Abstract
Free full text

Trafficking of endosomal Toll-like receptors
Abstract
Over the last decade we have learned much about nucleic acid recognition by the innate immune system and in particular by Toll-like receptors (TLRs). These receptors localize to endosomal compartments where they are poised to recognize microbial nucleic acids. Multiple regulatory mechanisms function to limit responses to self DNA or RNA, and breakdowns in these mechanisms can contribute to autoimmune or inflammatory disorders. In this review we discuss our current understanding of the cell biology of TLRs involved in nucleic acid recognition and how localization and trafficking of these receptors regulates their function.
Innate Immune Receptors and Pattern Recognition
The immune system utilizes a variety of receptors to detect potentially harmful microbes, such as bacteria, viruses and fungi. The adaptive immune system consists of millions of lymphocytes, each expressing a distinct antigen receptor generated through gene rearrangement, and provides an extremely broad repertoire of specificities. The innate immune system employs a distinct strategy of microbial recognition with a limited set of receptors expressed broadly on multiple cell types. Thus, compared to the adaptive immune system, the innate immune system sacrifices breadth for the ability to respond rapidly [1].
Over the last fifteen years, several families of innate immune receptors have been described, including Toll-like receptors (TLRs), RIG-I like receptors (RLRs), AIM2-like receptors (ALRs), Nod-like receptors (NLRs) and C-type Lectin receptors (CLRs) [2–6]. Because the specificities of innate receptors are germline-encoded, they have been shaped through evolution to recognize highly conserved features of microbes, also called pathogen associated molecular patterns (PAMPs) [1]. Examples of PAMPs are lipopolysaccharide (LPS) and lipoteichoic acid, both conserved features of broad bacterial classes [2] (Figure 1). Targeting PAMPs enables innate immune cells to recognize diverse microbes with relatively few receptors. Signals induced by innate receptors not only initiate rapid antimicrobial defenses but also control activation of adaptive immunity [7]. In this way the innate immune system provides key information about the nature of antigen; that is, activation of innate receptors determines when an immune response is appropriate.

TLRs are synthesized in the endoplasmic reticulum (ER), traffic to the Golgi, and ultimately localize to the cell surface or remain intracellular in endosomes or lysosomes. All TLRs have a horseshoe-like ectodomain structure and interact with their ligands as dimers. Surface resident TLRs 1, 2, 4, 5, and 6 recognize microbial ligands such as lipopolysaccharide (LPS), bacterial lipoproteins, and flagellin. Most endosomal TLRs recognize microbial derived nucleic acids. The ectodomains of these nucleic acid sensing TLRs undergo proteolytic processing in endosomes to generate functional receptors capable of signaling upon ligand recognition.
While innate immune recognition of microbes is critical for effective immunity, it is also important that the immune system avoid responses to self molecules that could lead to autoimmunity. This possibility may appear unlikely, based on the specificity of innate receptors for PAMPs, but there is now considerable evidence that recognition of self ligands by innate immune receptors contributes to many autoimmune or autoinflammatory disorders [8]. This concept is particularly well-established for members of the TLR family, a subset of which recognizes nucleic acids, which can be foreign or self-derived [8] (Figure 1). Numerous regulatory mechanisms exist to prevent the activation of TLRs by self molecules, but one major aspect appears to be tight control of receptor localization and trafficking. In this review we discuss recent findings that examine the cell biological mechanisms that regulate nucleic acid recognition by TLRs and the consequences when these mechanisms fail.
Nucleic acid recognition by TLRs
TLRs comprise a family of type I transmembrane proteins, each with an N-terminal ectodomain consisting of multiple leucine-rich repeat (LRR) domains involved in ligand binding, as well as a C-terminal cytosolic region containing a Toll/Interleukin-1 receptor (TIR) domain that mediates recruitment of signaling components [2]. Based on existing structures of TLR ectodomains, the activated, ligand-bound state appears to be a dimer [9–16]. TLRs utilize common signaling adaptor molecules, MyD88 and/or TRIF, to initiate signaling [2].
Viruses and bacteria typically enter cells through endocytic or phagocytic pathways, which can lead to their degradation and release of microbial nucleic acids [17] (Figure 1). Multiple TLRs recognize specific nucleic acid structures: TLR3 recognizes double stranded RNA (dsRNA) [18], TLR7 and TLR8 recognize single stranded RNA (ssRNA) [19–22] and TLR9 recognizes DNA [23]. Additionally, TLR13, until recently an orphan receptor, recognizes ssRNA [24–26]. For some of these TLRs, specific sequence motifs within RNA or DNA are required for optimal ligand binding, and there is evidence that the optimal motifs differ between species [27–33]. Activation of nucleic acid sensing TLRs triggers downstream signaling and activation of transcription factors such as NF-κB and IRFs to promote pro-inflammatory and anti-viral responses, respectively [2].
