Abstract
Free full text

The Adaptive Endoplasmic Reticulum Stress Response to Lipotoxicity in Progressive Human Nonalcoholic Fatty Liver Disease
Abstract
Nonalcoholic fatty liver disease (NAFLD) may progress from simple steatosis to severe, nonalcoholic steatohepatitis (NASH) in 7%–14% of the U.S. population through a second “hit” in the form of increased oxidative stress and inflammation. Endoplasmic reticulum (ER) stress signaling and the unfolded protein response (UPR) are triggered when high levels of lipids and misfolded proteins alter ER homeostasis creating a lipotoxic environment within NAFLD livers. The objective of this study was to determine the coordinate regulation of ER stress–associated genes in the progressive stages of human NAFLD. Human liver samples categorized as normal, steatosis, NASH (Fatty), and NASH (Not Fatty) were analyzed by individual Affymetrix GeneChip Human 1.0 ST microarrays, immunoblots, and immunohistochemistry. A gene set enrichment analysis was performed on autophagy, apoptosis, lipogenesis, and ER stress/UPR gene categories. An enrichment of downregulated genes in the ER stress–associated lipogenesis and ER stress/UPR gene categories was observed in NASH. Conversely, an enrichment of upregulated ER stress–associated genes for autophagy and apoptosis gene categories was observed in NASH. Protein expression of the adaptive liver response protein STC2 and the transcription factor X-box binding protein 1 spliced (XBP-1s) were significantly elevated among NASH samples, whereas other downstream ER stress proteins including CHOP, ATF4, and phosphorylated JNK and eIF2α were not significantly changed in disease progression. Increased nuclear accumulation of total XBP-1 protein was observed in steatosis and NASH livers. The findings reveal the presence of a coordinated, adaptive transcriptional response to hepatic ER stress in human NAFLD.
Nonalcoholic fatty liver disease (NAFLD) is a chronic disease associated with the activation of the endoplasmic reticulum (ER) stress response and the unfolded protein response (UPR) (Puri et al., 2008). The pathological progression of this disease has been described by the “2-hit” hypothesis, which states that an initial first “hit” in the form of accumulating lipids within hepatocytes results in the initial profile of simple steatosis (Day and James, 1998). NAFLD patients with steatosis can have a second “hit” in the form of increased oxidative stress and inflammation that results in the development of nonalcoholic steatohepatitis (NASH), a severe disease that is difficult to reverse in contrast to the previously benign stage of steatosis (Day and James, 1998). NASH patients may progress from NASH (Fatty) to NASH (Not Fatty), which is similar to the pathological profile of cirrhosis. Approximately 30%–40% of the U.S. population is estimated to be afflicted with some form of NAFLD, whereas 7%–14% is estimated to have progressed to NASH (Ali and Cusi, 2009; McCullough, 2006). The pathological features of the disease have been well characterized; however, the exact mechanism of the progression from steatosis to NASH is not known. The lipotoxic environment of NAFLD is promoted by an oversupply of lipids that directly influences ER homeostasis and ER stress activation (Leamy et al., 2013). The signaling pathways resulting from lipotoxic stress may lead to either an adaptive or a harmful UPR (Achard and Laybutt, 2012).
The UPR is classically triggered in response to an accumulation of misfolded or unfolded proteins. Current evidence in the literature indicates that increased lipotoxic stress in the form of saturated fatty acids may also trigger the ER stress response (Fuchs and Sanyal, 2012; Leamy et al., 2013). The 3 branches of the ER stress response are composed of the type I transmembrane inositol-requiring enzyme 1 (IRE1) branch, the PKR-like ER kinase (PERK), and the activating transcription factor 6 (ATF6) branch. These 3 pathways of the ER stress response are activated through the 3 transmembrane receptors (IRE1, PERK, and ATF6), which are normally kept in an inactive state by close association with the chaperone GRP78. These sensors initiate downstream transcription of ER stress–associated genes. The regulation of the ER stress response is dependent on the activation of the transcription factors: X-box binding protein 1 spliced (XBP-1s), ATF6, and ATF4 (Adachi et al., 2008; Uemura et al., 2009). XBP-1s is a crucial transcription factor of the ER stress response shown to directly regulate lipogenesis gene expression (Glimcher and Lee, 2009; Lee et al., 2008). XBP-1s has multiple functions in the regulation of lipogenesis and insulin resistance (Ning et al., 2011). The dual role of XBP-1s in the regulation of lipogenesis and the activation of IRE1 pathway targets makes it an important regulatory component in the study of lipotoxic ER stress. The transcription factor ATF4 has also been shown to participate in cytoprotective functions through upregulation of stanniocalcin 2 (STC2) in the PERK pathway (Fazio et al., 2011; Ito et al., 2004). The activation of ATF6 has been reported to suppress lipogenesis genes under conditions of glucose deficiency and ER stress (Zeng et al., 2004). Thus, ER stress signaling has an important role in the regulation of lipogenesis and cell protection.
Autophagy and apoptosis are cellular processes that attempt to resolve ER stress or initiate controlled cell death mechanisms (Amir and Czaja, 2011; Gentile et al., 2011; Patil and Walter, 2001). Lipophagy is a form of autophagy that participates in the regulation of lipid metabolism (Singh et al., 2009). Autophagy and apoptosis are distinct cellular processes with the same cytoprotective goals. ER stress pathway transcription factors regulate both autophagy and apoptosis (Acosta-Alvear et al., 2007; Alkhouri et al., 2011; Amir and Czaja, 2011; Mei et al., 2011; Thorburn, 2008). Autophagy and apoptosis may also modulate the transition from steatosis to NASH (Amir and Czaja, 2011; Rautou et al., 2010). The connecting events of cell death, cytoprotection, and lipid processing mechanisms are connected and regulated through ER stress signaling (Glimcher and Lee, 2009; Lee and Glimcher, 2009; Ren et al., 2012). Thus, the hypothesis of the current study is that ER stress–associated gene categories of the UPR, lipogenesis, autophagy, and apoptosis are coordinately altered with progression of NAFLD in an attempt to resolve hepatic lipotoxicity and stress.
MATERIALS AND METHODS
Human liver samples.
Human liver samples were acquired from the National Institutes of Health (NIH)–funded Liver Tissue and Cell Distribution System that is comprised of the following institutions: the University of Minnesota, University of Pittsburgh, and Virginia Commonwealth University. Postmortem liver tissue and liver biopsies were acquired and scored for diagnosis by an NIH pathologist using the NAFLD activity scoring system (Kleiner et al., 2005). According to this method, a sample is diagnosed as steatosis when greater than 10% lipid deposition was scored within hepatocytes, but there was no accompanying inflammation or fibrosis. NASH with fatty liver samples were scored when greater than 5% lipid deposition in the presence of inflammation and fibrosis was observed. NASH without fatty liver samples were distinguished from NASH with fatty liver samples by having less than 5% fat deposition and increased inflammation and fibrosis. Human liver tissue information on age, sex, and demographics has been previously published and described (Fisher et al., 2009; Lake et al., 2011). Histology images demonstrating each stage of the disease of the same samples utilized in this study have been previously published (Fisher et al., 2009; Hardwick et al., 2010). Liver tissue was utilized for total mRNA isolation and purification for application to individual Affymetrix GeneChip Human 1.0 ST microarrays as described previously (Lake et al., 2011). Quality of the mRNA was assessed as previously described and reported (Lake et al., 2011). A sample size of 19 normal, 10 steatosis, 9 NASH Fatty, and 7 NASH Not Fatty samples were utilized in the generation of the individual Affymetrix microarrays. Whole cell liver lysates were generated for Western blot analyses from individual liver tissue samples as described previously for protein analysis (Hardwick et al., 2011). Whole cell liver lysate (7 normal, 7 steatosis, 8 NASH Fatty, and 5 NASH Not Fatty) samples were utilized for western blotting. Total protein content was analyzed using the Pierce BCA (Thermo Fisher Scientific, Waltham, Massachusetts) protein quantitation assay.
