Abstract
Background
Proteoglycans are found on the cell surface and in the extracellular matrix, and serve as prime sites for interaction with signaling molecules. Proteoglycans help regulate pathways that control stem cell fate, and therefore represent an excellent tool to manipulate these pathways. Despite their importance, there is a dearth of data linking glycosaminoglycan structure within proteoglycans with stem cell differentiation.Methods
Human embryonic stem cell line WA09 (H9) was differentiated into early mesoderm and endoderm lineages, and the glycosaminoglycanomic changes accompanying these transitions were studied using transcript analysis, immunoblotting, immunofluorescence and disaccharide analysis.Results
Pluripotent H9 cell lumican had no glycosaminoglycan chains whereas in splanchnic mesoderm lumican was glycosaminoglycanated. H9 cells have primarily non-sulfated heparan sulfate chains. On differentiation towards splanchnic mesoderm and hepatic lineages N-sulfo group content increases. Differences in transcript expression of NDST1, HS6ST2 and HS6ST3, three heparan sulfate biosynthetic enzymes, within splanchnic mesoderm cells compared to H9 cells correlate to changes in glycosaminoglycan structure.Conclusions
Differentiation of embryonic stem cells markedly changes the proteoglycanome.General significance
The glycosaminoglycan biosynthetic pathway is complex and highly regulated, and therefore, understanding the details of this pathway should enable better control with the aim of directing stem cell differentiation.Free full text

Changes in glycosaminoglycan structure on differentiation of human embryonic stem cells towards mesoderm and endoderm lineages
Associated Data
Abstract
Background
Proteoglycans are found on the cell surface and in the extracellular matrix, and serve as prime sites for interaction with signaling molecules. Proteoglycans help regulate pathways that control stem cell fate, and therefore represent an excellent tool to manipulate these pathways. Despite their importance, there is a dearth of data linking glycosaminoglycan structure within proteoglycans with stem cell differentiation.
Methods
Human embryonic stem cell line WA09 (H9) was differentiated into early mesoderm and endoderm lineages, and the glycosaminoglycanomic changes accompanying these transitions were studied using transcript analysis, immunoblotting, immunofluorescence and disaccharide analysis.
Results
Pluripotent H9 cell lumican had no glycosaminoglycan chains whereas in splanchnic mesoderm lumican was glycosaminoglycanated. H9 cells have primarily non-sulfated heparan sulfate chains. On differentiation towards splanchnic mesoderm and hepatic lineages N-sulfo group content increases. Differences in transcript expression of NDST1, HS6ST2 and HS6ST3, three heparan sulfate biosynthetic enzymes, within splanchnic mesoderm cells compared to H9 cells correlate to changes in glycosaminoglycan structure.
Conclusions
Differentiation of embryonic stem cells markedly change the proteoglycanome
General Significance
The glycosaminoglycan biosynthetic pathway is complex and highly regulated, and therefore, understanding the details of this pathway should enable better control with the aim of directing stem cell differentiation.
1. Introduction
Embryonic stem cells (ESCs) have remarkable potential in translational medicine. ESCs might one day be used to replenish tissues that have been damaged in various diseases including Parkinson’s and diabetes, and in this way provide a direct clinical use. ESCs might also be indirectly used as targets for drug development and as models to study developmental processes. Their use in such studies is favored over using primary cell lines, immortalized tumor cells, or genetically transformed cells, since ESCs can be maintained in culture for extended periods of time and give rise to uniform and genetically normal cell populations. The distinctiveness of ESCs is inherent in their pluripotency, which imparts the ability to differentiate into cell types of all germ layers, the ectoderm, mesoderm and endoderm.
Controlled cell-extrinsic [1] and cell-intrinsic signals direct the pathways determining the ESC fate (differentiated versus non-differentiated). A tightly controlled network of transcription factors [2–4], interacting with the microRNA network [5–10], process information received from the extracellular environment and in turn regulate the expression of genes required for maintenance of pluripotency or drive differentiation towards a specific lineage.
Proteoglycans (PGs) primarily reside in the extracellular space, as cell membrane proteins and extracellular matrix (ECM) proteins. PGs consist of a protein core with glycosaminoglycan (GAG) chains attached [11]. PGs interact with chemokines, growth factors, and morphogens, and they are important for modulating signaling pathways such as FGF, Wnt, and BMP [12–17], which are important in determining stem cell fate. The principal activity of PGs has been associated with their GAG chains, although their core proteins can also display activity [18,19]. GAGs are linear polysaccharides consisting of repeating disaccharides and can be divided into four classes: heparan sulfate (HS)/heparin (HP), chondroitin sulfate (CS)/dermatan sulfate (DS), keratan sulfate (KS) and hyaluronan (HA). These classes differ in the structure of the repeating disaccharides and also in their function [11].
The role of diverse elements in stem cell fate determination, including transcription factors, microRNAs, and chromatin modifiers have been extensively studied (20), but the functions of PGs remain less clearly defined. There have been limited studies connecting PGs to stem cell fate [21,22]. Even fewer studies have been done linking GAG structure to stem cell commitment towards the various lineages [23,24]. Most studies on PGs have involved neural stem cells, satellite cells and hematopoetic stem cells [20].
In the current study, pluripotent human embryonic stem cells (H9) were differentiated into multi-potent splanchnic mesoderm, which has the capacity for differentiation into the major cardiovascular lineages [25]. H9 cells were also differentiated into early stages of hepatocytes. Changes in HS/HP and CS/DS chain compositions were examined to establish changes in the cellular glycosaminoglycanome accompanying differentiation towards splanchnic mesoderm and hepatic cell types. Changes in the transcript abundance for genes involved in the biosynthesis of GAGs and genes encoding PG core proteins were analyzed in undifferentiated H9 cells and differentiated Isl1+ (splanchnic mesoderm) cells to determine to what extent changes in GAG structures might be regulated at the gene level. Understanding the role of GAGs in the genesis of splanchnic mesoderm cells and hepatocytes should enable researchers to control these differentiation processes with the aim of utilizing those cells for regenerative medicine as well as drug development.
2. Materials and Methods
2.1 hESC H9 cell culture
The hESC line H9 (WiCell Research Institute, Inc, Madison, WI) was maintained on Matrigel coated cell culture dishes in complete mTeSR-1 media (Stem Cell Technologies, Vancouver, Canada) supplemented with 100 U/mL penicillin and streptomycin (Life Technologies, Grand Island, NY) and cultured at 37°C in a humidified atmosphere with 5% CO2. Cells were passaged every 5–6 days using collagenase IV (Life Technologies) to release cells from Matrigel.
2.2 hESC H9 differentiation
H9 hESCs were differentiated to splanchnic mesoderm by addition of BMP4 (100 ng/ml, R&D Systems) and Wnt3a (25 ng/ml, R&D Systems) for 4 days.
H9 differentiation towards hepatocytes was performed as described in the literature [26]. Briefly, H9 cells were primed towards definitive endoderm in RPMI 1640 medium (ATCC, Manassas, VA) supplemented with B27, Activin A and Wnt3a for 3 days. Hepatic differentiation was induced in KnockOut-DMEM-medium (ATCC) supplemented with DMSO and KnockOut-Serum replacement (Life Technologies) for 5 days. Hepatic maturation was continued for 9 days in L-15 medium (ATCC) containing hepatic growth factor (R&D Inc., Minneapolis, MN), oncostatin M (R&D Inc.) and 10% FBS (Life technologies).
