Abstract
Free full text

PlantTFDB 3.0: a portal for the functional and evolutionary study of plant transcription factors
Abstract
With the aim to provide a resource for functional and evolutionary study of plant transcription factors (TFs), we updated the plant TF database PlantTFDB to version 3.0 (http://planttfdb.cbi.pku.edu.cn). After refining the TF classification pipeline, we systematically identified 129 288 TFs from 83 species, of which 67 species have genome sequences, covering main lineages of green plants. Besides the abundant annotation provided in the previous version, we generated more annotations for identified TFs, including expression, regulation, interaction, conserved elements, phenotype information, expert-curated descriptions derived from UniProt, TAIR and NCBI GeneRIF, as well as references to provide clues for functional studies of TFs. To help identify evolutionary relationship among identified TFs, we assigned 69 450 TFs into 3924 orthologous groups, and constructed 9217 phylogenetic trees for TFs within the same families or same orthologous groups, respectively. In addition, we set up a TF prediction server in this version for users to identify TFs from their own sequences.
INTRODUCTION
Transcription factors (TFs) play key roles in plant development and stress response by temporarily and spatially regulating the transcription of their target genes. TFs are usually classified into different families based on their DNA-binding domains (DBDs). In 2000, Riechmann et al. (1) made the first attempt for the genome-wide analysis of TFs in Arabidopsis thaliana soon after the availability of its whole genome sequence. In the following years, several databases dedicated to identification and annotation of plant TFs became publicly available, either for multiple species, such as PlnTFDB (2), PlanTAPDB (3), GRASSIUS (4), LegumeTFDB (5), DATFAP (6) and TreeTFDB (7), or for individual organisms, such as AGRIS (8), RARTF (9), TOBFAC (10), SoyDB (11) and wDBTF (12). During the past 8 years, we have constructed three species-specific TF databases DATF (13), DRTF (14) and DPTF (15) for model organisms Arabidopsis, rice and poplar, as well as a comprehensive plant TF database (PlantTFDB) (16,17). The databases we constructed were accessed >10 million hits per year and were widely used for functional and evolutionary study of plant TFs, as well as for the prediction and annotation of TFs in newly sequenced genomes.
To meet requirements from our user community, we updated PlantTFDB to version 3.0 (http://planttfdb.cbi.pku.edu.cn/). In comparison with the previous two versions, PlantTFDB 3.0 covers more species and more TFs identified by the refined family assignment rules and improved prediction pipeline. In addition, new types of annotations were added, and phylogenetic trees and orthologous groups (OGs) were re-constructed. Finally, an online TF prediction server was set up (Table 1).
Table 1.
Comparison among the three versions of PlantTFDB
| PlantTFDB | Version 1.0 | Version 2.0 | Version 3.0 |
|---|---|---|---|
| Species | 22 | 49 | 83 |
| Species with genome sequences | 5 | 28 | 67 |
| Species without genome sequences | 17 | 21 | 16 |
| TF family | 64 | 58 | 58 |
| TF number | 26 402 | 53 574 | 129 288 |
| Annotation | |||
    Expert-curated description Expert-curated description | No | No | Yes |
    Expression Expression | Yes | Yes | Yes |
    Regulation Regulation | No | No | Yes |
    Interaction Interaction | No | No | Yes |
    Phenotype Phenotype | No | No | Yes |
    Reference Reference | Yes | Yes | Yes |
| Orthologous group | Yes | Yes | Yes |
| Phylogenetic tree | |||
    Family Family | No | Yes | Yes |
    Orthologous group Orthologous group | No | No | Yes |
| Web service | No | Yes | No |
| TF prediction server | No | No | Yes |
We believe that PlantTFDB 3.0 provides users with complete TF datasets, comprehensive annotations and useful analysis tools.
