Abstract
Free full text

Hijacking of leguminous nodulation signaling by the rhizobial type III secretion system
Significance
Root–nodule symbiosis between leguminous plants and rhizobia requires rhizobial Nod factors (NFs) and their leguminous receptors (NFRs). Here we show that symbiosis in the soybean rhizobium Bradyrhizobium elkanii is promoted by the type III secretion system (T3SS), which delivers virulence factors via pathogenic bacteria. Nodulation tests and expression analyses using mutants of both B. elkanii and soybean (Glycine max) revealed that rhizobial T3SS activates host nodulation signaling in the absence of NFs and NFRs. These results suggest that rhizobia have adopted a pathogenic system that stimulates their legume hosts to initiate symbiotic programs.
Abstract
Root–nodule symbiosis between leguminous plants and nitrogen-fixing bacteria (rhizobia) involves molecular communication between the two partners. Key components for the establishment of symbiosis are rhizobium-derived lipochitooligosaccharides (Nod factors; NFs) and their leguminous receptors (NFRs) that initiate nodule development and bacterial entry. Here we demonstrate that the soybean microsymbiont Bradyrhizobium elkanii uses the type III secretion system (T3SS), which is known for its delivery of virulence factors by pathogenic bacteria, to promote symbiosis. Intriguingly, wild-type B. elkanii, but not the T3SS-deficient mutant, was able to form nitrogen-fixing nodules on soybean nfr mutant En1282. Furthermore, even the NF-deficient B. elkanii mutant induced nodules unless T3SS genes were mutated. Transcriptional analysis revealed that expression of the soybean nodulation-specific genes ENOD40 and NIN was increased in the roots of En1282 inoculated with B. elkanii but not with its T3SS mutant, suggesting that T3SS activates host nodulation signaling by bypassing NF recognition. Root-hair curling and infection threads were not observed in the roots of En1282 inoculated with B. elkanii, indicating that T3SS is involved in crack entry or intercellular infection. These findings suggest that B. elkanii has adopted a pathogenic system for activating host symbiosis signaling to promote its infection.
Leguminous plants such as soybean and pea establish symbiosis with a group of soil bacteria called rhizobia. Rhizobia induce the development of root nodules in leguminous plants and fix atmospheric dinitrogen to ammonia, which can be effectively used by the host plants. Nitrogen-fixing nodules are formed as a consequence of molecular interactions between partners. For example, flavonoids excreted from host plant roots interact with the rhizobial transcriptional factor NodD to induce the expression of nodulation (nod) genes of rhizobia, which are required for the synthesis of lipochitooligosaccharides known as Nod factors (NFs) (1). The perception of NFs via cognate host receptors (NFRs) induces cascades of signal transduction that are required for root-hair infection and nodule organogenesis (2).
Besides NFs, several rhizobial traits have been reported to affect symbiosis with host legumes. Among them, the type III secretion system (T3SS) is known as an introducer of virulence factors in animal and plant pathogens (3). Pathogenic bacteria use the system to deliver so-called type III effector (T3E) proteins directly into host cells or the external environment, where they manipulate host cellular processes to promote pathogenicity (4). Some rhizobia have been shown to possess T3SSs that affect the ability of rhizobia to infect host legumes (5). A distinct characteristic of rhizobial T3SS is that its expression is induced by flavonoids derived from the host legume. Flavonoids induce NodD to activate the expression of ttsI, a transcriptional activator of the type III secretion gene cluster (tts). TtsI subsequently activates the tts gene cluster, and proteins are secreted via T3SS (6, 7). This cascade confines the expression of rhizobial T3SS during host-plant infection.
Previous studies have shown both positive and negative effects of rhizobial T3SSs on symbiosis. In the case of Mesorhizobium loti, a microsymbiont of Lotus spp., deletion of its tts genes results in a reduction of the number of nodules formed on the roots of Lotus corniculatus subsp. frondosus, whereas the number of nodules on the roots of Lotus halophilus significantly increases when inoculated with a T3SS-null mutant (8). Meanwhile, the T3Es NopL and NopP of NGR234 have positive effects on symbiosis with their host legumes (9). Both NopL and NopP are phosphorylated by plant protein kinases, and NopL can prevent the complete induction of pathogenesis-related defense proteins when expressed in Lotus japonicus roots (10). However, the molecular basis for T3SS-involved nodulation enhancement has not been fully elucidated.
Bradyrhizobium elkanii is a microsymbiont of leguminous hosts such as Glycine max and Arachis hypogea, and is used as a commercial inoculant for soybean in Brazil (11). We previously demonstrated that B. elkanii USDA61 nodulation on the soybean cultivar Clark-rj1 was T3SS-dependent (12). Clark-rj1 is genotypically characterized as rj1rj1, and the rj1 locus is a nonnodulation trait that was recently reported to represent a frameshift mutation leading to a truncated NFR1 (13). Clark-rj1 exhibits a nonnodulation phenotype as well as a Lotus nfr1 mutant (14) when inoculated with most rhizobia, with the exception of USDA61. We therefore speculated that B. elkanii T3SS can compensate for NF signaling downstream of NFRs in soybean. However, it remains to be determined whether T3SS-dependent nodulation occurs in other nfr mutant soybeans, and whether the induction of soybean symbiosis genes by B. elkanii T3SS is independent of NF signaling. The current study examined the involvement of B. elkanii T3SS in the nodulation and infection properties of an additional nfr1 mutant derived from the soybean cultivar Enrei, and compared it with the wild-type variant. We also investigated gene expression in soybean roots induced by B. elkanii T3SS, and compared this with conventional NF/NFR signaling.