2.3 Total RNA isolation, cDNA synthesis and qRT-PCR reactions
Four biological replicates of undifferentiated and differentiated H9 cell samples were harvested, flash frozen in liquid nitrogen and stored at −80°C until use. For measurement of PG-related gene expression levels, total RNA was isolated from cell lysates using the RNeasy Plus kit (Qiagen, Valencia, CA) and cDNA synthesis was performed using Superscript III First Strand Synthesis (Life Technologies) as previously described [27]. The qRT-PCR reactions were performed in triplicate for each gene analyzed. Cycling conditions and analysis of amplimer products were performed as previously described [23]. Briefly, reactions contained 1.25 μl of diluted cDNA template (1:10), 1.25 μL of primer pair mix (125 μM final concentration) and 2.5 μl iQ SYBR Green Supermix (BioRad, Hercules, CA) added to 96-well microtiter plates. Primers for the control gene, RPL4, were included on each plate to control for run variation and to normalize individual gene expression. Primer pairs for GAG-related genes were designed within a single exon [23,28] and primer design validated previously using the standard curve method [23,29].
2.4 Calculation of relative transcript abundance and statistical analysis
An average of the triplicate Ct values for each gene was determined and the standard deviation calculated. Samples were re-run if the standard deviation value was >0.5 Ct units. The logarithmic average Ct value for the control gene and each tested gene was converted to a linear value by the equation 2−Ct. Converted values were normalized to RPL4 by dividing individual gene value by control gene value. Normalized values were scaled so that genes below the level of detection were given a value of 1 × 10−6, and this value was used as the lower limit on histograms. A non-parametric Mann-Whitney test (GraphPad InStat3, v3.10) was used to determine statistically significant changes (p<0.05) in transcript abundance between undifferentiated and differentiated samples. In cases where no transcripts were detected in one of the two cell types, no p-values were determined. Fold change was calculated by dividing normalized values of tested genes in differentiated cells by those in undifferentiated H9 cells.
2.5 Protein isolation, quantification and immunoblotting
For total protein extraction, cells were lysed in Nonidet-P40 lysis buffer (Boston Bioproducts, Ashland, MA) on ice for 30 min in the presence of a cocktail of protease and phosphatase inhibitors (Thermo Fisher Scientific) which included AEBSF, aprotinin, bestatin, E-64, leupeptin, and pepstatin A. Protein concentration was determined using the BCA assay (Pierce). Approximately 40 μg of total protein was loaded and separated on a precast 4–20% gradient polyacrylamide gel. After transfer to a PVDF membrane, proteins of interest were detected using relevant primary and HRP-conjugated secondary antibodies followed by chemiluminescent exposure (Super Signal West Pico ECL substrate, Pierce) on high performance chemiluminescence film (GE Healthcare, Little Chalfont, UK, Amersham Hyperfilm ECL). Primary antibodies used were anti-γ-tubulin (T3320, Sigma Aldrich), anti-decorin (H00001634-B01P, Abnova, Walnut, CA), anti-lumican (H00004060-D01P, Abnova), anti-serglycin (H00005552-M03, Abnova,), anti-glypican 5 (sc-84278, Santa Cruz Biotechnology, Santa Cruz, CA), anti-Oct3/4 (611202, BD Biosciences, Franklin Lakes, NJ) and anti-nestin (611658, BD Biosciences).
2.6 Immunofluorescence
Cells were grown in Lab-Tek chamber slides. Media from cells was washed off and cells were fixed with 4% paraformaldehyde. Cells were blocked with DPBS-Triton 100-X solution supplemented with 5% bovine serum albumin (BSA). They were incubated overnight at 4°C with primary antibody diluted in Dulbecco’s phosphate-buffered saline (DPBS)-Triton X-100 solution supplemented with 1% BSA. The following day, after several washes with DPBS, cells were incubated with secondary antibody (goat anti-mouse Alexa Fluor 488, goat anti-rabbit Alexa Fluor 647, A11001, A21244, Life Technologies) diluted in DPBS-Triton X-100 solution supplemented with 1% BSA at room temperature for 1 h. After several washes with DPBS, cells were stained with Hoechst 33342 for 5 min at room temperature. After several washes, chambers were detached from slides, which were incubated overnight with ProLong Gold anti-fade reagent and covered with a cover slip. The following day slides were sealed and analyzed with a Zeiss 510 Meta multi-photon confocal microscope. Primary antibodies used were anti-Oct3/4 (611202, BD Biosciences, Franklin Lakes, NJ), anti-Isl (AF1837, R&D Systems, Minneapolis, MN), anti-AFP (WH0000174M1-100g, Sigma-Aldrich), anti-albumin (sc-271605, Santa Cruz Biotechnology), anti-HNF4α (sc-6556, Santa Cruz Biotechnology), anti-FOXA2 (AF2400, R&D Systems), and anti-HepPar1 (IR624, DAKO, Carpinteria, CA). Alexa Fluor 488 conjugated phalloidin stain was used for actin staining (Life Technologies, A12379).
2.7 Isolation and purification of total and cell surface associated GAGs
For total GAG recovery, H9, splanchnic mesoderm and premature hepatocyte cells were harvested, washed in phosphate buffered saline (PBS) three-times, and centrifuged to pellet cells. Isolation and purification of GAGs was performed as previously described with some modifications [27,30]. Specifically, cell pellets were re-suspended in 1 ml water and subjected to proteolysis at 55°C in 2 mg/ml actinase E (Kaken Biochemicals, Tokyo, Japan) for 20 h. After proteolysis, particulates were removed using a 0.22 μm syringe-top filter. Peptides were removed from the samples using Microcon Centrifugal Filter Units YM-10 (10 k MWCO, 15 ml, Vivascience, Ridgewood, NJ). Samples were collected from the top layer of the filtration membrane and lyophilized, then dissolved in 8 M urea containing 2% CHAPS (pH 8.3, Sigma-Aldrich). A Vivapure MINI Q H spin column was equilibrated with 200 μl of 8 M urea containing 2% CHAPS (pH 8.3). To remove any remaining proteins, the clarified, filtered samples were loaded onto and run through the Vivapure MINI Q H spin columns under centrifugal force (700 × g). The columns were then washed with 200 μl of 8 M urea containing 2% CHAPS at pH 8.3, followed by five washes with 200 μl of 200 mM NaCl. GAGs were released from the spin column by washing three times with 100 μl of 16% NaCl. GAGs were desalted with YM-10 spin columns. The GAGs were lyophilized and stored at room temperature for future use.
