Abstract
Free full text

Immunological and Virological Mechanisms of Vaccine-Mediated Protection Against SIV and HIV
Summary
A major challenge for the development of a highly effective AIDS vaccine is the identification of mechanisms of protective immunity. To address this question, we used a non-human primate challenge model with simian immunodeficiency virus (SIV). We show that antibodies to the SIV Envelope are necessary and sufficient to prevent infection. Moreover, sequencing of viruses from breakthrough infections revealed selective pressure against neutralization-sensitive viruses; we identified a two amino acid signature that alters antigenicity and confers neutralization resistance. A similar signature confers resistance of HIV-1 to neutralization by monoclonal antibodies against variable regions 1 and 2 (V1V2), suggesting that SIV and HIV share a fundamental mechanism of immune escape from vaccine- or naturally-elicited antibodies. These analyses provide insight into the limited efficacy seen in HIV vaccine trials.
Among the five human efficacy trials of HIV-1 vaccines, only one has shown some success in preventing HIV infection. In the RV144 trial, a combination viral vector and protein immunization achieved a modest 31% efficacy in a cohort of low risk adults in Thailand1. In-depth immunological correlates analysis suggested that specific antibody responses to the HIV-1 envelope variable regions 1 and 2 (V1V2) region correlated with protection while an IgA response showed a negative association2,3. Virus sequencing of the breakthrough infections in RV144 suggested a possible vaccine mediated selection pressure against certain virus variants4; the mechanism of immune pressure remains elusive, but may include elicitation of antibodies targeting V1V2 of those variants5. In contrast, the recent HVTN 505 trial, using a DNA-prime, recombinant adenovirus type 5 (rAd5) boost, was halted for futility with no vaccine efficacy6.
Infection of nonhuman primates with SIV represents the best available animal model for testing vaccine concepts for protecting against HIV infection, and mucosal challenge with SIV can be used to model human mucosal HIV exposure7. Several SIV challenge studies have shown partial protection from acquisition; in some cases, there has been an association to elicited antibodies, but a strong immunological mechanism or correlate has not been identified8–13. Here, we used a repetitive intra-rectal challenge using an SIV E660 challenge virus that was unmatched to the vaccines14. The E660 virus swarm is heterogeneous, comprising groups or clusters of viruses ranging from neutralization sensitive to resistant15. We reasoned that, in the absence of complete protection, the naturally occurring diversity of neutralization profiles would provide the most informative correlates analysis.
Our goals were to define cellular and humoral immune correlates of immunity, and to understand the mechanism leading to protection against SIV infection. Our immunogens included “T-cell mosaics” designed to optimize coverage of epitope diversity for cellular responses16,17. We designed a four arm study to define mechanisms of vaccine protection: (i) mosaic Gag; (ii) mosaic heterologous envelope (Env); (iii) heterologous Env based on a natural SIV mac239 sequence; and (iv) control vaccine. Our primary questions were: (1) Is Env immunization sufficient and/or necessary to provide protection against acquisition?; (2) Does Gag (alone) immunization provide any protection against acquisition?; and (3) Does the use of “T cell mosaic” Envs provide additional benefit over a natural Env sequence?
The number of acquisition endpoints in this study was similar to a large human efficacy study. We demonstrated that an Env-elicited immune response is necessary and sufficient to provide protection from acquisition. Importantly, by integrating immunological and virological analyses, we elucidated antibody mediated mechanisms of protection and discovered a fundamental mechanism of virus escape from antibody-mediated control, shared by SIV and HIV, that has broad implications for understanding vaccine mediated protection and potentially for vaccine design.
Vaccine Immunogenicity
80 Indian origin rhesus macaques were enrolled in a DNA prime, rAd5 boost immunization study. Animals were randomized into four groups of 20 based on TRIM5α alleles, gender, age, and weight. All animals received three shots of DNA at 4 week intervals, followed by rAd5 at week 3014. The control group received vectors that contained no inserts; the second group (“mosaic Gag”) received 2 SIV Gag mosaic immunogens17; the third group (“mosaic Env”) received 2 SIV Env mosaic immunogens (78% and 87% sequence identity to E543, a clone similar to E66016); and the fourth group (“mac239 Env”) received an immunogen encoding SIVmac239 Env (83% sequence identity to E543). Envelope sequences are shown in Supplementary Table 1, and sequence distances in Supplementary Table 2.
Vaccination elicited the expected cellular (Extended Fig 1) and humoral (Extended Fig 2) responses. Notably, compared to mac239 Env immunization, mosaic Env induced modestly lower and qualitatively different humoral responses (Extended Fig 2). Mapping of the antibody response to unglycosylated linear peptides (Extended Fig 2c) revealed that mac239 Env elicited a broader response than mosaic Env. Overall, immunization elicited mild neutralization activity against a limited set of viral strains (Extended Fig 2e–g).
SIV Challenge Outcome
To test vaccine efficacy against infectious challenge, we exposed animals weekly to intrarectal administration of E660 at a dose that infects ~30% of control animals per exposure14. Each animal was challenged up to 12 times or until it had detectable plasma viremia. Immunization with mac239 Env provided significant protection against acquisition, whereas mosaic Env immunization did not achieve significance (Fig 1a). There was no difference in acquisition between Gag-immunized animals and control animals. For protection against acquisition, vaccine efficacy (VE: the reduction in the rate of infection at each challenge)18,19 was 69% for mac239 Env (Fig 1d).

a, The fraction infected animals in each arm following each of 12 challenges is shown. Five animals in the mac239 Env arm and one animal in the mosaic Env arm remained uninfected after 12 challenges. b, For each arm, the geometric mean plasma viral load (RNA copies/ml) for infected animals is shown. Each animal is synchronized to its peak VL. Inset: expanded scale for the acute phase. c, The peak and set point plasma viral load distributions for all infected animals. d, The infection rate is the fraction of infections out of the total number of exposures; vaccine efficacy was calculated as described in the methods.
All infected animals that received active immunization showed 0.7–1.1 log decrease in peak viral load (VL) on average (Fig 1b,c; Extended Fig 3b). The best control of acute VL occurred in the mosaic Env arm, while the mosaic Gag arm showed the best long-term control (Fig 1D). We confirmed previous findings that animals with certain alleles of TRIM5α showed better innate control of infection and pathogenesis14 (Extended Fig 3d, e). Due to the stratification by TRIM5α alleles in our study, including this factor as a covariate in analyses does not impact our conclusions. All three vaccine arms showed protection against loss of CD4 cells (Extended Fig 3c). Thus, the mosaic Env constructs elicited effective T cell responses that protected against pathogenic effects of infection despite their inability to block acquisition.