Results
B. elkanii Nodulates nfr1 Mutant Soybeans in a T3SS-Dependent Manner.
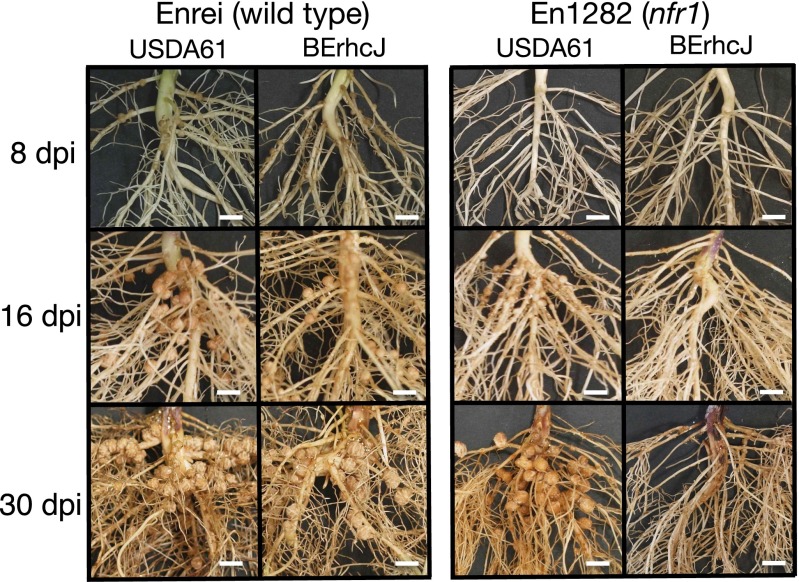
In B. elkanii, the rhcJ gene is in an operon encoding several T3SS components, and strain BErhcJ is a mutant defective for protein secretion by this system (12). In wild-type Enrei, the mutant BErhcJ induced 20–40% fewer nodules than wild-type B. elkanii (Fig. 1A). Nodule numbers derived by BEttsI, the mutant of transcriptional activator TtsI, also decreased to similar levels observed for BErhcJ. In En1282 (a mutant of Enrei), however, nodulation was observed in roots inoculated with USDA61, whereas few nodules were observed following inoculation with T3SS mutants or Bradyrhizobium japonicum USDA110 (Fig. 1B). In comparison with wild-type Enrei (Fig. 1A), En1282 formed significantly fewer nodules when inoculated with B. elkanii (Figs. 1B and and2).2). Notably, the nodulation timing was delayed in En1282; nodules became visible on the roots of Enrei 7–8 d after B. elkanii inoculation, compared with 14–16 d on the roots of En1282. Similar nodulation phenotypes were observed when cultivar Clark-rj1 was selected as the host plant (Fig. S1). These results demonstrate that B. elkanii USDA61 is capable of nodulating nfr1 mutant soybeans of two different cultivars.

Nodulation of G. max cv. Enrei (A) and En1282 (B) inoculated with wild-type and mutant strains of B. elkanii USDA61 and B. japonicum USDA110. Nodule numbers were counted 30 d after inoculation. Nodulation tests were performed at least three times, values shown are means obtained from at least 18 plants, and error bars indicate SDs. Statistical analyses (Student t test) were performed comparing mutants with the wild-type strain. *P < 0.05; **P < 0.01.
Synergistic Effects of NFs and T3SS Signaling on the Efficiency of Rhizobial Infection.
To explore further the importance of NFs in the interaction between B. elkanii and nfr1 soybean, we constructed the nodC mutant of B. elkanii, BEnodC, which is unable to synthesize NFs, and examined its nodulation phenotype in wild-type and nfr1 mutant soybeans. Contrary to expectations, BEnodC induced nodule formation on Enrei roots, although the nodule number was significantly reduced compared with that of the B. elkanii wild-type strain (Fig. 1A). The bacteria reisolated from nodules retained the same phenotype (Table S1). The nodule numbers of En1282 inoculated with BEnodC were roughly halved compared with those inoculated with the B. elkanii wild-type strain. The nodule numbers of Enrei and En1282 inoculated with BEnodC were not significantly different (Student t test P < 0.05). Almost all nodules induced by BEnodC on both Enrei and En1282 showed irregular or tumor-like morphology (Figs. 1B and 3 G and H). In En1282 inoculated with BEnodC, ~20–50% of nodules showed tumor-like structures that contained no infected cells (Fig. 3 H and L). However, when both nodC and ttsI were mutated (BEttsnod), the nodulation ability of B. elkanii was mostly abolished (Fig. 1 A and B). These results indicate that B. elkanii is capable of infecting and nodulating soybean in a T3SS-dependent manner in the absence of NFs, although the nodulation efficiency was much lower than NF-derived nodulation.