The use of nucleic acids to detect the presence of microbes may seem paradoxical due to the potential for inappropriate recognition of host nucleic acids. In fact, nucleic acids can be present in the extracellular milieu, either through non-apoptotic forms of cell death, secondary necrosis of uncleared apoptotic debris, or release of DNA by neutrophils during NETosis [34–36]. Inappropriate recognition of these host-derived nucleic acids can lead to autoimmune or autoinflammatory disorders, such as systemic lupus erythematosus (SLE) [37, 38]. While these potential risks are great, the emergence of nucleic acid recognition as a general strategy of innate immune recognition suggests an evolutionary benefit that outweighs the risk of responding to self nucleic acids. Therefore, mechanisms must exist that reduce the likelihood of responses to self nucleic acids. For example, nucleases present in the extracellular environment as well as within endosomes degrade free nucleic acids that could otherwise activate TLRs. Nucleic acids within viral particles or bacteria, however, are protected from these nucleases and can activate TLRs when released in protease-rich acidic compartments [17, 39]. A series of studies over the past several years support the idea that regulation of receptor trafficking and localization is a critical aspect of limiting responses to host nucleic acids. Examining how TLRs maintain proper self versus non-self discrimination provides an attractive opportunity to study cell biology while also providing mechanistic insights to autoimmune diseases.
Regulation of nucleic acid sensing TLRs
Despite sharing common structural and functional features, individual TLRs possess unique characteristics, including the subcellular compartments to which they localize. TLRs 1, 2, 4, 5, and 6 localize to the plasma membrane, while TLRs 3, 7, 8, 9, and likely 11, 12 and 13 localize within intracellular compartments [2, 26, 40, 41] (Figure 1). The localization of nucleic acid sensing TLRs within endosomes and lysosomes places these receptors in a strategic position to encounter nucleic acids released from engulfed microbes. Endosomal or lysosomal localization is also necessary for function and activation of this subset of receptors. For example, neutralizing the pH of endosomes and lysosomes with drugs such as bafilomycinA1 and chloroquine inhibits the activation of nucleic acid sensing TLRs [29, 42]. Additionally, initiation of anti-viral responses by TLRs is generally reserved for TLRs activated within endosomes where they activate transcription factors IRF3 or IRF7 to induce type I interferon (IFN) [43, 44].
Intracellular localization may also provide a mechanism to prevent recognition of self nucleic acids. The first evidence that the endosomal localization of TLRs is important for maintaining appropriate responses to nucleic acids came from studies of TLR9- and TLR7-dependent responses to internalized self DNA- or RNA-containing immune complexes. Normally, self nucleic acids do not stimulate TLR9 or TLR7 expressing cells; however, uptake through Fc receptors or B cell antigen receptors, or through association with anti-microbial peptide LL37 or secreted HMGB1, can efficiently deliver these self nucleic acid ligands to TLR-containing endosomes [45–51]. The mechanisms underlying this process are under active investigation, but, regardless of the precise regulatory details, these experiments highlight the danger associated with bypassing the compartmentalization of TLRs capable of nucleic acid recognition.
Unlike their surface localized counterparts, the ectodomains of TLRs 3, 7, and 9 are proteolytically processed in endosomal compartments by endosomal proteases such as cathepsins and asparagine endopeptidase in a step-wise manner [52–55] (Figure 1). This cleavage event separates an N-terminal portion of the ectodomain from the rest of the TLR. The resulting C-terminal fragment contains the truncated ectodomain, transmembrane, and cytosolic regions and acts as a functional, cleaved receptor. Surprisingly, while proteolytic processing is not required for ligand binding, it is necessary to initiate downstream signals because only the cleaved receptor is able to recruit MyD88, and inhibition of processing blocks receptor signaling [52]. Studies examining ligand binding by TLR9 suggest that a conformational change occurs in the ectodomain upon binding [56]. Ectodomain cleavage may facilitate this conformational change, which seems to reorient the cytosolic TIR domains and promote adaptor recruitment.