Microarray ER stress–associated genes.
Affymetrix GeneChip Human 1.0 ST Arrays (Affymetrix, La Jolla, California) were generated for each human liver sample. Array hybridization, washing, and scanning were performed according to manufacturer’s recommendations in the Affymetrix GeneChip Whole Transcript Sense Target Labeling protocol (Affymetrix). All microarray data processing, archiving, and analysis were generated by the Genomics Core Facility at the Arizona Cancer Center. NASH Fatty and NASH Not Fatty microarray data were combined in the analysis into 1 category designated as NASH due to the lack of global gene expression changes and mechanistic differences between the 2 categories as reported previously (Lake et al., 2011). Although morphologically the 2 stages of NASH are categorized by pathologists as separate pathologies with NASH Fatty occurring first in the change of progression and NASH Not Fatty occurring shortly after as the liver progresses to cirrhosis, no mechanistic changes in global gene expression occur between these 2 pathological designations. A total of 33 252 annotated and unannotated genes are represented for each sample. The data set is accessible at the ArrayExpress public repository for microarray data under the accession number E-MEXP-3291 (http://www.webcitation.org/5zyojNu7T).
Gene set enrichment analysis.
Autophagy, apoptosis, lipogenesis, and ER stress/UPR gene categories were tested for enrichment in either upregulation or downregulation using the human microarray data. Gene set enrichment tests were performed using the Generally Applicable Gene-set Enrichment (GAGE) analysis method as described in the literature (Luo et al., 2009). The GAGE method of gene enrichment analysis is appropriate for gene expression data of different sample sizes and experimental design and was employed to analyze the ER stress gene categories in the NAFLD human microarray data (Luo et al., 2009). Hierarchical clustering analysis was also performed on each of the ER stress–associated gene categories. Heat maps were generated to visualize clustering patterns of diagnosis groups and gene expression changes in the tested ER stress gene sets.
Gene set coexpression analysis.
A comparison of expression profiles was performed for all ER stress gene categories. The different categories of ER stress genes (ER stress/UPR, lipogenesis, apoptosis, and autophagy) were each analyzed for gene coexpression using correlation coefficients. Histograms showing the correlation of ER stress gene categories with each other in comparison with the overall background gene expression in the microarray data were generated for each category.
Protein expression analysis.
ER stress pathway proteins were analyzed by Western blot analysis using the human whole cell liver lysate samples. The samples (80 μg of total protein) were diluted in Laemmli buffer (Bio-Rad Laboratories, Hercules, California) and subjected to polyacrylamide gel electrophoresis using 10% SDS-polyacrylamide gels. Proteins were transferred to PVDF membranes overnight, blocked in 5% nonfat dry milk in TBST buffer, and incubated overnight in the following primary antibodies. Phosphorylated and total JNK and phosphorylated and total eIF2α (Cell Signaling Technology, Boston, Massachusetts), XBP-1s (Biolegend, San Diego, California), ATF4 (Santa Cruz Biotechnology Inc, Santa Cruz, California), STC2 (Abcam Inc, Cambridge, Massachusetts), and CHOP (Santa Cruz Biotechnology Inc). A nitrocellulose membrane was used for CHOP protein analysis rather than PVDF. Following primary antibody incubation, the blots were incubated with the corresponding horseradish peroxidase–tagged secondary antibodies (Santa Cruz Biotechnology Inc). The blots were washed and imaged with Femto West Super Signal chemiluminescent solution (ThermoScientific, Rockford, Illinois). Densitometric analysis was performed with ImageJ Software (NIH, Bethesda, Maryland) and proteins were normalized to total pan-Cadherin protein (Abcam Inc). Densitometric data were graphed and illustrated in Graphpad Prism 5 (GraphPad Software Inc, San Diego, California) with box-and-whisker plots.
Immunohistochemical staining of total XBP-1.
Normal (n = 5), steatotic (n = 5), NASH Fatty (n = 7), and NASH Not Fatty (n = 7) formalin fixed and paraffin-embedded (FFPE) human liver samples were stained for XBP-1 nuclear localization. Each sample was deparaffinized, followed by rehydration in an ethanol gradient. Antigen retrieval was performed with citrate-EDTA buffer (10mM citric acid, 2mM EDTA, 0.05% Tween 20, pH 6.2) in a microwave followed by a 0.3% (vol/vol) endogenous peroxidase block as previously described for FFPE human liver samples (Hardwick et al., 2011). All samples were incubated in total XBP-1 primary antibody (1:110, Abcam Inc) overnight at 4°C. Antibody binding was detected by the MACH4 kit (Biocare Medical, Concord, California), and color development was performed with Betazoid DAB (Biocare Medical) according to the manufacturer’s recommendation. All slides were imaged with a Nikon Eclipse E4000 microscope and a Sony Exwave DXC-390 camera. Hepatocyte nuclei positive for XBP-1 staining and hepatocyte nuclei negative for XBP-1 staining were quantified within a ×40 field of vision with a total of 10 fields assessed for each sample. The number of total hepatocytes was calculated from the positive and negative nuclei counts and was used to determine the percentage of positive nuclei within a field.
RESULTS
Gene Set Enrichment Analysis
Gene set enrichment analysis was performed on the ER stress gene categories of lipogenesis, apoptosis, autophagy, and ER stress/UPR acquired from the human NAFLD microarray data set. The lipogenesis and ER Stress/UPR gene categories were significantly enriched for downregulated genes in NASH liver samples (Table 1). Conversely, apoptosis and autophagy gene sets were significantly enriched for upregulated genes in NASH samples (Table 1). Autophagy gene expression was also enriched for upregulation in steatosis samples compared with normals. No significant changes in steatosis for any other gene category for either upregulation or downregulation were observed.