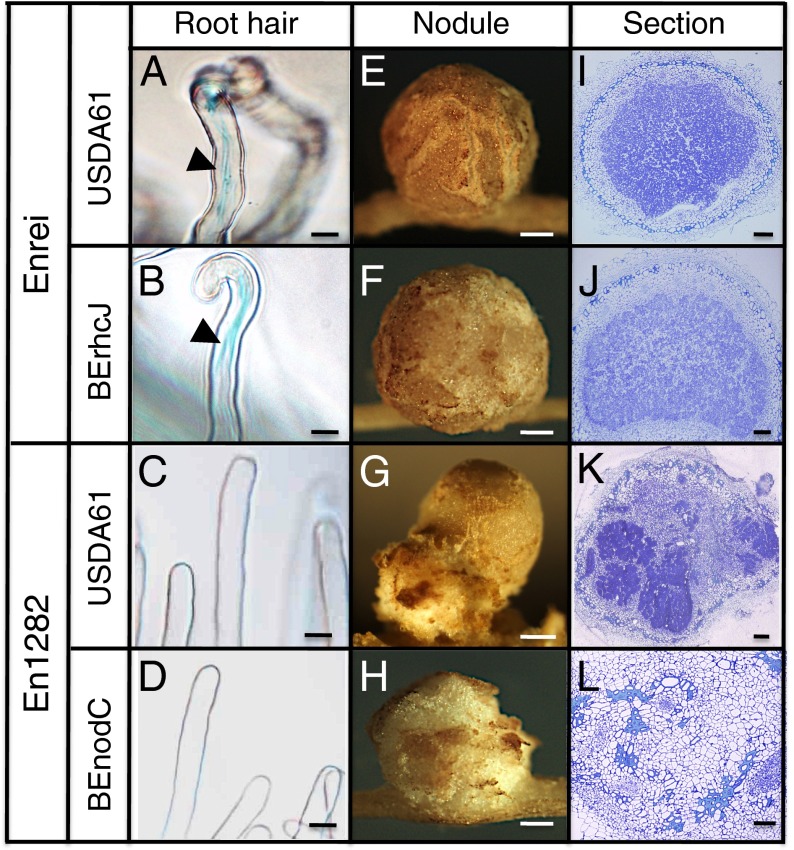
Root hairs and nodules of G. max cv. Enrei and nfr1 mutant En1282 inoculated with B. elkanii strains. (A) Root-hair curling and infection threads in Enrei inoculated with B. elkanii USDA61. (B) Root-hair curling and infection threads in Enrei inoculated with type III mutant BErhcJ. (C) Root hairs of nfr1 mutant En1282 inoculated with USDA61. (D) Root hairs of En1282 inoculated with BEnodC. (E) Nodule of Enrei inoculated with USDA61. (F) Nodule of Enrei inoculated with BErhcJ. (G) Nodule of En1282 inoculated with USDA61. (H) Nodule of En1282 inoculated with BEnodC. (I) Thin section of Enrei nodule infected by USDA61. (J) Thin section of Enrei nodule infected by BErhcJ. (K) Thin section of En1282 nodule infected by USDA61. (L) Thin section of En1282 nodule infected by BEnodC. [Scale bars, (A–D) 20 μm; (E–H) 200 μm; (I–L) 100 μm.]
Infective Properties of B. elkanii.
To investigate the infective properties of B. elkanii in the absence of NF signaling, we followed the infection process of B. elkanii strains on Enrei and En1282. In Enrei soybean, root-hair curling and infection threads were observed on roots inoculated with USDA61 and BErhcJ (Fig. 3 A and B), followed by the emergence of globular nodules containing infected cells (Fig. 3 E, F, I, and J). The numbers of infection threads formed on Enrei roots were 13.6 ± 3.9 (n = 5) with USDA61 and 9.6 ± 2.4 (n = 5) with BErhcJ. By contrast, root-hair curling and infection threads were absent on the roots of En1282 soybean inoculated with both USDA61 and BEnodC (Fig. 3 C and D). In En1282 inoculated with USDA61, about 20% of nodules showed irregular shapes and contained sectored, infected regions within which the infected cells were distributed unevenly in the central nodule zone (Fig. 3 G and K).
Dissection of NF- and T3SS-Derived Gene Expression in Soybean by Microarray Analysis.
Plant inoculation tests suggested that nodule formation in nfr1 soybeans is dependent on B. elkanii T3SS. To investigate the molecular basis for T3SS-dependent nodulation and distinguish it from NFR-dependent nodulation, we examined gene-expression profiles in soybean roots using microarray analysis. Comparative analysis of microarray data was performed to dissect soybean genes up- or down-regulated by NF or T3SS signaling (Fig. 4). To extract soybean genes up- or down-regulated by NFR signaling, we compared gene expression in the roots of Enrei and En1282 inoculated with BErhcJ (Fig. 4A). Eight days after inoculation, roots were sampled as suitable material for target RNA preparation. To detect soybean genes up- or down-regulated by signals derived by B. elkanii T3SS, we compared the gene expression of En1282 roots inoculated with USDA61 and BErhcJ (Fig. 4A).

Experimental design and summary of microarray analysis. (A) Experimental design. To extract soybean genes up- or down-regulated by Nod-factor signaling, we compared gene expression in the roots of Enrei and En1282 inoculated with BErhcJ. To detect soybean genes up- or down-regulated by the signals derived by B. elkanii T3SS, we compared the gene expression of En1282 roots inoculated with USDA61 and BErhcJ. (B) Summary of microarray analyses of genes up-regulated by NF and T3SS signaling. Soybean genes with signal intensities more than 2-fold that of uninfected roots were considered to be up-regulated. (C) Summary of microarray analyses of genes down-regulated by NF and T3SS signaling. Soybean genes with signal intensities less than 0.5-fold that of uninfected roots were considered to be down-regulated.
