Abstract
Free full text

PNAS Plus
Hierarchical recruitment of Plk4 and regulation of centriole biogenesis by two centrosomal scaffolds, Cep192 and Cep152
Significance
Found in most eukaryotic cells, a centriole is a cylindrically shaped subcellular structure that plays an important role in various cellular processes, including mitotic spindle formation and chromosome segregation. Centriole duplication occurs only once per cell cycle, thus ensuring accurate control of centriole numbers to maintain genomic integrity. Although a growing body of evidence suggests that a Ser/Thr protein kinase, polo-like kinase 4 (Plk4), is a key regulator of centriole duplication, how Plk4 is recruited to centrosomes remains largely unknown. Here we showed that Plk4 dynamically localizes to distinct subcentrosomal regions by interacting with two hierarchically regulated scaffolds, Cep192 and Cep152. Highlighting the importance of these interactions, mutational disruption of either one of these interactions was sufficient to cripple Plk4-dependent centriole biogenesis.
Abstract
Centrosomes play an important role in various cellular processes, including spindle formation and chromosome segregation. They are composed of two orthogonally arranged centrioles, whose duplication occurs only once per cell cycle. Accurate control of centriole numbers is essential for the maintenance of genomic integrity. Although it is well appreciated that polo-like kinase 4 (Plk4) plays a central role in centriole biogenesis, how it is recruited to centrosomes and whether this step is necessary for centriole biogenesis remain largely elusive. Here we showed that Plk4 localizes to distinct subcentrosomal regions in a temporally and spatially regulated manner, and that Cep192 and Cep152 serve as two distinct scaffolds that recruit Plk4 to centrosomes in a hierarchical order. Interestingly, Cep192 and Cep152 competitively interacted with the cryptic polo box of Plk4 through their homologous N-terminal sequences containing acidic-α-helix and N/Q-rich motifs. Consistent with these observations, the expression of either one of these N-terminal fragments was sufficient to delocalize Plk4 from centrosomes. Furthermore, loss of the Cep192- or Cep152-dependent interaction with Plk4 resulted in impaired centriole duplication that led to delayed cell proliferation. Thus, the spatiotemporal regulation of Plk4 localization by two hierarchical scaffolds, Cep192 and Cep152, is critical for centriole biogenesis.
The centrosome is the main microtubule-organizing center in mammalian cells that plays a central role in spindle formation and chromosome segregation during mitosis. Centrosomes are composed of two orthogonally arranged centrioles surrounded by an amorphous mass of electron-dense pericentriolar material (PCM). Centrioles duplicate precisely once per cell cycle and serve as platforms for the assembly of centrosomes, primary cilia, and flagella (1–4).
Centriole duplication is initiated by the assembly of a procentriole in early S phase. In Caenorhabditis elegans, a centrosomal scaffold protein, called Spd-2, is required for proper recruitment of a Ser/Thr kinase, Zyg-1 (5), to centrosomes, and this step in turn allows the recruitment of Sas-6, Sas-5, and Sas-4 to the site of procentriole assembly (6, 7). Sas6 plays a pivotal role in self-assembling a cartwheel-like structure at this site of the procentriole with Sas5 and Sas4 (8–12). In Drosophila, the overexpression of polo-like kinase 4 (Plk4; also called Sak), the Zyg-1 ortholog, is sufficient to induce centriole amplification, whereas the depletion of Plk4 disrupts centriole duplication (12, 13). Interestingly, however, Drosophila Spd-2 is dispensable for Plk4-mediated centriole duplication (14). Instead, another scaffold, Asterless, has been suggested to play a critical role in targeting Plk4 to centrosomes (15), hinting that the mechanism underlying Plk4 recruitment is distinct in different organisms.
Accumulated evidence in humans suggests that Plk4 is a functional ortholog of C. elegans Zyg-1 and Drosophila Plk4, and that it plays a key role in centriole duplication (16, 17). When overexpressed, Plk4 can induce multiple centriole precursors surrounding a single parental centriole, and centrosomally localized Plk4 appears to be required for this event (16). The cryptic polo box (CPB) present at the upstream of the C-terminal polo box (PB) (18) is necessary and sufficient for targeting Plk4 to centrosomes (16, 19). Interestingly, the CPB comprises two structurally related motifs and forms a homodimer (19) to interact with its binding targets. However, the molecular basis of how Plk4 binds to its targets and localizes to centrosomes remains largely elusive.
Studies have shown that Cep152, a human ortholog of Drosophila Asterless, interacts with Plk4 through the CPB (20, 21). However, the depletion of Cep152 does not significantly decrease the level of Plk4 at centrosomes. Recently, Sonnen et al. have shown that a C. elegans Spd-2 ortholog, Cep192, interacts with Plk4 and promotes the recruitment of Plk4 to centrosomes (22). Moreover, Cep192 binds to Cep152, and the depletion of both enhances the Plk4 localization defect (22). Based on these observations, Sonnen et al. proposed that Cep192 cooperates with Cep152 to properly recruit Plk4 to centrosomes and to promote centriole duplication (22).
In this study, we demonstrated that disrupting either the Cep192–Plk4 interaction or the Cep152–Plk4 interaction was sufficient to impair centriole duplication. We further showed that Plk4 dynamically localizes to different subcentrosomal regions in a cell cycle-specific manner, and that Cep192 functions at a point upstream of Cep152 to regulate Plk4 localization. Thus, we propose that the spatiotemporal regulation of Plk4 localization by two hierarchical scaffolds, Cep192 and Cep152, is critical for Plk4-dependent centriole biogenesis.
Results
Identification of Plk4 as a Cep192-Binding Protein.
Cep192 plays a critical role in PCM assembly and mitotic progression (23, 24). To better understand the function of Cep192, we isolated Cep192-binding proteins by carrying out tandem affinity purification followed by mass spectrometry analyses. Among the 15 candidates we obtained, Cep192 strongly interacted with Plk4, whereas several others interacted weakly with Plk4 (SI Appendix, Fig. S1A). We confirmed that Cep192 interacts efficiently with Plk4 under thymidine (S phase)- and nocodazole (M phase)-treated conditions (SI Appendix, Fig. S1B), suggesting that the interaction occurs as early as the S phase of the cell cycle.
The N-Terminal Region of Cep192 Binds to the CPB of Plk4.
To determine the region of Cep192 responsible for interacting with Plk4, we generated several GFP-fused Cep192 fragments and examined their capacity to bind to FLAG-tagged Plk4 by carrying out anti-GFP immunoprecipitation (SI Appendix, Fig. S2A). Results showed that an N-terminal Cep192 fragment (1–647) was sufficient to bind to Plk4. Subsequent deletion analyses revealed that the Cep192 (201–280) fragment possessed the full capacity to bind to Plk4 (SI Appendix, Fig. S2 B and C).
Next, we generated various Plk4-truncated forms to determine the region of Plk4 that is critical for Cep192 binding. We observed that the CPB of Plk4 and CPB-containing Plk4 forms (full-length and C), but not the kinase domain or the C-terminal PB, interacts with Cep192 (201–310; SI Appendix, Fig. S3A) or Cep192 (201–280; i.e., DN80, as detailed later; Fig. 1A). These interactions were confirmed by reverse immunoprecipitation analyses with anti-FLAG antibody (SI Appendix, Fig. S3 B and C). Thus, the CPB of Plk4 binds to the Cep192 (201–280) fragment.
