Abstract
Free full text

TGF-β: Duality of Function Between Tumor Prevention and Carcinogenesis
Abstract
Several mechanisms underlying tumor progression have remained elusive, particularly in relation to transforming growth factor beta (TGF-β). Although TGF-β initially inhibits epithelial growth, it appears to promote the progression of advanced tumors. Defects in normal TGF-β pathways partially explain this paradox, which can lead to a cascade of downstream events that drive multiple oncogenic pathways, manifesting as several key features of tumorigenesis (uncontrolled proliferation, loss of apoptosis, epithelial-to-mesenchymal transition, sustained angiogenesis, evasion of immune surveillance, and metastasis). Understanding the mechanisms of TGF-β dysregulation will likely reveal novel points of convergence between TGF-β and other pathways that can be specifically targeted for therapy.
Even the most lethal cancers are initially indolent and localized, yet they acquire the ability to invade both surrounding and distant tissues. Although several key components of this progression have been identified, a comprehensive understanding of these events remains unclear (1–3). Recent data suggest that a potentially vital step in this mechanism is aberration of transforming growth factor beta (TGF-β) signaling (4–6). Dysregulation of TGF-β pathways leads to extensive signal reprogramming, allowing cancer cells to hijack normal function to ensure their own survival.
TGF-β Cell Biology
TGF-β, comprising three isoforms, is a potent pleiotropic cytokine that regulates mammalian development, differentiation, and homeostasis in nearly all cell types and tissues. Knockout studies have revealed that each of the isoforms is essential for development. TGF-β1-null mice are viable for 2 weeks after birth, yet soon develop severe inflammatory lesions in multiple organs and a rapid wasting syndrome, culminating in death at 3 to 5 weeks of age (7). TGF-β2-null mice are predominantly perinatal lethal with the surviving pups developing cyanosis. These mice present with severe cardiac dysfunction in addition to defects in several other organs and are phenotypically distinct from knockouts of the other TGF-β isoforms (8). Lastly, TGF-β3-null mice present with severe cleft palates and begin gasping soon after birth. These mice are unable to suckle, become cyanotic, and die within 24 hours after birth (9).
Each of these three TGF-β isoforms is initially synthesized as a 75-kDa homodimer known as pro-TGF-β. Pro-TGF-β is then cleaved in the Golgi to form the mature TGF-β homodimer (10). These 25-kDa homodimers interact with latency-associated proteins to form the small latent complex (10–12). In the endoplasmic reticulum, a single latent TGF-β binding protein forms a disulfide bond with the TGF-β homodimer to form the large latent complex, allowing for targeted export to the extracellular matrix (11).
After export, the large latent complex interacts with fibronectin fibrils and heparin sulfate proteoglycans on the cell membrane. Eventually, the large latent complex localizes to fibrillin-rich microfibrils in the extracellular matrix, where it is stored until its activation (13,14). There, latent TGF-β is stored, where it remains biologically unavailable until its activation (10). Latent TGF-β is activated by several factors, including proteases (14,15), thrombospondin 1 (16), reactive oxygen species (17), and integrins (18,19). These factors release mature TGF-β by freeing it from the microfibril-bound large latent complex. This occurs through liberation from latency-associated proteins, degradation of latent TGF-β binding protein, or modification of latent complex conformation.
TGF-β Signaling Pathways
Once the ligand is activated, TGF-β signaling is mediated through SMAD and non-SMAD pathways to regulate transcription, translation, microRNA biogenesis, protein synthesis, and post-translational modifications (20–22). Although the downstream effects of TGF-β are heavily context dependent, its signaling is at least partially conserved in many cell types (23). In the canonical pathway, the TGF-β ligand binds to the type 2 TGF-β receptor (TGFBR2) that recruits the type 1 TGF-β receptor (TGFBR1). These receptors dimerize and autophosphorylate serine/threonine residues, allowing for the phosphorylation of SMAD2 and SMAD3 by TGFBR1. The now activated SMAD proteins dissociate from the SMAD anchor for receptor activation (SARA) protein, hetero-oligomerize with SMAD4, and translocate to the nucleus, interacting with myriad transcriptional coregulators and other factors to mediate target gene expression or repression (23,24) (Figure 1). There also exists a type 3 TGF-β receptor (TGFBR3 or betaglycan), a transmembrane proteoglycan that binds the TGF-β ligand, whose function is relatively unknown. Although TGFBR3 appears to lack a cytoplasmic signaling domain, it appears to have important roles in development, as well as in regulating TGFBR1 and TGFBR2 (25–27).