The microarray analysis results are summarized in Fig. 4 B and C. Soybean genes with signal intensities more than 2-fold and less than 0.5-fold that of uninfected roots were considered up- and down-regulated, respectively. We identified 173 and 991 genes potentially up- and down-regulated by NFR signaling (Table S2 and Dataset S1, respectively), and 29 and 36 genes potentially up- and down-regulated by T3SS signaling (Tables S3 and S4, respectively). The 173 genes potentially up-regulated by NF signaling include a number of known nodulins and symbiosis-related genes (Table S2). Of these, 16 are commonly up-regulated by T3SS signaling, including symbiosis-related genes such as root-nodule extensin, ENOD40, and nodule inception (NIN) (Table 1). The fold changes of signal intensity were not comparable between NF and T3SS signaling. Nodulins 21 and 26 were up-regulated only by NF signaling (Table S2), whereas T3SS signaling triggered the up-regulation of endo-β-1,4-glucanase, cyclin, and early nodulin GmN93 (Table S3).
Table 1.
G. max genes up-regulated or down-regulated by both Nod-factor and T3SS signaling
| ID no. | Similarity | NF pathway* | T3SS pathway† | ||
| Fold change | P value | Fold change | P value | ||
| Up-regulated | |||||
 BI316036.1 BI316036.1 | Root-nodule extensin | 217.69 | 0.0021 | 71.55 | 0.0063 |
 BI970567.1 BI970567.1 | G. max uncharacterized protein | 178.16 | 0.0007 | 2.36 | 0.0323 |
 BI971063.1 BI971063.1 | Subtilisin-like protease | 81.41 | 0.0048 | 6.45 | 0.0037 |
 BW679106.1 BW679106.1 | F-box/kelch-repeat protein | 45.95 | 0.0027 | 10.67 | 0.0021 |
 CO978759.1 CO978759.1 | CASP-like protein N24–like protein | 34.24 | 0.0015 | 4.63 | 0.0126 |
 CO985886.1 CO985886.1 | Gibberellin 3-β-dioxygenase 1–like protein | 30.45 | 0.0030 | 14.11 | 0.0117 |
 BW676134.1 BW676134.1 | G. max cDNA clone Gm-c1082-2286 | 17.90 | 0.0066 | 2.85 | 0.0044 |
 BW657022.1 BW657022.1 | Nodule inception protein (nin) | 15.73 | 0.0104 | 5.10 | 0.0135 |
 BI425123.1 BI425123.1 | Cytochrome P450 71A1-like protein | 15.10 | 0.0115 | 4.10 | 0.0028 |
 BW676262.1 BW676262.1 | ENOD40-2 | 10.90 | 0.0071 | 3.30 | 0.0336 |
 CF922378.1 CF922378.1 | Early nodulin GmN36a | 9.17 | 0.0081 | 3.33 | 0.0230 |
 AW830341.1 AW830341.1 | G. max hypothetical 17.8-kDa protein | 6.60 | 0.0213 | 7.60 | 0.0032 |
 BI969878.1 BI969878.1 | G. max uncharacterized protein | 6.03 | 0.0068 | 2.27 | 0.0081 |
 BE346992.1 BE346992.1 | G. max uncharacterized protein | 2.50 | 0.0346 | 2.13 | 0.0271 |
 BI426827.1 BI426827.1 | Indole-3-acetate O-methyltransferase 1-like–protein | 2.46 | 0.0017 | 2.31 | 0.0031 |
 BI967256.1 BI967256.1 | G. max uncharacterized protein | 2.17 | 0.0409 | 3.60 | 0.0019 |
| Down-regulated | |||||
 CO981522.1 CO981522.1 | Tryptophan aminotransferase-related protein | 0.05 | 0.000 | 0.15 | 0.043 |
 BQ080234.1 BQ080234.1 | Callose synthase 9-like, transcript variant | 0.09 | 0.002 | 0.28 | 0.012 |
 BM177253.1 BM177253.1 | G. max uncharacterized protein | 0.11 | 0.004 | 0.13 | 0.016 |
 CD405795.1 CD405795.1 | G. max mRNA induced by salicylic acid | 0.13 | 0.014 | 0.27 | 0.011 |
 BM892625.1 BM892625.1 | G. max uncharacterized protein | 0.20 | 0.021 | 0.20 | 0.042 |
 AW620254.1 AW620254.1 | 17.9-kDa class II heat shock protein | 0.20 | 0.003 | 0.48 | 0.043 |
 AW705695.1 AW705695.1 | Vicilin-like antimicrobial peptides 2-2–like protein | 0.26 | 0.007 | 0.46 | 0.025 |
 CA938283.1 CA938283.1 | Chlorophyll a-b–binding protein | 0.27 | 0.047 | 0.34 | 0.001 |
 CD391543.1 CD391543.1 | G. max uncharacterized protein | 0.31 | 0.027 | 0.41 | 0.041 |
 BE346566.1 BE346566.1 | Myb-related protein 306–like transcription factor | 0.33 | 0.046 | 0.38 | 0.048 |
 BG839502.1 BG839502.1 | NAC domain-containing protein 8–like protein | 0.37 | 0.016 | 0.49 | 0.045 |
 BW683725.1 BW683725.1 | G. max uncharacterized protein | 0.40 | 0.047 | 0.10 | 0.002 |
NF signaling down-regulated 991 genes, of which 12 were commonly down-regulated by T3SS signaling. These include tryptophan aminotransferase-related protein, callose synthase, and salicylic acid-inducing genes (Table 1). By contrast, down-regulation of 24 genes was identified only in T3SS signaling, including vacuolar processing enzyme 2, chalcone synthase, and vicilin-like antimicrobial peptides (Table S4).
T3SS-Dependent Expression of Symbiosis Genes ENOD40 and GmNIN in Soybean.