MATERIALS AND METHODS
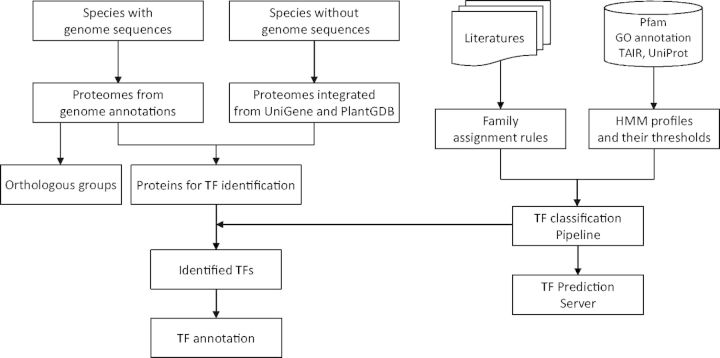
Figure 1 shows the main steps in the construction of PlantTFDB 3.0, including data integration, TF classification, TF annotation and construction of orthologous groups.
Sequence data
We downloaded protein sequences of 67 species with genome sequences from the Joint Genome Institute (JGI) and several other institutions engaged in plant genome sequencing and annotation projects (Supplementary Table S1). For 16 species without genome sequences, we downloaded their expressed sequence tag sequences from UniGene (18) and PlantGDB-assembled unique transcripts from PlantGDB (19), and then built reference proteome for each species (Supplementary Table S2) using a previous established pipeline (17).
Family assignment rules
TFs are usually classified into different families based on their DBDs. We used auxiliary and forbidden domains to distinguish complicated TF families with multiple signature domains. After a comprehensive literature review, we improved the family assignment rules described in the previous version (17) and arranged several families into superfamilies (Figure 2). We removed the forbidden domain Glyco_hydro_14 of the BES1 family, as recent studies demonstrated that BES1 family proteins with this domain also showed TF activity (20).
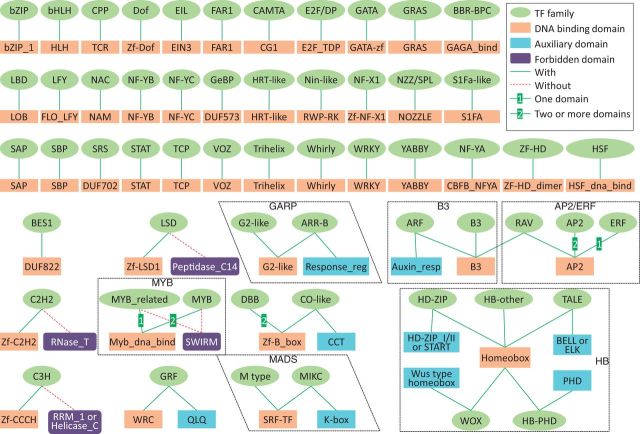
Refined family assignment rules used for TF identification and assignment. Green ellipses represent TF families and red rectangles represent DBDs. Blue rectangles denote auxiliary domains and purple rectangles denote forbidden domains. Green solid lines link families and DBDs or auxiliary domains and number ‘1’ or ‘2’ indicates number of DBDs. Red dash lines link families and forbidden domains. Families belonging to the same superfamily are arranged within rectangles or rhombi.
Prediction pipeline
We refined the TF prediction pipeline by updating the hidden Markov model (HMM) profiles used to identify TFs and adjusted their thresholds. We downloaded the latest version of HMM profiles from Pfam (version 27.0) (21) for most signature domains and built our own HMM profiles for the remaining domain that did not have available Pfam HMM profiles. We used HMMER 3.0 (22) to identify TFs and assigned them into different families according to the family assignment rules described earlier.