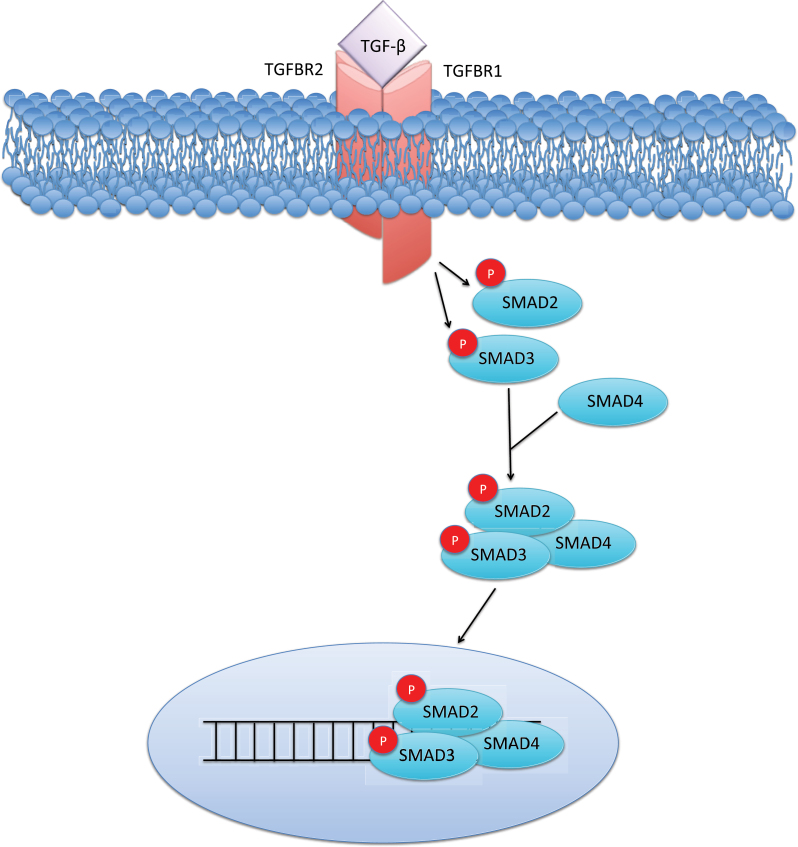
Canonical transforming growth factor β (TGF-β) signaling. The TGF-β ligand binds its type 2 transmembrane receptor (TGFBR2), recruiting the type 1 receptor (TGFBR1), leading to the phosphorylation of SMAD2 and SMAD3. This phosphorylation event allows for complexing with SMAD4 and nuclear translocation, leading to growth arrest in benign epithelial cells. P = phosphorylation.
TGF-β also signals through a number of non-SMAD pathways, including p38 MAPK, p42/p44 MAPK, c-Src, m-TOR, RhoA, RAS, PI3K/Akt, protein phosphatase 2A (PP2A)/p70s6K, and JNK MAPK (22,28–32). Additionally, two studies have linked translational regulation to the cytostatic program governed by TGF-β. The first mechanism involves transcriptional activation of the translation-inhibiting protein eukaryotic translation initiation factor 4E-binding protein 1 (4E-BP1) mediated by the SMAD signaling pathway (33), whereas the second relies on catalytic inactivation of the translation initiation factor eEF1A1 (eukaryotic elongation factor 1A1) by TGFBR1 (21).
Both SMAD-dependent signaling and SMAD-independent signaling play multiple roles in homeostasis, particularly in the growth and plasticity of epithelial cells. SMAD-dependent TGF-β signaling induces growth arrest through a number of mechanisms, including control over various cyclin-dependent kinase inhibitors (5,34–37). SMAD-independent mechanisms of TGF-β–induced apoptosis involve DAXX/HIPK2 and transforming growth factor β associated kinase (TAK1)/TRAF6–dependent p38/JNK activation (38,39).
MAPK Regulation of SMAD Signaling
TGF-β signaling is tightly regulated by mitogen-activated protein kinases (MAPKs). Although extracellular signal-regulated kinase (ERK) MAPK signaling is more commonly associated with receptor tyrosine kinases, it has been demonstrated that TGFBR1 also has tyrosine kinase activity. TGFBR1 directly phosphorylates tyrosine on ShcA, allowing for interaction with Grb2 and Sos. This leads to Ras activation and downstream ERK phosphorylation (40).
ERK and other MAPKs regulate the SMAD-dependent arm of TGF-β signaling through phosphorylation of SMAD2 and SMAD3 at specific amino acid residues. Together, these regions are known as the interdomain SMAD-linker region. ERK phosphorylation of the linker region occurs at Ser245/250/255 and Thr220 residues on SMAD2 and at Ser204/208 and Thr17 on SMAD3 and reportedly reduces SMAD2/3 signaling. In contrast, enhancement of SMAD signaling can occur through phosphorylation at various SMAD3 residues, including Thr8 by ERK, Ser204/208/213 by p38 MAPK/ROCK, or Ser208/213 by JNK MAPK. SMAD3 phosphorylation at Ser208/213 residues is common to advanced tumors and may play a role in the aberrant response to TGF-β in cancer (41).
TGF-β Paradox in Tumor Biology
TGF-β expression has been studied in nearly all epithelial cancers, including, prostate, breast, lung, colorectal, pancreatic, and skin cancers (42). Through these studies, it has become clear that TGF-β can function as both a tumor suppressor and a tumor promoter (4,43–45). In benign epithelia and many early-stage tumors, TGF-β is a potent inducer of growth arrest. However, in advanced tumors, TGF-β signaling pathways are severely dysregulated. Rather than inhibiting carcinogenesis, TGF-β promotes tumor growth and progression at late stages (4,5,42,43,46–48). This functional switch is known as the TGF-β paradox (44).
This paradox is reflected in the clinic, where in early stage cancers, levels of TGF-β are positively associated with a favorable prognosis. Yet in advanced tumors, levels of TGF-β in the tumor microenvironment are positively associated with tumor size, invasiveness, and dedifferentiation, making TGF-β a useful prognostic biomarker and predictor of recurrence after initial or failed therapy (42,47–49).
TGF-β Homeostatic Feedback
TGF-β is a key regulator of its own expression (50,51), and in some cancers, tumor progression corresponds to changes in TGF-β self-regulation, which can include downregulation or inactivation of the TGFBRs (52–54). This receptor repression may correspond with compensatory induction of the TGF-β ligand, which is further complicated by data suggesting that TGF-β can function as a cell-autonomous tumor promoter through noncanonical mechanisms (55).