Microarray analysis indicated that T3SS-derived signaling from B. elkanii activates several symbiosis-related genes, which are known to be expressed by the recognition of compatible NFs via host legume NFRs. To investigate further the importance of NFs and NFRs in the expression of these symbiosis genes, we compared their expression in the roots of Enrei and En1282 inoculated with T3SS and nodC mutants using real-time RT-PCR. Two well-characterized symbiosis genes, ENOD40 (15) and NIN (16), were selected as marker genes because their expression is induced early after NF perception (17, 18). In both Enrei and En1282 roots, relative expression levels of ENOD40 and NIN were significantly induced by the inoculation of USDA61 (Fig. 5). Deletion of the nodC gene decreased the up-regulation of these genes, but it remained significant compared with the mock treatment. However, the up-regulation of both genes was abolished by inoculation of the T3SS mutant BErhcJ on En1282. These results indicated that the expression of ENOD40 and NIN was induced by T3SS-derived signaling and that NF production enhances the expression even in roots of the nfr1 mutant En1282.
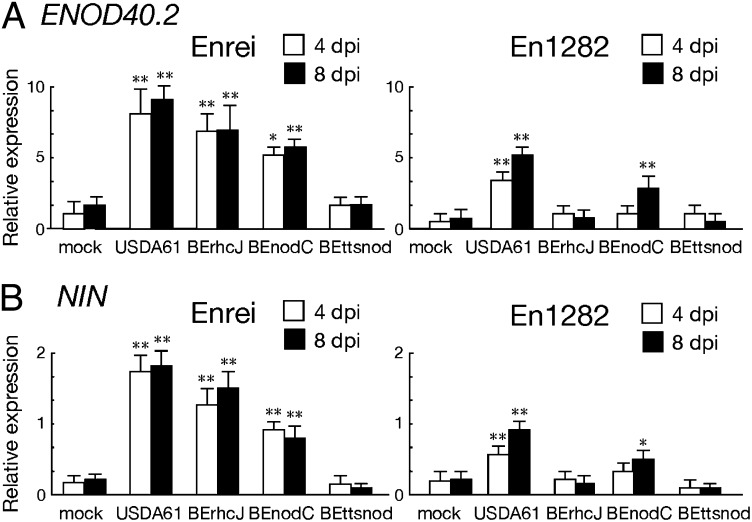
Real-time RT-PCR analysis of G. max ENOD40 and G. max NIN transcripts in roots of Enrei and En1282 inoculated with wild-type and mutants of B. elkanii USDA61. RNAs were isolated from the roots of Enrei and its nfr1 mutant En1282 4 and 8 d postinoculation. cDNA synthesis and real-time PCR of G. max ENOD40 (A) and G. max NIN (B) were performed with gene-specific primers (Table S5) as described in Materials and Methods. The values are means of triplicate measurements, and the error bars indicate SDs. Statistical analyses (Student t test) were performed comparing each inoculation with the mock treatment. *P < 0.05; **P < 0.01.
Discussion
The present study investigated the activation of soybean nodulation signaling by T3SS in B. elkanii, homologs of which have been widely observed in pathogenic bacteria, and which play crucial roles in infecting their animal and plant hosts. We demonstrated that the activation does not require the initial step of symbiotic molecular interaction involving rhizobium NF and host legume NFR via the observation that wild-type B. elkanii, but not the T3SS-deficient mutant, was able to form nitrogen-fixing nodules on soybean nfr1 mutants En1282 and Clark-rj1. Nodulation of these soybean mutants was observed by infecting the B. elkanii NF-deficient mutant BEnodC unless T3SS genes were mutated.
Microarray and RT-PCR analysis revealed that T3SS-derived signaling from B. elkanii can induce the expression of symbiosis genes ENOD40 and NIN in the absence of NFR1 and NF (Fig. 5 and Table 1). These symbiosis genes were previously thought to be up-regulated only following the recognition of NFs via associated NFRs. Although the mechanism for their activation by T3SS remains to be elucidated, recent findings from plant mutants that form nodules in the absence of NFs or rhizobia suggest a number of possibilities. The L. japonicus mutant spontaneous nodule formation 1 (snf1) carries a mutation in CCaMK that leads to autoactivation of its kinase domain and the development of spontaneous nodules in the absence of rhizobia (19). snf1 and other CCaMK gain-of-function mutations lead to the induction of nodulation genes such as ENOD11, resulting in the development of spontaneous nodules in the absence of either NFs or rhizobia (19). Similarly, the gain-of-function mutant in the cytokinin receptor LHK1 was shown to develop spontaneous nodules, concomitant with the expression of NIN in the absence of rhizobia (20, 21). Moreover, exogenous cytokinins induce the expression of early nodulin genes including ENOD40 and NIN in several legume species (22, 23). It remains to be determined whether B. elkanii T3SS activates CCaMK or LHK, or affects cytokinin levels in the nfr1 mutant soybean, leading to nodule development.