Transmitted founder analysis
Because E660 is a viral swarm with >3% sequence diversity, the number of transmitted founder (T/F) viruses can be determined by single genome amplification (SGA). For every infected animal, sequencing was done on plasma from the earliest time point with detectable plasma VL, one week after infection: thus, the inferred sequences represent the original infecting viruses7. Both Env arms showed a significant decrease in T/F variants (Fig 2a). From these data, an efficacy can be calculated by the reduction of T/F variants per challenge; theoretically, this value estimates VE for a very low (clinically-relevant) infectious dose. Immunization with mac239 Env reduced the number of T/F variants by 81%, and mosaic Env reduced T/F by 51% (Fig 2b).

a, The distribution of unique T/F viruses in the first virus-positive plasma sample is shown for all 80 animals. b, The average number of T/F viruses per exposure event was calculated. Here, vaccine efficacy (VE) is computed as the reduction in the number of T/F viruses (ns: p > 0.05). c, For each position in Env, the p value is shown for a permutation test comparing the fraction of viruses with the consensus amino acid in the Env T/F vs the control and Gag T/F. p values at positions 23, 45, and 47 remain significant after correction for multiple comparisons. d–f, Based on the sequence at positions 45 and 47, T/F viruses were divided into “TR” (45T+47R) and “A/K” (45A or 47K) viruses. d, Proportion of A/K viruses in the E660 challenge stock was measured by deep sequencing or by SGA, and among T/F in the immunization arms by SGA. A Fisher’s exact test was performed to determine the significance of the difference in A/K viruses compared to the Control+Gag arms (ns: p > 0.05). e,f, Cumulative infection probabilities by TR or A/K viruses was done using a non-parametric estimate for competing risks23; the VE and p-values are computed using likelihoods from a modified Hudgens and Gilbert leaky vaccine model18. Tick marks indicate censoring of animals solely infected by the other virus type (challenges 1–12), or remaining uninfected after 12 challenges.
Phylogenetic analysis using all complete Env sequences did not reveal an obvious clustering of T/F variants by vaccine arm. However, a strong “sieving” effect was discerned by examining individual amino acid variants. Over the Env coding sequence, the 133 T/F sequences showed variation at 63 sites (Supplementary Table 3); 20 positions in the cytoplasmic domain or with rare variation (<5 of 133 T/F) were excluded from further analysis. Among the remaining sites, we found significant differences in variant representation in the Env vaccinated arms compared to the control and Gag arms (Fig 2c; Extended Fig 4). The strongest effect was seen at positions 23, 45, and 47. The consensus T/F sequence at these positions (VTR) was found in a majority of T/F viruses in the control and Gag arms. In contrast, variant sequences (IAK) were significantly overrepresented in the Env immunized arms. Thus, immunization with Env sequences induced an immune response selecting against virions with the VTR signature.
Mechanism of Virus Selection in Vaccinees
In order to define the mechanism of vaccine-mediated selection against viral variants, we measured the neutralization profile of all 40 Env immunized animals against pseudo-typed viruses. CP3C-P-A8 (“CP3C” for brevity), a clone from the E660 swarm, is a neutralization-sensitive virus and has the amino acids VTR at positions 23, 45 and 47. CR54-PK-2A5 (“CR54”), another clone from the E660 swarm, is a neutralization resistant virus with IAK at these positions. Sera from immunized animals completely neutralized CP3C, with an inhibitory concentration potency (ICHM, defined as the dilution giving half-maximal inhibition) that varied 1000-fold (Fig 3a). In contrast, the same sera only achieved a maximum of ~50% inhibition of CR54. Importantly, this shows that CR54 is a heterogeneous population of virions despite being genetically clonal: half of the virions are easily neutralized by antisera, and half are completely resistant.
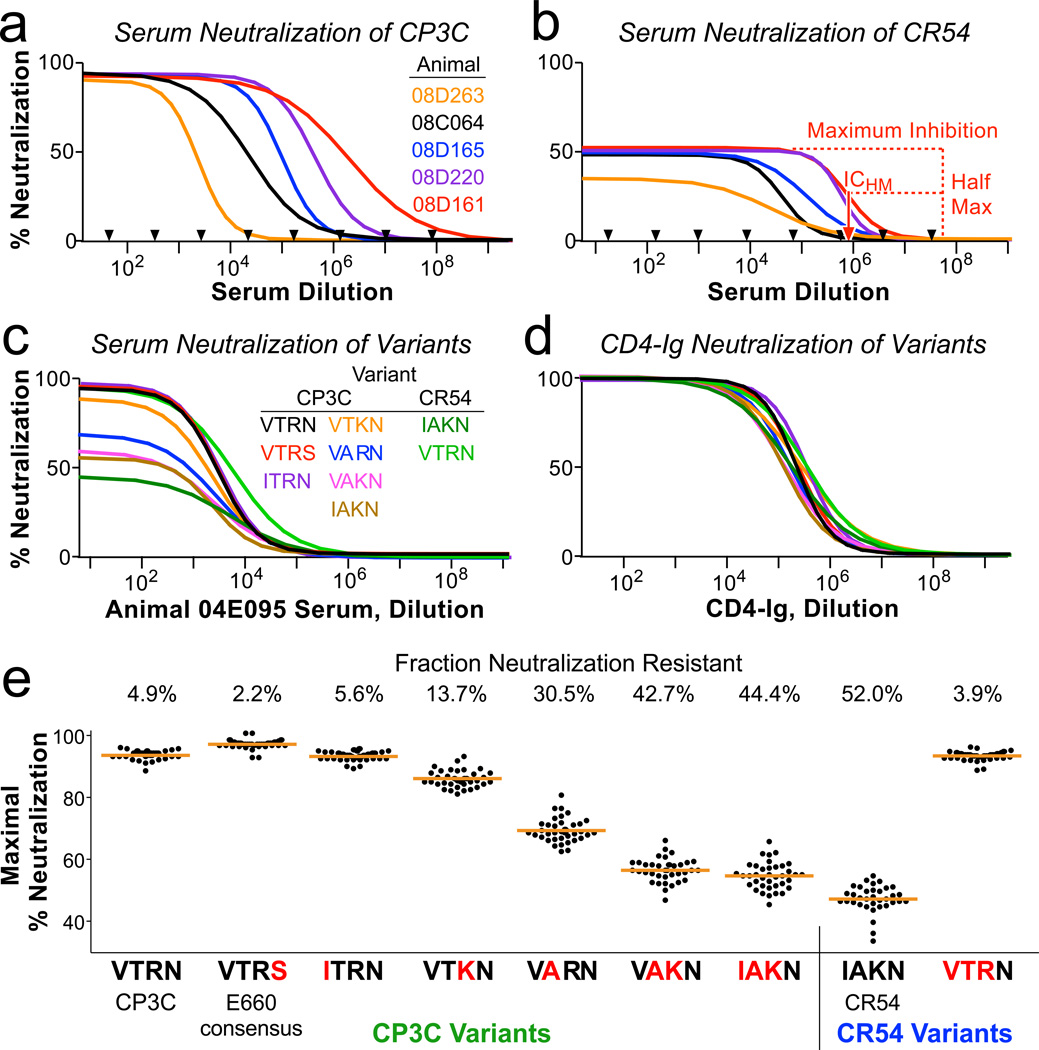
a,b, Neutralization curves of CP3C, a sensitive clone from E660 (a), and of CR54, a resistant clone from E660 (b), using dilutions of sera from five Env-immunized animals (selected to show the range of potency). Black arrows indicate which dilutions were tested in duplicate; curves represent non-linear least squares regressions of a four-parameter binding model. Nearly 100% of CP3C virions, but only 40–50% of CR54 virions, can be neutralized by immune sera. Red dashed lines show how ICHM is derived for animal 08D161. c,d, Neutralization curves of 9 viral variants using sera from one animal (c) or CD4-Ig (d). The parent virus into which mutations were made is listed, along with the amino acids at positions 23, 45, 47, and 70. e, All variants were assayed using serial dilutions of sera from all 40 Env-immunized animals. Shown is the maximum fraction of each virus that was neutralized (determined by regression analysis). Blue letters indicate amino acid substitutions compared to the parent virus. The numbers above the graphic indicate the mean resistant fraction for each virus.