Recent data suggest that a potential crux of the TGF-β paradox in prostate cancer may be the differential activation of MAP kinases, such as ERK, between benign and malignant cells. This action is believed to control synthesis of TGF-β in a homeostatic feedback mechanism. ERK activation through TGF-β signaling induces synthesis of the TGF-β ligand in prostate cells in vitro. However, when the level of TGF-β is elevated, benign prostate cells can repress ERK activation. It appears that this mechanism becomes perturbed in advanced disease, and the described negative feedback loop collapses, establishing a feed-forward signaling mechanism in its place and failing to suppress pathological levels of active ERK. In many tumor types, ERK is constitutively activated independent of the level of TGF-β, and, consequently, TGF-β may be continuously produced (51).
The effects of ERK activation on TGF-β signaling are not solely limited to TGF-β synthesis. TGF-β is a key regulator of gene methylation, a process dependent on ERK activation of DNA methyltransferase (DNMT). In prostate and ovarian cancer cells, TGF-β stimulates DNMT expression (52,56). Overexpression of TGF-β and/or DNMTs is associated with an aggressive phenotype and poor prognosis in prostate cancer (52,57). Among the many genes affected by DNA hypermethylation are TGFBR1 and TGFBR2 (52,54,56).
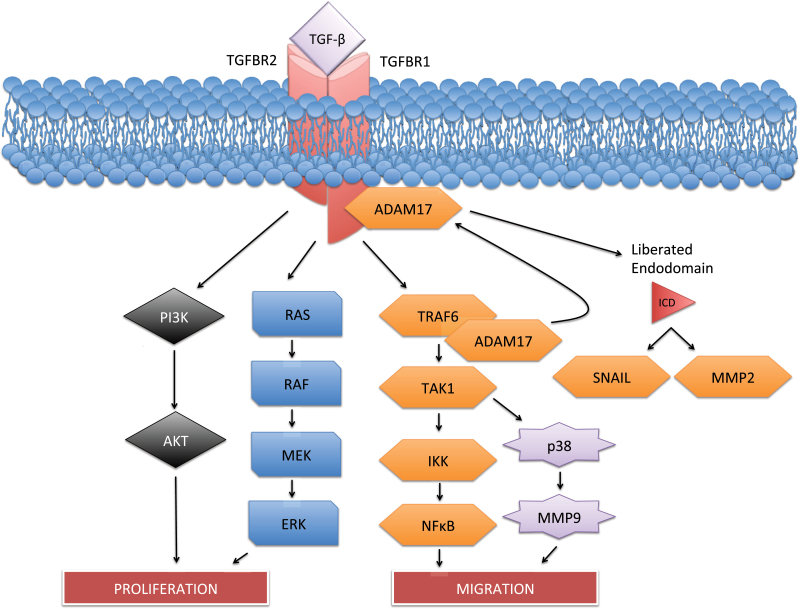
Although ERK may function as a key transcriptional inhibitor of TGFBR1 expression, TGFBR1 may also be repressed at the protein level. Also in prostate cancer cells, degradation of the functional TGFBR1 protein is dependent on a cascade involving tumor necrosis factor receptor associated factor (TRAF6), ADAM metallopeptidase domain 17 (ADAM17), and protein kinase C zeta (PKCζ). In response to TGF-β, TRAF6, a ubiquitin ligase, ubiquitinates TGFBR1, leading to its cleavage by ADAM17 and nuclear translocation of its liberated intracellular domain (ICD) (Figure 2) (58). Interestingly, ADAM17 targeting of TGFBR1 has been linked to ERK activation in nonprostate cells, resulting in shedding of the TGFBR1 ectodomain (59). This suggests ERK may be involved in both protein and transcriptional repression of the TGFBRs.
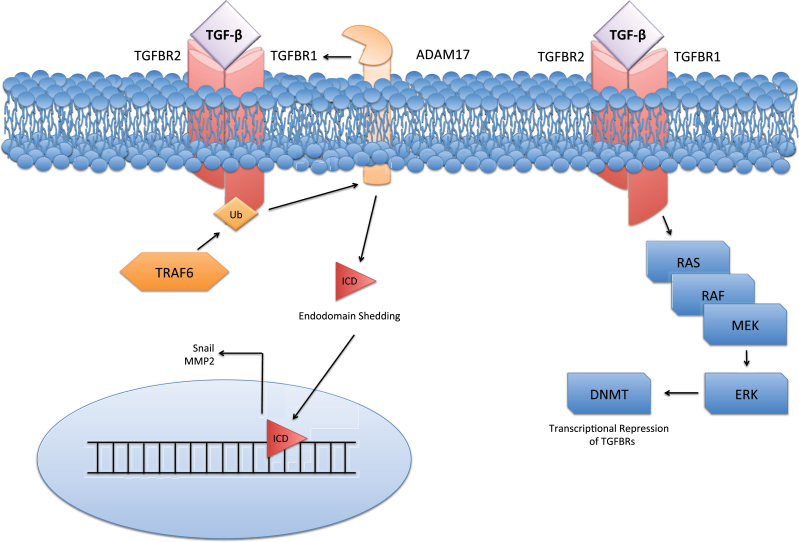
Mechanisms of transforming growth factor β receptor (TGFBR) repression. Repression of TGFBRs occurs at both the epigenetic and protein levels. It was recently discovered that the ubiquitination of TGFBR1 by tumor necrosis factor receptor associated factor 6 (TRAF6) leads to ADAM metallopeptidase domain 17 (ADAM17) mediated cleavage, liberating the TGFBR1 intracellular domain (ICD). The TGFBR1-ICD is then free to translocate to the nucleus and activate factors including Snail and matrix metalloproteinase 2 (MMP2). Additionally, epigenetic repression of the TGFBRs may be dependent on ERK activation of DNA methyltransferase (DNMT), which hypermethylates the TGFBR promoters, downregulating their expression.