Table 1
Gene Enrichment Tests of ER Stress–Associated Gene Categories
| Group | p Geometric Normal–Steatosis | p Geometric Normal–NASH | FDR q Value Normal–Steatosis | FDR q Value Normal–NASH | Set Size |
|---|---|---|---|---|---|
| Upregulation | |||||
 Autophagy Autophagy | 0.01 | 0.06 | 3.16e-05 | 0.001 | 240 |
 Apoptosis Apoptosis | 0.25 | 0.25 | 2.12 e-01 | 0.033 | 27 |
 ER stress ER stress | 0.31 | 0.94 | 9.73 e-01 | 1.00 | 98 |
 ER/UPR ER/UPR | 0.14 | 0.97 | 5.73e-01 | 1.00 | 150 |
 Lipogenesis Lipogenesis | 0.42 | 0.91 | 9.73e-01 | 1.00 | 49 |
 UPR UPR | 0.11 | 0.84 | 1.76e-01 | 1.00 | 52 |
| Downregulation | |||||
 ER/UPR ER/UPR | 0.10 | 0.001 | 1.00 | 9.68e-33 | 150 |
 UPR UPR | 0.28 | 0.002 | 1.00 | 3.37e-24 | 52 |
 Lipogenesis Lipogenesis | 0.12 | 0.002 | 0.16 | 2.87e-24 | 49 |
 ER stress ER stress | 0.14 | 0.02 | 0.55 | 1.25e-14 | 98 |
 Apoptosis Apoptosis | 0.49 | 0.66 | 1.00 | 1.00 | 27 |
 Autophagy Autophagy | 0.40 | 0.44 | 1.00 | 1.00 | 240 |
Notes. Gene set enrichment testing for upregulation or downregulation is shown. The gene categories for autophagy, apoptosis, ER stress, UPR, ER stress/UPR combined signaling genes, and lipogenesis genes were all tested individually. p Geometric represents the geometric mean of the individual p values from multiple single array based gene set tests. FDR q value represents the q value adjustment of the global p value using the Benjamini and Hochberg procedure. Set size represents the effective gene set size or number of genes. Abbreviation: FDR, false discovery rate.
Microarray Gene Expression
ER stress and downstream UPR genes from the human NAFLD microarray data set were analyzed in normal, steatosis, and NASH human microarray data sets. For the IRE1 signaling pathway, IRE1 and JNK mRNA were not significantly altered in NASH samples compared with normal. The mRNA levels of PERK were not significantly changed in samples diagnosed as NASH (Fig. 1). The gene expression for eIF2α and CHOP were decreased significantly in NASH compared with steatosis samples (Fig. 1). The gene for the transcription factor ATF6 exhibited a significant increase in gene expression in steatosis samples compared with normal samples but was unchanged in NASH samples. ATF4 was significantly decreased in NASH samples compared with both normal and steatosis samples. The gene designated for the transcription factor XBP-1 was significantly decreased in NASH relative to normal and steatosis samples (Fig. 1). The associated PERK pathway gene STC2 was significantly increased in NASH samples compared with steatosis samples (Fig. 1).
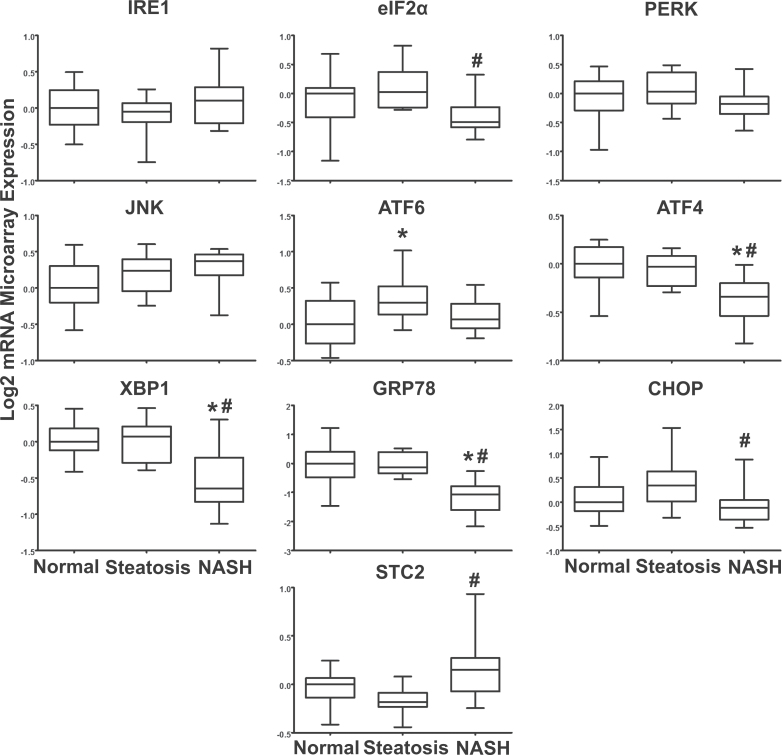
ER stress gene expression. The log transformed microarray data in human samples of NAFLD are shown. Array data represent 19 normal livers, 10 steatosis, and a total of 16 NASH samples, which were analyzed by 1-way ANOVA with post hoc Tukey testing. mRNA expression of ER stress genes was normalized to the median of the normal diagnosis group and is presented in box-and-whisker plots as the mean relative expression ± SE. Asterisk (*) represents a significant change from normal samples and pound sign (#) is representative of significance from steatosis (p ≤ .05). Abbreviations: ATF4, activating transcription factor 4; ATF6, activating transcription factor 6; ER, endoplasmic reticulum; IRE1, inositol-requiring enzyme 1; NAFLD, nonalcoholic fatty liver disease; NASH, nonalcoholic steatohepatitis; PERK, PKR-like ER kinase; STC2, stanniocalcin 2; XBP-1, X-box binding protein 1.
Hierarchical Clustering Analysis
Liver samples diagnosed as NASH predominantly clustered together for all of the ER stress–associated gene categories in a hierarchical clustering analysis (Fig. 2). ER stress/UPR, apoptosis, and autophagy gene set heat maps revealed this effect in NASH samples (Fig. 2). For each of the gene sets, steatosis and normal samples did not cluster and were randomly distributed throughout the heat maps (Fig. 2). The hierarchical clustering analysis demonstrates the capacity of the gene expression profiles for each ER stress–associated gene set to differentiate NASH from the other diagnosis groups. The clustering is indicative of a potential coordinate regulation of gene expression in NASH.