2.8 Enzymatic depolymerization of GAGs for HS/HP analysis
Isolated GAG samples were incubated with chondroitin lyase ABC (10 m-units, Seikagaku Corporation, Tokyo, Japan) and chondroitin lyase ACII (5 m-units, Seikagaku Corporation) at 37°C for 10 h and the enzymatic products were recovered by centrifugal filtration as described above, but at 13,000 × g. CS/DS disaccharides that passed through the filter were freeze-dried for future analysis. GAGs remaining in the retentate were collected by reversing the filter and spinning at 13,000 × g, followed by incubation with 10 m-units of heparin lyases I, II, and III at 37°C for 10 h. The products were recovered by centrifugal filtration and the HS/HP disaccharides collected and freeze-dried for LC-MS analysis. Cloning, overexpression in Escherichia coli, and purification of the recombinant heparin lyase I (EC 4.2.2.7), heparin lyase II (no EC assigned), and heparin lyase III (EC 4.2.2.8) from Flavobacterium heparinum were all performed as previously described [31,32].
2.9 LC-MS disaccharide composition analysis of HS/HP
LC-MS analyses were performed on an Agilent 1200 LC/MS instrument (Agilent Technologies, Inc. Wilmington, DE) equipped with a 6300 ion trap with two separate systems for the CS/DS disaccharide analysis and HS/HP disaccharide analysis. For HS/HP disaccharide analysis, eluent A was water/acetonitrile (85:15) v/v, and eluent B was water/acetonitrile (35:65) v/v. Both eluents contained 12 mM tributylamine (TrBA) and 38 mM NH4OAc with pH adjusted to 6.5 with acetic acid. The column temperature was maintained at 45°C. For disaccharide analysis, eluent A was used for 10 min, followed by a linear gradient from 10 to 40 min of 0 to 50% (v/v) eluent B at the flow rate of 100 μl/min. The electrospray interface was set in negative ionization mode with the skimmer potential and capillary exit of −40.0 V, with a temperature of 350°C to obtain maximum abundance of ions in a full-scan spectra (350 1500 Da, 10 full scans/s). Nitrogen was used as a drying gas (8 l/min) and nebulizing gas (40 psi) [33].
Quantification analysis of HS/HP disaccharides was performed using calibration curves constructed by separation of increasing amounts of unsaturated HS/HP disaccharide standards (2, 5, 10, 15, 20, 30, 50, and 100 ng per disaccharide). Unsaturated disaccharide standards of HS/HP (0S: ΔUA-GlcNAc, NS: ΔUA-GlcNS, 6S: ΔUA-GlcNAc6S, 2S: ΔUA2S-GlcNAc, NS2S: ΔUA2S-GlcNS, NS6S: ΔUA-GlcNS6S, 2S6S: ΔUA2S-GlcNAc6S, TriS: ΔUA2S-GlcNS6S) were obtained from Iduron (Manchester, UK). Linearity was assessed based on the amount of disaccharide and peak intensity in MS. All analyses were performed in triplicate.
3. Results
3.1 Experimental hESCs WA09 (H9) possess normal karyotype and express pluripotency markers
Pluripotent WA09 (H9) cells were grown on Matrigel and exhibited morphology typical of non-differentiated stem cells (Figure 1A). They were routinely karyotyped to ensure that no chromosomal abnormalities developed during stem cell maintenance (Figure 1B). Pluripotency was monitored by checking for the expression of relevant markers including Oct3/4, and Nestin. Our H9 cell line expressed Oct3/4, confirming their pluripotency (Figure 1C). Nestin, which is associated with early neural differentiation [34], was not expressed, further confirming pluripotency (Figure 1D).
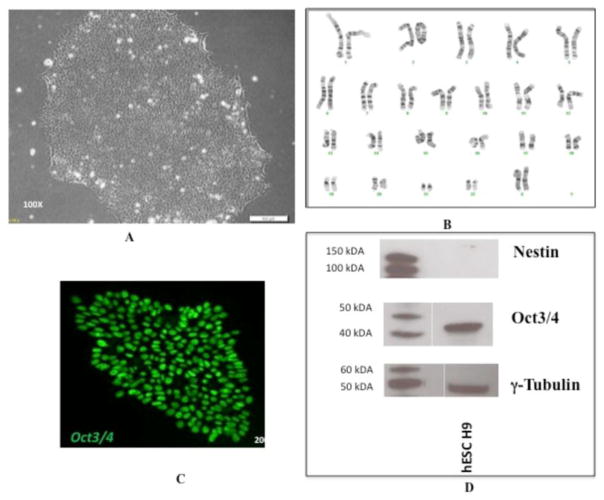
Expression of pluripotency markers and karyotyping of hESC cell line H9. (A) Pluripotent H9 cells express typical morphology and (B) normal karyotype1 when grown on matrigel substrate with mTeSR1 media. (C) They also express the pluripotency markers Oct3/4, as detected by immunocytochemistry and (D) do not express the early differentiation marker Nestin, as detected by immunoblotting. The lanes on the left show standard proteins with their molecular weights indicated and the lanes on the right shows hESC H9.
3.2 Differentiation of hESC line H9 towards splanchnic mesoderm cells
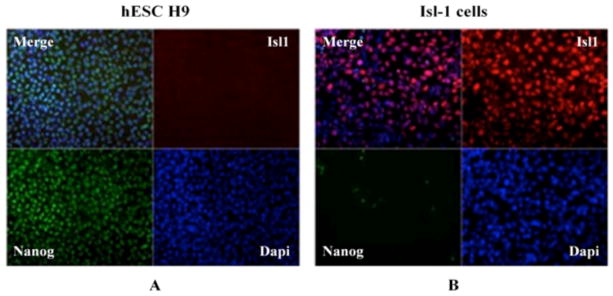
Differentiation of H9 cells towards Isl-1+ splanchnic mesoderm was monitored by checking the expression of pluripotency marker Nanog, as well as the early mesoderm marker, Isl-1 (Figure 2). The expression of the Nanog transcription factor was high in non-differentiated H9 cells, whereas it decreased upon differentiation. In contrast, pluripotent H9 cells did not express the Isl-1 marker, but an increased level of that protein was observed in mesoderm cells, confirming their differentiation. Co-localization of DAPI (nuclear stain) with expression of Nanog and Isl-1 markers confirms the transcription factor origins of those proteins.
3.3 Differentiation of the hESC line H9 towards hepatocyte cell type
During differentiation towards the hepatic lineage, hESCs traverse several stages, including specified hepatic cells, hepatoblasts and hepatocytes (Figure 3A). Each of these cell types vary in their morphological appearance and present typical protein marker expression. During differentiation, we observed several cell types in differentiated H9 cell populations, including those that morphologically resembled hepatocytes (Figure 3B). Pluripotent H9 cells had round cells packed tightly into colonies. During differentiation, polygonal shaped cells appeared, which resembled the morphology of hepatocytes.