Of particular interest are the nature and function of B. elkanii T3Es in activating soybean nodulation signaling. B. japonicum, a close relative of B. elkanii, also possesses a functional T3SS that is activated by soybean genistein, and secretes some proteins that are also secreted by B. elkanii (6, 24). However, this and other studies show that B. japonicum is unable to nodulate on either En1282 or Clark-rj1, suggesting that B. elkanii possesses a unique T3E(s) responsible for activation of nodulation signaling. Although we have not yet detected any B. elkanii unique T3E candidate genes, B. elkanii encodes several genes for T3E candidates, including nopL, nopM, and nopP, which have been reported to play either a positive or a negative role in symbiosis in other rhizobia (9, 25). Intriguingly, CCaMK and LHK1, mentioned above, are protein kinases that play pivotal roles in nodulation and should participate in phosphorylation systems (26). As a number of T3Es in plant pathogens are known to target host kinases (27), and the rhizobial T3Es NopL and NopP are phosphorylated in host cytoplasm (9), it is conceivable that some T3Es perturb the host phosphorylation state for nodulation.
Intriguingly, in the present study, the nodC mutant induced fewer nodules than wild-type B. elkanii on En1282 or Clark-rj1 soybean (Fig. 1 and Fig. S1), indicating that rhizobial NF signals even play a positive role in the nodule formation of the nfr1 mutant soybean. The positive effects of NFs on nfr1 soybean were also confirmed by expression analysis of ENOD40 and NIN, as the expression of both genes in the roots of En1282 was higher for USDA61 than BEnodC inoculation (Fig. 5). The NFRs belong to lysine motif (LysM) receptor-like kinases (LysM-RKs), and besides NFR1/NFR5 homologs that directly bind to NFs (28), legume plants are known to possess larger numbers of LysM-RKs than nonleguminous plants (18, 29). Together, these results suggest that rhizobial NFs are perceived by additional receptors in soybean roots, and that the derived signal and that from NFRs synergistically activate nodulation signaling.
Another intriguing observation was the lack of root-hair curling or infection-thread formation in the roots of En1282 inoculated with USDA61. This suggested that T3SS plays a role in alternative infection processes such as crack entry or intercellular infection, although further microscopic analysis will be necessary to elucidate the nature of the infection enhanced by T3SS. Crack-entry infection is well-studied in A. hypogea, one of the host plants of B. elkanii, although the nodulation to A. hypogea is likely to be NF-dependent (30). In the course of A. hypogea nodulation, neither root hairs nor cortical infection threads are formed. Instead, rhizobia invade through fissures in epidermal cells, and the intercellular rhizobia directly internalize cortical cells via an endocytosis-like process (30). Recently, photosynthetic Bradyrhizobium spp. lacking the canonical nod genes were shown to infect their host legume Aeschynomene spp. through gaps between axillary root hairs and invade the cortical cells by invagination of the host cell wall (31). Moreover, Madsen et al. reported that infection between M. loti nodC and L. japonicus nfr1-1 snf1 mutants involves intercellular infection with no infection threads in either root hairs or nodule tissues (26). Such ground-state infection occurs at a 20- to 100-fold lower frequency compared with normal root-hair invasion (26). Similarly, we observed limited but significant nodulation on Enrei and En1282 by the B. elkanii nodC mutant and no nodulation by the nodC-ttsI double mutant (Fig. 1). This suggests that B. elkanii T3SS can promote intercellular infection. In the presence of NFs and NFRs, however, this poorly efficient infection would be overridden by that accompanying root-hair infection threads. Although the details of the T3SS-related infection of B. elkanii remain to be elucidated, some insights might be gained by examining the T3SS-mediated Salmonella infection process. Salmonella spp. use T3Es to manipulate cellular processes in promoting their uptake and subsequent survival within vacuoles (32), which makes a fitting analogy with rhizobia being entrapped by root cortical cells via an endocytosis-like process that develops symbiosomes of infected cells in nodules.
Although the B. elkanii nodC mutant was shown to form nodules in the present study (Fig. 1), most of these nodules were poorly infected (Fig. 3). This suggests that concerted expression, rather than simple copossession, of T3SS and nodulation genes is required for the formation of effective nodules. Such a requirement might be why most known rhizobial T3SS genes are situated downstream of flavonoid-responsive signaling (6, 7). Host-derived flavonoid induction is found in most T3SS-harboring rhizobia, except for the β-rhizobium Cupriavidus taiwanensis, which incorporates a T3SS similar to that of the β-proteobacterium Burkholderia, and its expression is more likely to be glutamate-dependent. These findings suggest that the acquisition of T3SS from ancestor pathogenic bacteria has been adapted for the orchestrated expression of symbiotic genes during coevolution with host legume plants.
Another interesting aspect of rhizobial T3SS is that T3 effector proteins can cause restriction of nodulation. Our previous study revealed that soybean cultivar Hill, which carries the Rj4 gene and is known to restrict nodulation by B. elkanii USDA61, formed nodules when inoculated with type III knockout mutants of USDA61 (12). Although the Rj4 gene has not been identified, the soybean genes Rj2 and Rfg1 were recently identified as allelic genes encoding the Toll-interleukin receptor/nucleotide-binding site/leucine-rich repeat (TIR-NBS-LRR) class of plant resistance (R) proteins that restrict nodulation with B. japonicum USDA122 and Sinorhizobium fredii USDA257, respectively (33). Introduction of the Rfg1 allele of Williams 82 into Peking (genetically defined as rfg1/rfg1) abolished nodulation of S. fredii USDA257, whereas the T3SS knockout mutant of S. fredii USDA257 exhibited the ability to nodulate soybean carrying the Rfg1 allele (33). These findings imply that rhizobial T3Es, when recognized by cognate host R proteins, trigger a strong defense reaction (effector-triggered immunity) that halts rhizobial infection.