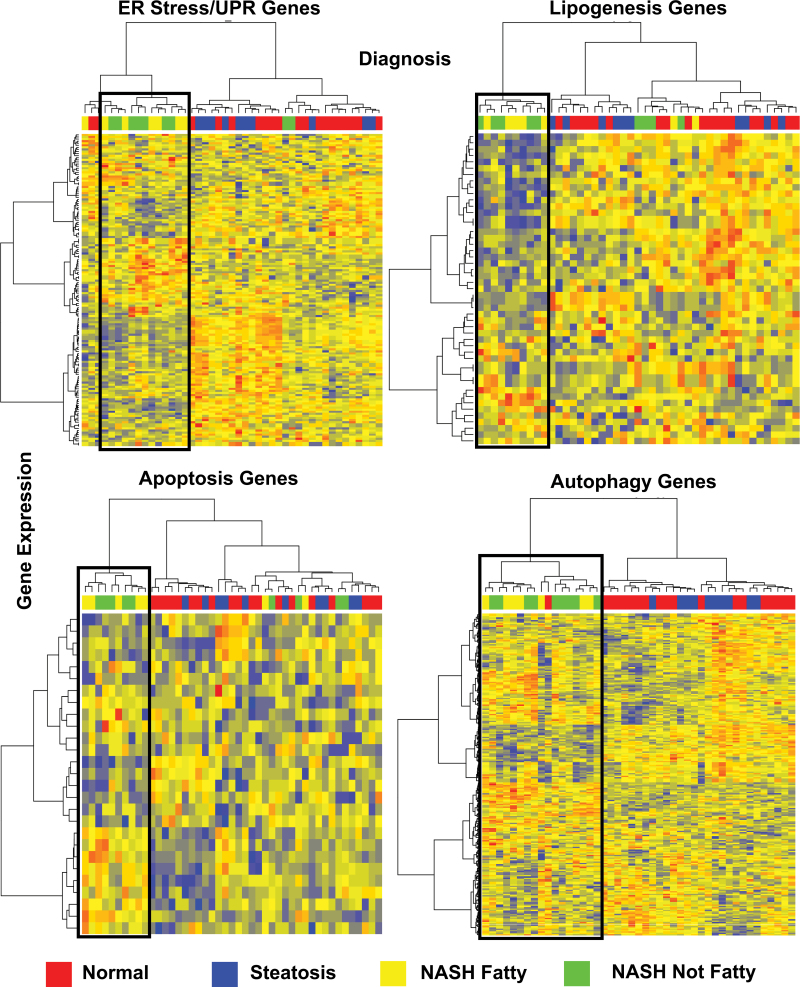
Heat maps of ER stress gene categories. Hierarchical clustering analysis of 4 separate ER stress gene categories (lipogenesis, autophagy, apoptosis, and ER stress/UPR genes) is shown. Diagnosis of samples is as follows: red = normal, blue = steatosis, yellow = NASH (Fatty), green = NASH (Not Fatty). Genes that are downregulated in the heat map are shown in shades of blue, whereas upregulated genes are in red. Genes with unaltered expression are represented in yellow. Boxes are centered over gene expression changes in NASH Fatty and NASH Not Fatty samples, which cluster together. Abbreviations: ER, endoplasmic reticulum; NASH, nonalcoholic steatohepatitis; UPR, unfolded protein response.
Immunoblot Analysis
Western blots for phosphorylated eIF2α, total eIF2α, phosphorylated JNK, total JNK, XBP-1s, CHOP, ATF4, and STC2 were performed to determine changes in protein levels using whole cell liver lysates of human samples diagnosed as normal, steatosis, NASH Fatty, and NASH Not Fatty (Figs. 3 and and4).4). The ratio of phosphorylated to total eIF2α and phosphorylated to total JNK did not significantly change in NASH and variability among liver samples is shown in the box-and-whisker plots (Fig. 4). The IRE1 pathway transcription factor XBP-1s and the PERK pathway product STC2 were significantly increased in NASH Not Fatty samples compared with normal, whereas XBP-1s was also significantly increased in NASH Fatty compared with normal samples (Fig. 4). This statistical increase is paralleled by the representative blots of the proteins (Fig. 3). ATF4 and CHOP protein expression was not significantly changed in NASH due to interindividual variability in the samples (Fig. 4) despite the strong expression changes in the representative blots (Fig. 3).
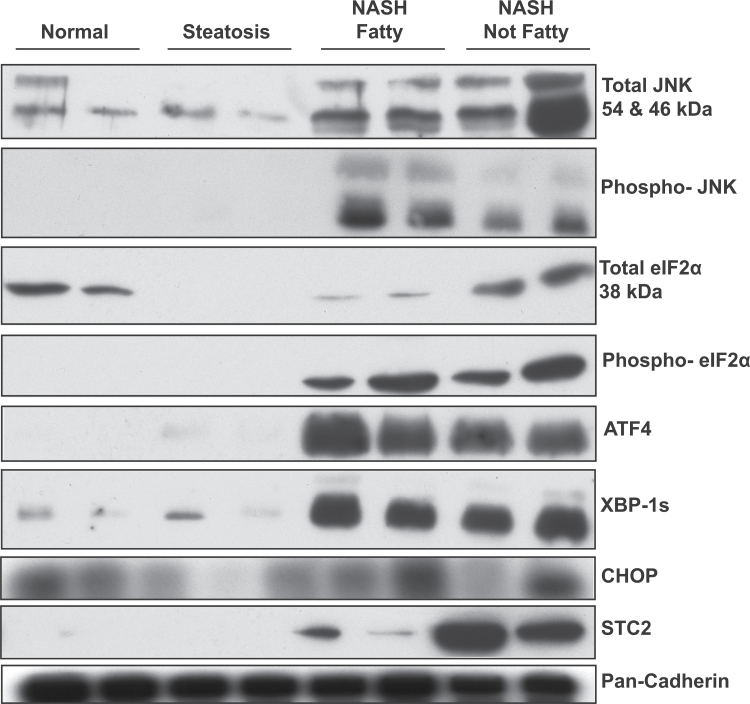
Representative immunoblots of ER stress proteins. Representative immunoblots of total JNK, phosphorylated JNK, total eIF2α, phosphorylated eIF2α, ATF4, XBP-1s, CHOP, STC2 are shown with pan-Cadherin control protein. Two samples per diagnosis group for normal, steatosis, NASH Fatty, and NASH Not Fatty are shown here as representative of all samples run in the protein expression analysis. Abbreviations: ATF4, activating transcription factor 4; ER, endoplasmic reticulum; NASH, nonalcoholic steatohepatitis; STC2, stanniocalcin 2; XBP-1s, X-box binding protein 1 spliced.
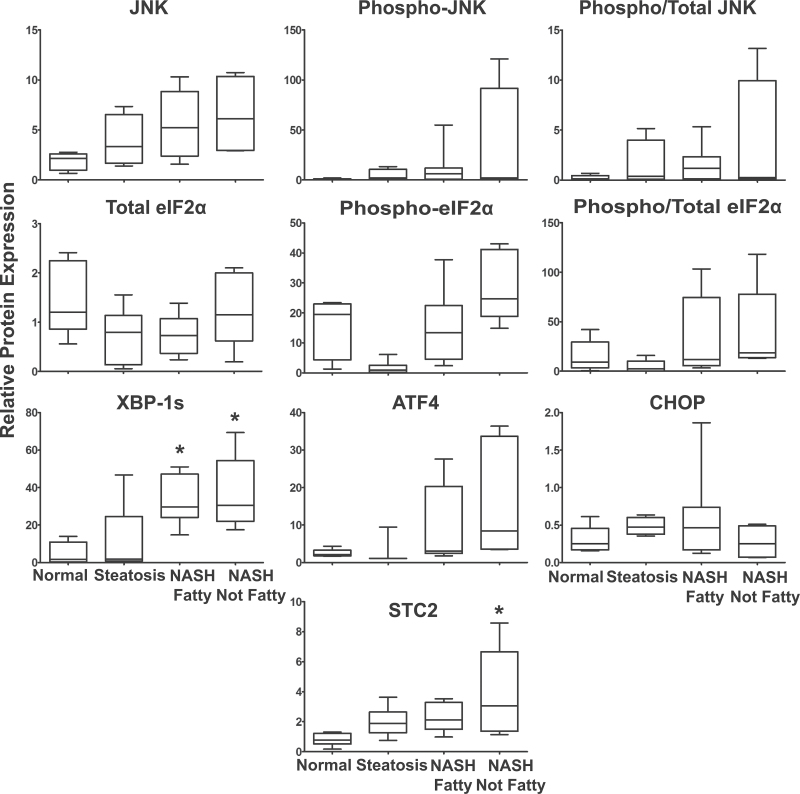
ER stress protein expression. Hepatic expression of selected ER stress proteins in normal (n = 7), steatosis (n = 7), NASH Fatty (n = 8), and NASH Not Fatty (n = 5) whole cell liver lysates. Densitometric analysis was performed and the output analyzed by 1-way ANOVA with post hoc Tukey testing. Data were normalized to total pan-Cadherin protein and are presented in box-and-whisker plots as the mean relative protein expression ± SE. Significance was determined by p ≤ .05. Asterisk (*) represents significance from normal samples. Abbreviations: ATF4, activating transcription factor 4; ER, endoplasmic reticulum; NASH, nonalcoholic steatohepatitis; STC2, stanniocalcin 2; XBP-1s,X-box binding protein 1 spliced.