We introduced variants of these four amino acids into CP3C and CR54 Env to test which might be responsible for modulating neutralization resistance. These amino acid variations did not change the potency of the antisera (ICHM varied <2-fold: Fig 3c and Extended Fig 5a), and did not change the sensitivity to neutralization by CD4-Ig (Fig 3d). However, variant sequences affected the fraction of neutralization-resistant virions.
Despite the wide breadth of epitopes targeted by vaccine-elicited antisera (Extended Fig 2), there was little variation in the fraction of each virus that was neutralization resistant (Fig 3e). Moreover, an identical neutralization-resistant fraction was observed for a panel of SIV monoclonal antibodies directed near the CD4 binding site or the V1V2 loop (Extended Fig 5b,c). Therefore, even clonally-derived virions are remarkably heterogeneous: a fraction are easily neutralized, and the remainder are completely resistant to antibody-based neutralization.
Generation of this resistant Env phenotype was favored by amino acid substitutions in the C1 region. By making point-mutations, we showed that the T45A and R47K mutations individually result in increased resistance. Together, changing these two amino acids converts the sensitive CP3C Env to a nearly fully-resistant phenotype, and the resistant CR54 to fully-sensitive. For parsimony in subsequent analyses, we divided E660 viruses into two categories: viruses with 45T and 47R (“TR”) that are putatively neutralization sensitive, and viruses with 45A or 47K (“A/K”) that should be generally resistant to vaccine-elicited sera.
Deep sequencing and SGA of Env genes showed that ~20% of the E660 challenge swarm were neutralization-resistant A/K viruses (Fig 2d). The same proportion was found among infecting T/F sequences in the control and Gag arms, demonstrating that there is no innate selection for or against A/K sequences. Furthermore, A/K infections resulted in the same peak and setpoint plasma VL, suggesting that these viruses are no more or less fit than TR viruses (Extended Fig 6). However, vaccine-elicited responses strongly selected against infection by TR viruses – such that, in the mac239 Env arm, the infrequent (neutralization-resistant) A/K variants comprised nearly 75% of T/F viruses.
We next computed the VE against A/K and TR viruses separately. The TR (sensitive) variants are highly susceptible to vaccine-mediated control, with a VE of 90% (Fig 2e). In contrast, the VE against A/K viruses did not reach significance (Fig 2f). Thus, the heterogeneous neutralization of even clonal SIV virions, programmed by C1 amino acid variations, represents a novel mechanism of immune escape from Env-specific antibodies.
Immune correlates of risk of infection
A panoply of cellular and humoral assays quantifying vaccine-elicited responses were performed at baseline, peak post boost, and pre-challenge time points. We found strong associations between several antibody responses and probability of infection, but no associations between T cell responses and delayed acquisition.
Given that the E660 swarm is comprised of both neutralization sensitive (TR) and resistant (A/K) genotypes, it made sense to analyze correlates in two ways: first, by including all infections, irrespective of variant; and second, by separating the two types of infections. Since the vaccine is largely ineffective against A/K viruses, pooling A/K-infected with TR-infected animals may mask potential correlates.
The data shown in Fig. 4 illustrate these analyses. Among all 40 Env-vaccinated animals, plasma IgG binding to CP3C gp120 Env at the time of challenge did not correlate significantly with time to infection (Fig 4a). In contrast, when we exclude animals who were infected solely with A/K viruses, we found a strong correlation with IgG binding to CP3C gp120 (Fig 4b), but not other Envs (Extended Fig 7a,b). We grouped all Env-immunized animals by those with an IgG response to CP3C above or below 570 (the median value, corresponding to an endpoint titer of ~1:1000). Animals with the higher response had a 75% lower rate of infection by TR viruses (Fig 4c).

a,b, Week 52 plasma IgG against the CP3C envelope is graphed against time to infection (uninfected animals were assigned a value of 13). No significant correlation was found when all infection events were considered (a); however, by excluding infected solely with A/K viruses, a strong predictive relationship is seen (b). The line is from a linear regression; statistics are based on Spearman correlation. c, Week 52 plasma IgG against the mac239 V1V2 is significantly associated with protection against TR viruses, and also against all viruses (Extended Fig 7B). d, Kaplan Meier (KM) analysis was performed by dividing the 40 Env-immunized animals in two equal groups based on the anti-CP3C IgG responses (median = 570). Animals remaining uninfected or infected solely with A/K viruses were censored as shown by vertical lines. e, KM analysis comparing Env-immunized animals with higher vs. lower week 32 serum activity against the CD4 binding site of envelope. f, KM analysis comparing Env-immunized animals with higher vs. lower week 52 against virus pseudotyped with a CP3C Env containing a T45A mutation (“VARN”), a sequence shared by E543. g, The mean response (upper) and proportion of responders (lower) against each linear peptide is shown for animals grouped by time to infection: 1–3 challenges (red) vs 4 or more challenges (blue). Green boxes highlight regions potentially associated with protection identified by a Fisher’s exact test; overlapping peptide numbers are in green, with sequences given in Supplementary Table 5. h, Average binding to the linear C3 peptides 119 and 120 correlates strongly with time to infection. i, KM analysis comparing Env-immunized animals with a positive response to C3 peptides to those with a negative response. j, KM analyses comparing all animals in the control and Gag arms (black), all Env-immunized animals having a CP3C IgG response <570 and a negative C3 peptides response (grey), and animals in either Env arm having either antibody response.
