was isolated and assayed for ACT1 and SUC2 transcripts by real-time RT-PCR. The results were normalized to ACT1 RNA and expressed relative to the value for the wild-type (WT) strain at 0 min. The experiments were repeated three times, and a typical time course is shown. (B) Closeup view of the values for the snf1Δ and snf1Δ tetO7-ACC1 mutants. (C) The indicated strains were grown in YPD medium to an A600 of 1.0 at 30°C. Total RNA was isolated and assayed for ACT1 and SUC2 transcripts by real-time RT-PCR. The results were normalized to ACT1 RNA and expressed relative to the value for the mig1Δ mutant strain. The experiments were repeated three times, and the results are shown as means ± standard deviations. Values that are statistically significantly different (P < 0.05) from those of the mig1Δ mutant strain are indicated by an asterisk, and values that are statistically significantly different (P < 0.05) from each other are indicated by a bracket and an asterisk.
Role of SNF1 in the regulation of acetyl-CoA homeostasis during metabolic transitions.
The availability of glucose affects the cellular level of acetyl-CoA and global histone acetylation (29, 66). To determine the role of SNF1 in the regulation of acetyl-CoA homeostasis during metabolic transitions, we subjected exponentially growing wild-type and snf1Δ mutant cells to glucose depletion and repletion. After 1 h of glucose depletion, the acetyl-CoA level increased 1.7-fold in wild-type cells (Fig. 10A). However, this increase was almost absent in snf1Δ mutant cells. The acetylation of bulk histones, as determined by Western blot analysis, did not change in wild-type and snf1Δ mutant cells (data not shown); however, the histone acetylation in the promoters of the SNF1-dependent REG2 and GAL4 genes increased significantly more in wild-type cells than in snf1Δ mutant cells (Fig. 10B). This increase is likely cotranscriptional, since SNF1 is required for the recruitment of SAGA to the SUC2, HXT2, and HXT4 promoters under glucose limitation (20, 67). We interpret these results as meaning that upon glucose depletion, the increased acetyl-CoA level supports targeted histone acetylation in the promoters of SNF1-dependent genes. As expected for SNF1-dependent genes, the expression of REG2 and GAL4 increased only in wild-type cells and not in snf1Δ mutant cells (Fig. 10C). Glucose refeeding also resulted in an increased acetyl-CoA level in wild-type cells (Fig. 10D). The increase was significantly attenuated and delayed in snf1Δ mutant cells. Glucose addition also did not affect global histone acetylation in wild-type and snf1Δ mutant cells (data not shown). This result differs from the addition of glucose to stationary-phase cells that depleted utilizable carbon sources from the medium (29). Stationary-phase cells have a very low global histone acetylation level, most likely because of the extremely low level of acetyl-CoA (68). Addition of glucose to these cells induces NuA4- and SAGA-dependent histone acetylation (29). In our experiment, glucose was added to cells only after 2 h of glucose depletion, which did not result in decreased acetyl-CoA and global histone acetylation levels. However, glucose addition induced histone acetylation in the promoters of the ribosomal protein genes RPS22B and RPS11B and increased the expression of these genes in wild-type cells. The acetylation of promoter histones and expression of RPS22B and RPS11B were significantly less pronounced in snf1Δ mutant cells (Fig. 10E and andF).F). The increase in the acetylation of promoter histones is likely mediated by NuA4, which is recruited to ribosomal protein genes under favorable nutritional conditions and thus promotes their expression (69). This corresponds to our results, which show increased acetylation of particularly histone H4 in the promoters of RPS22B and RPS11B (Fig. 10E). These results suggest that the increased level of acetyl-CoA is responsible for the increased expression of the ribosomal protein genes in wild-type cells and are consistent with the notion that the increased cellular level of acetyl-CoA promotes the acetylation of histones at growth genes (31, 70).