Immunohistochemical Staining of XBP-1 Protein
Immunohistochemical staining of total XBP-1 was performed on formalin fixed and paraffin embedded (FFPE) samples representing the entire spectrum of NAFLD to determine cellular localization (Fig. 5). Nuclear staining for XBP-1 was clearly visible in all stages of NAFLD with NASH (Fatty) and NASH (Not Fatty) being more prominent with 55.40% and 58.50% positive staining, respectively (Table 2). Nuclear translocation in normal samples was rarely seen (10.05%), and normal samples exhibited only faint staining (Table 2). Steatosis samples exhibited increased nuclear staining (48.40%) in contrast to normal samples (Table 2).
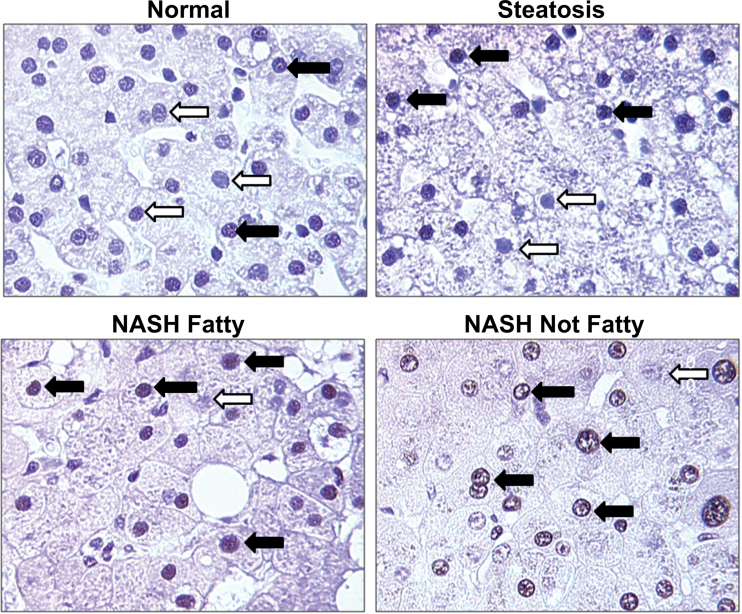
XBP-1 transcription factor localization in NAFLD. Representative images show total XBP-1 staining and nuclear localization with an antibody that is selective for both XBP-1s and XBP-1u. Normal, steatosis, NASH Fatty, and NASH Not Fatty FFPE tissues were stained and assessed. Closed arrows indicate nuclei positive for XBP-1, whereas open arrows indicate nuclei negative for XBP-1. Magnification: ×40. Abbreviations: FFPE, formalin fixed and paraffin embedded; NAFLD, nonalcoholic fatty liver disease; NASH, nonalcoholic steatohepatitis; XBP-1s, X-box binding protein 1 spliced; XBP-1u, X-box binding protein 1 unspliced.
Table 2
Average Percent Positive Values for Nuclear Staining With XBP-1 Protein
| Diagnosis | Average (+) Nuclei ± SD | Average (−) Nuclei ± SD | Average Total ± SD | Average Percent Positive ± SD |
|---|---|---|---|---|
| Normal | 11.0±5.8 | 101.8±26.9 | 112.9±27.6 | 10.0±5.9% |
| Steatosis | 44.3±4.8 | 46.9±4.3 | 91.2±7.3 | 48.4±3.2% |
| NASH Fatty | 44.0±9.1 | 35.1±7.6 | 79.1±15.8 | 55.4±3.1% |
| NASH Not Fatty | 39.5±12.1 | 27.1±8.6 | 66.6±20.2 | 58.5±4.5% |
Notes. FFPE human liver samples representing normal, steatosis, NASH Fatty, and NASH Not Fatty stages were stained and assessed for XBP-1 nuclear localization. The average percent positive values ± SD are shown for XBP-1 nuclear localization in each diagnosis category of normal (n = 5), steatosis (n = 5), NASH Fatty (n = 7), and NASH Not Fatty (n = 7).
DISCUSSION
The current study investigated the altered gene expression patterns in ER stress–associated gene categories of lipogenesis, autophagy, and apoptosis gene sets in hepatic samples of progressive human NAFLD. We identified novel patterns in the coordinate regulation of several ER stress–associated gene categories across the progression of human NAFLD from normal livers to simple steatosis (the first “hit”) and from steatosis to NASH (the second “hit”). The majority of gene expression changes for ER stress genes occurred in NASH and not steatosis compared with normal samples. These patterns included an enrichment of upregulated genes in autophagy and apoptosis categories, whereas in the lipogenesis and ER stress/UPR gene sets a downregulation enrichment of genes is present in NASH. The autophagy gene set represented the only gene category in the normal to steatosis comparison that was significantly enriched for gene expression upregulation. The enrichment profile of autophagy gene expression has implications for the promotion of cell survival and cell death mechanisms in steatosis and NASH. The survival versus cell death response is thought to be modulated by the accumulation of free fatty acids, a common occurrence in NAFLD (Mei et al., 2011). Saturated fatty acids are reported to suppress autophagy and promote apoptosis, whereas unsaturated fatty acids have been shown to induce autophagy (Mei et al., 2011). Thus, the upregulation of autophagy genes (Table 1) in our steatosis and NASH liver samples may represent an adaptive response of these livers to lipotoxicity. Autophagy is generally recognized as a cytoprotective process and the upregulation of autophagy genes represents an important aspect of the adaptive ER stress response. The adaptive response assists in the clearing of excess lipids from the liver in an attempt to relieve the lipotoxic stress (Amir and Czaja, 2011; Singh et al., 2009). Apoptosis is also clearly correlated with the severity of liver disease in NASH patients (Wieckowska et al., 2006). It is recognized as a mechanism that arises when the UPR and protective autophagy components fail to relieve ER stress (Pagliassotti, 2012). The activation of both lipoapoptosis and fatty acid–induced apoptosis is a concerted effort to combat lipotoxicity in NASH (Cusi, 2012; Malhi and Gores, 2008).