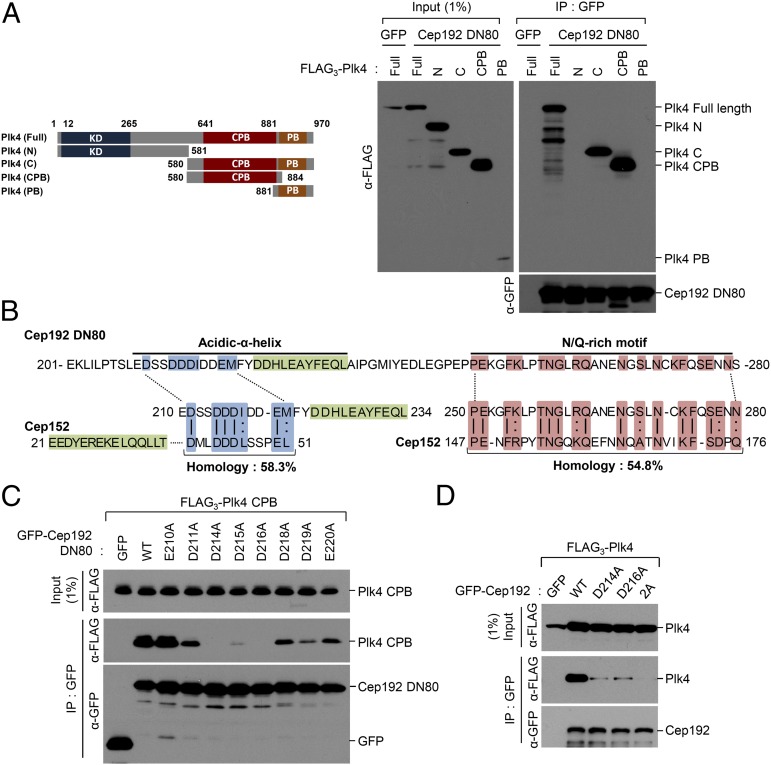
Analyses of the interaction between Plk4 and Cep192. (A) 293T cells cotransfected with the indicated constructs were immunoprecipitated and then immunoblotted. Schematic diagram (Left) denotes various Plk4 forms used in the experiment. Numbers indicate amino acid residues (KD, kinase domain). Cep192 DN80 is an 80-residue-long fragment (residues 201–280) from Cep192 that appears to be fully capable of binding to Plk4 CPB (SI Appendix, Figs. S2 and S3). The control GFP ligand is not shown in the anti-GFP panel because of its small molecular size. (B) The primary sequence of Cep192 DN80 (residues 201–280) was aligned with that of Cep152 DN217 (residues 1–217) (21) by BLAST. The N-terminal acidic region (green) and α-helix motif (blue) and the C-terminal N/Q-rich region (pink) are marked, and the identical (vertical line) and conserved (colon) residues are indicated. (C and D) 293T cells cotransfected with the truncated (C) or full-length (D) forms of Plk4 and Cep192 constructs were analyzed.
Mutations in the Acidic α-Helix Motif Disrupt the Cep192-Plk4 Interaction.
A previous report showed that the Cep152 (1–217) fragment efficiently binds to Plk4 (21). Comparative analysis of primary amino acid sequences between Cep192 (201–280) and Cep152 (1–217) revealed that both fragments contained an N-terminal acidic (D/E) region and α-helix (or α-helix and acidic region for 1–217) motif and a C-terminal N/Q-rich motif (Fig. 1B). Therefore, based on the presence of the D/E- and N/Q-rich regions in each sequence, we will refer to Cep192 (201–280) and Cep152 (1–217) hereafter as DN80 and DN217, respectively, for simplicity. As with the importance of both motifs for Plk4 binding, the deletion of the N-terminal D/E-enriched sequence (i.e., residues 210–220) greatly diminished the ability of Cep192 to interact with Plk4, whereas the deletion of the C-terminal N/Q-enriched sequence (i.e., residues 261–280) significantly (i.e., a few fold) weakened the interaction (SI Appendix, Fig. S2C).
In an attempt to determine residues critical for Plk4 binding, we mutated 16 single residues (eight residues each for the N- and the C-terminal motif) in Cep192 DN80, and then tested the resulting mutants for Plk4 CPB binding. Results showed that mutating D214 or D216 to A greatly crippled the CPB binding, whereas mutating D215 to A impaired it at a somewhat reduced level (Fig. 1C). On the contrary, single mutations in the N/Q-rich motif generally exhibited a moderate level of defect in Plk4 CPB binding (SI Appendix, Fig. S4). As expected, the full-length Cep192 (D214A D216A) double mutant (2A) failed to interact with the full-length Plk4, whereas each single mutant interacted with Plk4 somewhat weakly (Fig. 1D). Therefore, we used the Cep192 (2A) mutant to investigate the phenotypic consequence of the loss of the Cep192-Plk4 interaction below (see Fig. 5).

Loss of Cep192- or Cep152-dependent interaction with Plk4 induces defects in centriole duplication and cell proliferation. (A–D) U2OS cells stably expressing the indicated constructs were silenced for control luciferase (siGL), Cep192 (siCep192), or Cep152 (siCep152; SI Appendix, Fig. S10 shows details of the experimental procedure). (A) The resulting cells were used for immunoblotting analyses, and the membranes were stained with Coomassie Brilliant Blue (CBB). (B–D) The cells from A were immunostained for further analyses. (B) Relative fluorescence intensities for centrosome-localized Plk4 signals were quantified from three independent experiments (≥50 cells per experiment). Bars indicate mean ± SEM. (C and D) Number of cells with no detectable Cep192 (C) or Cep152 (D) signals was quantified from three independent experiments (≥200 cells per experiment). Bars indicate SD. (E) Number of centrosomal Sas6 signals per cell was quantified from three independent experiments (≥200 cells per experiment). Bars indicate SD. The actual confocal images used for quantification in B and E are provided in SI Appendix, Figs. S13 and S14. (F) The cells obtained in A were quantified 4 d after treatment with siRNAs. Data were obtained from three independent experiments (≥2,000 cells per experiment). Bars indicate SD. (G) Model illustrating the mechanism of how Cep192 and Cep152 promote Plk4 recruitment and centriole biogenesis. In early G1, Cep192 localizes to mother and daughter centrioles, whereas Cep152 localizes only to mother centriole. At this stage, Plk4 mainly associates with mother centriole. Soon after the recruitment of Plk4 to daughter centriole in late G1, Cep152 was amassed at this location (SI Appendix, Fig. S8B). These series of events in turn allow the recruitment of Sas6 to the site of procentriole assembly. As a result of a hierarchical assembly of Cep192 and Cep152, depleting Cep192 delocalizes Cep152 from centrioles, thus resulting in a greatly diminished Plk4 signals (two tiny red Plk4 dots) at centrosomes. On the contrary, depleting Cep152, which is present in centrosomes and cytosol, unleashes Plk4 from the Cep152 tether and allows freed Plk4 to be enriched (two larger red Plk4 dots) at Cep192-associated centrioles. In the absence of Cep192 or Cep152, Sas6 recruitment is disrupted.
Cep152 DN217 Binds to Plk4 CPB in a Manner Similar to That of Cep192 DN80.