Annotation pipeline
We used a pipeline comprising several packages to annotate identified TFs. Domain structure and GO annotation were predicted by InterProScan (version 4.8) (23). Cross-links to well-known resources were assigned to the best BLAST hits with maximal e-value 1e-10. Nuclear localization signals were predicted by PredictNLS (24). Other information such as expert-curated description, expression, regulation, conserved elements and references was collected from corresponding databases. Multiple sequence alignments (MSAs) for DBDs were constructed by HMM-guided method, and MSAs for full-length protein sequences were inferred by T-coffee (version 9.03) (25). Family trees across 83 species were inferred by FastTree (version 2.1.3) (26) with 100 resamplings. Family trees within each species were inferred by MrBayes (version 3.2.1) (27) based on the Dayhoff model for 50 000 generations. The Help page (http://planttfdb.cbi.pku.edu.cn/help_info.php#tfinfo) describes more detailed information on datasets and parameter settings.
Orthologous groups
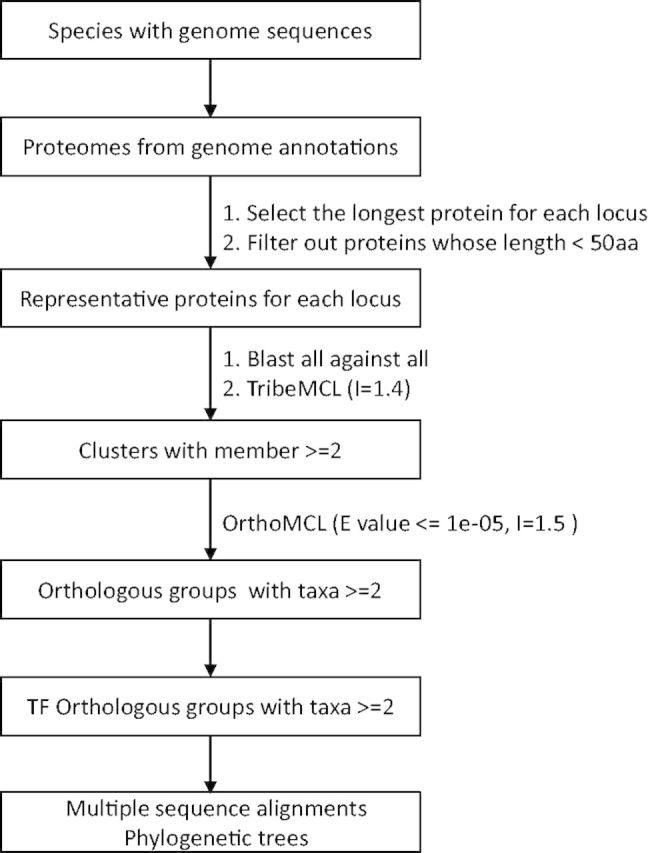
Orthologous groups were inferred using the following methods implemented as a pipeline of Plaza (Figure 3) (28).
First, we selected a representative gene model for each locus from 67 species with genome sequences and filtered out proteins if their lengths were <50 aa. Then we classified these proteins into clusters by TribeMCL (29). After that, proteins within the same cluster were assigned into orthologous groups by OrthoMCL (30). For TFs in the same orthologous group, MSAs were constructed by T-coffee and phylogenetic trees were inferred by MrBayes (27) with the same parameters described earlier.
RESULTS AND DISCUSSION
Genomic TF repertoires of green plants
Using the refined TF prediction pipeline, we identified 129 288 TFs (116 585 loci) from 2 691 496 proteins (2 437 666 loci) of 83 species (Table 2, Supplementary Tables S3 and S4).
Table 2.
Average number of TFs in different taxonomic lineages summarized from 67 species with genome sequences
| Lineage | Species | Gene | TF (%) | Family |
|---|---|---|---|---|
| Chlorophyta | 10 | 10 550 | 141 (1.34) | 35 |
| Bryophytaa | 1 | 32 273 | 1079 (3.34) | 53 |
| Lycopodiophytab | 1 | 22 271 | 665 (2.99) | 54 |
| Coniferophytac | 1 | 71 158 | 1851 (2.60) | 55 |
| Basal Magnoliophytad | 1 | 26 846 | 900 (3.35) | 58 |
| Monocot | 15 | 34 017 | 1701 (5.00) | 58 |
| Eudicot | 38 | 34 798 | 1861 (5.35) | 58 |
aPhyscomitrella patens.
bSelaginella moellendorffii.
cPicea abies.
dAmborella trichopoda.