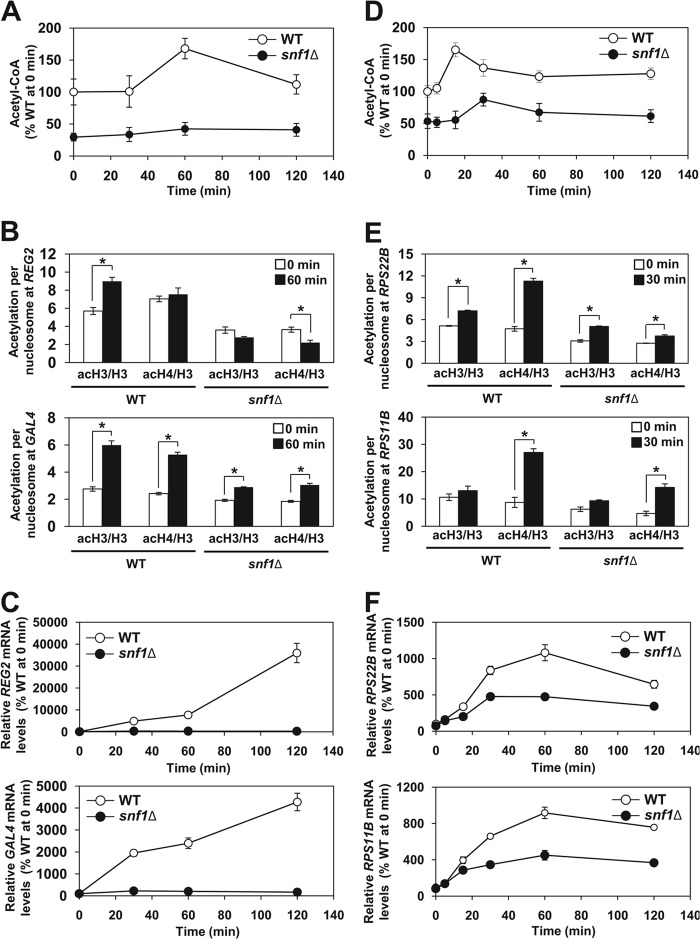
Glucose starvation and refeeding result in an SNF1-dependent increase in the acetyl-CoA level and increased histone acetylation in the promoters of the activated genes. (A, B, and C) Glucose starvation. (A) snf1Δ mutant cells fail to elevate the acetyl-CoA level during glucose starvation. The indicated strains were grown in YPD medium to an A600 of 1.0 at 30°C. The cells were harvested, washed, and resuspended in YEP medium containing 0.05% glucose. Samples were taken at the indicated time points, and acetyl-CoA was assayed in cell lysates. The concentration of acetyl-CoA in wild-type (WT) cells at 0 min was 1.8 nmol/107 cells. The experiments were repeated three times, and the results are shown as means ± standard deviations. (B) ChIP experiments were performed with antibodies against total histone H3 (H3), histone H3 acetylated at lysine 14 (acH3), and hyperacetylated histone H4 (acH4). Occupancies of H3, acH3, and acH4 were determined in the promoter regions of REG2 and GAL4. Acetylation per nucleosome was calculated as ratios of acH3 to total H3 and acH4 to total H3. (C) Total RNA was isolated and assayed for REG2 and GAL4 transcripts by real-time RT-PCR. The results were normalized to ACT1 RNA and expressed relative to the value for the wild-type strain at 0 min. The experiments were repeated three times, and the results are shown as means ± standard deviations. (D, E, and F) Glucose repletion. (D) The increase in the acetyl-CoA level upon glucose addition is attenuated in snf1Δ mutant cells. The indicated strains were grown in YPD medium to an A600 of 0.5 at 30°C. The cells were harvested, washed, and grown in YEP medium containing 0.05% glucose for 2 h. Glucose was added to 2%, and the cells were harvested at the indicated time points for acetyl-CoA assay. The concentration of acetyl-CoA in wild-type cells was 0.9 nmol/107 cells at 0 min. The experiments were repeated three times, and the results are shown as means ± standard deviations. (E) ChIP experiments were performed with antibodies against total histone H3 (H3), histone H3 acetylated at lysine 14 (acH3), and hyperacetylated histone H4 (acH4). Occupancies of H3, acH3, and acH4 were determined in the promoter regions of RPS22B and RPS11B. Acetylation per nucleosome was calculated as ratios of acH3 to total H3 and acH4 to total H3. (F) Total RNA was isolated and assayed for RPS22B and RPS11B transcripts by real-time RT-PCR. The results were normalized to ACT1 RNA and expressed relative to the value for the wild-type strain at 0 min. The experiments were repeated three times, and the results are shown as means ± standard deviations. (B and E) The experiments were repeated three times, and the results are shown as means ± standard deviations. Values that were statistically significantly different (P < 0.05) from each other are indicated by a bracket and an asterisk.