Despite the strong evidence supporting a functional requirement for ectodomain cleavage, several studies suggest that the N-terminal portion of the receptor plays a role in ligand binding and signaling. First, a single amino acid mutation in the N-terminus of TLR9 resulted in a signaling defect, suggesting a role for the N-terminus for activation [57]. Second, structural studies of TLR3 identified binding sites for polyI:C (a synthetic dsRNA ligand) in both N- and C-terminal regions [9, 58–60]. Third, recent evidence suggests that N-terminal and C-terminal fragments of TLR3 remain associated after proteolytic processing [53, 61]. Finally, a recent structure of human TLR8 bound to ligand revealed separated N- and C-terminal fragments making contacts with ligand [15]. Thus, it seems quite likely that both the N- and C-terminal fragments facilitate ligand binding. Whether the N-terminal contacts are absolutely required for receptor function remains controversial, as two reports show that TLR9 deficiency is rescued by expression of a pre-truncated receptor [53, 54].
Why did this complex regulatory step evolve? The requirement for proteolytic processing may serve as an additional safeguard against receptor activation at the cell surface, where self nucleic acids are more likely to be encountered [39]. Because the proteases required for processing are only active within endosomes and lysosomes, the requirement for ectodomain cleavage limits receptor function to these intracellular compartments. Mutation of TLR receptor domains in cells or mice has demonstrated the importance of receptor localization in self nucleic acid recognition. Redirecting TLR9 to the cell surface by swapping the transmembrane domains and cytosolic regions of TLR9 with that of TLR4, a surface localized TLR, enabled TLR9 to respond to vertebrate genomic DNA [62]. However, because signaling downstream of this chimeric receptor was mediated by the TLR4 cytosolic domain, these studies were limited to in vitro analyses of ligand recognition [62]. A separate study introduced mutations in the transmembrane domain of TLR9, creating a mutant receptor (TLR9TM-MUT) that localized to the cell surface and no longer required proteolytic processing for activation [63]. The more conservative mutations in this receptor enabled more relevant in vivo studies assessing the consequence of altered receptor localization and regulation. TLR9TM-MUT gained the ability to respond to vertebrate genomic DNA, and mice expressing TLR9TM-MUT developed systemic lethal inflammation. Thus, both intracellular sequestration and proteolytic cleavage of nucleic acid sensing TLRs appear to be necessary safeguards to ensure proper self versus non-self discrimination.
Trafficking from the ER
TLRs traffic via the conventional secretory pathway from the endoplasmic reticulum (ER) to the Golgi where they are sorted to the endosomal network. While it is understood that proper trafficking of nucleic acid sensing TLRs is a critical regulatory step for activation and self versus non-self discrimination, our understanding of how these receptors gain access to signaling compartments remains incomplete. A critical mode of regulation may be at the initial step of ER export, as alterations at this step may determine the level of functional receptor present in endolysosomes and thereby influence the threshold of receptor activation. Controlling this threshold could be highly relevant in autoimmunity. For example, simply overexpressing TLR7 is sufficient to cause autoimmune disease in mice [64–66].
UNC93B1 is an important regulatory factor that controls TLR trafficking from the ER [67]. Unc93b1 was originally identified through a forward genetic screen in mice where a mutation (H412R) resulted in a defect in the signaling of nucleic acid sensing TLRs 3, 7, and 9 but not surface localized TLRs [68]. Subsequent studies have focused on the mechanistic details of UNC93B1 function, which indicate that this ER-resident protein is required for proper trafficking of multiple TLRs [67, 69]. UNC93B1 facilitates incorporation of TLR9 into COPII vesicles, which transport protein cargo from the ER to Golgi [69] (Figure 2A). This finding opens the possibility that active transport mechanisms such as cargo selection by Sec24 proteins (part of the COPII machinery) may be involved in regulating ER export of TLRs. TLRs 3, 7, 11, 12 and 13 also require UNC93B1 to exit the ER [69], presumably via loading into COPII vesicles, although this mechanism has not been formally demonstrated for these TLRs. Further research will certainly focus on how the levels of individual TLR export are established and whether this ER trafficking step is regulated in response to external stimuli.