The increased number of species with genome sequences and the availability of a conifer genome (31) gave us the chance to show the genomic TF repertoires across green plants for the first time (Table 2, Supplementary Table S3). Compared with green alga, land plants have a large increase in the number of TF families, TFs and percentage of TFs in their genome, which might correlate with morphological complexity of land plants (32).
Comprehensive annotations for TFs
A database of well-annotated TFs may provide users with rich information as well as insightful clues for further study. In an attempt to construct a comprehensive knowledgebase for plant TFs, we collected expert-curated description, expression, regulation, mutation and phenotype data from various public resources and made annotations for identified TFs in PlantTFDB 3.0 (Table 3), in addition to abundant annotations provided in the previous two versions (16,17). By integrating information from Entrez Gene (33), UniProtKB (34), GeneRIF (33) and mined by ourselves, we added related references for TFs.
Table 3.
Summary of annotations for TFs in PlantTFDB 3.0
| Typea | Species | TF | Entry |
|---|---|---|---|
| Expert-curated description | 22 | 2128 | 6649 |
| Expression | |||
    UniGene UniGene | 44 | 44 862 | 45 239 |
    Microarray Microarray | 14 | 15 424 | 31 975 |
 Plant ontology Plant ontology | 5 | 6850 | 174 162 |
| Regulation | |||
 Binding site/matrix Binding site/matrix | 24 | 541 | 729 |
 ChIP-chip/ChIP-seq ChIP-chip/ChIP-seq | 1 | 54 | 75 |
 microRNA microRNA | 1 | 28 | 43 |
 Hormone Hormone | 1 | 417 | 803 |
| Interaction | 10 | 992 | 3101 |
| Conserved element | 2 | 3709 | 63 859 |
| Phenotype | 2 | 4704 | 147 684 |
| Reference | 59 | 5004 | 20 255 |
aNew types of annotations in this version are marked in bold.
Evolutionary conserved elements may work as transcriptional regulatory elements (35,36). Therefore, we collected these elements, which were identified based on the genome alignments of 9 crucifers (36) and 20 angiosperm plants (37), and added them into the current version, in addition to functional genomic annotations described earlier.
Orthologs usually have similar function and are widely used to explore functions of poorly studied proteins. To help users infer the functions of poorly studied TFs, we constructed MSAs and phylogenetic trees within the same family across 83 species, based on conserved DBDs. We further assigned 69 450 TFs into 3924 orthologous groups and constructed phylogenetic trees for each orthologous group. As an aid to decipher their evolutionary relationships, we also built trees for individual TF families within the same species. Hyperlinks to TF pages were added in the tree branches so that the users could browse them conveniently. The MSAs and phylogenetic trees in PlantTFDB 3.0 can be freely downloaded for further analyses. Direct links to TFs of A. thaliana, the best-studied model plant and the best-annotated species in PlantTFDB 3.0, were also generated for all TFs in other species.
TF prediction server
In recent years, the TF classification rules we constructed have been widely used to annotate TFs of newly sequenced genomes (38,39). In this regard, we set up a TF prediction server (http://planttfdb.cbi.pku.edu.cn/prediction.php) for users to identify TFs from their own protein sequences. As A. thaliana is the best-annotated species in PlantTFDB 3.0, links to the best hits in A. thaliana are provided for predicted TFs. Currently, users can upload up to 100 sequences and obtain results within a minute from our server.