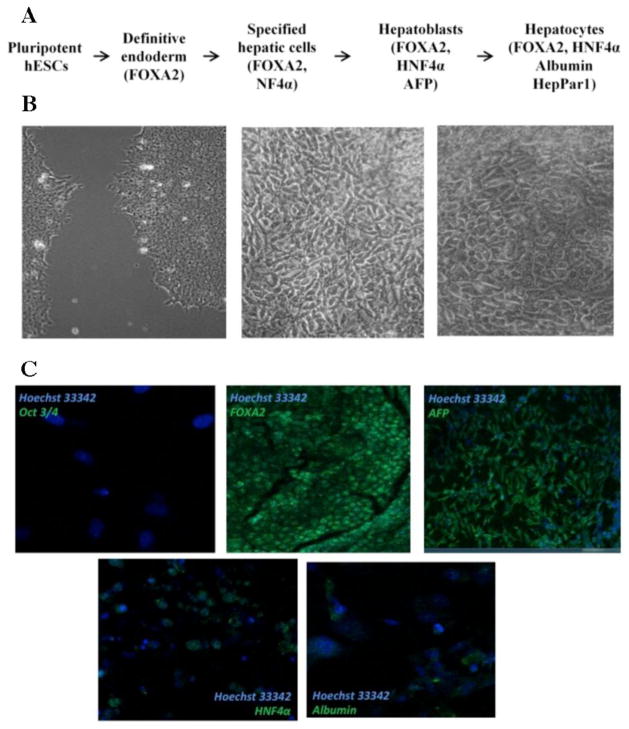
Differentiation of H9 cells towards the hepatic lineage. (A) Scheme of the stages observed during differentiation of hESCs towards hepatocytes and markers expressed at each stag. (B) Cell types observed during differentiation of H9 cells during differentiation towards hepatocytes: pluripotent H9 cells (left), cells resembling premature hepatocytes (middle), cells resembling mature hepatocytes (right). (C) Expression of hepatic markers during differentiation of H9 cells detected by immunocytochemistry.
Expression of the pluripotency marker Oct3/4 was not detected in a differentiated H9 cell population, demonstrating that they had lost pluripotency. Hepatocyte nuclear factor (HNF) 3β (FOXA2) is a transcription factor that regulates expression of α-fetoprotein (AFP) protein in the early stages of hepatic differentiation [35]. Both FOXA2 and AFP were expressed in our differentiated H9 cells, confirming that differentiation was primed towards an endodermal fate. HNF4α is a transcription factor that regulates the expression of albumin [36], one of the most abundant proteins in mature hepatocytes. Expression of both HNF4α and albumin were detected at low levels in differentiated H9 cells (Figure 3C). Expression of hepatic fate associated markers (HNF1α, HNF4α, and AFP) was also detected at the transcript level (Supplementary Figure 1).
HepG2 cells are hepatocellular carcinoma cells that are widely used as a model to study human hepatocytes, as propagation and maintenance of primary hepatocytes is challenging. Expression of hepatocyte-associated markers was tested in HepG2 cells to establish the baseline level of expression in mature hepatocytes (Supplementary Figure 2). The expression of proteins found in mature hepatocytes such as HNF4α and albumin was high in HepG2 cells. When the expression of hepatic markers in differentiated H9 cells and HepG2 cells were compared, it was clear that H9 cells had differentiated into immature hepatocytes. The expression of the mature hepatocyte markers HNF4α and albumin were lower in H9 cells differentiated to hepatic lineage than in HepG2 cells. In addition, H9 cells differentiated into the hepatic lineage did not secret fibrinogen, a protein associated with mature hepatocytes (data not shown).
3.4 Expression of GAG core proteins in splanchnic mesoderm cells
GAGs are synthesized on core proteins, which can carry KS, CS/DS and HS/HP. The primary function of the core protein is typically associated with the specific GAG chain present [17]. With this in mind, we assessed the relative transcript abundances of GAG core proteins by qRT-PCR in H9 cells and splanchnic mesoderm cells. The most striking changes in transcript abundance between the two cell populations were for genes encoding lumican, decorin, serglycin and glypican-5. Transcripts for lumican increased >500-fold in splanchnic mesoderm cells compared to H9 cells. Decorin transcripts were only detected in the differentiated cell population yielding an increase of >200-fold. Transcript levels for Aggrecan, Procollagen 1Xa2 and glypican-5 were elevated in splanchnic mesoderm cells 11-fold, 6.5-fold and 15-fold, respectively. Smaller (< 5-fold), but statistically significant, increases in transcript abundances were identified for Fibromodulin, Biglycan, Thrombomodulin, Syndecan-2, Syndecan-3, Syndecan-4, and Glypican 1. Serglycin transcripts were only detected in H9 ES resulting in a 10-fold decrease in abundance compared to differentiated Isl1+ cells, although the overall expression level of serglycin in Isl1+ cells was on the lower end of the detectable range.. Statistically significant decreases in transcript abundance were also observed for Brevican (−3.4-fold), CD74 (−1.8-fold), Syndecan 1 (−2.4-fold) and Glypican 2 (−5-fold). Finally, there were no statistically significant changes between the two cell populations detected in the transcript levels for versican, neurocan, NG2, beuroglycan, bamacan, epican (CD44), perlecan, agrin, glypican-3, glypican-4 and glypican-6 (Figure 4 A).
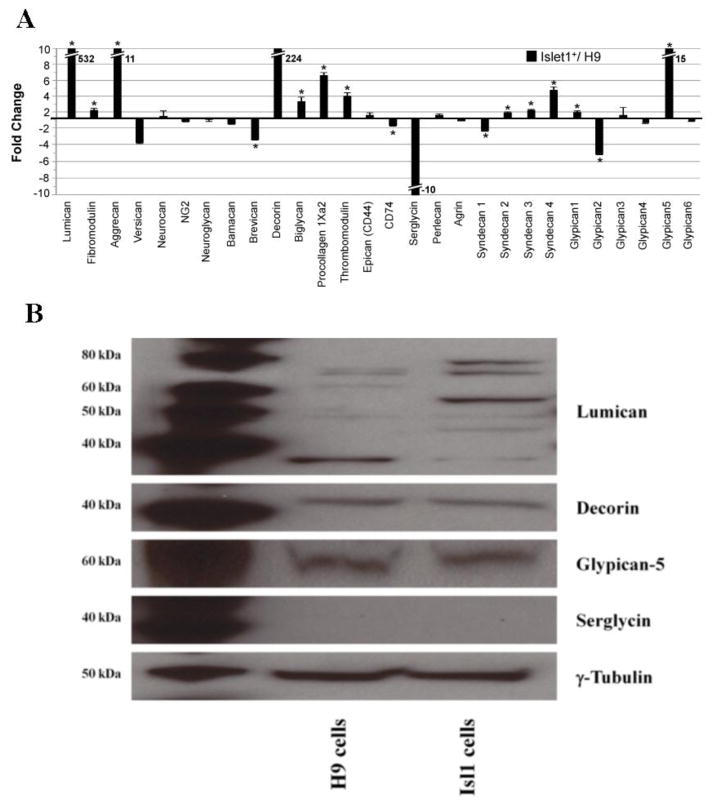
Gene transcript and protein expression of GAG core proteins in H9 and splanchnic mesoderm cells. (A) Fold change of CS/DS and HS/HP core proteins detected by qRT-PCR and normalized to RPL4. The center line indicates a value of 1 (no fold-change). Positive fold change values indicate that the transcript for a given gene is more abundant in the differentiated Isl1+ cells than in H9 cells, while negative fold change indicates that the transcript is more abundant in the undifferentiated H9 cells than the differentiated population. An asterisk (*) indicates a statistically significant change (p<0.05). Hash marks are used to indicate an off-scale value and the fold change is indicated beside the histogram bar. (B) Change in protein expression level of lumican, decorin, glypican-5 and serglyin in hESC line H9 and splanchnic mesoderm cells by Western blotting. The lane on the left shows the positive controls.