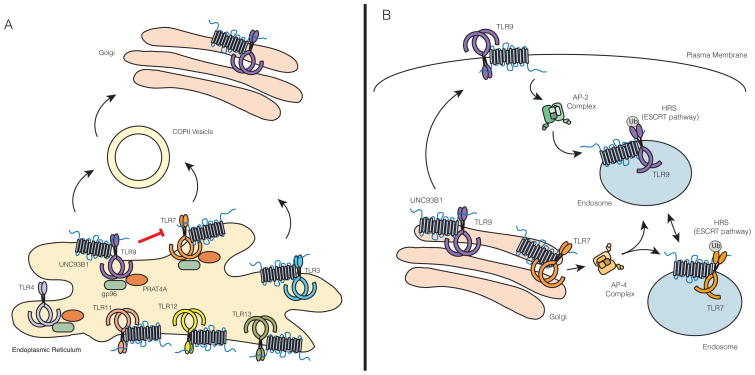
(A) Regulation at the Endoplasmic Reticulum (ER). ER export is an important mode of regulation for controlling levels of functional TLRs in the cell. TLRs 3, 7, 8, 9, 11, 12, and 13 require UNC93B1 to exit the ER. UNC93B1 is necessary for TLR9 incorporation into COPII vesicles. There is evidence for direct competition between TLR7 and TLR9 for UNC93B1, as the presence of TLR9 inhibits trafficking of TLR7. To what extent other TLRs compete for UNC93B1 is still an open question. For most TLRs, with the exception of TLR3, trafficking from the ER also requires the folding chaperones gp96 and PRAT4A. (B) Trafficking routes from Golgi to endosome. Upon reaching the Golgi, nucleic acid sensing TLRs are sorted to endosomes. UNC93B1 traffics with nucleic acid sensing TLRs to endosomes, but the role for UNC93B1 in mediating TLR trafficking from Golgi to endosomes differs for individual TLRs. While TLR9 requires UNC93B1 to recruit the AP-2 complex at the cell surface to mediate endocytosis of the UNC93B1/TLR9 complex, TLR7 can directly recruit the AP-4 complex and mediate its own trafficking to endosomes. The trafficking pathways for other endosomal TLRs have not been directly investigated. TLR9, and presumably TLR7, are ubiqutinated and recognized by the HRS/ESCRT pathway to maintain receptor stability in lysosomes.
The determinants of UNC93B1 specificity for TLRs, especially discrimination between individual TLRs, remain poorly defined. While the UNC93B1-H412R mutant fails to interact with TLRs, this mutation likely destabilizes the protein rather than disrupts specific interactions [70]. Analyses of chimeric TLRs have implicated TLR transmembrane domains as critical for UNC93B1 association [70]. Furthermore, the interaction between UNC93B1 and TLRs in the ER is a necessary step for ER export, since TLR9 cannot exit the ER in the presence of UNC93B1-H412R [69]. A recent study has demonstrated the importance of acidic residues (D812 and E813) juxtaposed to the transmembrane region of TLR9 for association with UNC93B1 [71]. While these residues alone are not sufficient to mediate interaction with UNC93B1, they are necessary for binding and subsequent UNC93B1-dependent trafficking. Similar residues were identified in TLR3; however, other UNC93B1 dependent TLRs have yet to be examined and the complementary residues on UNC93B1 have yet to be identified [71]. Previous studies have also described roles for other regions within TLRs for proper trafficking. For example, the linker region, a short amino acid sequence between the transmembrane and TIR domains, is important for the proper trafficking of TLR3, while the transmembrane domain of TLR7 is predominantly responsible for its proper trafficking [72, 73]. Additionally, TLR9 trafficking was shown to be dependent on both its transmembrane and cytosolic regions [74–76]. These studies did not examine in detail whether these regions have any functional relationship with UNC93B1, and so, it will be interesting to identify the regions of TLRs important for both UNC93B1-dependent and UNC93B1-independent trafficking.
A number of additional factors regulate TLR exit from the ER, but these proteins generally function as folding chaperones [77] (Figure 2A). Glycoprotein 96 (gp96), a paralogue of heat-shock protein 90 (HSP90), is involved in the folding of many surface proteins, including integrins and TLRs in the ER [78]. Protein associated with TLR4 A (PRAT4A), also known as CNPY3, is more specifically required for TLR folding, but, unlike UNC93B1, does not discriminate between intracellular or surface TLRs [79]. Gp96 and CNPY3 seem to work together to coordinate the folding of all TLRs, with the exception of TLR3 [80]. Whether these factors coordinate with UNC93B1 for TLR trafficking or whether additional factors exist that control TLR trafficking remain unanswered questions.
Trafficking beyond the ER
After export from the ER, nucleic acid sensing TLRs pass through the Golgi before they are sorted to endosomal compartments [52, 55, 81]. The molecular details underlying these sorting events remain poorly defined, although recent studies have identified some of the key players. Surprisingly, UNC93B1 remains associated with TLRs after ER exit and plays a role in these later trafficking events [67, 69]. The first mechanistic demonstration of a role for UNC93B1 function beyond ER export was revealed through identification of a critical YxxΦ motif in the C terminal region of UNC93B1. Mutating this motif (Y539A) disrupted the direct interaction between UNC93B1 and AP-2, an adaptor protein complex that facilitates endocytosis from the cell surface. In cells expressing this UNC93B1 mutant, TLR9 fails to reach endosomes and accumulates at the plasma membrane [69]. This surprising finding indicates that TLR9 traffics via the cell surface en route to endosomes, further justifying the requirement for proteolytic processing as a mechanism to prevent activation at the cell surface (Figure 2B).