Cumulatively, these results indicate that both glucose starvation and refeeding result in a SNF1-dependent temporary acetyl-CoA level increase, which correlates with the increased targeted acetylation of histones in the promoters of the activated genes. This occurs in addition to the globally higher histone acetylation level in wild-type cells than in snf1Δ mutant cells. The mechanism responsible for the temporarily increased level of acetyl-CoA upon glucose depletion or repletion likely involves the activation of SNF1 and SNF1-mediated inhibition of Acc1p. However, our results illustrated in Fig. 7 suggest a possible contribution from a second mechanism that is triggered during glucose starvation and involves SNF1-dependent derepression of ACS1.
DISCUSSION
Acetyl-CoA is a substrate for HATs, and metabolic pathways that regulate the homeostasis of acetyl-CoA in the nucleocytosolic compartment affect the global acetylation of chromatin histones (28, 30–32). Therefore, nucleocytosolic acetyl-CoA is the link between cellular energy and carbon metabolism and between histone acetylation and chromatin regulation (29, 31, 32, 71). In yeast, glycolytically produced pyruvate is converted to acetaldehyde and subsequently to acetate in the cytosol (42). Nucleocytosolic Acs2p then converts acetate to acetyl-CoA (28). In mammalian cells, the nucleocytosolic enzyme ATP-citrate lyase (ACL) is the major source of acetyl-CoA for histone acetylation (30). ACL generates acetyl-CoA in the nucleocytosolic compartment from glucose-derived citrate, and glucose availability affects histone acetylation in an ACL-dependent manner.
Since nucleocytosolic acetyl-CoA is also used for the de novo synthesis of fatty acids (27), histone acetylation and synthesis of fatty acids compete for the same acetyl-CoA pool. The first and rate-limiting reaction in the de novo synthesis of fatty acids is carboxylation of acetyl-CoA to form malonyl-CoA, catalyzed by ACC (27). We have shown that lowering the flux of acetyl-CoA through the fatty acid synthesis pathway by reducing ACC1 expression results in globally increased histone acetylation and altered transcriptional regulation (32). Since ACC is phosphorylated and inhibited by Snf1p/AMPK in both yeast and mammalian cells (23–26), we hypothesized that SNF1 contributes to the regulation of homeostasis of acetyl-CoA and is one of the factors that set the level of global untargeted acetylation of histones. The decreased level of acetyl-CoA and increased level of malonyl-CoA are consistent with the higher enzymatic activity of Acc1p (25, 26) and increased conversion of acetyl-CoA to malonyl-CoA in snf1Δ mutant cells (Fig. 1A and andB).B). Regulation of the global level of acetylation of chromatin histones thus represents another, previously unrecognized, mechanism by which SNF1 regulates transcription. The role of SNF1 in the regulation of global histone acetylation is also supported by the synthetic genetic interactions between snf1Δ and mutations in the SAGA and NuA4 HATs and by the partial suppression of the temperature sensitivity of snf1Δ mutant cells by the hda1Δ and rpd3Δ mutations (Fig. 5). Histone hypoacetylation also likely contributes to the stress sensitivity of snf1Δ mutant cells by preventing efficient and timely transcriptional responses to different stresses. Several phenotypes of snf1Δ mutant cells are at least partially suppressed by the introduction of the tetO7-ACC1 allele. Cells expressing the tetO7-ACC1 allele display reduced Acc1p enzymatic activity (32) and elevated acetyl-CoA levels (Fig. 1A). The tetO7-ACC1 allele elevates acetyl-CoA levels and global histone acetylation in snf1Δ mutant cells and suppresses temperature sensitivity, alkaline pH sensitivity, and hydroxyurea sensitivity. In addition to the tetO7-ACC1 allele, we found that the low level of acetyl-CoA, histone hypoacetylation, and stress sensitivity of snf1Δ mutant cells can be suppressed by derepression of the acetyl-CoA synthetase ACS1. Deletion of MIG1 derepresses ACS1, which is normally repressed by glucose, and results in the increased synthesis of acetyl-CoA (Fig. 7). This result provides additional evidence that some of the phenotypes and transcriptional defects of snf1Δ mutant cells are due to their low cellular acetyl-CoA level and global histone hypoacetylation.