TGFBR1 degradation by ADAM17 desensitizes cells to the growth inhibitory effects of TGF-β, and nuclear translocation of the liberated TGFBR1-ICD is involved in the upregulation of other oncogenic factors (58,59). The effects of TRAF6 extend well beyond TGFBR1 degradation and include JNK and NF-kB activation, partially explaining the link between these cascades and TGF-β (60). Mass cleavage coupled with repression of the TGFBR1 transcripts is a potential explanation of the repression of TGFBRs seen in some advanced diseases and warrants future study. Together, these observations provide a partial mechanism through which sensitivity to TGF-β becomes reduced in tumor cells (Figure 2). Yet, because TAK1/p38 signaling can have proapoptotic effects in the epithelia (61), the tumor suppressive effects of this cascade must be carefully weighed against its seemingly tumor-promoting effects to determine its relevance as a clinical target.
Inhibitory SMAD Signaling
As discussed, the TGF-β ligand signals through transmembrane receptors, leading to phosphorylation of the downstream SMAD proteins. These canonical effectors can be classified as R-SMADs (SMAD2/3) and the co-SMAD (SMAD4). However, there are also inhibitor SMADs (I-SMADs; SMAD6/7) that function to negatively regulate R-SMAD activation and nuclear translocation. Overexpression of the I-SMADs, particularly SMAD7, is common to pathologies in which TGF-β signaling is perturbed (62). SMAD7 normally antagonizes TGF-β signaling through a negative feedback loop, mediating the crosstalk between TGF-β and other signaling pathways. In response to TGF-β signaling events, SMAD7 is recruited to TGFBR1, blocking SMAD2/3 phosphorylation and its downstream signaling (63).
Additionally, SMAD7 indirectly represses TGF-β signaling by targeting TGFBR1, TGFBR2, and other SMAD transcription factors for proteasomal degradation (64,65). A key mechanism of TGFBR repression by SMAD7 is the recruitment of the HECT type E3 ubiquitin ligases Smurf1 and Smurf2, which ubiquitinate TGFBR1 and TGFBR2 (66). Ubiquitination of the TGFBRs can lead to either clathrin-dependent internalization into EEA1-positive endosome recycling or internalization by the caveolar pathway and receptor proteasome degradation (67–69). It was recently discovered that receptor internalization was enhanced by the TGF-β coreceptor, CD109 (67), making CD109 a novel target for therapies aiming to preserve the effects of TGF-β.
Phosphatase Regulation of TGF-β Signaling
The above data illustrate how some tumors may become less responsive to the antigrowth signals of TGF-β in the cancer epithelia; however, this model does not explain how TGF-β promotes tumor progression at later stages of disease. One candidate protein that may be at least partially responsible for these paradoxical events in some cancers is protein phosphatase 2a (PP2A). PP2A is synthesized in the cytoplasm and can be recruited by activated TGFBR1 to the cell membrane through a mechanism that is not completely understood. Once there, it can function as a regulator of TGF-β receptor stability and activity (70), as well as a context-specific inhibitor to TAK1 and ERK activation (51,71,72).
In prostate cancer cells, it is reported that PP2A can be recruited by activated TGFBR1 depending on the level of TGF-β available to the target cells. In response to a high dose of TGF-β, a sufficient quantity of PP2A is recruited by activated TGFBR1, resulting in inhibition of ERK. Yet at a low dose of TGF-β, because of limited quantity of activated TGFBR1, the quantity of PP2A available is restricted, and ERK remains activated. In malignant prostate cells, downregulation of TGFBR1 is much more pronounced, limiting recruitment of PP2A to the membrane. This results in constitutive ERK activation and induction of the TGF-β ligand (Figure 3). In other words, PP2A is recruited by TGFBR1 and may serve as a barrier to the SMAD-independent arm of TGF-β signaling while also protecting against the surge of TGF-β secreted by advanced tumors (51). Thus reduced expression of TGFBR1 may cause downstream preference of mitogenic pathways in response to TGF-β or desensitization to its antiproliferative effects.