A genome-wide RNAi screen to search for genes involved in TLR7 and TLR9 activation identified the hepatocyte growth factor-regulated tyrosine kinase substrate (HRS), a component of the ESCRT pathway, as a key factor in TLR7 and TLR9 trafficking [82]. The canonical ESCRT pathway is involved in receptor downregulation from the cell surface and receptor trafficking to multi-vesicular bodies for degradation in lysosomes. However, TLR9 and TLR7 seem to adopt the use of a non-canonical ESCRT pathway in which ESCRT is used for sorting or recycling rather than degradation [82]. TLR9, and presumably TLR7, is ubiquitinated by an as yet unidentified E3 ubiquitin ligase in a post-ER compartment. This ubiquitylation step enables recognition by HRS and trafficking to multi-vesicular bodies in order to maintain residence in lysosomal compartments [82] (Figure 2B).
Differential trafficking of nucleic acid sensing TLRs by UNC93B1
Differential regulation of TLR7 and TLR9 is of particular interest because of the role for these receptors in SLE. TLR7 and TLR9 can recognize nucleic acid-protein complexes and contribute to disease pathology in SLE-like mouse models, such as MRL/lpr [8]. In this model, disease is ameliorated by a loss of TLR7 expression but exacerbated by a loss of TLR9 [83]. Recent studies in mice deficient in both TLR7 and TLR9 show that loss of TLR7 can rescue the exacerbated disease seen in TLR9 deficient mice [84]. This study, along with in vitro studies showing the inhibitory effect of TLR9 on TLR7 function through physical interaction, suggests that TLR9 regulates the levels of TLR7 [85].
There is accumulating evidence that UNC93B1 mediates differential trafficking of TLR7 and TLR9, potentially at multiple levels. A point mutation (D34A) in the N terminus of UNC93B1 enhanced TLR7 responses, while concomitantly suppressing TLR9 responses [86]. Further examination of cells expressing UNC93B1-D34A revealed an increase in interaction with TLR7 and simultaneous decrease with TLR9 resulting in increased endosomal TLR7 and decreased TLR9. The study also observed enhanced interaction between UNC93B1-D34A and TLR8 as well as TLR13, which share specificity for ssRNA [22, 24, 25, 86]. Thus, the N-terminal portion of UNC93B1 appears to be involved in the selectivity of this chaperone for distinct TLR family members, and the D34A residue in particular is required for interaction with TLR9. Furthermore, mice expressing Unc93b1 mutant D34A alleles develop a systemic lethal inflammation [87]. This outcome is the result of enhanced TLR7 responses with a corresponding diminishment in TLR9 responses. Moreover, the disease is entirely TLR7-dependent, consistent with the finding that overexpression of TLR7 can cause a SLE-like disease [64–66, 87]. These observations support a model in which competition for UNC93B1 binding by individual TLRs, presumably in the ER, determines their rate of export, and aberrations in this regulatory step can lead to disease.
Further evidence for differential regulation of endosomal TLR trafficking comes from analysis of post-ER sorting by UNC93B1. While TLR9 utilizes UNC93B1 to interact with AP-2, TLR7 trafficking is unexpectedly independent of this pathway. Instead, TLR7 appears to directly recruit AP-4, a distinct sorting complex known to mediate direct transport between the trans Golgi network and endosomes [69] (Figure 2B). Again, these results argue that TLR binding to UNC93B1 must be mutually exclusive, at least for the TLRs that utilize these distinct trafficking pathways.
Altogether, these findings suggest that the use of distinct trafficking machinery may enable differential regulation of these two receptors thereby explaining the differential involvement of TLR7 and TLR9 in SLE. Moreover, these mechanisms may serve to deliver TLRs to distinct functional compartments, a possibility that we discuss in the following section.
Type I interferon production from specialized endosomes
Another layer of complexity in endosomal TLR biology is revealed by analyses of gene induction. A major conceptual breakthrough was achieved by Taniguchi’s group when they demonstrated that the signals leading to the transcriptional induction of two classes of genes (proinflammatory cytokines versus type I IFN) originate from different types of endosomes [44]. This observation has led to speculation that endosomal TLRs populate heterogeneous endosomes possessing distinct signaling capabilities.
