Abstract
Free full text

SIRT1 is highly expressed in brain metastasis tissues of non-small cell lung cancer (NSCLC) and in positive regulation of NSCLC cell migration
Associated Data
Abstract
Brain metastases are a frequent and ongoing major complication of non-small cell lung cancer (NSCLC). To deepen our understanding to the underlying mechanisms by which NSCLC cells metastasize to brain and hence to improve the therapy, a high throughput RNAi screening with shRNA library of 153 epigenetic genes was subjected to A549, a NSCLC cell line with high migration ability, to examine the effects of these genes on cell migration by wound-healing assay. The screening results showed that knockdown of 2 genes (KDM5B and SIRT1) dramatically and specifically inhibits A549 migration but not affects the proliferation, which was subsequently confirmed through transwell migration assay. Furthermore, SIRT1 is found to be highly expressed in brain metastasis tissues of NSCLC, compared to the NSCLC tissues, suggesting that SIRT1 may play roles in brain metastasis of NSCLC. The relationship between SIRT1 expression and cell migration ability was further investigated in three NSCLC cell lines and the result indicated that SIRT1 expression is tightly correlated with cell migration ability. Collectively, our work provides potential biomarker and therapeutic target for brain metastasis of NSCLC.
Introduction
Brain metastases are a frequent and ongoing major complication of non-small cell lung cancer (NSCLC). Among NSCLC patients, approximately 20% to 40% will develop brain metastases at some point [1,2]. The prognosis is generally poor and available standard therapeutic options provide a limited improvement in local control, overall survival and symptom relief. The median survival for untreated patients is 1-2 months, which may be extended to 6 months with surgery, radiotherapy, and chemotherapy [3-5]. The emerging targeted therapies, such as EGFR tyrosine kinase inhibitor (TKI), erlotinib and gefitinib, and anti-VEGF Bevacizumab treatment, have been proved to be promising therapy for these patients in some clinical trials [6,7]. However, hemorrhage of brain metastasis from non-small cell lung cancer post targeted therapies [8,9] suggests that these therapies need further safety evaluation in more patients. Therefore, it is urgent to deepen our understanding of the underlying mechanism by which NSCLC cells metastasize to brain and hence to improve the therapy.
Cancer metastasis is a multistep process during which tumor cells, responding to different intrinsic and extrinsic stimuli, detach from the primary tumor mass, invade the contiguous stroma, migrate over a long distance, and colonize distant organs. In this complex process, reciprocal regulation of tumor cells and the microenvironment leads to the consequence of metastasis. Except that EGFR mutations [10], αv-Integrin isoform [11], CXCR4/CXCL12 [12], E-cadherin [13], S100B [14] and MMP2 expression [15] in NSCLC cells and junctional adhesion molecules and microRNAs [16] in brain [17] are involved into brain metastases of NSCLC, growing evidences suggest that epigenetic regulation plays vital roles in cancer metastasis.
Epigenetic regulation, including genomic DNA methylation, histone modifications, chromatin remodeling and microRNAs regulation, implicates in tumorigenesis and cancer metastasis. It is proposed that epigenetic control of epithelial-to-mesenchymal transition (EMT) is also critical for the conversion of early stage tumors into invasive malignancies and therefore contributes to cancer metastasis [18]. Li et al reported that silencing of Wnt5a during colon cancer metastasis involves histone modifications, including lower levels of acetylated histone H3, H4 and H3K4me2 and higher levels of H3K27me3 in the promoter region [19]. Carmona et al found that DNA methylation-associated transcriptional silencing of CDH11, an inhibitor of tumor growth, motility and dissemination, occurred in the corresponding lymph node metastases of melanoma and head and neck cancer cells but not in the primary tumors [20]. Wu et al found epigenetic inactivation of the canonical Wnt antagonist secreted frizzled-related protein 1 by promoter hypermethylation in hepatocellular carcinoma cells contributes to metastasis in HCC progression [21]. A reciprocal repression between ZEB1 and members of the miR-200 family promotes EMT and invasion in cancer cells [22] and the ZEB1/miR-200 feedback loop controls Notch signalling in cancer cells [23]. All these suggest that epigenetic regulation may dynamically control the expression of key genes involved in tumor progress and contribute to cancer metastasis.