We observed that the two previously identified N-terminal fragments (1–512 and 1–217) of Cep152 (20, 21) interacted with the full-length Plk4 or Plk4 CPB as efficiently as the Cep192 DN80 fragment did (SI Appendix, Fig. S5 A and B). Interestingly, the Cep152 (27–180) fragment lacking both the N-terminal 26 and the C-terminal 37 residues from the Cep152 (1–217) fragment (i.e., DN217 for simplicity), completely disrupted the binding (SI Appendix, Fig. S5). As Cep152 (27–180) contains the entire C-terminal N/Q motif, its inability to interact with Plk4 could be the consequence of truncating the N-terminal α-helix of the α-helix and acidic motif.
As several mutations in the acidic α-helix motif of DN80 crippled the DN80–Plk4 CPB interaction, we generated analogous mutations in the corresponding region of DN217. Among them, we found that a mutation of D43 or D44 to A greatly impaired the DN217-Plk4 CPB interaction, whereas a mutation of D40 to A crippled the interaction at a slightly reduced level (Fig. 2A). Consistent with these observations, the full-length Cep152 (D43A D44A) double mutant (2A) exhibited a significantly compromised ability to bind to the full-length Plk4 (Fig. 2B), suggesting that the mode of the Cep152–Plk4 interaction is similar to that of the Cep192–Plk4 interaction.
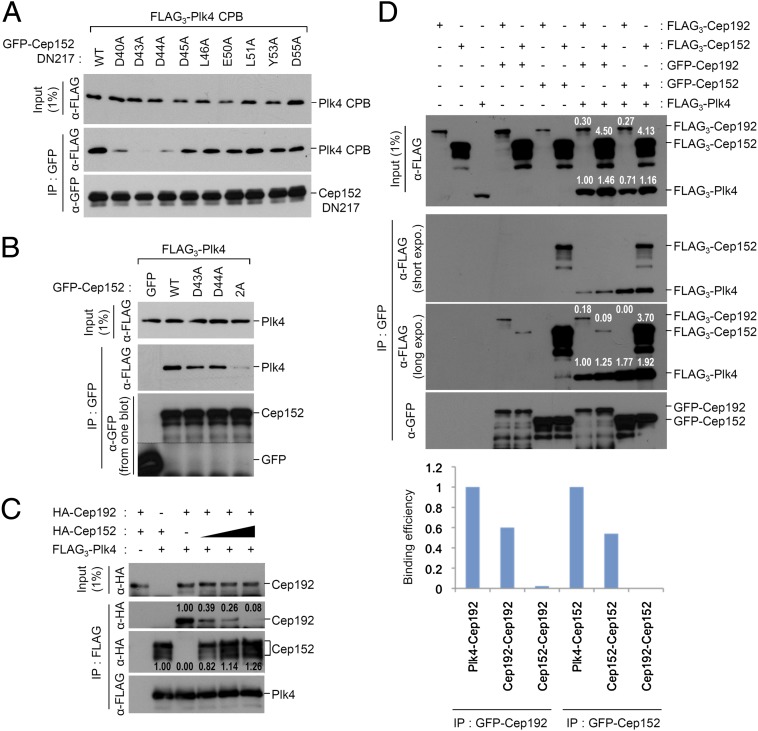
Cep152 contains a binding motif analogous to that of Cep192, and competes against the latter in Plk4 binding. (A and B) 293T cells cotransfected with the indicated constructs were analyzed. (C) 293T cells expressing FLAG3-Plk4, HA-Cep192, or increasing amounts (black right-angled triangle) of HA-Cep152 were mixed, as indicated (plus symbols), and subjected to coimmunoprecipitation analyses. Numbers indicate relative signal intensities. (D) For Cep152 and Cep192 expression, 293T cells were singly transfected or cotransfected with the indicated FLAG- or GFP-tagged constructs. The resulting total lysates expressing Cep192, Cep152, or both were mixed with the same amounts of other total lysates expressing either control vector or FLAG3-Plk4, and then the combined lysates were subjected to coimmunoprecipitation analyses. Numbers indicate relative expression levels in the input and amounts of coimmunoprecipitated proteins. The graph shows calculated levels of interactions based on the signal intensities quantified from the blots.
Cep192 and Cep152 Bind to Plk4 CPB in a Competitive Fashion.
The observed similarities in the binding mode of DN80 or DN217 to Plk4 CPB predict that Cep192 and Cep152 may compete with each other to bind to Plk4. To test this possibility, we mixed cell lysates expressing varying amounts of Cep152 with the second lysates expressing a constant level of Cep192 (the expression of Cep192 in different amounts was not feasible because its overexpression was cytotoxic). Under these conditions, as the level of Cep152 bound to Plk4 progressively increased, the level of Cep192 gradually decreased (Fig. 2C), suggesting that Cep192 and Cep152 bind to the same or an overlapping region of Plk4 in a competitive manner.
The competitive binding nature of Cep192 and Cep152 to Plk4 suggests that Cep192 and Cep152 may not form a heterodimeric/multimeric complex between the two proteins, as suggested previously (22), but they may function independently of each other. To closely examine this possibility, we performed coimmunoprecipitation analyses by using cotransfected 293T cells. Under the conditions in which Plk4 is not coexpressed, the immunoprecipitation of GFP-tagged Cep192 or Cep152 efficiently coprecipitated FLAG-tagged Cep192 or Cep152, respectively (Fig. 2D, lanes 4 and 7). In contrast, the immunoprecipitation of GFP-Cep192 weakly coprecipitated FLAG-Cep152, whereas the immunoprecipitation of GFP-Cep152 failed to coprecipitate a detectable amount of FLAG-192 (Fig. 2D, lanes 5 and 6). Plk4 coexpression did not appear to alter the interaction capacity of these two proteins (Fig. 2D, last 4 lanes). Therefore, as both Cep192 and Cep152 bind well to Plk4, we estimated the efficiencies of their inter- and intramolecular interactions by using their binding levels to Plk4 as internal controls. Results showed that Cep192 and Cep152 homodimerize/multimerize at a rate of ~60% and ~54% of the corresponding Cep192–Plk4 and Cep152–Plk4 interactions (Fig. 2D) (detailed calculations are provided in Materials and Methods). In stark contrast, the immunoprecipitation of GFP-Cep192 coprecipitated Cep152 very weakly (~2% of the Cep192–Plk4 interaction), whereas the immunoprecipitation of GFP-Cep152 failed to coprecipitate Cep192 at a detectable level (Fig. 2D). Taken together, these findings suggest that Cep192 and Cep152 may likely function as individual homodimers/multimers, but not as a heterodimer/multimer made of the two proteins.
An Intact DN80 or DN217 Motif is Necessary for Proper Recruitment of Plk4 to Centrosomes.
To determine the functional relevance of the Cep192 DN80- or Cep152 DN217-dependent interaction with Plk4, we first examined whether GST-fused DN80 or DN217 binds to endogenous Plk4 by using an antibody generated against the C-terminal domain of Plk4 (SI Appendix, Fig. S6). Results showed that DN80 and DN217, but not their respective Plk4-binding–defective 2A mutants, were capable of precipitating endogenous Plk4 (Fig. 3A), although the former appeared to be a few fold less efficient than the latter. Thus, an intact DN80 or DN217 motif is necessary and sufficient to bind to endogenous Plk4.