Despite the enrichment of apoptosis genes among upregulated genes in NASH samples, the protein phosphorylation levels of JNK did not significantly change although significant variability in protein expression was present (Figs. 3 and and4).4). Similarly, the phosphorylation status of eIF2α did not significantly change. The interindividual variability in these liver samples may contribute to this observation. In contrast, other studies have shown significant elevation of JNK and eIF2α phosphorylation levels in livers diagnosed as NASH although interindividual variability was also present (Puri et al., 2008). Protein expression of CHOP and ATF4 were not significantly changed across the progression of NAFLD. This finding parallels results observed in another study of CHOP and ATF4 protein expression (Puri et al., 2008).
The liver is the predominant organ in the body delegated to de novo lipogenesis. Any modification to lipid metabolism mechanisms by disease prevalence or lipotoxicity may cause significant reorganization of normal lipid detoxifying processes (Lee and Glimcher, 2009). In the current study, we show a coordinated transcriptional downregulation of lipogenesis genes together with ER stress/UPR genes. Lipid accumulation and lipotoxicity are known to positively regulate the ER stress response through a circular cycle that leads to further toxic insults upon the liver (Glimcher and Lee, 2009). These results may represent an adaptive attempt during NASH to reduce transcriptional expression of de novo lipogenesis genes to alleviate further lipotoxic stress and pathological damage.
The transcription factor that connects lipogenesis to the UPR is XBP-1s (Glimcher and Lee, 2009). In the current study, we show an elevation of XBP-1s protein and increased nuclear localization of unspliced and spliced XBP-1 in NASH liver samples that strongly supports a functional increase of active XBP-1s (Figs. 3 and and4).4). Increased XBP-1s protein has also been demonstrated in a nondiabetic mouse model of NAFLD (Rinella et al., 2011). The mRNA expression of total XBP-1 in our microarray data does not parallel the elevated protein levels of XBP-1s (Fig. 1), which may suggest the presence of a negative feedback mechanism acting to decrease the transcriptional production of XBP-1. Mechanisms of negative feedback for XBP-1 have been explored by previous studies (Yoshida et al., 2006). Although XBP-1 mRNA levels are clearly downregulated with respect to gene expression in NASH samples, our microarray data do not differentiate between XBP-1u and XBP-1s. From the accumulated evidence of our immunoblots (Fig. 3) and nuclear localization staining (Fig. 5) in NASH, we speculate that functional XBP-1s is expressed at high levels in NASH despite the changes in gene expression. Other studies have analyzed the mRNA and protein expression of XBP-1s using in vitro and in vivo models and more recently in humans (Park et al., 2010; Puri et al., 2008). In contrast, decreased levels of XBP-1s protein and increased levels of unspliced XBP-1 mRNA were reported for these liver samples categorized as NASH (Puri et al., 2008). In support of our observation that XBP-1s protein is increased in NASH, immunohistochemical staining of total XBP-1 revealed that it is increasingly localized to the nucleus in steatosis and NASH (Table 2, Fig. 5). The mechanism for XBP-1s translocation to the nucleus has been demonstrated in the literature to occur with p85 chaperoned translocation (Park et al., 2010). In a study of ob/ob insulin deficient mice, the lack of p85 binding to XBP-1s resulted in inhibited XBP-1s translocation and a diminished chaperone response and metabolic dysregulation (Park et al., 2010). Even more interesting, increased levels of insulin, a symptom of many NAFLD patients, were responsible for enhancing the nuclear concentration of XBP-1s (Park et al., 2010). The increased XBP-1 nuclear localization in our steatosis and NASH liver samples suggests that similar mechanisms of XBP-1s translocation and stabilization may be occurring in human NAFLD.
An analysis of the PERK pathway in our NAFLD liver samples revealed the upregulation of an important cytoprotective downstream component: STC2. STC2 mRNA and protein were significantly upregulated in NASH (Not Fatty) samples (Figs. 1 and and4).4). STC2 may have an important role in the initiation of adaptive mechanisms given previous accounts of its cytoprotective properties during disease (Fazio et al., 2011; Ito et al., 2004). The upregulation of STC2 has been previously established to occur through the PERK pathway of ER stress signaling in a rodent model of pancreatitis (Fazio et al., 2011). The regulation of STC2 is proposed to be under the control of the transcription factor ATF4, which is induced by phosphorylated eIF2α (Fazio et al., 2011). The mRNA expression of STC2 increased significantly in NASH, but ATF4 was significantly decreased in NASH overall (Fig. 1). Protein levels of ATF4 were not significantly altered with progression of NAFLD (Fig. 3).
Altered transcriptomic and translational expression of XBP-1s and STC2 may have a relevant role in the initiation of the adaptive ER stress response to NAFLD progression. Although it is clear that the machinery that drives the transcription of the ER stress response is activated in NASH, it is important to note that the various gene categories are not necessarily regulated as part of that response, but rather encode genes that are categorized by their function. These genes that are associated with ER stress response signaling are not solely activated by ER stress and have many different mechanisms of regulation in the literature. Therefore, the opposing regulation of the potentially protective autophagy and apoptosis gene categories compared with the downregulation of the lipogenesis and UPR gene categories represents an interesting avenue of hepatic regulation in NASH. We have shown adaptive and potential cytoprotective mechanisms in NASH in an attempt to resolve hepatic lipotoxicity. Overall, the patterns in gene expression alterations and mechanistic changes of important regulatory proteins during NASH contribute new information to the field of ER stress mechanisms in humans with progressive NAFLD.
SUPPLEMENTARY DATA
Supplementary data are available online at http://toxsci.oxfordjournals.org/.
FUNDING
National Institute of Health Grants (DK068039;, AI083927;, ES006694;, HD06248); National Institute of Environmental Health Science Toxicology Training Grant (ES007091); Academy of Sciences of the Czech Republic (AVOZ50510513); Liver Tissue Cell Distribution System, National Institute of Health Contract (NO1-DK-7-0004/HHSN267200700004C).
ACKNOWLEDGMENTS
We sincerely thank the NIH-sponsored Liver Tissue Cell Distribution System members for their help in the acquisition of the human liver tissue samples at the University of Minnesota, Virginia Commonwealth University, and the University of Pittsburgh.