The clearest example of distinct TLR-dependent gene induction between cell types comes from analyses of different subsets of dendritic cells and macrophages. Plasmacytoid dendritic cells (pDCs) produce large amounts of type I IFN in response to TLR7 and TLR9 engagement while other innate immune cell types, such as conventional dendritic cells (cDCs) and macrophages, produce very little type I IFN in comparison. Initially, this difference was attributed to high basal expression of IRF7 and distinct signaling pathways by pDCs, but subsequent work has demonstrated that the cell biology of pDCs may also be a key feature [44, 88–91]. This possibility was first suggested by the finding that different classes of synthetic oligodinucleotide (ODNs) elicit very different downstream signaling outcomes in pDCs [44, 91–93]. For example, CpG-A ODN, which aggregate to form large complexes, strongly induce type I IFN, while CpG-B ODN, which remain monomeric, instead favor a pro-inflammatory response characterized by TNFα, IL-6 and IL-12p40 production. Remarkably, this phenomenon is the result of differential trafficking of the ODNs to endosomal compartments, where TLR9 signaling leads to activation of distinct transcription factors [44] (Figure 3). CpG-A preferentially traffics to early endosomes where TLR9 signaling recruits the transcription factor IRF7 and leads to a type I IFN response [44]. In contrast, CpG-B preferentially traffics to late endosomes where TLR9 signaling activates transcription factor NF-κB, resulting in a proinflammatory cytokine response [44] (Figure 3). Why CpG-A and CpG-B localize to different endosomal compartments remains unclear. Studies suggest that the structural conformations of the ODNs determine their distinct localization. Consistent with this model is the observation that complexing CpG-B with a cationic lipid DOTAP alters the localization of CpG-B to an early endosomal compartment in pDCs resulting in type I IFN production [44]. Whether sorting to different compartments occurs upon engagement of endocytic receptors at the surface or additional sorting receptors in the endocytic pathway remains unclear. Altogether, the location of TLR9 and ligand engagement determines the specific cytokine outcome in pDCs. It is also clear that these distinct endosomal signaling compartments exist in cDCs and macrophages. While CpG-A alone can promote a proinflammatory response in these cells, it does not induce type I IFN unless complexed with the liposomal transfection reagent DOTAP [44]. Thus, although some cell biological mechanisms may be unique to pDCs, it is clear that functionally distinct signaling compartments exist in other innate immune cells.
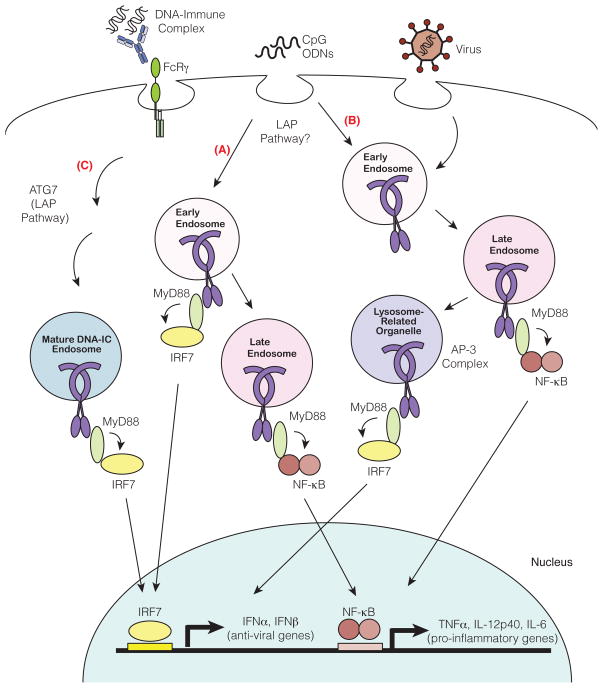
Nucleic acids encounter TLRs in endosomes after uptake from the extracellular milieu. Depending on the method of delivery into the cell (e.g. in an immune complex, as oligonucleotides, or encapsulated in a viral particle), nucleic acids may reach distinct endosomes with unique signaling properties. Early and late endosomes are spatially and temporally distinct from one another. TLR9 signaling from late endosomes leads to activation of NF-κB transcription factors and induction of pro-inflammatory cytokines (TNFα, IL-12p40 and IL-6) (A and B). TLR9 signaling from a distinct population of endosomes leads recruitment of IRF and induction of type I IFN (IFNα, IFNβ). One model suggests that this specialized compartment is simply early endosomes (A). Two other compartments capable of type I IFN induction have been described, and it remains unclear whether these three compartments are, in fact, distinct. The AP-3 complex regulates TLR9 trafficking to mature lysosome related organelles (LROs), compartments distinct from endosomes and lysosomes and capable of initiating type I interferon signaling (B). Second, a compartment for DNA-immune complex (DNA-IC)-dependent IFNα signaling requires components of the autophagy pathway through a mechanism termed LC3 associated phagocytosis (LAP). DNA-IC compartments mature using solely the LAP pathway and not the AP-3 dependent pathway (C). In contrast, CpG also utilizes autophagy pathways (mediated by ATG5 and ATG7) but also requires AP-3 for type I IFN signaling. While much of characterization of these compartments has focused on TLR9, it is likely that TLR7 populates similarly diverse and specialized endosomes.