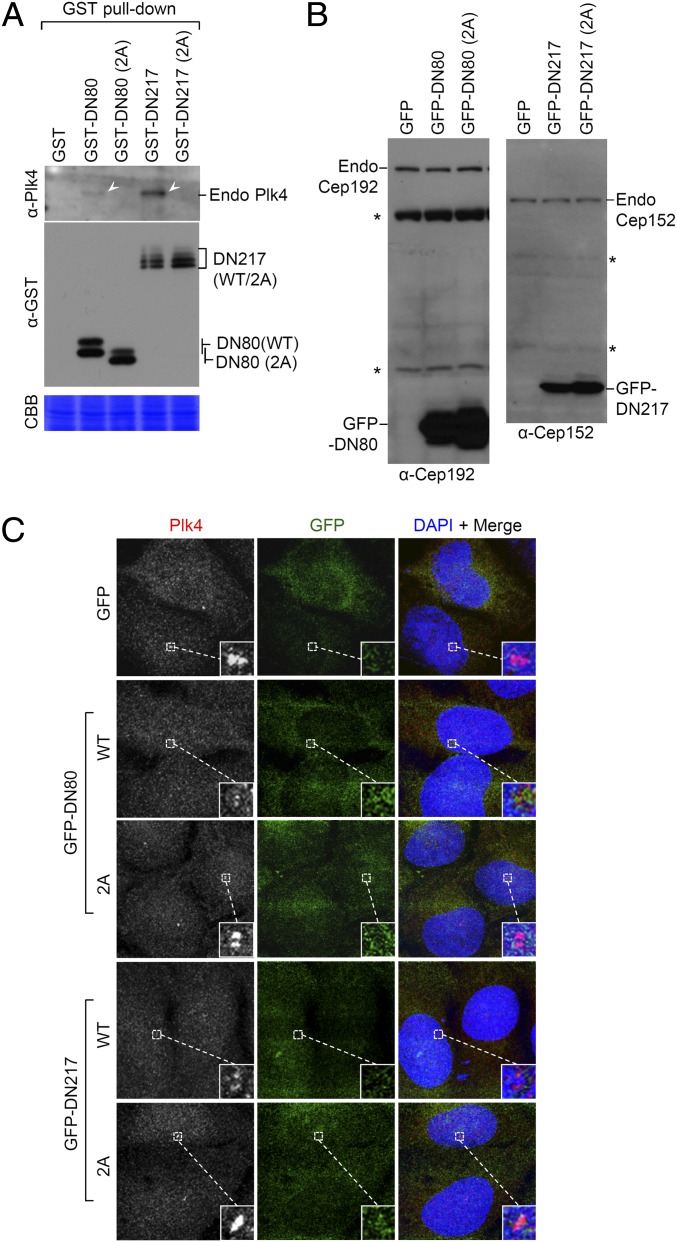
Expression of Cep192 DN80 or Cep152 DN217 is sufficient to alter subcellular localization of Plk4. (A) 293T cells transfected with the indicated constructs were subjected to GST pull-down assays. Arrowheads mark endogenous Plk4 coprecipitated with GST-DN80 or GST-DN217, but not with their respective 2A mutant or control GST (not shown because of small molecular size). (B and C) U2OS cells stably expressing the indicated GFP-DN80 or GFP-DN217 constructs were subjected to immunoblotting analyses (B) using antibodies that detect the epitope commonly present in the GFP-fused form and its respective endogenous protein. The cells were also immunostained (C) to determine the level of Plk4 at the centrosomes. Asterisks in (B) mark nonspecific cross-reacting proteins.
We then investigated whether the DN80 or DN217 motif was capable of altering Plk4 function in vivo by expressing GFP-fused DN80 (WT or 2A) or DN217 (WT or 2A) a few to several fold higher than its respective endogenous Cep192 or Cep152 (Fig. 3B). GFP-DN80 and GFP-DN217 exhibited diffused signals in cytosol (Fig. 3C). Under these conditions, ectopically expressed GFP-DN80 or GFP-DN217, but not the DN80 (2A) or DN217 (2A) mutant, effectively delocalized Plk4 from centrosomes (Fig. 3C). These observations suggest that expressing GFP-DN80 or GFP-DN217 is sufficient to interfere with endogenous Plk4 localization through direct interaction with Plk4 CPB.
Plk4 Dynamically Localizes to Centrioles in a Cell Cycle-Specific Manner.
To investigate the role of Cep192 and Cep152 in the localization of Plk4, we first investigated the cell cycle-dependent subcentriolar localization of Plk4 by using HeLa cells expressing Centrin2 fused with GFP. To this end, we harvested synchronized mitotic cells by mechanical shake-off, plated them directly onto coverslips, and then prepared cells in the G1, G1/S, and S phases of the cell cycle (less than 7 h, 10 h, and 14 h after shake-off, respectively). The cells were then immunostained and analyzed by 3D structured illumination microscopy (SIM). Immediately after mitotic exit, most cells exhibited a strong Plk4 dot-like signal associated with a proximal part of brighter mother centrioles (judged by the brightness of the Centrin2-GFP signal; Fig. 4A, first row). At the 3-h time point, a dot-like Plk4 appearance was replaced with a ring-like Plk4 signal surrounding the proximal part of mother centrioles, and, occasionally, daughter centrioles as well (Fig. 4A, second row; and Fig. 4B, G1 cell). The ring-like Plk4 signal persisted until the centrioles entered duplication (~10 h after shake-off). At the 10-h time point, ~50% of the cells exhibited full or partial ring-like Plk4 signals (Fig. 4A, third row; and Fig. 4B, late G1 cell), whereas the rest of the cells showed a dot-like Plk4 appearance mostly in colocalization with the Sas6 signal (Fig. 4A, fourth row), which was indicative of the initiation of procentriole formation. Interestingly, however, Sas6 was not detectable on a small fraction of centrioles with a dot-like Plk4 signal (Fig. 4A, third row), indicating that the transition of Plk4 localization from a ring-like to a dot-like appearance occurs before Sas6 recruitment. The existence of various patterns of Plk4 fluorescence signals suggests that dynamic changes occur in Plk4 localization around the time of procentriole formation. At 14 h after shake-off, the centrosomes in the majority of cells displayed prominent two Plk4 dot signals closely colocalized with two Sas6 dot signals (detected as purple dots), and these signals were found adjacent to the weaker Centrin2-GFP signals of the procentrioles (Fig. 4A, fifth row; Fig. 4B, S phase; and SI Appendix, Fig. S7), in accordance with expected localization of Plk4 within a diplosome. These observations suggest that Plk4 relocalizes to the site of future procentriole formation at the late G1 phase, and this step is important for recruiting Sas6 to this site.