REFERENCES
- Achard C. S., Laybutt D. R. (2012). Lipid-induced endoplasmic reticulum stress in liver cells results in two distinct outcomes: Adaptation with enhanced insulin signaling or insulin resistance. Endocrinology 153, 2164–2177 [Abstract] [Google Scholar]
- Acosta-Alvear D., Zhou Y., Blais A., Tsikitis M., Lents N. H., Arias C., Lennon C. J., Kluger Y., Dynlacht B. D. (2007). XBP1 controls diverse cell type- and condition-specific transcriptional regulatory networks. Mol. Cell 27, 53–66 [Abstract] [Google Scholar]
- Adachi Y., Yamamoto K., Okada T., Yoshida H., Harada A., Mori K. (2008). ATF6 is a transcription factor specializing in the regulation of quality control proteins in the endoplasmic reticulum. Cell Struct. Funct. 33, 75–89 [Abstract] [Google Scholar]
- Ali R., Cusi K. (2009). New diagnostic and treatment approaches in non-alcoholic fatty liver disease (NAFLD). Ann. Med. 41, 265–278 [Abstract] [Google Scholar]
- Alkhouri N., Carter-Kent C., Feldstein A. E. (2011). Apoptosis in nonalcoholic fatty liver disease: Diagnostic and therapeutic implications. Expert Rev. Gastroenterol. Hepatol. 5, 201–212 [Europe PMC free article] [Abstract] [Google Scholar]
- Amir M., Czaja M. J. (2011). Autophagy in nonalcoholic steatohepatitis. Expert Rev. Gastroenterol. Hepatol. 5, 159–166 [Europe PMC free article] [Abstract] [Google Scholar]
- Cusi K. (2012). Role of obesity and lipotoxicity in the development of nonalcoholic steatohepatitis: Pathophysiology and clinical implications. Gastroenterology 142, 711–725 [Abstract] [Google Scholar]
- Day C. P., James O. F. (1998). Steatohepatitis: A tale of two “hits”? Gastroenterology 114, 842–845 [Abstract] [Google Scholar]
- Fazio E. N., Dimattia G. E., Chadi S. A., Kernohan K. D., Pin C. L. (2011). Stanniocalcin 2 alters PERK signalling and reduces cellular injury during cerulein induced pancreatitis in mice. BMC Cell Biol. 12, 17. [Europe PMC free article] [Abstract] [Google Scholar]
- Fisher C. D., Lickteig A. J., Augustine L. M., Ranger-Moore J., Jackson J. P., Ferguson S. S., Cherrington N. J. (2009). Hepatic cytochrome P450 enzyme alterations in humans with progressive stages of nonalcoholic fatty liver disease. Drug Metab. Dispos. 37, 2087–2094 [Europe PMC free article] [Abstract] [Google Scholar]
- Fuchs M., Sanyal A. J. (2012). Lipotoxicity in NASH. J. Hepatol. 56, 291–293 [Abstract] [Google Scholar]
- Gentile C. L., Frye M., Pagliassotti M. J. (2011). Endoplasmic reticulum stress and the unfolded protein response in nonalcoholic fatty liver disease. Antioxid. Redox Signal. 15, 505–521 [Europe PMC free article] [Abstract] [Google Scholar]
- Glimcher L. H., Lee A. H. (2009). From sugar to fat: How the transcription factor XBP1 regulates hepatic lipogenesis. Ann. N.Y. Acad. Sci. 1173, E2–E9 [Europe PMC free article] [Abstract] [Google Scholar]
- Hardwick R. N., Fisher C. D., Canet M. J., Lake A. D., Cherrington N. J. (2010). Diversity in antioxidant response enzymes in progressive stages of human nonalcoholic fatty liver disease. Drug Metab. Dispos. 38, 2293–2301 [Europe PMC free article] [Abstract] [Google Scholar]
- Hardwick R. N., Fisher C. D., Canet M. J., Scheffer G. L., Cherrington N. J. (2011). Variations in ATP-binding cassette transporter regulation during the progression of human nonalcoholic fatty liver disease. Drug Metab. Dispos. 39, 2395–2402 [Europe PMC free article] [Abstract] [Google Scholar]
- Ito D., Walker J. R., Thompson C. S., Moroz I., Lin W., Veselits M. L., Hakim A. M., Fienberg A. A., Thinakaran G. (2004). Characterization of stanniocalcin 2, a novel target of the mammalian unfolded protein response with cytoprotective properties. Mol. Cell. Biol. 24, 9456–9469 [Europe PMC free article] [Abstract] [Google Scholar]
- Kleiner D. E., Brunt E. M., Van Natta M., Behling C., Contos M. J., Cummings O. W., Ferrell L. D., Liu Y. C., Torbenson M. S., Unalp-Arida A., et al. (2005). Design and validation of a histological scoring system for nonalcoholic fatty liver disease. Hepatology 41, 1313–1321 [Abstract] [Google Scholar]
- Lake A. D., Novak P., Fisher C. D., Jackson J. P., Hardwick R. N., Billheimer D. D., Klimecki W. T., Cherrington N. J. (2011). Analysis of global and absorption, distribution, metabolism, and elimination gene expression in the progressive stages of human nonalcoholic fatty liver disease. Drug Metab. Dispos. 39, 1954–1960 [Europe PMC free article] [Abstract] [Google Scholar]
- Leamy A. K., Egnatchik R. A., Young J. D. (2013). Molecular mechanisms and the role of saturated fatty acids in the progression of non-alcoholic fatty liver disease. Prog. Lipid Res. 52, 165–174 [Europe PMC free article] [Abstract] [Google Scholar]
- Lee A. H., Glimcher L. H. (2009). Intersection of the unfolded protein response and hepatic lipid metabolism. Cell. Mol. Life Sci. 66, 2835–2850 [Europe PMC free article] [Abstract] [Google Scholar]
- Lee A. H., Scapa E. F., Cohen D. E., Glimcher L. H. (2008). Regulation of hepatic lipogenesis by the transcription factor XBP1. Science 320, 1492–1496 [Europe PMC free article] [Abstract] [Google Scholar]
- Luo W., Friedman M. S., Shedden K., Hankenson K. D., Woolf P. J. (2009). GAGE: Generally applicable gene set enrichment for pathway analysis. BMC Bioinformatics 10, 161. [Europe PMC free article] [Abstract] [Google Scholar]
- Malhi H., Gores G. J. (2008). Molecular mechanisms of lipotoxicity in nonalcoholic fatty liver disease. Semin. Liver Dis. 28, 360–369 [Europe PMC free article] [Abstract] [Google Scholar]
- McCullough A. J. (2006). Pathophysiology of nonalcoholic steatohepatitis. J. Clin. Gastroenterol. 40, S17–S29 [Abstract] [Google Scholar]
- Mei S., Ni H. M., Manley S., Bockus A., Kassel K. M., Luyendyk J. P., Copple B. L., Ding W. X. (2011). Differential roles of unsaturated and saturated fatty acids on autophagy and apoptosis in hepatocytes. J. Pharmacol. Exp. Ther. 339, 487–498 [Europe PMC free article] [Abstract] [Google Scholar]
- Ning J., Hong T., Ward A., Pi J., Liu Z., Liu H. Y., Cao W. (2011). Constitutive role for IRE1α-XBP1 signaling pathway in the insulin-mediated hepatic lipogenic program. Endocrinology 152, 2247–2255 [Europe PMC free article] [Abstract] [Google Scholar]