Yet, there is not systemic evaluation of epigenetic enzymes on NSCLC migration until now. To this end, an RNAi screening with epigenetics genes was applied to A549, a NSCLC cell line with high migration ability, to systematically investigate the roles of these genes in NSCLC cells migration. Wound-healing experiments showed that the knockdown of 2 genes dramatically inhibits A549 migration, which was further validated through transwell migration experiments. And SIRT1 is also found to highly express in tissues of brain metastasis of NSCLC, compared to the NSCLC tissues, suggesting that SIRT1 may play roles in brain metastasis of NSCLC. Our results might propose a potential therapeutic target of brain metastases in NSCLC.
Materials and methods
Cell culture
NSCLC cell lines (A549, NCI-H460, 95C, 95D and U1752) were maintained in Dulbecco’s modified Eagle’s medium (DMEM) or RMPI 1640 medium supplemented with 10% fetal bovine serum (FBS) (Hyclone), penicillin (100 IU/ml) and Streptomycin (100 μg/ml) (Life Technologies) at 37°C and 5% CO2.
Construction of shRNA lentiviral library and lentivirus infection
A customized lentiviral shRNA library targeting 153 epigenetic genes (see Supplementary Table 1A) was constructed into lentivirus vector (Invitrogen, BLOCK-iT™ Lentiviral RNAi Expression System, K4944-00), shRNA lentivirus were packaged and titered in HEK293T cells according to the manufacturer’s protocol. Each gene in this library contained two pools of four distinct shRNA species targeting different positions of the target transcript. A549 cells were infected by shRNA lentivirus at MOI of 5 in the presence of polybrene (8 μg/ml).
RNAi screening
A549 cells (2500 per well) were plated in 96 well plates and infected 24 h later with shRNA library, screened as 306 pools of shRNAs per pool in independent triplicates in the primary screening. 96 h after infection, wound-healing experiments were performed to investigate the migration ability following shRNA treatments. Those genes that inhibited cell migration by higher than 20% were designated as positive hits. And then most positive hits were validated by secondary RNAi screening.
In vitro wound-healing assay
Cells in 96-well plates were transfected with shRNA lentiviral libraries. 96 h later, a 100-μL pipette tip was used to scribe a line across the cell monolayer, medium was then changed to DMEM containing 0~1% FBS. The relative migrated distances were measured using a fluorescence microspore 24 h later, compared to the NC group.
Cell migration assay
Migration assays were performed in Transwells (Corning, Inc.) (8.0-μm pore size). For migration assay, 96 h post transduction of lentiviral vectors, 20,000~10,000 cells (according to the cell types) in serum-free medium were added to the upper wells. Media containing 10% FBS were added to the lower wells. Cells that migrated through the filter after 24 h were stained and counted by phase microscopy.
Tissue samples
Twelve brain metastasis tissues of NSCLC were obtained from the Department of Neurosurgery, Huashan Hospital (Shanghai, China) for quantitative real-time PCR (qPCR) analysis. All tissue samples were obtained from the surgical removal and immediately snap frozen in liquid nitrogen and stored in liquid nitrogen until use. In addition, two primary NSCLC tissues and three normal lung tissues were collected for qPCR analysis to investigate the expression profi le of SIRT1. The study was approved by the ethics committee of Huashan Hospital. 100 mg or so tissues were homogenized in 1 ml Trizol (Life technologies), the total RNA was extracted according to the manufacturer’s protocol.
Quantitative real-time PCR (qPCR)
Total RNA was synthesized to cDNA using PrimeScript RT reagent kit with gDNA Eraser (Takara, RR074A) for RT-PCR with mixture of oligo-dT and Random Primer (9 mer). The primers used for qPCR validation were list in Supplementary Table 2. Real-time qPCR was performed on CFX-96 (Bio-lab), with endogenous control hActb. Gene expression was calculated relative to expression of hActb endogenous control and adjusted relative to expression in negative control cells (for gene silencing efficacy detection) or in normal tissue 1 (for SIRT1 expression in tissues) or in A549 (for SIRT1 expression in NSCLC cell lines).