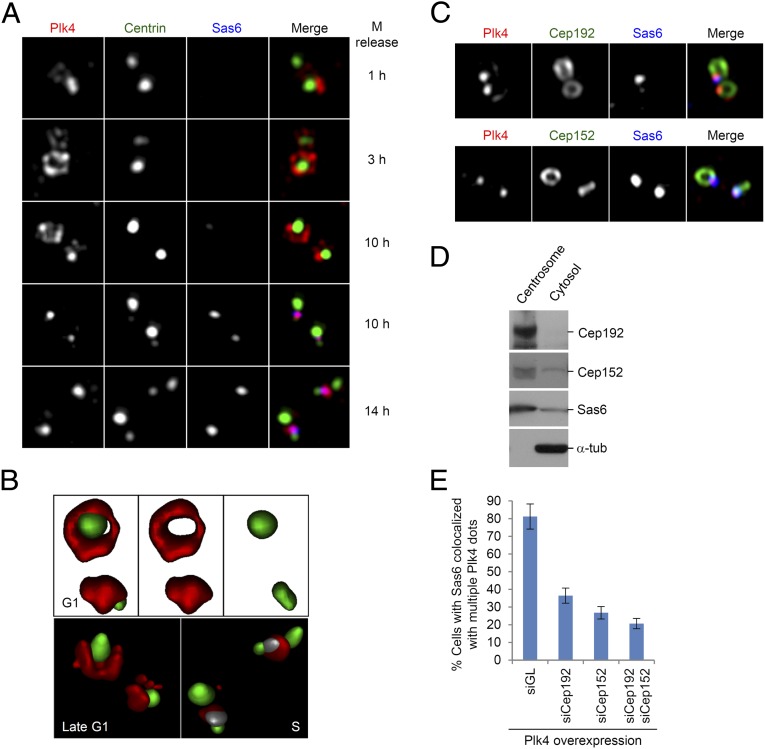
The role of Cep192 and Cep152 in the subcentrosomal localization and function of Plk4. (A–C) Three-dimensional SIM was carried out to examine the patterns of subcentrosomal localizations of Plk4. (A) Mitotic HeLa cells expressing Centrin2-GFP were prepared by mechanical shake-off, and were then cultured in prewarmed medium for the indicated lengths of time before being subjected to immunostaining analysis. Represented images obtained at each time point are shown. Note the presence of a ring-like Plk4 signal surrounding the mother centriole at the 3-h time point. Pink dots at the 10-h (the fourth row) and 14-h time points indicate colocalized Plk4 (red) and Sas6 (blue) signals at procentrioles. (B) Surface rendering of Plk4 (red), Centrin2-GFP (green), and Sas6 (pseudocolored in gray) signals for selected images from A shows dynamic subcentrosomal localization of Plk4 at different stages of the cell cycle. Note that, at the onset of procentriole assembly, Sas6 (gray) is almost completely colocalized with Plk4 (red) and is located close to two nascent Centrin (green) signals from daughter centrioles. (C) Asynchronously growing U2OS cells were immunostained, and representative images of Plk4 colocalized with Cep192 or Cep152 are provided. Additional images showing the localization patterns of Plk4 in relation to those of Cep192 or Cep152 are provided in SI Appendix, Fig. S8. (D) Enriched centrosome and corresponding cytosol fractions were prepared from HeLa cells arrested in S phase, and then subjected to immunoblotting analyses. (E) U2OS stable cells expressing a tetracycline-inducible GFP-Plk4 were transfected with the indicated siRNAs, and were fixed and immunostained. Cells that exhibit Sas6 signals colocalized with multiple Plk4 dot signals were quantified. Bars indicate SD.
Next, we examined whether Plk4 colocalizes with Cep192 and Cep152, as cells proceed through the cell cycle. We observed that Cep192 localizes along the cylindrical centrioles during the G1 and S phases, and then spreads out to expanding PCM at the late stages of the cell cycle (SI Appendix, Fig. S8A). As previously demonstrated (22), Cep152 exhibited a short cylindrical distribution at the proximal part of centrioles (SI Appendix, Fig. S8B). Plk4 clearly colocalized with Cep192 or Cep152, albeit with only a small fraction of their signals (Fig. 4C and SI Appendix, Fig. S8), thus supporting the finding that Plk4 interacts with the latter proteins (Figs. 1 and and22).
Cep192 and Cep152 Hierarchically Regulate the Recruitment of Plk4 to Centrosomes.
To examine whether the two scaffolds, Cep192 and Cep152, are tightly associated with centrosomes or whether they are also present in cytosol, we carried out immunoblotting analysis by using the isolated centrosomal fraction and its respective cytosolic fraction from S-phase HeLa cells. As expected, Cep192 was found to be solely associated with the centrosomal fraction (Fig. 4D). However, Cep152 was present not only in the centrosomal fraction but also in the cytosolic fraction (Fig. 4D). Considering that the isolated centrosomes that we used were highly enriched, a substantial level of the Cep152–Plk4 complex may exist in the cytosol.
We then investigated whether Cep192 and Cep152 are required for proper localization and function of Plk4 by using U2OS cells silenced for the control luciferase (siGL), Cep192 (siCep192) or Cep152 (siCep152) alone, or both (siCep192 siCep152). Under the conditions in which Cep192 and Cep152 were efficiently depleted to a similar degree (SI Appendix, Fig. S9A), we performed immunostaining analysis to examine the level of centrosome-localized Plk4 signal (SI Appendix, Fig. S9B). Consistent with the role of Cep192 in the recruitment of Plk4, depleting Cep192 diminished the level of Plk4 to ~53% of the control siGL cells (SI Appendix, Fig. S9C). Surprisingly, however, depleting Cep152 did not decrease Plk4 signals at centrosomes, but rather greatly (~2.4-fold) increased them. Interestingly, codepletion of Cep192 and Cep152 not only annihilated the effect of Cep152 depletion on Plk4 localization but also further diminished the level of Plk4 signals at centrosomes to ~38% of the control siGL cells (SI Appendix, Fig. S9C). As depleting either Cep192 or Cep152 does not alter the total amount of the other protein (SI Appendix, Fig. S9A), these results suggest that Cep192 and Cep152 play rather discrete but interconnected roles in Plk4 localization, and that Cep192 functions at a point upstream of Cep152 in this process.
Both Cep192 and Cep152 Are Required for Proper Recruitment of Sas6 to Centrosomes.
To further investigate the role of Cep192 and Cep152 in centriole biogenesis, the cells obtained from SI Appendix, Fig. S9A, were immunostained with a Sas6 antibody (SI Appendix, Fig. S9B). Results showed that ~71% of the cells silenced for the control siGL exhibited one or two Sas6 dots, whereas ~29% of these cells showed no detectable Sas6 signals (SI Appendix, Fig. S9D). Under these conditions, depleting Cep192 increased the fraction of cells with no Sas6 signals to ~62%, whereas depleting Cep152 increased this population to ~73%. Codepletion of Cep192 and Cep152 slightly increased the cells with no Sas6 to ~80% (SI Appendix, Fig. S9D). Although a modest increase (8–9%) in mitotic population was apparent for siCep192 and siCep152 cells, these cells did not exhibit a delayed G1 and S progression (SI Appendix, Fig. S9E), suggesting that the drastically reduced level of Sas6 signals in the siCep192 or siCep152 cells is not a result of an altered cell cycle. The requirement of Cep192 and Cep152 in recruiting Sas6 to centrosomes was confirmed by a second experiment that used cells overexpressing Plk4 (Fig. 4E), a condition that is sufficient to give rise to overduplicated centrioles (16, 17). Given that the depletion of Cep192 or Cep152 induces opposing effects on the level of centrosome-localized Plk4 (SI Appendix, Fig. S9C), the positive roles of Cep192 and Cep152 in the Sas6 localization are somewhat unexpected (as detailed later).