Correlation with time to infection was also observed for plasma antibody avidity (Extended Fig 7e), CD4-binding site activity (Fig 4e), and neutralization of some viral strains (Fig 4f). These data suggest that the quality of the antibody response is important. Thus, we investigated binding to specific regions within the Env.
By comparing peptide binding data for animals grouped by time to infection (Fig 4g), we identified four linear epitopes possibly associated with protection. There was a strong association between the breadth amongst these four epitopes and time to infection (Extended Fig 8a–c). In contrast, there was no significant association with the breadth of response across all Env epitopes (Extended Fig 8d). Thus, both breadth and magnitude of the response to selected epitopes are strong correlates of protection from acquisition.
The response to C3 (peptides 119+120) was the most significantly associated with protection, whether all viruses (Extended Fig 8e,f) or just TR viruses were considered (Fig 4h,i). This epitope corresponds to the α2 helix of Env and was identified as a neutralization target in HIV-120,21. In a multivariable model, both IgG to CP3C (p = 0.004) and binding to the C3 peptides (p = 0.02) provided independent prediction of time to infection. We thus compared animals that had neither (i) a response to the C3 peptides, nor (ii) IgG to CP3C (n = 12, combining both Env arms), to animals with either response (mac239 Env: n = 19/20; mosaic Env: n = 9/20). For animals with neither antibody response, the rate of infection (12 infections in 27 exposures, 44%), and the proportion of infections with only A/K viruses (3/12, 25%) was not different from the control (unvaccinated) or Gag arms. In contrast, animals with either antibody response were primarily infected with resistant A/K viruses, and the VE was >90% against TR viruses (Fig 4j).
V1V2 and vaccine-specific sieving
In the human RV144 trial, antibody binding to HIV V1V2 was a primary correlate of protection against infection. Similarly, antibody to the SIV V1V2 predicted protection against infection (Fig 4c). The mosaic and mac239 immunogen sequences varied significantly in this region (Supplentary Table 1), and consequently elicited somewhat different antibody responses (Fig 5a). To determine if these responses are associated with sieving, we analyzed variation in T/F sequences (as in Fig 3), after censoring vaccine-nonresponsive A/K viruses. This analysis revealed a strong selection associated with position 162 (Fig 5b). The mosaic immunization completely selected against TR viruses with 162N (0/14 viruses, compared to 17/34 in the control; Fig 5c). In contrast, the mac239 immunogen may have selected against the other variant (162S), although there were too few TR virus infections in this group to reach significance. Amongst A/K virus infections, there was no significant difference in representation of the 162N/S variants in across vaccine arms. These data show that selection against V1V2 sequences by the SIV vaccines is limited to neutralization-sensitive viruses, and, within those, selection is vaccine sequence-specific and thus not broad.
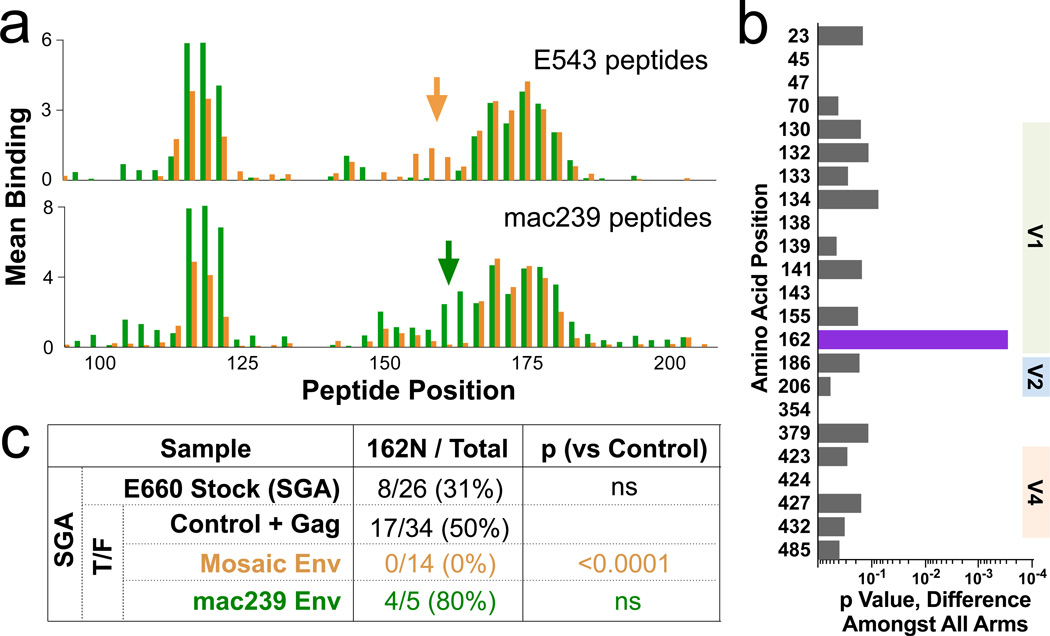
a, The binding of plasma from all 40 Env-immunized animals to linear 15mer peptides spanning the V1V2 region of either E543 (top) or mac239 (bottom) was measured; bars represent the average binding for the 20 mosaic- (orange) or the 20 mac239- (green) immunized animals. Arrows indicate an area of V1V2 showing vaccine-specific responses, encompassing amino acids 154–170. b, Sieving analysis was done as in Fig 2b, but after excluding neutralization-resistant A/K viruses. The only significant association with immunization arm was at position 162. c, Representation of 162N (vs 162S) as determined by SGA for TR viruses in the swarm or in T/F. Note that all immunogens encode 162N, so selection is likely mediated against a neighboring epitope; this epitope is found only in (one of) the mosaic immunogens, and occurs in linkage disequilibrium with 162N in the E660 swarm.
Antibody escape mechanism in HIV
To assess whether our findings extend to HIV, we measured the inhibition of 51 distinct HIV-1 envelope pseudotyped viruses by the V1V2-specific monoclonal antibodies PG9 and PG16. As we saw for neutralization resistant A/K SIV viruses, neutralization of some clonal HIV strains was incomplete; i.e. a fraction of virions could not be neutralized (Fig 6a). We examine the influence of sequence variation of these HIV envelopes on the fraction of neutralization-resistant virus (Fig 6b); the most significant association was at position 47, with 47R viruses being sensitive (Fig 6c). Sequence alignment with SIV envelope shows that position 47 in HIV is in a similar area of C1 as is position 47 in SIV (Fig 6d); the similar signature (arginine vs lysine) suggests that a common mechanism of neutralization escape may be shared by SIV and HIV.