Hypomorphic ACC1 mutations were isolated by genetic screening for suppressors of inositol auxotrophy caused by low INO1 gene expression in snf1Δ mutant cells (26). The inositol auxotrophy of snf1Δ mutant cells was suppressed by the tetO7-ACC1 allele, by inhibiting the enzymatic activity of Acc1p with the ACC inhibitor soraphen A or by inhibiting Acc1p by providing long-chain fatty acids in the medium (26). The ACC1 gene was also recovered by genetic screening for mutations that allow constitutive expression of the PHO5 gene in medium with a high phosphate concentration (72). Both groups concluded that it is a metabolite produced or utilized in fatty acid biosynthesis that affects INO1 or PHO5 transcription (26, 72). Our results suggest that this metabolite is acetyl-CoA, which regulates global histone acetylation. The reason that the hypomorphic ACC1 mutations were recovered in both genetic selections is the fact that INO1 and PHO5 are regulated by global histone acetylation (44, 73).
Histones are not the only proteins that are acetylated. Many acetylated nonchromatin proteins were identified in yeast and mammals, and protein acetylation appears as widespread as protein phosphorylation (38, 48, 74). Many of the acetylated proteins are located in the nucleus or cytosol, and their acetylation is regulated by the concentration of acetyl-CoA in the nucleocytosolic compartment (32). The regulatory role of nucleocytosolic acetyl-CoA thus likely extends beyond histone acetylation and chromatin-dependent processes. We showed that in addition to histones, the attenuated acetyl-CoA level in snf1Δ mutant cells affects the acetylation status of Swi4p (Fig. 4) and that the defect in the transcription of the histone genes in snf1Δ mutant cells is due not only to the hypoacetylation of histones but also to the hypoacetylation of Swi4p (Fig. 8F and andG).G). Regulation of Swi4p acetylation and transcription of SBF-dependent genes by the cellular concentration of acetyl-CoA likely endow the cells with an additional mechanism linking their metabolic state with cell cycle progression and growth. Sip2p, one of the regulatory β subunits of the SNF1 complex, is also acetylated (48). The level of Sip2p acetylation depends on the nucleocytosolic level of acetyl-CoA and is increased in tetO7-ACC1 cells (32) and decreased in snf1Δ mutant cells (Fig. 4). The acetylation of Sip2p increases its interaction and inhibition of Snf1p (49). It is tempting to speculate that Sip2p acetylation and Snf1p inhibition form a regulatory loop with Acc1p that contributes to the regulation of acetyl-CoA homeostasis (Fig. 11). An increased acetyl-CoA level would promote Sip2p acetylation and inhibition of Snf1p. Decreased Snf1p activity would result in lower Snf1p-mediated phosphorylation and inhibition of Acc1p and thus in increased conversion of acetyl-CoA to malonyl-CoA. On the other hand, a decreased acetyl-CoA level would result in Sip2p hypoacetylation and increased Snf1p activity. This, in turn, would lead to increased phosphorylation and inhibition of Acc1p and decreased conversion of acetyl-CoA to malonyl-CoA (Fig. 11). This homeostatic mechanism would maintain the nucleocytosolic level of acetyl-CoA within certain limits and would prevent gross hypoacetylation or hyperacetylation of chromatin histones, a condition that might alter the regulation of chromatin-based processes, such as transcription or DNA replication and repair. However, regulation of nucleocytosolic acetyl-CoA homeostasis is probably more complex and involves additional mechanisms. For example, transcription of ACC1 is decreased in gcn5 cells (75) and in cells expressing a nonacetylatable version of histone H4 (H4K5,8,12,16R) (63). Decreased histone acetylation because of a low nucleocytosolic acetyl-CoA concentration would thus diminish ACC1 transcription and likely also the conversion of acetyl-CoA to malonyl-CoA, which would elevate the acetyl-CoA level.