Defining the molecular features of specialized endosomes capable of type I IFN signaling has been an area of intense study. Several studies have implicated AP-3, an adaptor complex that facilitates trafficking from endosomes to lysosome related organelles (LROs), as a critical mechanism for TLR7- and TLR9-dependent type I IFN responses [94, 95]. AP-3 was shown to interact directly with TLR9 and facilitate trafficking from endosome compartments marked by VAMP3 to specialized LRO compartments marked by LAMP2 [94] (Figure 3). Additionally, AP-3 can associate with IRF7 and TRAF3, perhaps linking these signaling components of the type I IFN signaling pathway to LROs [94]. PDCs from Ap3b1-deficient mice fail to produce type I IFN in response to TLR7 and TLR9 ligands, but there remains some disagreement about the selectivity of this pDC defect. One study reported that Ap3b1-deficient pDCs produce normal levels of other cytokines like IL-12p40 [94] while another group reported that all TLR7- or TLR9-dependent cytokine production is absent in mutant cells [95]. The extent of the defect is not a minor detail, as a complete block of TLR7 and TLR9 signaling would imply that AP-3 plays a more general role in TLR trafficking, at least in pDCs. The potential role for LROs in TLR9 mediated type I IFN signaling is difficult to reconcile with previous studies identifying early endosomes as the key compartment, as these two organelles are quite distinct. Nevertheless, additional support for the importance of LROs in TLR9 and TLR7 signaling comes from the finding that mutations in additional genes required for LRO biogenesis (BLOC-1 and BLOC-2) also impact TLR9 and TLR7 function in pDCs [95].
A third specialized compartment, possibly distinct from early endosomes and/or LROs, has been described for the TLR9-dependent type I IFN response to DNA-antibody immune complexes (DNA-IC) by pDCs. Uptake of these complexes induces a non-canonical autophagy pathway called LC3-associated phagocytosis (LAP) [96]. TLR9 and UNC93B1 are recruited to endosomal compartments after DNA-IC uptake, but the nature of these compartments is influenced by components of the LAP pathway [96]. In Atg7-deficient pDCs, DNA-IC compartments acquire the early maturation marker VAMP3 but fail to recruit maturation markers, LAMP1 or LAMP2, and consequently fail to produce IFNa in response to DNA-IC [96]. Thus, the LAP pathway appears to be involved in trafficking of DNA-IC to an IFNα signaling compartment (Figure 3). Interestingly, DNA-IC compartments are distinct from AP-3-dependent LROs since the IFNα response induced by DNA-IC stimulation is normal in Ap3b1-deficient pDCs [96]. Although it is tempting to classify the CpG and DNA-IC type I IFN pathways as completely distinct, it is important to note that autophagy is also involved in CpG-dependent IFNα signaling in pDCs. Both Atg7- and Atg5-deficient pDCs show some reduction in CpG-dependent IFNα signaling [96] [97]. Nevertheless, the differential dependence on AP-3 by DNA-ICs and CpG supports the idea that distinct mechanisms exist to mediate responses to different types of cargo.
As is clear from the results discussed here, the field is grappling with the apparent heterogeneity of endosomal compartments from which TLR signaling can initiate. Future studies will likely establish the components necessary for building and maintaining the compartments responsible for differential signaling by TLRs as well as clarify whether there are differences in the composition of these compartments between specialized cell types. Understanding the precise mechanism by which differential signaling is initiated from distinct endosomes via a common signaling adaptor, MyD88, is critical as well.