MTS cell proliferation assay
The tetrazolium compound (MTS) cell proliferation assay is a quantitative colorimetric assay for mammalian cell survival and proliferation. A549 cells (4×103) were grown in 100 μl of culture medium containing serum per well in a 96-well plate. After 24 h, the cells were treated by shSIRT1-AD or shSIRT1-EH lentivirus for 96 h (shSIRT1-AD is a shRNAs pool composed of shSIRT1-A, shSIRT1-B, shSIRT1-C and shSIRT1-D; shSIRT1-EH is a shRNAs pool composed of shSIRT1-E, shSIRT1-F, shSIRT1-G and shSIRT1-H). Every treatment for each cell line was triplicate in the same experiment. Then 20 μl of MTS (CellTiter 96 AQueous One Solution Reagent; Promega) was added to each well for 2 h at 37°C. After incubation, the absorbance was read at a wavelength of 490 nm according to the manufacturer’s protocol.
Results
RNAi screening in A549 cells for migration-related epigenetic genes
Primary screening with shRNA library of 153 epigenetic genes through wound-healing assay showed that 25 epigenetic genes positively regulate the NSCLC cell migration ability. Knockdown of these genes significantly (p<0.05) reduced migration ability of A549 cells (Figure 1A). Interestingly, the other 26 epigenetic genes were involved in the negative regulation of A549 cells migration (cell migration ability was increased by higher than 20% following shRNA treatments, p<0.05, see Supplementary Table 1B), such as HDAC6. Silence of HDAC6 increased A549 migration ability by 24% or 40% (two pools of HDAC6 shRNAs induced A549 migration by different extents). In total, 1/3 of epigenetic genes in our library were associated with the regulation of A549 cells migration. A secondary screening was performed for those 25 positive hits to validate their roles in cell migration and 5 genes were found to be positive in these two cycles of screening (Figure 1C). Among these 5 genes, ALKBH5 is a mammalian RNA demethylase that impacts RNA metabolism [24], KDM5B is a histone demethylase, PRDM14 may possess histone methyltransferase activity, PRMT1 encodes a member of the protein arginine N-methyltransferase (PRMT) family, while the functions of human SIRT1 have not yet been determined; however, yeast sirtuin proteins are known to regulate epigenetic gene silencing.
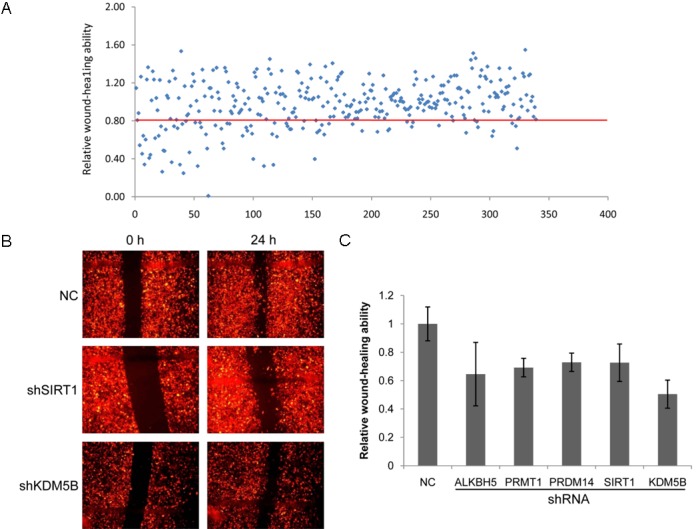
A high throughput screening with shRNAs library of 153 epigenetic genes. A: Primary screening. A549 cells were treated with shRNA lentiviral library for 96 h, and then wound-healing assay was performed to examine the effects of these genes on cell migration ability. There are two pools of shRNAs for each gene; every pool of some gene is comprised of four distinct shRNA species targeted to this gene. Those genes that cell migration was decreased by higher than 20% following knockdown of this gene (the dots below the red line) and p<0.05 were designed as positive hits. B: Wound-healing pictures of two positive hits (KDM5B and SIRT1) in the secondary screening. C: The histogram of positive hits in the secondary screening. Every treatment was triplicate in the same experiment. Bars represent the standard errors.