SNF1/AMPK is a master regulator of metabolism (2, 3). Intracellular acetyl-CoA reflects the metabolic state of the cell and, by promoting histone acetylation, connects metabolism with transcriptional regulation (31). This study suggests that by regulating acetyl-CoA homeostasis and histone acetylation, SNF1 is equipped with another, previously unrecognized, mechanism that relays information about the cellular metabolic state to the transcriptional machinery.
ACKNOWLEDGMENTS
We thank B. Andrews, K. M. Arndt, M. Carlson, M. H. Kuo, L. F. Pemberton, M. C. Schmidt, D. J. Stillman, T. Tsukiyama, F. Winston, and H. Zhu for strains and plasmids and members of the Vancura lab and I. Vancurova for helpful comments.
This work was supported by grant GM106324 from the National Institutes of Health to A.V.
REFERENCES
Articles from Molecular and Cellular Biology are provided here courtesy of Taylor & Francis
Full text links
Read article at publisher's site: https://doi.org/10.1128/mcb.00198-13
Read article for free, from open access legal sources, via Unpaywall:
https://mcb.asm.org/content/mcb/33/23/4701.full.pdf
Citations & impact
Impact metrics
Citations of article over time
Alternative metrics
Smart citations by scite.ai
Explore citation contexts and check if this article has been
supported or disputed.
https://scite.ai/reports/10.1128/mcb.00198-13
Article citations
Advances in S. cerevisiae Engineering for Xylose Fermentation and Biofuel Production: Balancing Growth, Metabolism, and Defense.
J Fungi (Basel), 9(8):786, 26 Jul 2023
Cited by: 0 articles | PMID: 37623557 | PMCID: PMC10455348
Review Free full text in Europe PMC
Perturbed fatty-acid metabolism is linked to localized chromatin hyperacetylation, increased stress-response gene expression and resistance to oxidative stress.
PLoS Genet, 19(1):e1010582, 10 Jan 2023
Cited by: 3 articles | PMID: 36626368 | PMCID: PMC9870116
The sound of silence: Transgene silencing in mammalian cell engineering.
Cell Syst, 13(12):950-973, 01 Dec 2022
Cited by: 31 articles | PMID: 36549273 | PMCID: PMC9880859
Review Free full text in Europe PMC
The polyHIS Tract of Yeast AMPK Coordinates Carbon Metabolism with Iron Availability.
Int J Mol Sci, 24(2):1368, 10 Jan 2023
Cited by: 4 articles | PMID: 36674878 | PMCID: PMC9863760
Multi-omics Data Reveal the Effect of Sodium Butyrate on Gene Expression and Protein Modification in Streptomyces.
Genomics Proteomics Bioinformatics, 21(6):1149-1162, 15 Sep 2022
Cited by: 2 articles | PMID: 36115661 | PMCID: PMC11082262
Go to all (49) article citations
Data
Similar Articles
To arrive at the top five similar articles we use a word-weighted algorithm to compare words from the Title and Abstract of each citation.
Activation of AMP-activated Protein Kinase by Metformin Induces Protein Acetylation in Prostate and Ovarian Cancer Cells.
J Biol Chem, 291(48):25154-25166, 12 Oct 2016
Cited by: 54 articles | PMID: 27733682 | PMCID: PMC5122782
Yeast phospholipase C is required for normal acetyl-CoA homeostasis and global histone acetylation.
J Biol Chem, 288(39):27986-27998, 02 Aug 2013
Cited by: 10 articles | PMID: 23913687 | PMCID: PMC3784712
Acetyl-CoA carboxylase regulates global histone acetylation.
J Biol Chem, 287(28):23865-23876, 11 May 2012
Cited by: 130 articles | PMID: 22580297 | PMCID: PMC3390662
Reciprocal Regulation of AMPK/SNF1 and Protein Acetylation.
Int J Mol Sci, 19(11):E3314, 25 Oct 2018
Cited by: 23 articles | PMID: 30366365 | PMCID: PMC6274705
Review Free full text in Europe PMC
Funding
Funders who supported this work.
NIGMS NIH HHS (2)
Grant ID: GM106324
Grant ID: R15 GM106324