In conclusion, rhizobia use a pathogenic secretion system for symbiotic purposes. B. elkanii T3SS hijacks soybean symbiosis genes in the absence of canonical NFs and NFRs that are known to be dispensable for the initiation of nodulation. We propose a model of nodulation signaling and modulation by the rhizobial type III secretion system in Fig. 6. Rhizobial T3SS is likely to promote intercellular infection, which can be superimposed by more efficient root-hair infection mediated by canonical NF and NFR signaling. Our efforts are now being directed toward the identification of the B. elkanii effector protein that hijacks soybean symbiosis signaling, to acquire a better understanding of the evolutionary aspects of interactions between endosymbionts and their eukaryotic hosts.
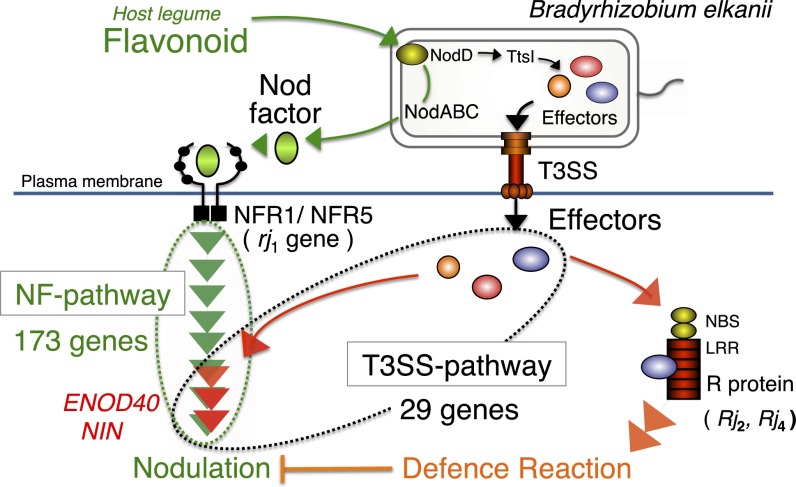
Model of nodulation signaling and modulation by the rhizobial type III secretion system. A host plant-derived flavonoid induces the production of Nod factors in rhizobia. Recognition of NFs by NF receptors triggers a signaling cascade leading to nodulation. The flavonoid also induces rhizobial T3SS, which injects effector proteins into host cells. One effector modulates nodulation signaling toward nodulation, whereas another is recognized by the host defense system, which is capable of aborting the nodulation process.
Materials and Methods
Bacterial Strains and Growth Conditions.
The bacterial strains used in this study are listed in Table S5. B. elkanii cultures were grown at 28 °C in arabinose–gluconate medium (34) or peptone–salts–yeast extract medium (35) supplemented with appropriate antibiotics (50 µg/mL polymyxin, 100 µg/mL tetracycline, 100 µg/mL kanamycin). Escherichia coli strains were grown at 37 °C in Luria–Bertani medium supplemented with appropriate antibiotics (12.5 µg/mL tetracycline, 100 µg/mL ampicillin, 50 µg/mL kanamycin).
Construction of B. elkanii Mutants.
Disruption of ttsI in B. elkanii was performed using a previously described method (9). Briefly, ttsI-containing fragments were amplified by PCR using primers ttsI_F and ttsI_R (Table S6) from the USDA61 genome. The resulting PCR product was cloned into the SmaI site of pK18 mobsacB (36) to form pK18MSBttsI. The aadA gene containing 45-bp extensions homologous to regions flanking the ttsI gene was PCR-amplified using pKST001Rtr (37) as the template and primers ttsI_U and ttsI_L (Table S6). The resulting PCR products were electroporated into E. coli DH10B harboring plasmids pK18MSBttsI and pKD46 (38) and then integrated into the cosmid via lambda red-mediated recombination. The plasmid derivatives with deletion–insertion loci were then transferred to B. elkanii USDA61 by conjugation. The deletion mutant BEttsI was obtained as a result of homologous recombination. Disruption of nodC was performed using a similar method to that for ttsI, except that pS18mobsacB was used rather than pK18mobsacB, and pKD13 (38) was used rather than pKST001Rtr to disrupt the nodC gene by a kanamycin-resistant gene (kan). The ttsI and nodC double-deletion mutants were obtained by introducing pS18MSBnodC into BEttsI by conjugation and selection of strains with both kanamycin and streptomycin resistance.
Plant Assays.
The soybean cultivars used were G. max (L.) Merr. cultivar Clark-rj1, Enrei (wild-type nodulating cultivar), and En1282 (nonnodulating mutant derived from Enrei by ethyl methanesulfonate mutagenesis) (39). En1282 was mutated in the NFR1 gene, which encodes an LysM-type receptor kinase involved in NF recognition (40). As far as we are aware, no backcross experiments have been performed with En1282. Soybean seeds were surface-sterilized and germinated as described by Krause et al. (6). One day after transplantation, all seedlings were inoculated with 1 mL of 107 cells per mL of wild-type or mutant strains of B. elkanii USDA61 or sterilized water (mock treatment). Plants were cultivated in a growth chamber at 25 °C and 70% humidity under a daytime/nighttime regimen of 16/8 h.
RNA Extraction from Soybean Roots and Microarray Analysis.