Validation of positive genes by transwell migration assay and selection of effective shRNA species by qPCR
To further validate the roles of positive genes in A549 cells migration, transwell migration assay was performed. The 4 distinct shRNA species comprised of each positive pool for KDM5B and SIRT1 were constructed, respectively, and infected A549 cells. The transwell migration results showed that knockdown of KDM5B and SIRT1 by more than 2 single shRNA species really suppressed the migration ability of A549 cells (Figure 2A, ,2B),2B), suggesting these inhibition is not caused by off-target effect [25].

SIRT1 and KDM5B silencing by shRNAs specifically inhibits the migration ability of NSCLC A549 cells. A: Transwell migration assay for SIRT1 or KDM5B knockdown. SIRT1 or KDM5B knockdown (by three distinct shRNA species of each gene, respectively, for 96 h) cells were seeded into the upper well, 24 h later the migrated cells were stained and photographed. For eah shRNA specie, two repeats were performed. Two fields of each repeat were list in the figure. B: The histogram of migration ability for KDM5B or SIRT1 knockdown. C: Gene silencing by shRNA was assayed by qPCR. Bars represent the standard errors.
KDM5B and SIRT1 silencing by shRNAs was confirmed by quantitative PCR (Figure 2C). The two KDM5B/SIRT1-targeting shRNAs that caused the most significant effects on transwell migration ability were also shown to cause the most significant silencing (Figure 2B, ,2C2C).
Collectively, KDM5B and SIRT1 silencing by shRNAs specifically inhibits the migration ability of NSCLC A549 cells.
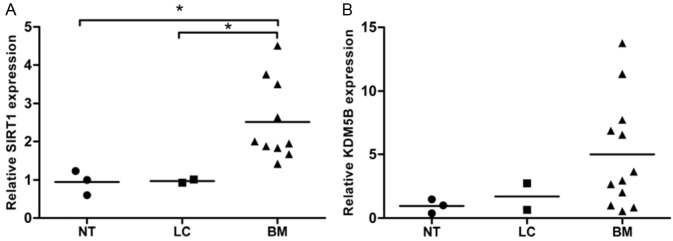
SIRT1 is highly expressed in brain metastasis tissues of NSCLC patients
To evaluate the clinical importance of KDM5B and SIRT1 for NSCLC metastasis, we examined the mRNA expression of these two genes in NSCLC. 2 NSCLC tissues and corresponding normal tissues and 12 brain metastasis tissues of NSCLC patients were collected. qPCR results showed that SIRT1 expression in brain metastasis tissues of NSCLC patients was significantly increased, compared to that in NSCLC tissues and corresponding normal tissues (Figure 3A). However, there was not marked difference in KDM5B expression between these 3 types of tissues (Figure 3B). This suggests that high expression of SIRT1 is highly correlated to the brain metastasis of NSCLC in clinic.
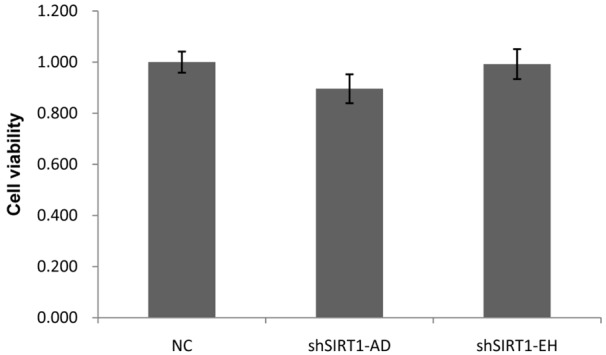
SIRT1 knockdown does not affect A549 cell proliferation
SIRT1 is proposed to be involved in proliferation of cancer cells [26,27], so we investigated cell viability following SIRT1 knockdown (Figure 4). The results showed that A549 cell proliferation was not significantly changed after SIRT1 silencing, suggesting that the effect of SIRT1 on cell migration above mentioned is not caused by anti-proliferation of SIRT1 knockdown.