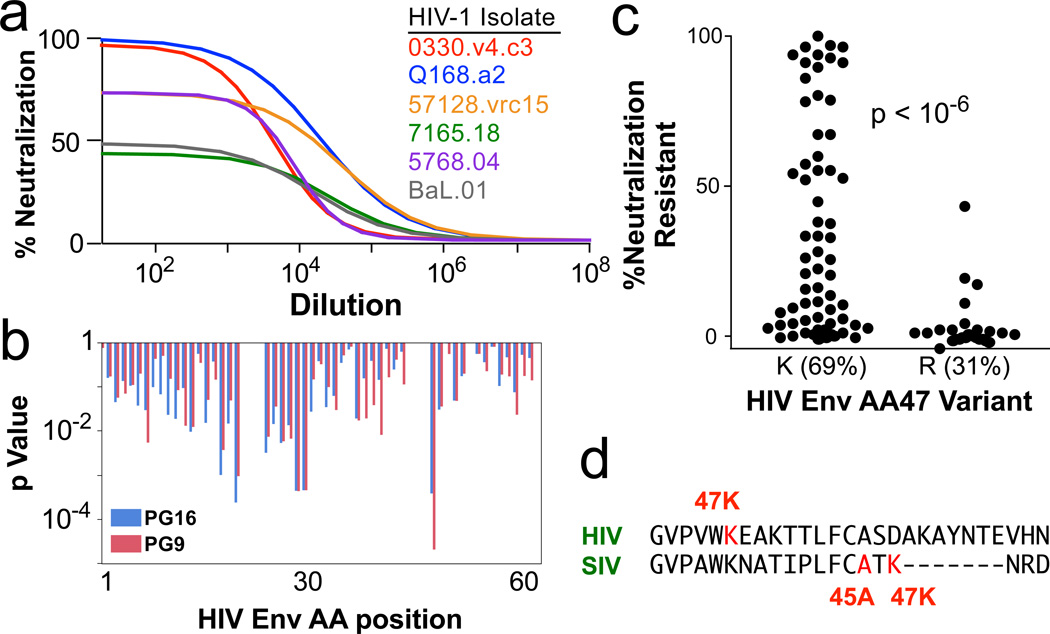
a, Neutralization profiles of 51 different HIV-1 strains by the V1V2 antibodies PG9 and PG16 were determined. Example curves of PG16 on six viruses are shown. As for SIV A/K viruses, a variable fraction of each clonal virus is completely neutralization resistant; the remainder is highly sensitive. b, The influence of variants at each position in envelope on the fraction of neutralization resistant virus is shown as a p value from a Fisher’s test; shown is the C1 region. c, The most significant association for all positions in Env was amino acid 47. The distribution of the fraction of neutralization resistant virus is shown for the two variants, 47K and 47R. d, Alignment of SIV and HIV Env proteins in the middle of the C1 region highlighting the positions of the neutralization signatures.
Discussion
Immune correlate studies that interrogate both virus sequences and immune responses can provide key insights on mechanisms of protection from HIV-1 acquisition. Using a nonhuman primate model with a number of acquisition endpoints similar to large human efficacy studies, we demonstrated that an Env-elicited immune response is necessary and sufficient to provide protection from acquisition. We identified antibody-based correlates including responses to several epitopes. In our study, SIV Env T cell mosaic immunogens elicited more effective T cell responses, but less effective antibody responses. With respect to the virus, we identified a strong sieving effect of Env immunization, selecting for minor variants in the challenge swarm. And finally, we identify a sequence signature in the SIV Env, possibly shared by HIV, that programs the neutralization phenotype of the viruses through a mechanism affecting the entire antigenic surface of the protein.
Among our three vaccine groups, there was no association between protection from infection and protection from pathogenesis (e.g., VL control). This suggests that humoral responses that effectively block acquisition are not necessarily correlated with cellular responses that control pathogenesis. Furthermore, we show that the Env-induced CTL suppressed acute viremia better than Gag CTL, but suppressed chronic viremia less effectively (Fig 1b,d). Our data also show that vaccination resulted in reduced T/F viruses in breakthrough infections. This suggests that the primary mechanism of protection is by lowering the effective infectious dose, i.e., in vivo neutralization.
Analysis of the sequences of breakthrough viruses revealed an amino acid signature, in the C1 region of Env, of viruses more likely to escape this neutralization. By creating point mutations that interconverted the neutralization profile of well-characterized viral envelopes, we defined a minimal two amino acid signature at positions 45 and 47 (TR vs A/K). Importantly, introduction of the A/K signature resulted in a fraction of clonally-derived Env proteins having a “global” antigenic change. This was manifest as resistance to polyclonal sera from dozens of animals, as well as resistance to monoclonal antibodies directed to the CD4 binding site or the V1V2 loops. Thus, the mechanism of resistance likely includes post-transcriptional modification, such as alternative glycosylation or folding, capable of masking the majority of epitopes on the viral Env.
We identified a hierarchy within this neutralization escape mechanism. This phenotype can occur for only a specific domain of the Env, such as for V1V2-directed antibodies against SIV (Extended Fig 5b) and HIV (Fig 6). This probably occurs through alternative glycosylation pathways restricted to this site. Resistance can also be global, affecting virtually any epitope, as we show for the SIV envelope (Extended Fig 5a). A hierarchy was observed in vivo, in that sieving at the V1V2 domain was only observed in viruses lacking the global resistance phenotype (i.e., in TR but not A/K viruses).
The observation that C1 amino acid variations can lead to alternative Env structures is consistent with data from Del Prete et al.22, who found that a single amino acid substitution could confer co-receptor dual tropism on mac239. Strikingly, the mutation responsible for the altered structure at the distant V3 loop was 47E – i.e., within the signature we identified as conferring altered antigenicity upon SIV Env.
It is notable that all viruses in the mac251 swarm contain the resistant A/K signature. This may account for the weak correlation with vaccine-induced antibody in previous studies11,14. It is likely that the resistant Env form can be neutralized by antibodies targeting “sites of vulnerability” (i.e., rare epitopes conserved across all structures); for SIV, as it is in HIV21, one of these may be the α2 helix. Antibody responses to this peptide were not only highly correlated with protection against TR viruses (Fig 4G), but also showed a trend for protection against infection with the A/K viruses (p = 0.07). Likewise, the Ig-CD4 molecule fully neutralized A/K viruses, suggesting that an appropriately-targeted antibody to the CD4 binding site could have a similar effect.
By restricting our correlates analysis to exclude infections resulting from neutralization resistant viruses (which are insensitive to the vaccine responses), we identified multiple strong correlates of risk of infection. All of these correlates derive from antibody measures, and include the magnitude of binding, the avidity of binding, and the breadth to selected epitopes of the SIV envelope. The importance of taking into account the virology is underscored by our analysis of the mosaic arm: despite this arm not achieving statistically significant protection overall (Fig 1), we could identify active immune mechanisms (Fig 4) as well as identify a mosaic immunogen-specific sieving effect in V1V2 (Fig 5).