Protein expression of lumican, decorin, serglycin, and glypican-5 was examined by immunoblotting to assess variation in their levels in splanchnic mesoderm cells compared to H9 cells. Our results showed that splanchnic mesoderm cells primarily express the GAGylated form of lumican, whereas pluripotent H9 cells express only the core protein (~ 39 kDa). The size of lumican with GAG chains ranges from ~ 50 kDa to ~ 80 kDa. No changes were detected, by Western blot analysis, in the levels of decorin or glypican-5 in H9 and splanchnic mesoderm cells. Serglycin was not detectable by Western blotting in either the H9 or the splanchnic mesoderm (Figure 4 B).
3.5 Change in the transcript abundance of genes encoding HS/HP chain initiation, elongation and modification enzymes in splanchnic mesoderm cells compared to H9 cells
HS/HP chains are synthesized on a core of carrier proteins, to which a tetrasaccharide linkage region is attached. This linkage region is constructed through the action of several enzymes, including xylosyl transferases (XYLT1 and XYLT2), β4-galactosyltransferase (β4GALT7), β3-galactosyltransferase (β3GALT6), and β3-glucuronosyltransferase (β3GAT3). Further elongation of the HS/HP chain is carried out by α-N-acetylglucosaminyltransferases (EXTL2, EXTL3) and α-N-acetylglucosaminyl-β-glucuronosyl transferases (EXT1, EXT2) (Figure 5). Modest changes (under ~ 5-fold) in transcript levels were observed for all genes involved in chain initiation and elongation steps of HS/HP biosynthesis (Table 1).
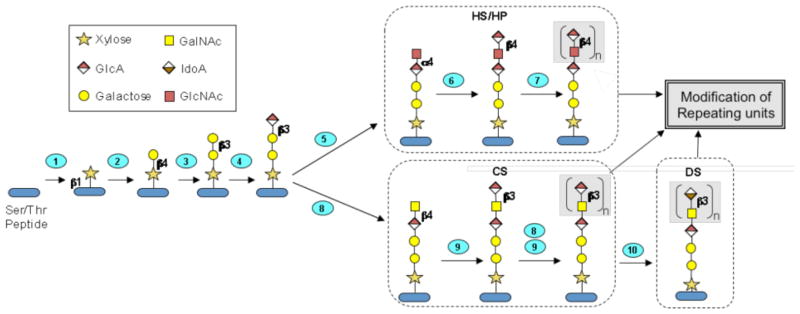
Graphic diagram of GAG core tetrasaccharide biosynthesis and HS chain polymerization. The Ser/Thr-containing polypeptide core protein is glycosylated and modified through ten enzymatic reactions occurring in the endoplasmic reticulum and Golgi. Numbered steps correspond to the reactions catalyzed by enzymes encoded by the following genes (1): XYLT1, XYLT2, (2): B4GALT7, (3): B3GALT6, (4): B3GALT3, (5): EXTL2, EXTL3, (6): EXT1, EXT2, (7): EXT1, EXT2, EXTL1, EXTL3. (8): CSGALNACT1, CSGALNACT2, (9): CHSY1, CHPF1, CHSY3, CHPF2, and (10): DSE1, DSE2.
Table 1
Fold change in transcript abundance of heparan sulfate/heparin chain initiation, elongation and modification genes in splanchnic mesoderm cells compared to H9 cells.
| Gene | Fold change* | Gene | Fold change* | Gene | Fold change* |
|---|---|---|---|---|---|
| XYLT1 | −1.2 | EXTL3 | −1.7 | HS6ST3 | 4.9** |
| XYLT2 | −2.1** | NDST1 | 3.7** | HS3ST1 | 14.1** |
| B4GALT7 | 1.0 | NDST2 | 1.0 | HS3ST2 | −7.2** |
| B3GALT6 | 1.5 | NDST3 | 1.0 | HS3ST3A1 | 1.5 |
| B3GAT3 | 1.3** | NDST4 | −1.1 | HS3ST3B1 | 1.0 |
| EXTL2 | −1.4 | GLCE | 2.3** | HS3ST4 | −1.1 |
| EXT1 | 3.6** | HS2ST1 | 1.1 | HS3ST5 | −2.1 |
| EXT2 | 1.1 | HS6ST1 | −1.2 | HS3ST6 | −111.7** |
| EXTL1 | 3.6** | HS6ST2 | 3.8** |
Nascent HS/HP (heparosan) goes through a series of modifications, including epimerization of uronic acids and introduction of sulfo groups at multiple locations along the polysaccharide chain. These reactions are catalyzed by N-deacetylase-N-sulfotransferases (NDSTs), C5-epimerase (GLCE), 2-O-sulfotransferase (HS2ST1), 6-O-sulfotransferases (HS6STs) and 3-O-sulfotransferases (HS3STs) (Figure 6). Many of these enzymes have multiple isozymes with different spatial and temporal distributions [37]. Slight, but statitistically significant, <5-fold increases were observed for transcripts for NDST1, GLCE, HS6ST2, and HS6ST3 in splanchnic mesoderm cells compared to H9 cells. The greatest increase was observed in the transcript level of HS3ST1, which was elevated ~ 14-fold in splanchnic mesoderm cells compared to H9 cells. We noted only minor decreases in transcript levels for NDST4, HS6ST1, HS3ST4 and HS3ST5 in splanchnic mesoderm cells as compared to H9 cells. Two genes displayed statistically significant decreases in transcript abundance. Transcripts for HS3ST2 were reduced ~ 14-fold and HS3ST6 transcripts were reduced ~112-fold in splanchnic mesoderm cells compared to undifferentiated H9 cells. No significant changes in transcript abundance were observed for NDST2, NDST3, HS2ST1, HS3ST3A1 and HS3ST3B1 on differentiation of H9 cells into splanchnic mesoderm cells (Table 1).
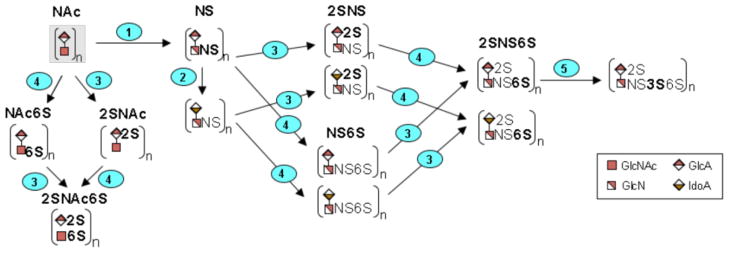
Graphic diagram of the modification of the repeating units in the HS/HP chain. Numbers on steps correspond to the reactions catalyzed by enzymes encoded by the following genes (1): NDST1, NDST2, NDST3, NDST4, (2): GLCE, (3): HST2T1, (4): HS6ST1, HS6ST2, HS6ST3, and (5): HS3ST1, HS3ST2, HS3ST3A1, HS3ST3B1, HS3ST4, HS3ST5, HS3ST6. While the initial, unsulfated substrate is believed to be modified by enzymes 1–5, in that order, the figure includes all other permutations that are possible to achieve the final product, HP/HS.