Interaction of the Acidic α-Helix Motif of Cep192 or Cep152 with Plk4 Is Critical for Normal Plk4 Localization to Centrosomes.
We next investigated whether the interaction of the acidic α-helix motif of Cep192 or Cep152 with Plk4 CPB is required for proper recruitment of Plk4 to centrosomes. To this end, U2OS cells expressing WT or a Plk4 binding-defective 2A mutant form of the RNAi-insensitive Cep192-sil or Cep152-sil allele (bearing no epitope tag was necessary to increase the functionality of exogenously expressed proteins) were treated with siCep192 or siCep152, respectively (detailed experimental procedure is shown in SI Appendix, Fig. S10). Under the conditions in which endogenous Cep192 or Cep152 was efficiently depleted, their respective exogenous proteins were RNAi-insensitive (Cep192) or less RNAi-sensitive (Cep152; Fig. 5A). Proper centrosomal localization of all the exogenously expressed proteins was verified by confocal analyses (SI Appendix, Fig. S11).
In control siGL cells, we observed that the expression of Cep192 or Cep152, but not its respective 2A mutant, significantly (26–29%) enhanced the level of Plk4 localized at centrosomes (SI Appendix, Fig. S12), demonstrating that Cep192 and Cep152 are capable of recruiting Plk4 to centrosomes. Consistent with these findings, depletion of Cep192 significantly lowered the level of Plk4 at centrosomes, whereas expression of Cep192 WT, but not its 2A mutant, rescued the Plk4 localization defect in the siCep192 cells (Fig. 5B; actual image is shown in SI Appendix, Fig. S13A). However, in line with the results shown in SI Appendix, Fig. S9C, Cep152 depletion in control vector-expressing cells greatly (~2.9-fold) increased the level of centrosome-associated Plk4. In addition, the cells expressing Cep152 WT, but not its respective 2A mutant, restored the level of Plk4 back to the level observed with the siGL cells expressing the control vector (Fig. 5B; actual image is shown in SI Appendix, Fig. S13B).
Cep192 Is Required for Proper Localization of Cep152.
Cep192 and Cep152 localize at the periphery of the outer wall of centrioles and exhibit toroid-like distributions with different diameters (25). Thus, we examined whether depleting one of these proteins would alter the localization of the other. In good agreement with previous observations (22), we found that depleting Cep152 did not influence Cep192 localization, whereas depleting Cep192 significantly delocalized Cep152 from centrosomes (Fig. 5 C and D). Furthermore, the expression of Cep192 effectively restored the Cep152 localization defect in the siCep192 cells, whereas the expression of the Plk4-binding-defective Cep192 (2A) mutant rescued it but somewhat less efficiently (Fig. 5D). These observations suggest that Cep192 and Cep152 are hierarchically organized in such a way that the recruitment of Cep152 requires the presence of the Cep192 toroid around the outer wall of centrioles.
Loss of Either the Cep192- or Cep152-Dependent Interaction with Plk4 Impairs Sas6 Recruitment and Cell Proliferation.
To investigate whether the Cep192- or Cep152-dependent interaction with Plk4 is important for centriole biogenesis, we used the cells in Fig. 5A to examine the level of Sas6 recruited to procentrioles. In control siGL cells expressing the vector, ~72% of cells exhibited one or two Sas6 signals (Fig. 5E; actual image is shown in SI Appendix, Fig. S14). Under these conditions, depleting Cep192 or Cep152 substantially increased the population of cells with no detectable Sas6 signals (72% or 82%, respectively). Expressing Cep192 or Cep152, but not its respective 2A mutant, efficiently rescued the Sas6 recruitment defect in these cells (Fig. 5E; actual image is shown in SI Appendix, Fig. S14). Notably, the presence of a near-normal level of Cep152 in the Cep192 (2A)-expressing cells in Fig. 5D suggests that the Sas6 recruitment defect associated with the Cep192 (2A)-expressing cells is likely Cep152 independent. As expected, if the Cep192- or Cep152-dependent interaction with Plk4 was critical to promoting centriole biogenesis, the Plk4-binding–defective Cep192 (2A) or Cep152 (2A) mutant failed to rescue the cell-proliferation defect associated with the depletion of Cep192 or Cep152, respectively (Fig. 5F). This finding is in good agreement with the observation that depletion of Cep192 or ep152 leads to abnormal spindle formation (20, 21, 24), and induces a mild but significant level of mitotic arrest (SI Appendix, Fig. S9E).
Discussion
Discovery of a Binding Motif of Plk4 CPB.
Although Cep152 has been shown to bind to Plk4 (20, 21), whether and, if so, how it promotes Plk4 recruitment to centrosomes remain unclear. Here, we showed that Cep192, which is required for proper PCM expansion and spindle assembly (23, 24), also plays a critical role in Plk4 recruitment through direct interaction. We further showed that a fragment present in the N-terminal region of Cep192 or Cep152 (DN80 or DN217, respectively) was sufficient to bind to the CPB of Plk4. This finding was largely in agreement with a previous report (22).
Comparative analyses of the primary sequences of DN80 and DN217 revealed the presence of acidic (D/E)-α-helix [or α-helix-acidic (D/E) region for DN217] and N/Q-rich motifs (Fig. 1B). The acidic α-helix motif appeared to be crucial for Plk4 binding, whereas the N/Q-rich motif was important but not decisive for the interaction (Figs. 1C and and2A2A and SI Appendix, Figs. S2 and S4). The significance of the α-helix for Plk4 binding was demonstrated by the observation that Cep152 (27–180) containing a truncated α-helix motif failed to bind to CPB (SI Appendix, Fig. S5). Notably, the α-helices neighboring the two homologous D/E-enriched motifs in DN80 and DN217 are dissimilar in their sequences. These observations led us to speculate that the D/E-dependent electrostatic interactions may serve as a high-affinity anchor, whereas the α-helix may provide specificity for the interaction. As the α-helices in DN80 and DN217 are positioned at the opposite ends of their D/E-enriched motifs, they may bind to Plk4 CPB in opposing directions. Determining the cocrystal structures of Plk4 CPB in complex with DN80 or DN217 may likely be important for providing new insights into the binding nature of these complexes.
How Cep192 and Cep152 localize around the cylindrical centriole structure and serve as centrosomal scaffolds for Plk4 recruitment remains to be further investigated. Interestingly, although Cep192 and Cep152 did not significantly interact with each other, each appears to efficiently interact with its own molecules in a manner that does not require Plk4 (Fig. 2D). These findings suggest that Cep192 and Cep152 may form two biochemically distinct homodimeric/multimeric complexes, even in the absence of Plk4 binding. In this regard, it will be interesting to investigate whether the homodimerization/multimerization of Cep192 or Cep152 is an important step for localizing these centrosomal scaffolds to centrosomes, and/or whether this step is a prerequisite event for Plk4 recruitment to this location.
Dynamic Localization of Plk4 During the Cell Cycle.