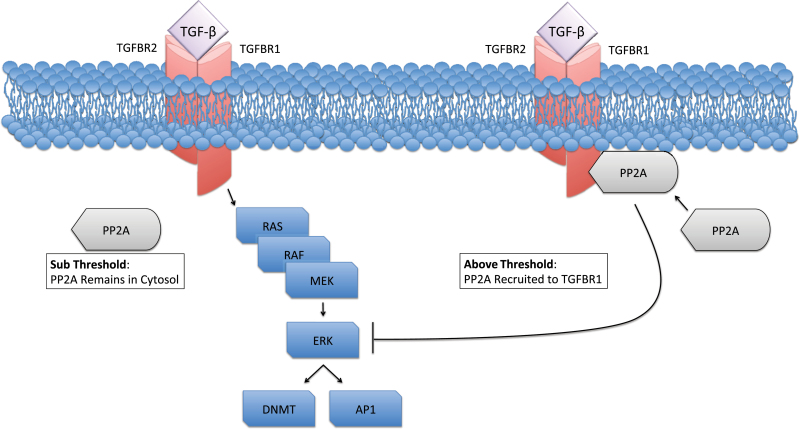
Transforming growth factor β (TGF-β) control over ERK MAPK. It is hypothesized that in some cells, there is a reciprocal control mechanism involving ERK and the transforming growth factor β receptors (TGFBRs). ERK activation leads to DNA methyltransferase (DNMT) and activator protein (AP1) expression, which is believed to lead to epigenetic repression of the TGFBRs and upregulation of the TGF-β ligand. In some contexts, TGF-β suppresses ERK activation. A key step in this process appears to be recruitment of protein phosphatase 2A (PP2A) to the cell membrane by TGFBR1. In the proposed model, once a sufficient quantity of PP2A is recruited, it serves as a barrier to ERK activation, where DNMT and AP1 expression are not induced, and the canonical SMAD arm of the TGF-βcascade is favored.
The many interactions between TGF-β and ERK are extremely complex, and this PP2A-mediated phenomenon has not been sufficiently explored in other cell types. As mentioned, it has also been demonstrated that TGFBR1 can directly lead to ERK activation (40) and is a necessary in vivo activator of ERK in the pancreas (73). This suggests that ERK may also have roles in TGF-β–induced growth suppression. Further study is needed to understand the cell-specific contributions of ERK and PP2A to the TGF-β paradox.
Roles of TGF-β in Tumor Progression
The SMAD-independent arm of the TGF-β cascade contextually activates several oncogenic factors. Additionally, the increased expression of SMAD7 in some cancers represses the SMAD-dependent arm of the TGF-β pathway, further silencing a key growth inhibitory mechanism of TGF-β signaling (74–76). In the proposed model, nearly all tumor-suppressive functions of TGF-β are lost and aberrant TGF-β signaling has a multitude of physiological ramifications promoting tumorigenesis and progression, which will be detailed in subsequent sections of this review.
TGF-β Control Over Proliferation and Loss of Apoptotic Response
In benign epithelial cells, TGF-β is generally considered an antiproliferative and proapoptotic signal (23). A key step in TGF-β dysregulation is the loss of this response. In advanced disease, TGF-β can have prosurvival/antiapoptotic effects (5,42,44,46,77). In the benign context, TGF-β regulates epithelial proliferation and apoptosis through a number of mechanisms. Among the most dominant and well known is SMAD control over c-Myc, p21, and cyclin D (34,36,78), although the effects of TGF-β can also be SMAD independent (28). Other pathways include PP2A dephosphorylation of p70S6K leading to cell cycle arrest (79), SMAD-dependent activation of BIM leading to compromised mitochondrial integrity (80), and the aforementioned TAK1/TRAF6 and DAXX/HIPK2 pathways leading to JNK-mediated apoptosis.
The SMAD complex has been shown to recognize the p15INK4b and p21CIP21 promoters, increasing expression of these genes, and stabilizing additional cyclin-dependent kinase inhibitors through other mechanisms (5,35,36). As TGF-β signaling is usurped in cancer, the balance between SMAD-dependent and SMAD-independent signaling likely shifts. In this context, TGF-β fails to induce the aforementioned cyclin-dependent kinase inhibitors, and cell proliferation is unchecked. Thus, increased presence of SMAD-inhibitory factors facilitates this process through blockade of the SMAD-independent arm of TGF-β growth inhibition.
The summation of these events leads to a prosurvival response to TGF-β by cancer cells. But, in this context, not only does TGF-β fail to inhibit growth, it also fails to induce apoptosis. TGF-β induction of apoptosis is also regulated by the direct interaction between SMAD3 and Akt (81). Akt, a target of TGF-β signaling (77,82,83) commonly overactivated in cancer (84), can actively repress SMAD3-induced apoptosis (81,85). The apoptotic response may also be affected by another target of TGF-β signaling, vimentin (86). Vimentin sequesters the tumor suppressor p53 in other models until its cleavage by caspase 4 (87), although this interaction has yet to be explored in cancer cells. Repression of p53 function may allow for the accumulation of mutations and further loss of the apoptotic response to TGF-β. Vimentin has a more prominent role in epithelial-to-mesenchymal transition (EMT)/metastasis and will be discussed below. Additionally, Src is a known regulator of death-associated protein kinase (DAPK) (88), a protein implicated in TGF-β–induced apoptosis of benign cells (89). The increased presence of Src in many cancers may contextually lead to repression of DAPK, potentially crippling the apoptotic response to TGF-β.
In contrast with its canonical role in regulating epithelial cell proliferation, TGF-β is considered a progrowth signal with respect to the mesenchyme. Classically, in response to injury, the influx of granulocytes, platelets, leukocytes, and additional parenchymal cells increases the presence of TGF-β at the site of the wound (90,91). TGF-β then induces fibroblast proliferation, myofibroblast differentiation, and remodeling of the extracellular matrix (90–94). Therefore, TGF-β is heavily implicated in a variety of fibrous diseases (91) and may serve as an attractive therapeutic target in desmoplastic tumors should the tumor epithelia be desensitized to the growth inhibitory effects of TGF-β. Additionally, in some tumors, TGF-β may even induce proliferation while failing to elicit an apoptotic response (95). Should the tumor environment establish a feed-forward loop resulting in uncontrolled expression of TGF-β, aberrant signaling events may continuously promote tumor growth at the level of the cancer epithelia.