3.6 Change in the expression of CS/DS chain elongation and modification enzymes in splanchnic mesoderm cells compared to H9 cells
The CS/DS and HS/HP biosynthetic pathways share a common initial pathway but these diverge at linkage region biosynthesis (Figure 5). CS/DS linkage region biosynthesis and extension is accomplished by the activity of β-4-N-acetylgalactosamine transferases (CSGALNACTs) and chondroitin synthases (CHSY1, CHPF1, CHPF2, CHSY3) (Figure 7). Dermatan sulfate epimerases (DSE1 and DSE2) convert glucuronic acid (GlcA) to iduronic acid (IdoA) in DS biosynthesis. Sulfation patterns characteristic of CS and DS are achieved by activity of chondroitin 4-O-sulfotransferases (CHST11 and CHST12), a dermatan 4-O-sulfotransferase (D4ST1), chondroitin uronosyl sulfotransferase (UST), chondroitin 6-O-sulfotransferases (CHST3 and CHST7), and N-acetylgalactosamine 4S, 6S transferase (CHST15). Transcript levels for CSGALNACT1,, CHFP1 and CHPF2 all showed increased transcript levels with the greatest increase observed for CSGALNACT1 (~ 69-fold). No significant change was detected in transcript levels of CSGALNACT2, CHSY1, CHPF, and CHSY3. Statistically significant decreases for transcript abundances for both DSE1 (−1.3-fold) and DSE2 (−4.1-fold) were determined in differentiated cells compared to H9 cells. Transcript analysis of the genes involved in CS/DS chain modification showed no change (D4ST1) or small (< 5-fold), but statistically significant changes upon differentiation of H9 cells into splanchnic mesoderm cells (Table 2).
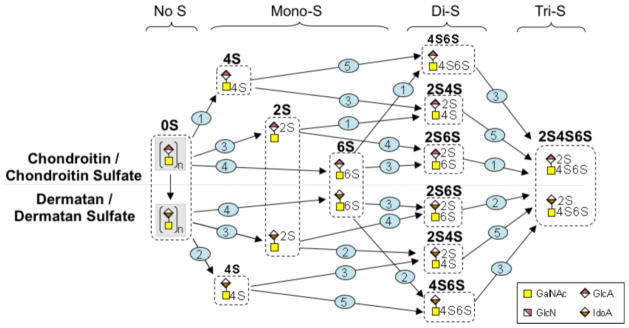
Graphic diagram of the modification of the repeating units in the CS/DS chain. Numbers on steps correspond to the reactions catalyzed by enzymes encoded by the following genes (1): CHST11, CHST12, (2): D4ST1, (3): UST, (4): CHST3, CHST7, and (5): CHST15. The figure includes all permutations that are possible to achieve the final product, CS/DS.
Table 2
Fold change in transcript abundance of chondroitin sulfate/dermatan sulfate biosynthetic genes in splanchnic mesoderm cells compared to H9 cells.
| Gene | Fold change* | Gene | Fold change* | Gene | Fold change* |
|---|---|---|---|---|---|
| CSGALNACT1 | 68.5** | CHPF2 | 3.1** | D4ST1 | 1.0 |
| CSGALNACT2 | −1.2 | DSE1 | −1.3** | UST | −1.6** |
| CHSY1 | 1.2 | DSE2 | −4.1** | CHST3 | 2.5** |
| CHPF1 | 2.0** | CHST11 | 1.8 | CHST7 | −1.4 |
| CHSY3 | 1.4 | CHST12 | 1.3** | CHST15 | 2.7** |
3.7 Structural changes in HS/HP GAGs in H9 cells differentiated into splanchnic mesoderm cells and premature hepatocytes
HS/HP chain structure was investigated in pluripotent H9 cells, splanchnic mesoderm cells and premature hepatocytes. In pluripotent H9 cells, the majority of cell-associated disaccharides were non-sulfated 0S disaccharides (75.5%). NS, 6S, NS2S and TriS disaccharides were also detected in H9 cells, but in far lower amounts (5.8%, 4.6%, 9.3%, and 5.1%, respectively). The 2S, NS6S and 2S6S disaccharides were not detected in H9 cells (Table 3). In splanchnic mesoderm cells, NS (31.8%), and 0S (42.1%) comprised the most substantial fraction of disaccharides. The 6S (5%), 2S (0.6%), NS6S (7.2%), NS2S (7.5%), and TriS (6%) disaccharides were also found in splanchnic mesoderm cells, whereas 2S6S structured disaccharides were not detected. In premature hepatocytes, the majority of disaccharides were comprised of 0S (49.1%), followed by NS6S (27.9%), NS (16.5%), 6S (6%), and 2S (0.4%). NS2S, 2S6S and TriS were not detected in premature hepatocytes.
Table 3
Distribution of heparan sulfate/heparin disaccharides in differentiated H9 cells.
| Sample | HS/HP disaccharide composition
| |||||||
|---|---|---|---|---|---|---|---|---|
| 0S | NS | 6S | 2S | NS6S | NS2S | 2S6S | TriS | |
| H9 cells
| 75.5 | 5.8 | 4.6 | 0 | 0 | 9.3 | 0 | 5.1 |
| Splanchnic mesoderm
| 42.1 | 31.8 | 5.0 | 0.6 | 7.2 | 7.5 | 0 | 6.0 |
| Immature hepatocytes | 49.1 | 16.5 | 6.0 | 0.4 | 27.9 | 0 | 0 | 0 |
3.8 Structural changes in CS/DS GAGs in H9 cells differentiated into splanchnic mesoderm cells and premature hepatocytes
CS/DS disaccharide structures were analyzed to establish changes in CS/DS accompanying differentiation of H9 cells towards splanchnic mesoderm and premature hepatic cells (Table 4). In pluripotent H9 cells the majority of CS/DS chains were comprised of 4S disaccharides (87.4%) followed by 6S containing structure (9.8%). Other disaccharides were also detected but in smaller amounts (0S (0.6%), 2S6S (1.3%), 2S4S (1.0%), 4S6S (0.4%). The 2S and TriS disaccharides were not detected in H9 cells. Upon differentiation towards splanchnic mesoderm cells the ratio of disaccharides changed, the majority becoming 6S (56.4%), followed by 4S (30.1%) and 0S (12.7). No additional disaccharides were detected in splanchnic mesoderm cells. CS/DS chains isolated from immature hepatocytes more closely resembled the structure of CS/DS from H9 cells. The major CS/DS disaccharide from premature hepatocytes was 4S (89.1%), followed by 6S (5.1%) and 0S (3.1%). The 2S4S and 4S6S were also detected but in smaller amounts (1.2% and 1.5%, respectively). The 2S, 2S6S, and TriS containing disaccharides were not detected in immature hepatocytes.