Earlier studies reported a dot- or ring-like Plk4 signal localized at or near the Cep152 toroid (22, 26, 27). Here, we closely examined the patterns of Plk4 localization during cell cycle progression from G1 to S. Our results showed that, in early G1, Plk4 associates with mother centrioles and then assumes a ring-like structure surrounding the proximal part of the centrioles (Fig. 4A and SI Appendix, Fig. S7). Sometimes, ring-like structures around mother and daughter centrioles were evident (SI Appendix, Fig. S8). Around the time of procentriole formation, Plk4 ring signals appeared to be replaced with two discrete dot signals, in which each dot localized to the site of future centriole assembly adjacent to each existing centriole (mother and daughter centrioles). The formation of Plk4 dot signals slightly preceded Sas6 recruitment, and both Plk4 and Sas6 signals were sharply colocalized with each other (Fig. 4 A and B). These findings suggest that subcentrosomal localization of Plk4 is temporally and spatially regulated during the early stages of centriole biogenesis. Remarkably, although Cep152 and Cep192 can directly interact with Plk4, Plk4 localizes only to a small, confined area within Cep192 or Cep152 toroids (SI Appendix, Fig. S8), strongly indicating the existence of an additional layer(s) of regulatory mechanism that restricts Plk4 localization to these locations. Further investigation on how Plk4 dynamically relocalizes from the surroundings of mother and daughter centrioles to the sites of future procentriole assembly will likely be a key step in better understanding the mechanism underlying the recruitment of Sas6 and its downstream components to this location.
It is interesting to note that, in advance of Plk4 recruitment, Cep192 localized to almost the entire length of mother and daughter centrioles (SI Appendix, Fig. S8A). On the contrary, Cep152 localized to the proximal end of mother centrioles before centriole duplication, and then localized to the same end of daughter centrioles apparently after Plk4 recruitment to this structure (SI Appendix, Fig. S8B, arrows). Hence, Cep192 is likely critical for early recruitment of Plk4 and subsequent assembly of the Cep152 toroid around Cep192-loaded centrioles. Following these events, Sas6 is recruited to daughter centrioles in a fashion that is almost completely colocalized with Plk4 (Fig. 4 A–C). Given that Cep192 and Cep152 are the two major scaffolds that are important for Plk4 localization (as detailed later), differential localization of these two scaffolds within centrosomes may provide mechanistic grounds for cell cycle stage-specific Plk4 localization.
Cep192 and Cep152: Two Hierarchically Assembled Centriolar Scaffolds Critical for Plk4 Recruitment.
Recent positional mapping analyses of various PCM components revealed that the PCM is a highly organized architecture comprising multiple layers of concentric toroidal structures, and that the Cep192 toroid is placed adjacent to the inner face of the Cep152 toroid (25, 28). Sonnen et al. showed that Cep192 interacts with Cep152, and that depleting both Cep192 and Cep152, but not Cep192 or Cep152 alone, greatly impairs Plk4 localization to centrosomes and centriole duplication (22). Based on these observations, they proposed a model in which Cep192 and Cep152 cooperate to recruit Plk4 to centrosomes and promote centriole duplication (22).
On the contrary, we found that Cep192 and Cep152 do not significantly interact with each other, but do interact with Plk4 in a competitive manner (Fig. 2 C and D and SI Appendix, Fig. S5C). These apparent differences in binding could be in part attributable to different buffer conditions used for washing [150 mM NaCl for this study vs. 300 mM NaCl for Sonnen et al. (22)]. Because Cep192 forms a distinct ring structure significantly smaller in diameter than Cep152 (25), Cep192 and Cep152 may not significantly interact with each other. As expected, if they functioned as important scaffolds for Plk4 binding, depleting Cep192 significantly disrupted Plk4 localization to centrosomes and impaired the recruitment of downstream components such as Sas6 (SI Appendix, Fig. S9 B–D). To our surprise, however, depleting Cep152 rather dramatically increased the level of centrosomal Plk4 because of the presence of Cep192, but failed to recruit Sas6 (SI Appendix, Fig. S9 B and C). Similar results were obtained with cells expressing a Plk4-binding–defective Cep192 (2A) or Cep152 (2A) mutant under the respective Cep192- or Cep152-depleted conditions (Fig. 5B). The inability of Cep152-depleted cells to recruit Sas6 suggests that the accumulation of Cep192-bound Plk4 alone is not sufficient to induce centriole biogenesis, and that the formation of the Cep152–Plk4 complex is critically required for the recruitment of downstream components important for centriole biogenesis. Consistent with this opinion, we observed that Cep152, but not Cep192, interacted with Cep135 (SI Appendix, Fig. S15), a component vital for proper procentriole assembly (29, 30). Thus, Plk4 localization to centrosomes is regulated through direct interactions with its two scaffolds, Cep192 and Cep152.
How, then, can the depletion of Cep152, but not Cep192, lead to an increased level of centrosome-localized Plk4 signal? One possibility is that Cep152 is present not only at centrosomes but also in cytosol, and that depletion of Cep152 unleashes Plk4 from the Cep152 tether and permits the resulting free Plk4 to bind to the Cep192 scaffold localized at centrosomes (Fig. 5G). In support of this model, whereas Cep192 was found to be present in the centrosomal fraction, Cep152 was present in the centrosomal and cytosolic fractions (Fig. 4D). Furthermore, as Cep192 and Cep152 toroids are hierarchically assembled in such a way that Cep192 is required for the centrosomal localization of Cep152 (Fig. 5 C and D), Cep192 depletion not only delocalizes Cep192-bound Plk4 but also cripples Cep152-bound Plk4, thus obliterating the increased Plk4 signal observed in the Cep152-depleted cells (SI Appendix, Fig. S9 B and C). This observation suggests that Cep192 functions as an inner platform that allows the recruitment of outer PCM components. In addition, as Plk4 is at a low level at M phase centrosomes (Fig. S6C) and coexpression of Cep192 stabilizes Plk4 (SI Appendix, Fig. S2 A and B and S3A), Cep192 may also serve as a centrosomal scaffold in early G1 that safeguards Plk4 from degradation.
It is notable that the siCep192 siCep152 doubly depleted cells showed a slightly enhanced defect in the recruitment of Plk4 and Sas6 compared with the siCep192 or the siCep152 cells (Fig. 4E and SI Appendix, Fig. S9 C and D). This could be because of the consequence of further loss of any remaining Plk4 or Sas6 in the Cep192- or Cep152-depleted cells by an additional reciprocal depletion of Cep152 or Cep192, respectively. In addition, the Cep192 (2A)-expressing cells in Fig. 5D exhibited a severely impaired ability to recruit Sas6, even in the presence of a near-normal level of centrosome-associated Cep152. This defect could be attributable to the diminished level of Plk4 in Cep192 (2A) cells (Fig. 5B), thus failing to generate a sufficient level of the Cep152–Plk4 complex to induce proper procentriole assembly.
Our hierarchical model of Cep192- and Cep152-dependent Plk4 recruitment predicts that Cep192 and Cep152 function at two different layers: Cep192 may play an initial role in the recruitment of Plk4 to centrioles, whereas Cep152 requires a Cep192-generated toroidal structure to help promote Plk4 localization and interaction with its downstream target(s) at this location. Thus, we propose that not only the assemblies of Cep192 and Cep152 toroids but also the functions of these proteins could be hierarchically regulated.
Implication of Cep192- and Cep152-Dependent Plk4 Interactions in Centriole Biogenesis and Cell Proliferation.