Our study provides insight into the possible reasons for the failure of HVTN505 and the limited protection in RV1441. Vaccination using our specific SIV Env expression vectors generated an antibody response ineffective against specific variants and protected against the subset of neutralization sensitive viral variants (Fig 2e). Based on data here, we hypothesize that HVTN505 failed due to an inability to elicit antisera that completely neutralized circulating HIV-1 strains, which are primarily neutralization-resistant. In contrast, the moderate success of RV144 suggests that antibodies were elicited that could neutralize some viruses circulating in that cohort; these sensitive viruses were susceptible to the vaccine-matched V1V2, leading to sieving. In any case, it will be critical apply integrated analyses to HIV vaccine trials similar to what we did for this SIV study: i.e., to clone and determine the neutralization profile of T/F viruses in the placebo arms (defining resistance of the circulating strains) and the active arms (to determine if the vaccine selectively blocked a subset of viruses), to optimally assess factors associated with vaccine-mediated protection.
Deciding which vaccine products to advance into large, expensive efficacy trials is difficult and complex. It is reasonable to postulate that any highly efficacious candidate will need to elicit antibodies targeting universal sites of vulnerability (i.e., epitopes shared by the heterogeneous forms of even clonal virions), or to separately elicit antibodies targeting each structural form. Thus, understanding the biophysical basis for this viral heterogeneity will be crucial for designing vaccines capable of completely blocking HIV.
In conclusion, we identified a sequence signature of the SIV Env that distinguishes broadly neutralization-resistant viruses. By taking this signature of T/F viruses from break-through infections into account, we found multiple strong correlates of risk of against infection, all based on antigen-specific antibody measurements – even for a vaccine arm that did not, upon initial analysis, reach statistically-significant protection. We found that this signature, while likely not unique, is shared by SIV and HIV, and may underlie a fundamental mechanism of immune escape in both vaccinated and naturally-infected subjects. Finally, our combined virological and immunological analyses provide insight into the biology of vaccine-mediated control, and lay a foundation for analysis and advancement of future HIV vaccines.
Methods
Animals
Animals were handled in accordance with the standards of the American Association for the Accreditation of Laboratory Animal Care (AAALAC) and meet NIH standards as set forth in the Guidelines for Care and Use of Laboratory Animals. The animal protocol, VRC 10–332, was approved by the Vaccine Research Center IACUC. All animals were Indian-origin rhesus macaques, male or female, approximately 3–5 years of age. Animals selected for the study were negative for MHC class I alleles Mamu-A01, -B08, and -B1725. Animals were typed by PCR for Trim5α alleles, and categorized as having 0, 1, or 2 restrictive alleles26. 80 animals were randomized into four arms based on the following criteria: Trim5α allele category, gender, and weight. Blood was collected at regular intervals (weekly or biweekly). PBMC were prepared; a small number were reserved for phenotyping for absolute cell counts, and the remainder were viably cryopreserved (in fetal bovine serum containing 10% DMSO and stored in liquid phase nitrogen until analysis). Plasma was frozen at −80C for virological and serological analysis. Sample size (n=20 per arm) was chosen to have an 80% probability to detect a vaccine efficacy of 50%18.
Immunization
The design of the mosaic immunogens has been previously described16. Briefly, an input data set was assembled to include all available SIVsmm complete genome sequences that were either directly isolated from sooty mangabeys or (in a small number of cases) had been minimally passaged in tissue culture. So that viral challenges could reasonably approximate real-world post-vaccination exposure to unknown virus strains, we specifically excluded from the mosaic sequence input all sequences from the mac251 lineage (including mac239) as well as isolates of (and closely related sequences to) smE660. Mosaic sequence cocktails were generated sequentially, so that a single-sequence mosaic was generated first, and a second sequence was subsequently generated as the best complement to the first27,28. Coverage values of potential T-cell epitopes (as amino acid 9-mers) have been published16. Mosaic coding sequences were introduced into the same DNA and rAd5 backbones as the mac239 Env. All rAd5 vectors were produced by GenVec (Rockville, MD). The mosaic Env were given as gp160 in both the DNA prime and the rAd5 boost. The mac239 natural Env immunogens (DNA and rAd5) are identical to what was previously used14, and was given as a gp140 in the DNA prime, and a gp145 in the rAd5 boost.
Functionality of all immunogens (mac239 and mosaic, Env and Gag) was confirmed by multiple assays. Expression of Env and Gag from DNA and rAd5 vectors in 293 cells in tissue culture was assessed by Western blot analysis, and was found to be comparable. Immunogenicity of each vector/insert combination was confirmed in mouse studies.
Primates were immunized as previously described14: a total of four times; 4 mg of DNA was given intramuscularly (IM) at weeks 0, 4, and 8 and 1010 particles of rAd5 was given IM at week 28. The two Gag mosaic immunogens were mixed prior to administration, as were the two Env mosaic immunogens.
SIV Challenge
Animals were challenged weekly with a dose of smE660 previously shown to infect unvaccinated animals approximately 30% per exposure, as previously described14. Weekly challenges were initiated at week 53 (6 months after rAd5 boost), and were halted when an animal became PCR positive for viral RNA, or after 12 exposures. There was no statistically significant change in the rate of infection within any group over time, indicating that infection was stochastic and there was no selection for innately-resistant animals.
Assays
All immunological and virological assays performed for correlation analyses were performed by investigators blind as to group assignment and challenge outcome.
Virology
To quantify SIV viral load, viral RNA from plasma was isolated using a Qiagen (Valencia, CA) QIAsymphony Virus/Bacteria Midi kit on Qiagen’s automated sample preparation platform, the QIAsymphony SP. Viral RNA from 500uL of plasma was eluted into 60uL of elution buffer. All subsequent reactions were setup using Qiagen’s automated PCR setup platform, the QIAgility. 25uL of viral RNA was annealed to a target specific reverse primer 5’- CACTAG GTGTCT CTG CACTAT CTG TTT TG -3’ then reverse transcribed using SuperScript III RT (Invitrogen, Foster City, CA) and PCR nucleotides (Roche, Indianapolis, IN) along with RNAse Out (Invitrogen) using an optimized version of the manufacturer’s protocol. Resulting cDNA was treated with RNAse H (Applied Biosystems, Foster City, CA) according to manufacturer’s protocol. 10uL of cDNA was then used to setup a real-time PCR using Gene Expression Mastermix (Applied Biosystems) along with target specific labeled probe 5’- /56-FAM/CTT CCT CAGTGT GTT TCA CTT TCT CTT CTG CG/3BHQ_1/ -3’ and forward 5’- GTC TGC GTC ATCTGGTGC ATT C -3’ and reverse primers 5’- CACTAG GTGTCT CTG CACTAT CTG TTT TG -3’ (custom synthesis by Integrated DNA Technologies, Coralville, IA). Real-time PCR was performed on an Applied Biosystems StepOne Plus platform using the standard curve protocol. The RNA standard was transcribed from the pSP72 vector containing the first 731 bp of the SIVmac239- or SIVsmE660-gag gene using the Megascript T7 kit (Ambion Inc, Foster City, CA), quantitated by optical density (OD), and serially diluted to generate a standard curve. The quality of the RNA standard was assessed using an Agilent Bioanalyzer with RNA Nano 6000 chips (Agilent Inc., Santa Clara, CA). The sensitivity of this assay has been shown to be 250 copies/mL.