Induction of EMT, Dedifferentiation, and Migration in Advanced Disease by TGF-β
In more-advanced disease stages, TGF-β induces EMT in cancer cells (96,97). EMT is marked by the loss of E-cadherin and the expression of mesenchymal proteins such as vimentin, fibronectin, and N-cadherin, facilitating the invasion process and worsening prognosis (98,99). In cancer cells, the repression of E-cadherin and the induction of vimentin, matrix-metalloproteinases (MMPs), and other pro-EMT factors can be driven by TGF-β (44,86,96,97,100).
The loss of E-cadherin has been linked to TGF-β signaling in cancer cells through both SMAD-dependent and -independent mechanisms (12,101–103). In pancreatic cancer, TGF-β is sufficient to induce tyrosine phosphorylation of α-catenin and the downstream Wnt target β-catenin in a SMAD-independent manner. This action is dependent on PI3K/Akt and destabilizes the E-cadherin adhesion complex (103). Since E-cadherin represses the PI3K/Akt pathway [104], the initial loss of E-cadherin may lead to further destabilizing events, leading to loss of cell–cell adhesion.
Alternatively, in epithelial cell lines, TGF-β can induce expression of the transcription factor Snail, recruiting the Sin3A/HDAC1/HDAC2 complex, which leads to deacetylation of the E-cadherin promoter, thus repressing transcription (101). SMAD-independent activation of Snail is a result of Ras/ERK activation (12,102) and the nuclear translocation of the TGFBR1-ICD (58). This suggests crosstalk between the SMAD-dependent and SMAD-independent arms of TGF-β repression of E-cadherin in cancer cells. Although the mechanism remains incomplete and deserves further study, in the tumor setting of aberrant TGF-β signaling, TGF-β actively represses E-cadherin and promotes EMT (12,101–103).
TGF-β and Wnt pathways also appear to converge at vimentin. Vimentin is a mesenchymal type III intermediate filament believed to facilitate the transport of β1 integrin by dynein/kinesis motor proteins toward the (+) end of microtubules. In vitro, inhibition of vimentin is sufficient to partially reverse EMT in prostate cancer cell lines. Cells previously expressing a more mesenchymal phenotype were successfully induced to form organized, epithelial-like structures. Downregulation of vimentin in cancer cells is also sufficient to slow tumor growth in vivo (99,105).
A prior study suggested that TGF-β upregulation of vimentin in prostate cancer is mediated by NF-κB and TAK1 (86). Both NF-κB and TAK1 are regulated by PP2A in other models (71,106), offering one possible explanation of TGF-β induction of EMT and metastasis in advanced disease. However, the same evidence suggests the relationship between PP2A and NF-κB is complex and requires further study. It has also been suggested that β-catenin has a role in transactivation of vimentin (107). These two proposed mechanisms are not mutually exclusive because there is likely significant crosstalk between multiple oncogenic pathways contributing to disease progression.
Evidence also suggests that EMT may also lead to the emergence of cancer cells presenting stemlike characteristics, often referred to as cancer stem cells (CSCs). Compelling evidence implicating TGF-β–induced EMT in the generation and maintenance of CSCs is that either TGF-β signals or overexpression of EMT-associated transcription factors (Snail or Twist) in benign mammary epithelium leads to acquisition of a CD44Hi/CD24low CSC phenotype. Additionally, TGF-β–induced CD44Hi/CD24low cells formed 40 times more mammospheres in vitro than their untreated counterparts, suggesting they have an increased capacity for self-renewal (108,109).
TGF-β was also demonstrated to regulate expression of the CSC marker CD133 in hepatic epithelial cells (110). This mechanism appears to be dependent on TGF-β inhibition of DNMTs, although evidence also suggests that the SMAD-dependent arm of the TGF-β pathway may be involved. TGF-β–induced CD133+ cells induced more aggressive tumors in xenograft models (110), and these CD133+ cells have been demonstrated to have resistance to chemotherapy and TGF-β–induced apoptosis (111).
In addition to inducing structural changes associated with EMT and tumorigenic properties generated through acquisition of a CSC phenotype, TGF-β also upregulates MMPs to promote invasion through proteolytic degradation and remodeling of the extracellular matrix (112). In the tumor context, TGF-β upregulates expression of MMPs through at least two mechanisms. One mechanism is dependent on the TGFBR1-ICD, shown in prostate cancer, and the other is dependent on p38, demonstrated in breast cancer (58,100). Despite rapid ERK activation by TGF-β, ERK activation was not required for the expression of MMPs in breast cancer cells (100).
As discussed, in the tumor microenvironment, TGF-β has a positive association with both EMT and acquisition of a CSC phenotype. TGF-β may also promote metastasis by interrupting cell–cell adhesion (12,101,103), upregulating vimentin and MMPs (58,86,100), and attracting new vasculature. Yet once the cancer epithelium invades these blood vessels, they require the means to extravasate and establish secondary tumors. It has been suggested that after extravasation cells undergo a redifferentiation program known as mesenchymal-to-epithelial transition (MET) allowing for integration into ectopic tissues (113,114). Although control of the MET program is not completely understood, evidence suggests the TGF-β–related factors of the bone morphogenetic protein (BMP) family may be involved. Treatment with BMP7 neutralized TGF-β–induced EMT during development and in human esophageal carcinoma cells. BMP7 also induced melanoma cells to take a more epithelial morphology (115).