Soybean roots were sampled, immediately frozen in liquid nitrogen, and ground to a powder. RNA was prepared from 100 mg powdered roots using the Qiagen RNeasy Plant Mini Kit according to the manufacturer’s instructions. All RNA samples (0.5 µg) were labeled with cyanine-3 (Cy3) fluorescent dye using the Agilent Quick Amp Labeling Kit. Cy3-labeled cRNA (1.65 µg) was fragmented and hybridized to the Agilent G. max (soybean) Oligo Microarray 4 × 44K (array ID no. 016772) for 17 h at 65 °C in a rotating oven. After hybridization, microarrays were washed and scanned on the Agilent DNA Microarray Scanner (G2505B) and the scanned images were analyzed with Agilent Feature Extraction software (version 9.5.1). The data were then subjected to 75 percentile normalization and log2 transformation using the analysis tool Subio Basic Plug-in. The microarray data discussed in this publication have been deposited in the National Center for Biotechnology Information’s Gene Expression Omnibus (GEO) and are accessible through GEO Series accession no. GSE38520.
Real-Time RT-PCR Analysis.
cDNA was synthesized in a final volume of 20 µL from 400 ng total RNA using SuperScript III reverse transcriptase (New England BioLabs) and a random primer (Invitrogen) according to the manufacturers’ protocols. After 10-fold dilution, cDNA was subjected to real-time PCR analyses in a final volume of 20 µL using KAPA SYBR Fast qPCR Master Mix following the manufacturer’s protocol. All primer sets used in the expression analysis are listed in Table S6.
Microscopy.
Soybean nodules were fixed with 2% (vol/vol) glutaraldehyde in phosphate saline buffer at 4 °C overnight. Nodules were dehydrated in an ethanol series and embedded in LR white acrylic resin (Electron Microscopy Sciences). Semithin sections (1-μm) were obtained using a microtome (Leica Microsystems), stained with 1% (wt/vol) aqueous toluidine blue solution, and examined with a BX51 Olympus light microscope.
Acknowledgments
We thank K. Minamisawa (Tohoku University) for kindly providing soybean seeds and Dr. Satoshi Nakaba (Tokyo University of Agriculture and Technology) for technical assistance in preparing nodule sections. We are grateful to H. Kouchi and H. Imaizumi-Anraku (National Institute of Agrobiological Sciences, Japan) and M. Göttfert (Dresden University of Technology) for helpful discussions and critical reading of the manuscript. This research was supported in part by Grants-in-Aid for Young Scientists (to S.O.; Grants 21780056 and 23780068) and for Scientific Research on Innovative Areas (to K.S.; Grant 24113512).
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
Data deposition: The microarray data reported in this paper have been deposited in the Gene Expression Omnibus (GEO) database, www.ncbi.nlm.nih.gov/geo (accession no. GSE38520).
This article contains supporting information online at www.pnas.org/lookup/suppl/10.1073/pnas.1302360110/-/DCSupplemental.
References
Articles from Proceedings of the National Academy of Sciences of the United States of America are provided here courtesy of National Academy of Sciences
Full text links
Read article at publisher's site: https://doi.org/10.1073/pnas.1302360110
Read article for free, from open access legal sources, via Unpaywall:
https://www.pnas.org/content/pnas/110/42/17131.full.pdf
Citations & impact
Impact metrics
Citations of article over time
Alternative metrics
Article citations
Atypical rhizobia trigger nodulation and pathogenesis on the same legume hosts.
Nat Commun, 15(1):9246, 26 Oct 2024
Cited by: 0 articles | PMID: 39461961 | PMCID: PMC11513132
NopP2 effector of Bradyrhizobium elkanii USDA61 is a determinant of nodulation in Vigna radiata cultivars.
Sci Rep, 14(1):24541, 19 Oct 2024
Cited by: 0 articles | PMID: 39424841 | PMCID: PMC11489812
Transcriptomes of soybean roots and nodules inoculated with Sinorhizobium fredii with NopP and NopI variants.
Sci Data, 11(1):1146, 18 Oct 2024
Cited by: 0 articles | PMID: 39424807 | PMCID: PMC11489703
The type III effector NopL interacts with GmREM1a and GmNFR5 to promote symbiosis in soybean.
Nat Commun, 15(1):5852, 12 Jul 2024
Cited by: 0 articles | PMID: 38992018 | PMCID: PMC11239682
Comparative genomic and transcriptomic analyses provide new insight into symbiotic host specificity.
iScience, 27(7):110207, 06 Jun 2024
Cited by: 0 articles | PMID: 38984200 | PMCID: PMC11231455
Go to all (123) article citations
Data
Data behind the article
This data has been text mined from the article, or deposited into data resources.
BioStudies: supplemental material and supporting data
GEO - Gene Expression Omnibus
- (2 citations) GEO - GSE38520
Similar Articles
To arrive at the top five similar articles we use a word-weighted algorithm to compare words from the Title and Abstract of each citation.
Effector-Triggered Immunity Determines Host Genotype-Specific Incompatibility in Legume-Rhizobium Symbiosis.
Plant Cell Physiol, 57(8):1791-1800, 03 Jun 2016
Cited by: 53 articles | PMID: 27373538
Rhizobia use a pathogenic-like effector to hijack leguminous nodulation signalling.
Sci Rep, 11(1):2034, 21 Jan 2021
Cited by: 23 articles | PMID: 33479414 | PMCID: PMC7820406
Roles of flavonoids and the transcriptional regulator TtsI in the activation of the type III secretion system of Bradyrhizobium elkanii SEMIA587.
Microbiology (Reading), 157(pt 3):627-635, 25 Nov 2010
Cited by: 7 articles | PMID: 21109563
Type III effector provides a novel symbiotic pathway in legume-rhizobia symbiosis.
Biosci Biotechnol Biochem, 87(1):28-37, 01 Dec 2022
Cited by: 4 articles | PMID: 36367542
Review