The number of transmitted/founder (T/F) variants was determined by single genome amplification (SGA) of the full-length envelope gene as previously described7. The number of sequences analyzed per animal was 21.2 ± 4.8 (mean ± sd), with a range of 10–38. There was no difference by group.
All 1,629 sequences are deposited in GenBank under accession numbers KF602252-KF603880.
SIV Envelope Constructs
Sequences of the CP3C-P-A8 envelope (referred to in this paper as “CP3C” for brevity) and CR54-PK-2A5 (“CR54”) are shown in Supplementary Table 1. These sequences were used to produce protein for binding assays as well as pseudotyped viruses. Mutations were designed into each virus to create individual amino acid variants as listed in Fig 4; the relevant portion of the envelope, encompassing the C1 region, is shown aligned in Supplementary Table 4. SIV Env mutant plasmids were generated by site-directed mutagenesis by GeneImmune Biotechnology (Rockville, MD).
Immunology
Intracellular cytokine staining for antigen-specific responses was performed using a qualified assay as described29. Cells were stimulated with overlapping 15mer peptides from Gag or Env from mac239 or smE543 (a clone similar to smE660). Data are shown for stimulations with E543 peptides. For breadth analysis, IFNγ ELIspots were performed as described30, using pools of 10 peptides from each protein.
Raw peptide microarray data (PepStar) were pre-processed and normalized as previously described31. Responses to peptides from mac239 or E543 were measured; data are shown for E543 only, except in Fig 5a where data from both sets are shown. For each peptide, the mean binding from 10 control animals was subtracted from the value for each vaccinated animal. The distribution of resulting values was used to define a cut-off value of 1.2 for positivity: a large fraction of peptide responses constituted a near-normal distribution centered on 0 (after background subtraction); the 10th percentile of this distribution was −1.2; thus, +1.2 is an estimate of the 90th percentile of a completely negative response. For breadth analysis, positive responses to partially overlapping peptides were considered to comprise a single epitope.
SIV-specific humoral IgG and IgA levels were evaluated by a standardized antibody binding multiplex assay as previously described24,32. IgA levels were low and are shown only as MFI for the lowest dilution tested. IgG levels are shown as MFI AUC (area under the curve) computed over the dilutions in the linear range of the assay. Avidity was quantified by Surface Plasmon Resonance (SPR) as previously described2,33.
Viral neutralization assays
Neutralization was evaluated using three distinct assays. (1) Plasma neutralization of viral replication in PBMC was performed as previously described14. (2) Env-pseudovirus neutralization was measured using single-round-of-infection SIV Env-pseudoviruses with TZM-bl target cells stably expressing high levels of CD4 and the co-receptors CCR5 and CXCR434,35. Tat-regulated luciferase gene expression was quantified to determine the reduction in virus infection. Neutralization curves were fit by nonlinear least squares regression, and the 50% inhibitory concentrations (ICHM) was computed as the antibody concentration required to achieve 50% of maximal inhibition. (3) Replication competent SIV was used to infect TZM-bl cells as above, with cloned or uncloned swarm SIVs essentially as described36. Briefly, neutralization assays were performed with serial dilutions of heat-inactivated (56C, 1 hr) samples. Diluted samples were pre-incubated with virus (~150,000 relative light unit equivalents) for 1 hr at 37C before addition of cells. Following 48 hr incubation, cells were lysed and luciferase activity determined using a microtiter plate luminometer and BriteLite Plus Reagent (Perkin Elmer). Neutralization titers are the reciprocal sample dilution or concentration (for sCD4) at which relative luminescence units (RLU) were reduced by 50% compared to RLU in virus control wells after subtraction of background RLU in cell control wells.
CD4 binding inhibition by sera was determined as described37 with the following modifications. Plate-bound CP3C Env was incubated with or without a 1:5 dilution of plasma at 37°C for 1 hr. After washing, wells were incubated with 50µg/ml CD4-Ig-Biotin at 37°C for 1 hr. Plates were washed to remove excess CD4-Ig-Biotin and incubated with streptavidin horseradish peroxidase at 37C for 1 hr. Inhibition was calculated as the fraction of the signal in wells with plasma to those without.
Statistics
The analyses presented here used a variety of techniques. Comparisons of continuous endpoints between groups were based on t-tests and Analysis of Variance, log-transformed when appropriate (e.g. viral load). Comparisons of groups with respect to number of challenges until infection used the discrete time survival model assuming a leaky vaccine effect18. A comparison of the goodness-of-fit of possible models showed that the likelihood of the leaky model performed better than the all-or-none model (and the null hypothesis), and performed similarly to a model that allowed both types of effects18. For the cumulative incidence of A/K vs. TR viruses, we used nonparametric estimates that allowed for competing risks23. VE against each virus type was computed by modifying the Hudgens and Gilbert leaky vaccine model18 to account for two infection types, with p-values computed from likelihood ratio tests. We did not formally test for heterogeneity of per-exposure probability. However, two pieces of evidence support lack of heterogeneity. First, the leaky model is a better fit than the all-or-none model (which should be sensitive to one type of heterogeneity). Second, we evaluated the risk of infection as a function of challenge number, and found no statistically significant change over time in any group.
Associations between immunological measurements and number of challenges were based on similar models, with continuous predictors dichotomized at the median. Immunological predictors that showed some association using this method were investigated further using Cox Proportional Hazard models, both univariate and multivariate. All correlations were based on Spearman’s rho (nonparametric), in order to handle variables with censored readings below or above assay limits, as well as to include uninfected animals in correlations based on number of infections. P values reported are not corrected for the number of comparisons between multiple immunological assays and outcome, except as noted.
In this paper we identified several potential correlates of risk of infection38,39 that deserve further investigation and confirmation. Since these are not measureable in unvaccinated animals, the correlations with time to infection we report might be related to either differential effects of vaccines or to unspecified differences between the immune systems of the animals.
For sieve analysis, only Env positions with at least five variants amongst all sequences were considered. The distribution of variants at those positions passing this minimum threshold was compared between groups using a permutation-based version of the Fisher’s exact test, using 10,000 permutations. P values reported in Fig 2C and Extended Fig 4 are not corrected for multiple comparisons, but positions 23, 45, and 47 (in both analyses) remain significant (p ≤ 0.002) after such correction. Similarly, position 162 in Fig 5b is significant after correction for multiple comparisons.