Plk4 haploinsufficiency has been shown to cause mitotic abnormalities, such as aberrant spindle formation, chromosome missegregation, and aneuploidy, which ultimately lead to the spontaneous generation of cancers in mice (31). These results suggest that a proper level of Plk4 is important for normal mitotic progression and genomic stability. Hence, our finding that Cep192 and Cep152 are the two major centrosomal scaffolds that regulate the recruitment of Plk4 to centrosomes is significant. More importantly, we have demonstrated that the impairment of the interaction between the CPB of Plk4 and either one of the acidic α-helix motifs of the two scaffolds is sufficient to induce a defect in the recruitment of Sas6, a critical component of centriole biogenesis, and to ultimately cause a diminished rate of cell proliferation (Fig. 5 E and F). Consistent with our observations, depletion of Cep192 or Cep152 has been shown to induce a defect in centriole duplication followed by segregation in mitosis, as evidenced by the appearance of centrosomes with single centrioles rather than pairs and monopolar spindles with a single centrosome (20, 21, 24). As expected, these defects are similar to those associated with the depletion of Plk4 (16). Moreover, similar centrosomal and spindle defects were observed in Cep152-deficient Seckel fibroblasts that show an aneuploidy in ~14% of the population (32). Therefore, further investigation on the mechanism of how Cep192 and Cep152 intricately regulate the function of Plk4 will likely be an important step to better understand the mechanism underlying abnormal centrosome duplication and genomic instability in human cancers.
Materials and Methods
Plasmid Construction, Cell Culture, and Lentivirus Production.
Detailed information on plasmid construction, cell culture, and lentivirus production is provided in SI Appendix, SI Materials and Methods.
Antibodies.
Rabbit polyclonal antibodies were generated against bacterially expressed His-Cep192 (1–647), His-Cep152 (491–810), His-Plk4 (580–970), or a synthetic Cep152 N-terminal peptide (QQLLTDLPHDMLDDDL; residues 31–46) at Young In Frontier (Seoul, South Korea). Affinity purification of these antibodies was carried out by using GST-Cep192 (1–647), GST-Cep152 (491–810), or GST-Plk4 (580–970) that was immobilized to Affi-gel 15 (Bio-Rad Laboratories), or using C-(CH2)6-QQLLTDLPHDMLDDDL that was immobilized to SulfoLink coupling resins (Pierce). An antibody detecting only the Cep192 (201–280) region was prepared by performing affinity purification of anti–His-Cep192 (1–647) serum with bead-immobilized GST-Cep192 (201–280). This antibody was used to comparatively determine the expression level of GFP-DN80 over its endogenous Cep192. The Cep152 N-terminal peptide antibody was used to determine the relative expression level of GFP-DN217 over its endogenous Cep152. For immunostaining analysis, the anti-Cep192 (1–647) or anti-Cep152 (491–810) antibody was conjugated with Alexa Fluor 488 by using the Alexa Fluor 488 protein labeling kit (Molecular Probes). All other primary antibodies used in this study were purchased from the companies listed in SI Appendix, Table S2.
Immunoprecipitation, Immunoblotting, and Quantification.
Immunoprecipitation was carried out as previously described (33). Immunostaining and signal quantification were performed essentially as previously described (34). Alexa Fluor 488 (green)-conjugated anti-Cep192 (rabbit) or anti-Cep152 (rabbit) antibody was used to avoid cross-reactivities with another rabbit antibody. Further information is provided in SI Appendix, SI Materials and Methods.
Three-Dimensional SIM.
To obtain higher-resolution images of the centrosomes, the cells in various stages of the cell cycle were imaged on an ELYRA S1 Super resolution microscope (Zeiss) equipped with Zeiss Alpha Plan-Apo 100× 1.46 oil objective, 405 nm/488 nm/561 nm/640 nm laser illumination, and standard excitation and emission filter sets. Sections were acquired at 0.1 μm z steps, and images were reconstructed and aligned by ZEN software (Zeiss). ImageJ was used to obtain maximal-intensity projections. We used NIS-Elements software (Nikon) to create a surface rendering of the reconstructed stacks for Fig. 4B.
Acknowledgments
This work was supported in part by National Cancer Institute Intramural Grants (to K.S.L. and J.L.), Korea Basic Science Institute Research Grant T33418 (to J.K.B.), and the World Class Institute (WCI 2009-002) Program funded by Ministry of Science, ICT, and Future Planning of Korea (to B.Y.K.).
Footnotes
The authors declare no conflict of interest.
This article contains supporting information online at www.pnas.org/lookup/suppl/10.1073/pnas.1319656110/-/DCSupplemental.
References
Articles from Proceedings of the National Academy of Sciences of the United States of America are provided here courtesy of National Academy of Sciences
Full text links
Read article at publisher's site: https://doi.org/10.1073/pnas.1319656110
Read article for free, from open access legal sources, via Unpaywall:
https://www.pnas.org/content/pnas/110/50/E4849.full.pdf
Citations & impact
Impact metrics
Citations of article over time
Alternative metrics
Article citations
CRL4DCAF1 ubiquitin ligase regulates PLK4 protein levels to prevent premature centriole duplication.
Life Sci Alliance, 7(6):e202402668, 15 Mar 2024
Cited by: 1 article | PMID: 38490717 | PMCID: PMC10942865
Centrosome amplification and aneuploidy driven by the HIV-1-induced Vpr•VprBP•Plk4 complex in CD4+ T cells.
Nat Commun, 15(1):2017, 05 Mar 2024
Cited by: 3 articles | PMID: 38443376 | PMCID: PMC10914751
Prolonged overexpression of PLK4 leads to formation of centriole rosette clusters that are connected via canonical centrosome linker proteins.
Sci Rep, 14(1):4370, 22 Feb 2024
Cited by: 1 article | PMID: 38388511 | PMCID: PMC10883960
Novel Variants of CEP152 in a Case of Compound-Heterozygous Inheritance of Epilepsy.
Glob Med Genet, 11(1):20-24, 16 Jan 2024
Cited by: 0 articles | PMID: 38229970 | PMCID: PMC10791487
A simple Turing reaction-diffusion model explains how PLK4 breaks symmetry during centriole duplication and assembly.
PLoS Biol, 21(11):e3002391, 20 Nov 2023
Cited by: 1 article | PMID: 37983248
Go to all (130) article citations
Data
Data behind the article
This data has been text mined from the article, or deposited into data resources.
BioStudies: supplemental material and supporting data
Similar Articles
To arrive at the top five similar articles we use a word-weighted algorithm to compare words from the Title and Abstract of each citation.
Human Cep192 and Cep152 cooperate in Plk4 recruitment and centriole duplication.
J Cell Sci, 126(pt 14):3223-3233, 02 May 2013
Cited by: 149 articles | PMID: 23641073
Molecular basis for unidirectional scaffold switching of human Plk4 in centriole biogenesis.
Nat Struct Mol Biol, 21(8):696-703, 29 Jun 2014
Cited by: 70 articles | PMID: 24997597 | PMCID: PMC4125498
Cep152 interacts with Plk4 and is required for centriole duplication.
J Cell Biol, 191(4):721-729, 08 Nov 2010
Cited by: 183 articles | PMID: 21059850 | PMCID: PMC2983069
Role of Polo-like Kinases Plk1 and Plk4 in the Initiation of Centriole Duplication-Impact on Cancer.
Cells, 11(5):786, 24 Feb 2022
Cited by: 7 articles | PMID: 35269408 | PMCID: PMC8908989
Review Free full text in Europe PMC
Funding
Funders who supported this work.