- Pagliassotti M. J. (2012). Endoplasmic reticulum stress in nonalcoholic fatty liver disease. Annu. Rev. Nutr. 32, 17–33 [Abstract] [Google Scholar]
- Park S. W., Zhou Y., Lee J., Lu A., Sun C., Chung J., Ueki K., Ozcan U. (2010). The regulatory subunits of PI3K, p85alpha and p85beta, interact with XBP-1 and increase its nuclear translocation. Nat. Med. 16, 429–437 [Europe PMC free article] [Abstract] [Google Scholar]
- Patil C., Walter P. (2001). Intracellular signaling from the endoplasmic reticulum to the nucleus: The unfolded protein response in yeast and mammals. Curr. Opin. Cell Biol. 13, 349–355 [Abstract] [Google Scholar]
- Puri P., Mirshahi F., Cheung O., Natarajan R., Maher J. W., Kellum J. M., Sanyal A. J. (2008). Activation and dysregulation of the unfolded protein response in nonalcoholic fatty liver disease. Gastroenterology 134, 568–576 [Abstract] [Google Scholar]
- Rautou P. E., Mansouri A., Lebrec D., Durand F., Valla D., Moreau R. (2010). Autophagy in liver diseases. J. Hepatol. 53, 1123–1134 [Abstract] [Google Scholar]
- Ren L. P., Chan S. M., Zeng X. Y., Laybutt D. R., Iseli T. J., Sun R. Q., Kraegen E. W., Cooney G. J., Turner N., Ye J. M. (2012). Differing endoplasmic reticulum stress response to excess lipogenesis versus lipid oversupply in relation to hepatic steatosis and insulin resistance. PLoS One 7, e30816. [Europe PMC free article] [Abstract] [Google Scholar]
- Rinella M. E., Siddiqui M. S., Gardikiotes K., Gottstein J., Elias M., Green R. M. (2011). Dysregulation of the unfolded protein response in db/db mice with diet-induced steatohepatitis. Hepatology 54, 1600–1609 [Europe PMC free article] [Abstract] [Google Scholar]
- Singh R., Kaushik S., Wang Y., Xiang Y., Novak I., Komatsu M., Tanaka K., Cuervo A. M., Czaja M. J. (2009). Autophagy regulates lipid metabolism. Nature 458, 1131–1135 [Europe PMC free article] [Abstract] [Google Scholar]
- Thorburn A. (2008). Apoptosis and autophagy: Regulatory connections between two supposedly different processes. Apoptosis 13, 1–9 [Europe PMC free article] [Abstract] [Google Scholar]
- Uemura A., Oku M., Mori K., Yoshida H. (2009). Unconventional splicing of XBP1 mRNA occurs in the cytoplasm during the mammalian unfolded protein response. J. Cell Sci. 122(Pt 16), 2877–2886 [Abstract] [Google Scholar]
- Wieckowska A., Zein N. N., Yerian L. M., Lopez A. R., McCullough A. J., Feldstein A. E. (2006). In vivo assessment of liver cell apoptosis as a novel biomarker of disease severity in nonalcoholic fatty liver disease. Hepatology 44, 27–33 [Abstract] [Google Scholar]
- Yoshida H., Oku M., Suzuki M., Mori K. (2006). pXBP1(U) encoded in XBP1 pre-mRNA negatively regulates unfolded protein response activator pXBP1(S) in mammalian ER stress response. J. Cell Biol. 172, 565–575 [Europe PMC free article] [Abstract] [Google Scholar]
- Zeng L., Lu M., Mori K., Luo S., Lee A. S., Zhu Y., Shyy J. Y. (2004). ATF6 modulates SREBP2-mediated lipogenesis. EMBO J. 23, 950–958 [Europe PMC free article] [Abstract] [Google Scholar]
Articles from Toxicological Sciences are provided here courtesy of Oxford University Press
Full text links
Read article at publisher's site: https://doi.org/10.1093/toxsci/kft230
Read article for free, from open access legal sources, via Unpaywall:
https://europepmc.org/articles/pmc3871931?pdf=render
Citations & impact
Impact metrics
Citations of article over time
Smart citations by scite.ai
Explore citation contexts and check if this article has been
supported or disputed.
https://scite.ai/reports/10.1093/toxsci/kft230
Article citations
Autophagy alterations in obesity, type 2 diabetes, and metabolic dysfunction-associated steatotic liver disease: the evidence from human studies.
Intern Emerg Med, 19(5):1473-1491, 06 Jul 2024
Cited by: 1 article | PMID: 38971910 | PMCID: PMC11364608
Review Free full text in Europe PMC
A genetic mouse model of lean-NAFLD unveils sexual dimorphism in the liver-heart axis.
Commun Biol, 7(1):356, 22 Mar 2024
Cited by: 1 article | PMID: 38519536 | PMCID: PMC10959946
Mapping Interindividual Variability of Toxicodynamics Using High-Throughput Transcriptomics and Primary Human Hepatocytes from Fifty Donors.
Environ Health Perspect, 132(3):37005, 18 Mar 2024
Cited by: 0 articles | PMID: 38498338 | PMCID: PMC10947137
Glycemic Control Is Associated with Histological Findings of Nonalcoholic Fatty Liver Disease.
Diabetes Metab J, 48(3):440-448, 02 Feb 2024
Cited by: 0 articles | PMID: 38310878 | PMCID: PMC11140399
miR-12135 ameliorates liver fibrosis accompanied with the downregulation of integrin subunit alpha 11.
iScience, 27(1):108730, 14 Dec 2023
Cited by: 0 articles | PMID: 38235326 | PMCID: PMC10792239
Go to all (80) article citations
Data
Data behind the article
This data has been text mined from the article, or deposited into data resources.
BioStudies: supplemental material and supporting data
Functional Genomics Experiments
- (1 citation) ArrayExpress - E-MEXP-3291
Similar Articles
To arrive at the top five similar articles we use a word-weighted algorithm to compare words from the Title and Abstract of each citation.
Fibroblast growth factor 21 is regulated by the IRE1α-XBP1 branch of the unfolded protein response and counteracts endoplasmic reticulum stress-induced hepatic steatosis.
J Biol Chem, 289(43):29751-29765, 28 Aug 2014
Cited by: 114 articles | PMID: 25170079 | PMCID: PMC4207989
The unfolded protein response regulates hepatic autophagy by sXBP1-mediated activation of TFEB.
Autophagy, 17(8):1841-1855, 15 Jul 2020
Cited by: 41 articles | PMID: 32597296 | PMCID: PMC8386593
Gene regulatory network of unfolded protein response genes in endoplasmic reticulum stress.
Cell Stress Chaperones, 18(1):11-23, 18 Jul 2012
Cited by: 53 articles | PMID: 22802018 | PMCID: PMC3508129
Role of endoplasmic reticulum stress in the pathogenesis of nonalcoholic fatty liver disease.
World J Gastroenterol, 20(7):1768-1776, 01 Feb 2014
Cited by: 142 articles | PMID: 24587654 | PMCID: PMC3930975
Review Free full text in Europe PMC
Funding
Funders who supported this work.
NIAID NIH HHS (1)
Grant ID: AI083927
NICHD NIH HHS (2)
Grant ID: R01 HD062489
Grant ID: HD06248
NIDDK NIH HHS (2)
Grant ID: DK068039
Grant ID: N01-DK70004/HHSN267200700004C
NIEHS NIH HHS (4)
Grant ID: ES007091
Grant ID: ES006694
Grant ID: P30 ES006694
Grant ID: T32 ES007091