Table 4
Distribution of chondroitin sulfate/dermatan sulfate disaccharides in differentiated H9 cells.
| Sample | CS/DS disaccharide composition
| |||||||
|---|---|---|---|---|---|---|---|---|
| 0S | 6S | 4S | 2S | 2S6S | 2S4S | 4S6S | TriS | |
| H9 cells
| 0.6 | 9.8 | 87.4 | 0 | 1.3 | 1.0 | 0.4 | 0 |
| Splanchnic mesoderm
| 12.7 | 56.4 | 30.1 | 0 | 0 | 0 | 0 | 0 |
| Immature hepatocytes | 3.1 | 5.1 | 89.1 | 0 | 0 | 1.2 | 1.5 | 0 |
4. Discussion
Stem cells possess the potential to influence a vast range of important disciplines, but this prospective value is blunted by the necessity to define and harness advanced methods for controlling their differentiation. This challenge requires a thorough understanding of intrinsic and extrinsic signals that regulate stem cell fate. Although intrinsic components have garnered the majority of attention, the impact of the extrinsic components on cell signaling remains in its infancy. Extrinsic signals from the stem cell niche are essential in the direction of cell state. PGs are primarily associated with the extracellular environment and are indispensable components of the niche. Therefore, it is imperative to establish the role they play in stem cell biology. The importance of HSPGs with regards to cell exit from the pluripotency state has been demonstrated, as cells missing HS fail to differentiate [24]. ESCs express simple HS having N-sulfo groups [24]. Upon differentiation towards embryoid bodies, the level of 6-O-sulfo groups increases [23], and differentiation towards a neuronal fate is accompanied by elevated levels of N-sulfo, 2-O-sulfo, and 6-O-sulfo groups [24]. The introduction of 3-O-sulfo groups into HS has been shown to be important for transitioning of pluripotent mouse SCs to mouse epiblast stem cells [38]. In the current study, we differentiated the hESC line H9 into splanchnic mesoderm cells and premature hepatocytes to establish the glycosaminoglycanomic modifications accompanying cell differentiation processes. Comparison of the cell population differentiated towards hepatic lineage with the HepG2 cell line, using immunostaining with relevant markers, suggested that the H9 cells had differentiated into immature hepatocytes with a small fraction of mature hepatocytes. We used these cells along with splanchnic mesoderm cells to undertake our glycosaminoglycanomic analyses.
The HS biosynthetic pathway has been extensively characterized and the enzymes involved are well established [39]. Since HS biosynthetic enzymes are responsible for initiation, polymerization and modification of HS chains and the core proteins that carry those chains, the expression levels of genes encoding those enzymes and core proteins were inspected. Among HS core proteins, the most significant change was observed in the transcript levels of serglycin and glypican-5. Glypicans are cell surface associated PGs and are connected to the external side of the membrane through the GPI anchor. The role of glypicans in development and morphogenesis has been defined. They are known to play a role in the regulation of FGF, Hedgehog, Wnt, and BMP signaling primarily through their HS chains [46]. We did not detect a change in the protein expression levels of decorin or glypican-5 in cell based samples. Nevertheless, this does not preclude the possibility of observing such a change in cell culture media, which could account for the lack of correlation between cell associated protein and transcript abundance. This stands to reason because both decorin and glypican-5 are extracellular PGs that are often shed into the media, a possibility that was not investigated directly in this study. Serglycin is a PG that carries heparin chains and is found in connective tissue mast cells [47]. It has been proposed that serglycin might be involved in neurogenesis [48]. Even though we observed a change in serglycin transcript level, we could not correlate this change at the protein level by Western blotting. This contradiction could be explained by the fact that baseline protein expression of serglycin was too low to detect, and so a further drop in expression would remain undetectable, which coincides with the overall low transcript level for serglycin in differentiated Isl1+ cells.
Modification of HS/HP chains by biosynthetic enzymes is important for creating domains along the chains that are required for interaction with various signaling molecules. Although there were no sizeable changes in transcript levels of HS/HP chain initiation and elongation genes upon differentiation of H9 cells into splanchnic mesoderm cells, the abundances of HS/HP chain modification transcripts were affected. The most prevalent change was observed in the level of HS3ST1. There are seven isozymes of HS3ST, which have unique spatial and temporal distribution. The transcript of HS3ST1 increased, whereas the transcript level of HS3ST2 decreased in splanchnic mesoderm cells compared to H9 cells. HS3ST1 is found in kidney, brain and heart, whereas HS3ST2 was reported only in brain, which coincides with the increase in HS3ST1 transcripts in the cardiac progenitor Isl1+ line. These enzymes are responsible for introducing 3-O-sulfo groups into the HS/HP chain and these are important for interaction with proteins, such as antithrombin [37] and are also critical for differentiation of mouse ESCs through Fas signaling [49]. Due to the lack of available enzymes that can cut the HS/HP chains to yield 3-O-sulfo group containing disaccharides, we were unable to use LC-MS to verify whether the change in HS3ST transcript levels leads to a change in HS/HP structure. NDST1 is ubiquitously expressed while NDST3 is found in brain, kidney and liver [37]. While not as dramatic as H3ST1, we observed increases in expression for NDST1, GLCE, HS6ST2 and HS6ST3 transcripts. This result is echoed by the increase of NS and NS6S disaccharides detected by LC-MS.
We observed that the structure of HS/HP chains was modified upon differentiation towards different lineages. The most prominent change was the drop in the level of non-sulfated disaccharides and increase in N-sulfo group containing structures in both splanchnic mesoderm and early hepatic cells compared to H9 cells. Another significant change was the appearance of NS6S sulfated disaccharides upon loss of pluripotency, as this structure was observed only in differentiated cells and not in H9 cells. When compared to one another, splanchnic mesoderm cells closely resemble H9 cells in disaccharide composition, although the ratio of different disaccharides is distinctive in both cell types. The disaccharide composition and the ratio of different disaccharides show the greatest differences in premature hepatocytes when compared to H9 cells. The NS2S and TriS disaccharide were not found in premature hepatocytes, but two additional disaccharide structures, 2S and NS6S, appeared. This finding assigns premature hepatocytes a unique HS/HP disaccharide composition pattern. Taken together, we can surmise that H9 cells principally utilize non-sulfated HS/HP, and as development progresses, the HS/HP structure gains complexity through the introduction of sulfo groups at various positions in the HS/HP chains. The greater similarity of splanchnic mesoderm cells to H9 cells than premature hepatocytes to H9 cells can be explained by the fact that splanchnic mesoderm cells are mesendodermal cells that possess the ability to differentiate into either endodermal or mesodermal lineages, and appear earlier in development than early hepatic cells. As cells progress towards more committed cell types, such as premature hepatocytes, HS structural changes become more prominent.