Further direction
We have updated our PlantTFDB to version 3.0, which provides TF repertoires across the main lineages of green plants. The knowledge we collected, the OGs and phylogenetic trees we inferred are useful resources for further exploration of the physiological function and evolutionary relationship of TFs. We will continue to work on this project to refine the family assignment rules and the prediction pipeline, and collect more type of useful information for identified TFs in the future.
FUNDING
National Natural Science Foundation of China [31071160;, 31171242]; China High-Tech Program [2006AA02Z334;, 2012AA020409]; China National Key Basic Research Program [2011CBA01102]; China National Outstanding Youth Talents Program; China National Science and Technology Infrastructure Program [2009FY120100]. Funding for open access charge: State Key Laboratory of Protein and Plant Gene Research.
Conflict of interest statement. None declared.
ACKNOWLEDGEMENTS
The authors thank Joint Genome Institute for the genome annotation of four unpublished species, AGD for Amborella trichopoda, ICGC for Citrus clementina and AGI for three rice species. They also thank their users for their suggestions and comments. They specially thank Ying Dillaha for her language editing of the manuscript.
REFERENCES
Articles from Nucleic Acids Research are provided here courtesy of Oxford University Press
Full text links
Read article at publisher's site: https://doi.org/10.1093/nar/gkt1016
Read article for free, from open access legal sources, via Unpaywall:
https://academic.oup.com/nar/article-pdf/42/D1/D1182/16949620/gkt1016.pdf
Citations & impact
Impact metrics
Article citations
Specific binding between Arabidopsis thaliana phytochrome-interacting factor 3 (AtPIF3) bHLH and G-box originated prior to embryophyte emergence.
BMC Plant Biol, 24(1):1060, 11 Nov 2024
Cited by: 0 articles | PMID: 39523297 | PMCID: PMC11552376
Integrated hormone and transcriptome profiles provide insight into the pericarp differential development mechanism between Mandarin 'Shatangju' and 'Chunhongtangju'.
Front Plant Sci, 15:1461316, 10 Oct 2024
Cited by: 0 articles | PMID: 39450074 | PMCID: PMC11499144
The Critical Role of Phenylpropanoid Biosynthesis Pathway in Lily Resistance Against Gray Mold.
Int J Mol Sci, 25(20):11068, 15 Oct 2024
Cited by: 0 articles | PMID: 39456848 | PMCID: PMC11507431
Chromosome-scale genome assembly of Astragalus membranaceus using PacBio and Hi-C technologies.
Sci Data, 11(1):1071, 02 Oct 2024
Cited by: 0 articles | PMID: 39358417 | PMCID: PMC11446949
Integrative Analysis of Transcriptome and Metabolome Reveals the Pivotal Role of the NAM Family Genes in <i>Oncidium hybridum</i> Lodd. Pseudobulb Growth.
Int J Mol Sci, 25(19):10355, 26 Sep 2024
Cited by: 0 articles | PMID: 39408686 | PMCID: PMC11476975
Go to all (507) article citations
Other citations
Similar Articles
To arrive at the top five similar articles we use a word-weighted algorithm to compare words from the Title and Abstract of each citation.
PlantTFDB 2.0: update and improvement of the comprehensive plant transcription factor database.
Nucleic Acids Res, 39(database issue):D1114-7, 18 Nov 2010
Cited by: 184 articles | PMID: 21097470 | PMCID: PMC3013715
PlantTFDB: a comprehensive plant transcription factor database.
Nucleic Acids Res, 36(database issue):D966-9, 12 Oct 2007
Cited by: 138 articles | PMID: 17933783 | PMCID: PMC2238823
Computational identification of plant transcription factors and the construction of the PlantTFDB database.
Methods Mol Biol, 674:351-368, 01 Jan 2010
Cited by: 9 articles | PMID: 20827602
Review: WRKY transcription factors: Understanding the functional divergence.
Plant Sci, 334:111770, 13 Jun 2023
Cited by: 14 articles | PMID: 37321304
Review