As expected from our stratification, Trim5α alleles were found to have no impact on the conclusions of vaccine effects on protection; as follows: Analysis of the discrete time-to-infection model using only Trim5α-resistant animals did not change VE. Cox proportional hazard modeling of time-to-infection by group did not change when Trim5α was included as a covariate. Similarly, the importance of virus type infection (A/K vs TR) was unaffected by inclusion of Trim5α. Finally, immunological correlates analyses (prediction of time-to-infection by antibody measures) did not change when Trim5α was added as a covariate. The number of animals in each arm (out of 20) with homozygous Trim5α-sensitive alleles was 8 (control), 9 (mosaic Env), 7 (mosaic Gag), and 8 (mosaic Gag).
Acknowledgments
We are indebted to the following individuals: Wei Shi, Lan Wu, Sung-Youl Ko, Lingshu Wang, Wing-Pui Kong for immunogen construction; Mitzi M. Donaldson, Shing-Fen Kao, David Quinn, Joshua Owuor, Katherine Denison, Harikrishnan Balachandran, Corinne Luedemann, William T. Williams, Glenn Overman, Aaron Deal, Christie Brinkley, and Laura Racz for technical assistance with immunology assays; Alida Ault for assistance managing NHP studies; Shelby O’Connor for deep-sequencing data; Mike Seaman for providing plasmids encoding E660 envelopes; Francine McCutcheon, Julie Overbaugh, Jerome Kim, and David Montefiori for HIV-1 strains; and Peter Gilbert for advice with using the Aalen and Johansen model. This work was supported by the Intramural Research Program of the Vaccine Research Center, NIAID, NIH; by NIH contracts HHSN261200800001E (BFK, WG) and HHSN27201100016C (DCM); by NIH grant AI100645 (BTK, WF); and by the Bill and Melinda Gates Foundation grant OPP1032317.
Footnotes
Online Content. Additional Methods and Extended Data display items are available in the online version of the paper.
Supplementary Information is available in the online version of the paper.
Author Contributions: M.R., R.A.S., R.A.K., G.J.N., N.L.L., S.S.R., and J.R.M. designed and supervised the study. W.F., Z.Y., B.T.K., and G.J.N. designed and manufactured immunogens. J.M.T., N.L.L., and S.S.R. supervised nonhuman primate procedures. M.R., B.F.K., K.E.F., A.P.B., L.V.M., K.E.S., B.W., R.T.B., R.G., G.F., M.A., T.N.D., D.C.M., G.D.T., R.A.K., and J.R.M. supervised assays and performed primary data analysis. S.D.S., R.D.M., H.C.W., L.V.M., L.S., K.E.S., and B.W. performed assays. M.R., W.G., and M.N. aggregated data and performed statistical analysis. M.R. and J.R.M. wrote the manuscript.
T/F sequences are deposited in GenBank under accession numbers KF602252-KF603880.
The authors declare no competing financial interests.
References
Full text links
Read article at publisher's site: https://doi.org/10.1038/nature12893
Read article for free, from open access legal sources, via Unpaywall:
https://www.ncbi.nlm.nih.gov/pmc/articles/PMC3946913
Citations & impact
Impact metrics
Citations of article over time
Alternative metrics
Article citations
HIV Vaccine Development at a Crossroads: New B and T Cell Approaches.
Vaccines (Basel), 12(9):1043, 12 Sep 2024
Cited by: 0 articles | PMID: 39340073 | PMCID: PMC11435826
Review Free full text in Europe PMC
Viral vector delivered immunogen focuses HIV-1 antibody specificity and increases durability of the circulating antibody recall response.
PLoS Pathog, 19(5):e1011359, 31 May 2023
Cited by: 1 article | PMID: 37256916 | PMCID: PMC10284421
Trivalent mosaic or consensus HIV immunogens prime humoral and broader cellular immune responses in adults.
J Clin Invest, 133(4):e163338, 15 Feb 2023
Cited by: 5 articles | PMID: 36787249 | PMCID: PMC9927951
Stabilized HIV-1 envelope immunization induces neutralizing antibodies to the CD4bs and protects macaques against mucosal infection.
Sci Transl Med, 14(661):eabo5598, 07 Sep 2022
Cited by: 14 articles | PMID: 36070369 | PMCID: PMC10034035
Cryo-EM structures of prefusion SIV envelope trimer.
Nat Struct Mol Biol, 29(11):1080-1091, 07 Nov 2022
Cited by: 7 articles | PMID: 36344847 | PMCID: PMC10606957
Go to all (108) article citations
Data
Data behind the article
This data has been text mined from the article, or deposited into data resources.
BioStudies: supplemental material and supporting data
Nucleotide Sequences (2)
- (2 citations) ENA - KF603880
- (2 citations) ENA - KF602252
Similar Articles
To arrive at the top five similar articles we use a word-weighted algorithm to compare words from the Title and Abstract of each citation.
Breakthrough Virus Neutralization Resistance as a Correlate of Protection in a Nonhuman Primate Heterologous Simian Immunodeficiency Virus Vaccine Challenge Study.
J Virol, 89(24):12388-12400, 30 Sep 2015
Cited by: 9 articles | PMID: 26423953 | PMCID: PMC4665252
Low-dose penile SIVmac251 exposure of rhesus macaques infected with adenovirus type 5 (Ad5) and then immunized with a replication-defective Ad5-based SIV gag/pol/nef vaccine recapitulates the results of the phase IIb step trial of a similar HIV-1 vaccine.
J Virol, 86(4):2239-2250, 07 Dec 2011
Cited by: 74 articles | PMID: 22156519 | PMCID: PMC3302390
Rationally Designed Vaccines Targeting the V2 Region of HIV-1 gp120 Induce a Focused, Cross-Clade-Reactive, Biologically Functional Antibody Response.
J Virol, 90(24):10993-11006, 28 Nov 2016
Cited by: 30 articles | PMID: 27630234 | PMCID: PMC5126359
Vaccine-induced V1V2-specific antibodies control and or protect against infection with HIV, SIV and SHIV.
Curr Opin HIV AIDS, 14(4):309-317, 01 Jul 2019
Cited by: 22 articles | PMID: 30994501 | PMCID: PMC6542703
Review Free full text in Europe PMC
Funding
Funders who supported this work.
Intramural NIH HHS (2)
Grant ID: Z99 AI999999
Grant ID: ZIA AI005019-12
NCI NIH HHS (2)
Grant ID: HHSN261200800001E
Grant ID: HHSN261200800001C
NIAID NIH HHS (2)
Grant ID: UM1 AI100645
Grant ID: AI100645
PHS HHS (1)
Grant ID: HHSN27201100016C