The most significant change among CS core proteins was observed in the transcript levels of lumican and decorin. Lumican and decorin are small leucine-rich proteoglycans that can carry CS, DS and KS chains. The molecular weight of lumican is known to be heterogeneous, and it is initially translated as a precursor protein of ~ 50 kDa. However, in adult cells it is observed to be ~ 37 kDa, and this corresponds to the non-GAGylated form of lumican. Since it does have several potential glycosylation sites, it is also observed with a ladder of higher molecular weights [40]. This has been previously reported to occur during corneal development where it begins as a non-GAGylated protein, and as development progresses it is observed in the GAGylated form [41]. This is consistent with results observed from lumican expression upon differentiation of pluripotent stem cells towards splanchnic mesoderm cells. H9 cells predominantly utilize the non-GAGylated form of lumican, whereas in splanchnic mesoderm cells, the principal portion of lumican is a PG and this change in structure correlates well with the increased transcript level observed upon differentiation. Since KS is the major GAG that lumican carries, we speculate that KS is required for differentiation towards splanchnic mesoderm cells. Unfortunately, there is little information available on the role of KS in developmental processes, apart from those implicating its role in embryo implantation [42] and its inhibitory effect on neurite growth [43]. The increase in the transcript level of lumican quantified by qRT-PCR correlated well at the level of translation, which was assessed by immunoblotting. Decorin was shown to be important in the homeostasis regulation of satellite cells, which are committed muscle cell progenitors, through competition with TGF-β for TGF-β receptors [44]. Decorin expression has been shown to increase upon differentiation of the teratocarcinoma cell line NCCIT towards a neural lineage when treated with retinoic acid [45]. No correlation was observed between transcript abundance and protein levels for decorin. In fact, no transcripts could be detected for decorin in the undifferentiated cell population, which suggests that perhaps a minute amount of transcript is required for successful decorin translation. Variations observed in transcript and protein levels may be due to a number of factors, including, but not limited to mRNA and protein stability (RNA < protein), speed of generation of mRNA and protein (RNA < protein) and cell regulatory events which include post-transcriptional, translational and protein degradation regulation (RNA > protein).
The expression of CS/DS biosynthetic gene transcripts was not dramatically affected upon differentiation of H9 cells into splanchnic mesoderm cells, with the exception of CSGALNACT1 (involved in linkage region formation). Despite the lack of changes in transcript levels, we detected changes in the structure of CS/DS chains isolated from splanchnic mesoderm cells. While the majority of disaccharides were comprised of 4S and 6S containing structures, the ratio between those types of disaccharides also changed. CS/DS chains isolated from splanchnic mesoderm cells contained more non-sulfated structures compared to pluripotent H9 cells. There was not much change detected in the structure of CS/DS chains isolated from premature hepatocytes. This variation may be because the premature hepatocytes are derived from the endoderm while the splanchnic mesoderm cells are derived from the mesoderm. In general, we may conclude that the most significant changes were associated with HS/HP chains upon differentiating H9 cells into splanchnic mesoderm and premature hepatic cells.
The knowledge gained from a greater understanding of the processes leading to differentiation towards hepatocytes and splanchnic mesoderm cells is not limited to advancement of the treatment of damaged tissues, but can also be useful in drug discovery. The ability of hepatocytes to detoxify numerous compounds makes them invaluable for the homeostatic upkeep of organisms. This feature of hepatocytes can be harnessed in vitro for drug toxicology studies. Control of stem cell fate, coupled with high throughput platforms [50], can open the door for parallel toxicological screening of drugs and drug candidates on hepatocytes. Understanding the processes that lead to differentiation towards splanchnic mesoderm cells, and further towards cardiac progenitor cells, might one day be useful for patients with heart damage. Future work will be directed towards defining the underlying mechanisms by which the various GAG structures influence differentiation pathway decision making of stem cell fate.
Acknowledgments
The authors are grateful to Eric R. Gamache for proofreading and offering helpful suggestions for this manuscript. We also would like to acknowledge Dr. Hope E. Stansfield for general help and comments. This study was supported by New York State Department of Health-Empire State Stem Cell Board [grant number C024334] and the National Institutes of Health [grant numbers ES020903, P41RR018502 and P01GM085354].
Appendix A. Supplementary data
Supplementary data to this article can be found online at …
Footnotes
1Karyotyping of WA09 cells was done by Cell Line Genetics Inc., Madison, WI.
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
Full text links
Read article at publisher's site: https://doi.org/10.1016/j.bbagen.2014.01.007
Read article for free, from open access legal sources, via Unpaywall:
https://europepmc.org/articles/pmc4004645?pdf=render
Citations & impact
Impact metrics
Article citations
Genomic Diversity of the Retinta Breed Derived from Two Ancestral Bovine Lineages.
Vet Sci, 11(6):247, 30 May 2024
Cited by: 0 articles | PMID: 38921994 | PMCID: PMC11209511
Glycan Modifications as Regulators of Stem Cell Fate.
Biology (Basel), 13(2):76, 26 Jan 2024
Cited by: 3 articles | PMID: 38392295 | PMCID: PMC10886185
Review Free full text in Europe PMC
Dynamic Changes in Heparan Sulfate Nanostructure in Human Pluripotent Stem Cell Differentiation.
ACS Nano, 17(8):7207-7218, 12 Apr 2023
Cited by: 2 articles | PMID: 37042659 | PMCID: PMC11439449
The aberrant cancer metabolic gene carbohydrate sulfotransferase 11 promotes non-small cell lung cancer cell metastasis via dysregulation of ceruloplasmin and intracellular iron balance.
Transl Oncol, 25:101508, 16 Aug 2022
Cited by: 6 articles | PMID: 35985204 | PMCID: PMC9418604
CHST15 gene germline mutation is associated with the development of familial myeloproliferative neoplasms and higher transformation risk.
Cell Death Dis, 13(7):586, 07 Jul 2022
Cited by: 9 articles | PMID: 35798703 | PMCID: PMC9263130
Go to all (23) article citations
Data
Data behind the article
This data has been text mined from the article, or deposited into data resources.
BioStudies: supplemental material and supporting data
Similar Articles
To arrive at the top five similar articles we use a word-weighted algorithm to compare words from the Title and Abstract of each citation.
Micropatterning of human embryonic stem cells dissects the mesoderm and endoderm lineages.
Stem Cell Res, 2(2):155-162, 03 Dec 2008
Cited by: 59 articles | PMID: 19383420
Regulated Nodal signaling promotes differentiation of the definitive endoderm and mesoderm from ES cells.
J Cell Sci, 120(pt 12):2078-2090, 29 May 2007
Cited by: 44 articles | PMID: 17535850
Undersulfation of heparan sulfate restricts differentiation potential of mouse embryonic stem cells.
J Biol Chem, 287(14):10853-10862, 01 Feb 2012
Cited by: 42 articles | PMID: 22298785 | PMCID: PMC3322844
Hepatocyte differentiation: from the endoderm and beyond.
Curr Opin Genet Dev, 11(5):568-574, 01 Oct 2001
Cited by: 110 articles | PMID: 11532400
Review
Funding
Funders who supported this work.
NCRR NIH HHS (2)
Grant ID: P41 RR018502
Grant ID: P41RR018502
NHLBI NIH HHS (2)
Grant ID: R01 HL062244
Grant ID: R01 HL096972
NIEHS NIH HHS (2)
Grant ID: ES020903
Grant ID: R01 ES020903
NIGMS NIH HHS (4)
Grant ID: P01 GM085354
Grant ID: R01 GM038060
Grant ID: P41 GM103490
Grant ID: P01GM085354
National Institutes of Health (3)
Grant ID: ES020903
Grant ID: P41RR018502
Grant ID: P01GM085354
New York State Department of Health—Empire State Stem Cell Board (1)
Grant ID: C024334