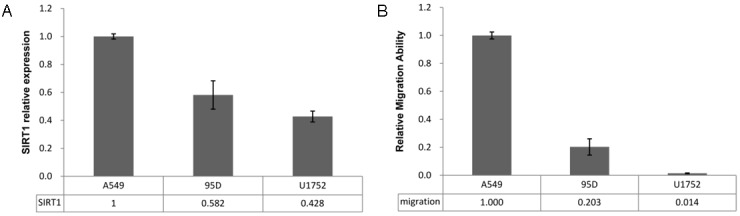
SIRT1 expression level is correlated to migration ability of NSCLC cell lines
And then the relationship between SIRT1 expression and cell migration ability was investigated in 3 NSCLC cell lines (A549, 95D and U1752). The results showed that Sirt1 expression was decreased gradually according to the following order: A549, 95D and U1752 (Figure 5A), and the cell migration ability was decreased stepwise according to the same order (Figure 5B). There are positive correspondences between these two data sets. Yet, due to limited cell line number, this correlation is still need to be further examined in more NSCLC cell lines and with functional confirmation by establishing over-expression stable cell lines. This indicates that SIRT1 expression is positively correlated to the migration ability of NSCLC cell lines.
Discussion
Brain metastasis of NSCLC is a great challenge for approximately 1/3 NSCLC patients, it is urgent to broaden and deepen our understandings on the mechanisms by which NSCLC cells metastasize to brain. As the first step, it is of great interest to find genes related to the migration of NSCLC cells in vitro.
Growing evidence suggsets that epigenetic regulation implicates in tumorigenesis and metastasis. But the systemic investigation of whole epigenetic genes in metastasis regulation has not been reported. In present work, a high throughput RNAi screening with shRNA library of 153 epigenetic genes was applied to A549, a NSCLC cell line with high migration ability.
Primary screening found that 1/3 or so epigenetic genes are involved in positive or negative regulation of A549 migration. This suggests an all-around regulation was carried out by epigenetic genes, some genes promote migration, and others inhibit migration, cell migration ability is controlled in a balance manner. Genetic variations in tumor cells or stimuli from microenvironment induce or inactivate some genes of this system and hence break this balance and contribute to the alteration on migration ability. So, it is the genetic variations in tumor cells or stimuli from microenvironment that is worthy of further studies to find the underlying reasons for alterations of migration ability in future.
Secondary screening found that 5 epigenetic genes were in positive regulation on A549 migration. 2 out of these 5 genes, KDM5B and SIRT1, were further validated through transwell migration assay and the gene silencing of shRNA was verified by qPCR. Knockdown of KDM5B and SIRT1 was found to specifically inhibit A549 migration. Subsequent experiments showed that SIRT1 was highly expressed in brain metastasis tissues of NSCLC patients, compared to that in NSCLC tissues and the corresponding normal tissues, suggesting the clinical importance of SIRT1 in brain metastasis of NSCLC.
The class III histone deacetylase SIRT1 is a nicotinamide adenine dinucleotide (NAD(+))-dependent deacetylase, SIRT1 is reported to be overexpressed and catalytically activated in a number of human cancers and implicated in tumorigenesis and metastasis of several types of cancer, including gastric carcinoma, pancreatic cancer, breast cancer, prostate cancer, colorectal adenocarcinoma and NSCLC. Cha et al reported in 2009 that SIRT1 expression is significantly correlated with lymph node metastasis and tumor invasion and is associated with poor prognosis of gastric carcinoma [28]. Noh et al reported SIRT1 expression is associated with lymph node metastasis and advanced tumor invasion [29]. Several studies suggest that SIRT1 can induce epithelial-mesenchymal transition (EMT) and invasion and hence promote cancer metastasis, silencing of SIRT1 inhibit cancer cell migration or invasion [30-32], which is consistent with our data. However, there have been conflicting data supporting whether SIRT1 act as a tumour promoter or as a tumour suppressor. It is proposed that SIRT1 suppresses EMT and hence inhibit cancer cell migration or invasion, the downregulation of SIRT1 expression plays roles in metastasis of ovarian cancer, lung cancer and colorectal adenocarcinoma [33-35]. The reason for this inconsistency may be in that cancer cell migration/invasion is probably controlled by a complex network comprised of many epigenetic genes, while SIRT1 is just one member of this network. Furthermore, SIRT1 is under control of some microRNAs, such as miR-204 [36], miR-520c and miR-373 [37], and miR-22 [38]. Therefore, it is necessary to reveal the upstream regulators and the downstream effectors of SIRT1 and to find out the other migration-regulated genes in further study.