However, as with the effects of the TGF-β ligands, the function of BMPs are also varied. Blockade of BMP signaling through insertion of a dominant negative form of the BMP receptor 2 (BMPR2) accelerated mammary tumor progression and invasive capability (116). Similarly, administration of BMP7 induced senescence in prostate cancer bone CSCs in a BMPR2-dependent manner (117).
Yet BMP9 induced EMT in hepatocellular carcinoma cells, and nuclear localization of pSMAD1, a downstream target of BMP signaling, was positively associated with expression of Snail and inversely associated with expression of E-cadherin (118). Additionally, blockade of BMP2 in metastatic colon cancer cells induced an MET event (115). These data suggest that, as with the three TGF-β ligands, the roles of BMPs in tumor progression are highly varied, and a similar conundrum lies in identifying and targeting only their tumor-promoting aspects.
Contradictory Roles of TGF-β in Sustained Angiogenesis
Sustained angiogenesis is a crucial step in tumor progression (119). Newly formed blood vessels allow for bulk nutrient and waste exchange and provide an entry point for metastatic cancer cells (119,120). In a number of studies, TGF-β is implicated in this process, although its exact contributions to angiogenesis are not fully understood. Tumor-derived TGF-β impacts the cancer epithelia as well as proximate stromal and endothelial cells (ECs), the combination of which have the potential for proangiogenic consequences (121). Although the contributions of the stroma to angiogenesis are vital, they are beyond the scope of this article. For an excellent review of the roles of TGF-β in the reactive stroma, see De Wever et al. (122).
Within many cancer cells, TGF-β is sufficient to upregulate the expression of vascular endothelial growth factor (VEGF), attracting neighboring ECs and promoting angiogenesis (123,124). A recent study showed TGF-β regulation of VEGF expression in breast cancer cells to be complex and, at least in part, SMAD dependent (123). SMAD-dependent signaling appears to at least initially repress VEGF expression (121). However, other studies suggest SMAD3 is overexpressed in some advanced disease and is associated with angiogenesis (125). Much remains to be elucidated regarding this mechanism, although in the clinic, TGF-β expression is positively and substantially associated with VEGF expression and angiogenesis in late-stage tumors (126).
Once secreted, TGF-β regulates the proliferation and migration of ECs, which has a multifaceted effect on EC function. TGF-β can bind both type 1 TGF-β superfamily receptors, activin receptor-like kinase 1 (ACVRL1), and activin receptor-like kinase 5 (ALK5). These receptors have antagonistic effects on EC function, and the balance of ALK1 vs ALK5 signaling dictates the overall effect of TGF-β on ECs. ECs undergo a biphasic response to TGF-β in vitro. In low concentrations, TGF-β promotes proliferation, migration, and expression of MMPs in endothelial cells through ALK1 activation of SMAD1/5, resulting in Id1 transcription. Interestingly, in high concentrations, the canonical SMAD2/3 pathway is activated by ALK5, inducing the expression of plasminogen activator inhibitor and fibronectin, thereby impeding angiogenesis (Figure 4). This suggests that any antiangiogenic effects of TGF-β are being overridden in advanced disease (121,127).
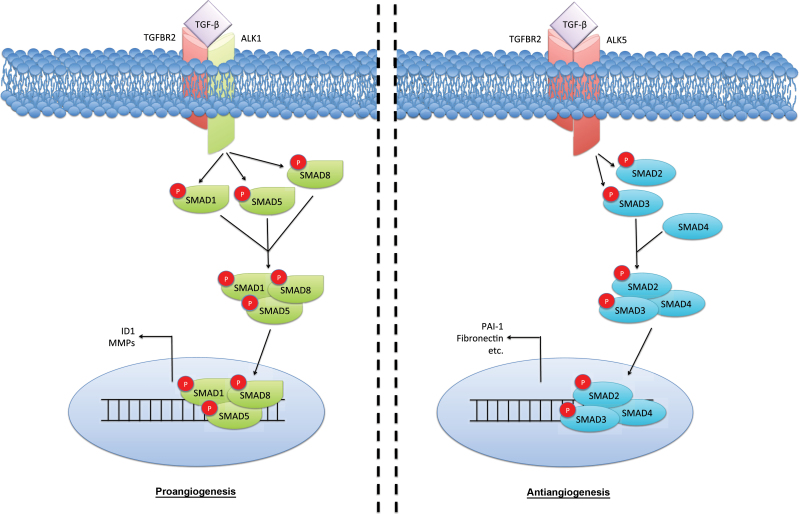
Transforming growth factor β (TGF-β) in endothelial cell function. The effects of TGF-β on the vascular endothelia are complex and seemingly biphasic. Low TGF-β concentrations appear to activate the activin receptor-like kinase 1 (ALK1)/SMAD1/5 pathway, stimulating endothelial cell (EC) proliferation and migration associated with higher levels of ID1. Yet, at higher levels, TGF-β activates its canonical SMAD2/3 pathway in a transforming growth factor β receptor 1 (TGFBR1; also known as ALK5)–dependent manner to induce antiangiogenic factors plasminiogen activator inhibitor-1 (PAI-1) and fibronectin. P = phosphorylation.
Although the mechanism is incomplete, the end physiological result is clear: TGF-β levels are positively associated with angiogenesis in the late-stage tumor microenvironment (126). Tumor-derived TGF-β therefore further sustains angiogenesis, facilitating nutrient exchange and metastasis that results in tumor progression.