Concluding Remarks
The trafficking and localization of TLRs has emerged as a primary mechanism to facilitate self versus non-self discrimination as well as induction of distinct signal transduction. Here, we have described our current understanding of the complex cell biology that regulates these receptors, but it is clear that additional components will likely be identified. Indeed, while we are beginning to understand the basic pathways that deliver TLRs to endosomal compartments, we still lack mechanistic details for many of the sorting steps (Outstanding Questions box). Moreover, it remains entirely unclear if or how these pathways are modulated by external signals such as inflammatory or anti-inflammatory mediators. A second major challenge is to characterize the specialized compartments associated with TLR signaling. For example, as we describe above, there is evidence that TLRs populate endosomes from which distinct signaling pathways are initiated, but the features responsible for these specialized signaling properties remain unclear. Finally, comparing TLR trafficking between different cell types may be another fruitful direction for future work. Much of our current understanding of TLR localization comes from the study of macrophages or cell lines, but the specialized aspects of TLR cell biology are often best studied in primary cells, such as B cells and pDCs. In general, tools to study TLR cell biology in primary cells are lacking, so the scientific community must invest in the development of high quality antibodies or transgenic mice expressing TLRs with epitope tags. Perhaps recent advances in genome engineering will accelerate progress toward this goal [98, 99].
These lines of research will not only enhance our understanding of basic cell biology but also provide insight into the mechanisms that contribute to autoimmune diseases driven by inappropriate TLR activation. Describing the molecular pathways that regulate TLR function may also reveal new therapeutic opportunities. Efforts are underway to develop inhibitors of TLRs to treat autoimmune disorders, but these have primarily focused on inhibition of ligand binding. As we learn more about the pathways controlling TLR trafficking and localization, it is possible that strategies aimed at blocking these pathways will broaden therapeutic opportunities.
Acknowledgments
We thank our colleagues for helpful discussions. Work in the Barton Lab is supported by grants from the NIH, the Lupus Research Institute, and the Burroughs Wellcome Fund.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
Full text links
Read article at publisher's site: https://doi.org/10.1016/j.tcb.2013.12.002
Read article for free, from open access legal sources, via Unpaywall:
https://escholarship.org/content/qt82n5w8zg/qt82n5w8zg.pdf?t=rsqrge
Citations & impact
Impact metrics
Article citations
Phospholipid peroxidation in macrophage confers tumor resistance by suppressing phagocytic capability towards ferroptotic cells.
Cell Death Differ, 31(9):1184-1201, 05 Aug 2024
Cited by: 0 articles | PMID: 39103535
Stage-specific expression of Toll-like receptors in the seminiferous epithelium of mouse testis.
Histochem Cell Biol, 162(4):323-335, 31 Jul 2024
Cited by: 0 articles | PMID: 39085445 | PMCID: PMC11364606
Cholesterol Efflux Decreases TLR4-Target Gene Expression in Cultured Macrophages Exposed to T. brucei Ghosts.
Microorganisms, 12(8):1730, 22 Aug 2024
Cited by: 0 articles | PMID: 39203572 | PMCID: PMC11357207
PTEN acts as a crucial inflammatory checkpoint controlling TLR9/IL-6 axis in B cells.
iScience, 27(7):110388, 26 Jun 2024
Cited by: 0 articles | PMID: 39092178 | PMCID: PMC11292540
Toll-like receptors and integrins crosstalk.
Front Immunol, 15:1403764, 10 Jun 2024
Cited by: 0 articles | PMID: 38915411 | PMCID: PMC11194410
Review Free full text in Europe PMC
Go to all (120) article citations
Similar Articles
To arrive at the top five similar articles we use a word-weighted algorithm to compare words from the Title and Abstract of each citation.
The Chaperone UNC93B1 Regulates Toll-like Receptor Stability Independently of Endosomal TLR Transport.
Immunity, 48(5):911-922.e7, 01 May 2018
Cited by: 67 articles | PMID: 29768176 | PMCID: PMC6482051
UNC93B1 mediates differential trafficking of endosomal TLRs.
Elife, 2:e00291, 19 Feb 2013
Cited by: 167 articles | PMID: 23426999 | PMCID: PMC3576711
Cell Surface Expression of Endosomal Toll-Like Receptors-A Necessity or a Superfluous Duplication?
Front Immunol, 11:620972, 01 Feb 2021
Cited by: 22 articles | PMID: 33597952 | PMCID: PMC7882679
Review Free full text in Europe PMC
Dynamic control of nucleic-acid-sensing Toll-like receptors by the endosomal compartment.
Int Immunol, 33(12):835-840, 01 Nov 2021
Cited by: 2 articles | PMID: 34223897
Review
Funding
Funders who supported this work.
NIAID NIH HHS (2)
Grant ID: P01 AI063302
Grant ID: R01 AI072429
 and
and 