Taken together, our high throughput RNAi screening showed that many epigenetic genes are involved in migration regulation, SIRT1 is associated with positive regulation of A549 cells migration and SIRT1 is highly expressed in brain metastasis tissues of NSCLC. SIRT1 is a drugable target, which might do benefit to patients in clinical therapy combination with SIRT1 specific inhibitor.
Acknowledgements
This work was granted by the knowledge innovation project of Chinese Academy of Sciences (No. KSCX2-YW-R-71).
References
Articles from International Journal of Clinical and Experimental Pathology are provided here courtesy of e-Century Publishing Corporation
Citations & impact
Impact metrics
Citations of article over time
Article citations
The role of SIRT1 in autophagy and drug resistance: unveiling new targets and potential biomarkers in cancer therapy.
Front Pharmacol, 15:1469830, 30 Sep 2024
Cited by: 0 articles | PMID: 39403142 | PMCID: PMC11471651
Review Free full text in Europe PMC
Emerging role of sirtuins in non‑small cell lung cancer (Review).
Oncol Rep, 52(4):127, 02 Aug 2024
Cited by: 0 articles | PMID: 39092574 | PMCID: PMC11304160
Review Free full text in Europe PMC
The impact of vitamin D on cancer: A mini review.
J Steroid Biochem Mol Biol, 231:106308, 11 Apr 2023
Cited by: 17 articles | PMID: 37054849
Review
SIRT1 Expression Is a Promising Prognostic Biomarker in Esophageal Squamous Cell Carcinoma: A Systematic Review and Meta-analysis.
Cancer Diagn Progn, 2(2):126-133, 03 Mar 2022
Cited by: 4 articles | PMID: 35399170 | PMCID: PMC8962800
Review Free full text in Europe PMC
MicroRNA-326 impairs chemotherapy resistance in non small cell lung cancer by suppressing histone deacetylase SIRT1-mediated HIF1α and elevating VEGFA.
Bioengineered, 13(3):5685-5699, 01 Mar 2022
Cited by: 11 articles | PMID: 34696659 | PMCID: PMC8973918
Go to all (42) article citations
Data
Data behind the article
This data has been text mined from the article, or deposited into data resources.
BioStudies: supplemental material and supporting data
Similar Articles
To arrive at the top five similar articles we use a word-weighted algorithm to compare words from the Title and Abstract of each citation.
MicroRNA-330-3p promotes cell invasion and metastasis in non-small cell lung cancer through GRIA3 by activating MAPK/ERK signaling pathway.
J Hematol Oncol, 10(1):125, 19 Jun 2017
Cited by: 52 articles | PMID: 28629431 | PMCID: PMC5477161
Histone demethylase JARID1B/KDM5B promotes aggressiveness of non-small cell lung cancer and serves as a good prognostic predictor.
Clin Epigenetics, 10(1):107, 09 Aug 2018
Cited by: 34 articles | PMID: 30092824 | PMCID: PMC6085612
SIRT1 promotes metastasis of human osteosarcoma cells.
Oncotarget, 7(48):79654-79669, 01 Nov 2016
Cited by: 20 articles | PMID: 27793039 | PMCID: PMC5346743
A Screening of Epigenetic Therapeutic Targets for Non-Small Cell Lung Cancer Reveals PADI4 and KDM6B as Promising Candidates.
Int J Mol Sci, 23(19):11911, 07 Oct 2022
Cited by: 1 article | PMID: 36233212 | PMCID: PMC9570250
Review Free full text in Europe PMC