TGF-β Upregulation and Evasion of Immune Surveillance
Another late-stage obligate step to the cancer cell is selective suppression of the immune system. It is well established that cytotoxic lymphocytes, such as natural killer (NK) and T cells, directly destroy tumor cells. TGF-β plays a crucial role in the repression of the immune system, as attested by the gross autoimmunity developed in TGF-β1 null mice (128). Among cells that comprise the immune system, T cells are regarded as the main target cells of immunosuppressive activity of TGF-β because the selective and specific ablation of TGFBR2 in T cells leads to similar autoimmunity as observed in TGF-β1 null animals (129). TGF-β signaling in T cells represses both their inflammatory and cytotoxic differentiation programs. In the absence of TGF-β signaling, T lymphocytes rapidly acquire high levels of Fas-L and granzymeB, two main functional components for and markers of cytotoxicity (130,131). In line with the repressor role for TGF-β on T lymphocytes, T cells expressing a constitutive active form of TGFBR1 remain refractory to full activation (132).
In addition to impairing T-cell effector function, TGF-β has been proposed as a potent inducer of regulatory T cells (Tregs). Tregs are capable of repressing both the cytotoxic and inflammatory function of effector T cells. Both development and function of regulatory cells are under the control of the forkhead family transcription factor Foxp3 (133). Interestingly, TGF-β signaling in T cells participates in the expression and the stabilization of Foxp3 (134–136). This may be relevant clinically because enrichment of CD4+CD25+Foxp3+ Tregs in human tumors is associated with poor prognosis in breast cancer patients (137,138).
It was previously demonstrated that Foxp3 expression is induced by TGF-β inactivation of ERK/DNMT (57). Because TGF-β is secreted by the tumor cell at increasingly high concentrations, it induces Foxp3 expression in cytotoxic T cells, helper T cells, and their precursors, rendering them inactive. In other words, TGF-β provides the tumor with a cloaking mechanism, facilitating evasion of immune system surveillance (57,132,139–143).
Several studies suggest that TGF-β directly represses immune activity in the tumor setting, confirming the importance of TGF-β signaling in T cells in controlling tumor growth. In one such study, CD8+ T cells were rendered TGF-β insensitive by a dominant-negative TGFBR2. Adoptive transfer of these cells resulted in a fivefold increase in tumor-killing activity compared with adoptive transfer of tumor-reactive yet still TGF-β–sensitive CD8+ T cells. This result illustrates the pivotal role of TGF-β in tumor evasion of the immune system in cancer. TGF-β renders advanced tumors progressively less recognizable to the surveillance program. Resultant protection of the cancer cell from cytotoxic cells allows for the autocrine effects of TGF-β to take effect, leading to further dysregulation both in the cancer cell and the tumor microenvironment (142,143).
It should be noted that in addition to its role in evasion of immune surveillance, TGF-β also governs the T helper 17 (Th17) differentiation program. The mechanism of Th17 fating is complex and not fully understood. For a more thorough explanation of these events, see Hatton (144).
Mouse Models of TGF-β
Mouse models have been integral in illustrating and investigating the dual roles of TGF-β during carcinogenesis. There has been a marked enhancement of our understanding of how TGF-β can actually facilitate tumor progression and subsequent metastatic dissemination, including its tumor-suppressive effect on normal and hyperplastic tissues (as previously discussed). Particular attention will be devoted to underlining the tumor promoter role of TGF-β and its reliance on both cell-autonomous effects (on the epithelia) and nonautonomous effects in the tumor microenvironment (particularly on the different cell populations composing the stroma).
One of the first pieces of compelling in vivo evidence supporting the dual role of TGF-β during carcinogenesis was provided by genetically engineered mice presenting impaired TGF-β signaling targeted to the mammary glands. Several transgenes were used: a constitutively active TGF-β (TGF-β1S223/225), a constitutively active type 1 TGF-β receptor (TGFBR2CA), dominant negative type 2 TGF-β receptor (TGFBR2DN), and type 2 TGF-β knockout receptor (TGFBR2null). These genetic alterations were specifically targeted to mammary gland epithelium using regulatory elements from the mouse mammary tumor virus (MMTV) or whey acidic protein (WAP) genes. These transgenes were not independently sufficient to induce neoplastic lesions in the mammary gland as reported in many other organs. The only visible defects were an abnormal development of the branching ducts before puberty and impaired gland involution after lactation. These genetic perturbations of TGF-β signaling were later combined with known oncogenes in mammary gland predisposition cancer models. The most commonly used oncogenes are Her-2/neu (erbB-2), polyoma virus middle T antigen (PyVmT), and transforming growth factor alpha (TGFα) (145–151).
In each of these models, a decreased incidence and a slow progression of the primary mammary tumor was observed when TGF-β signaling was activated (TGF-β1S223/225 or TGFBR1CA). Opposite effects on the primary mammary tumor were observed when TGF-β signaling was inhibited (TGFBR2DN or TGFBR2null). Altogether, these models were a clear example of the tumor-suppressive role of TGF-β.
Close examination of progressing tumors in these models revealed that TGF-β has different roles during tumor initiation (number of primary tumors and latency to onset) and tumor progression (local invasion and metastasis). For example, a TGFBR2-neutralizing antibody compromised the formation of metastases induced by PyVmT, indicating that TGF-β has a prometastatic effect (146). This was the first observation in favor of a protumoral effect of TGF-β, which was further validated by transgenic mouse models, as demonstrated by inhibition of TGF-β, using TGFBR2DN, reducing the aggressiveness of MMTV-TGFα tumors (148) and the number of metastases arising from MMTV-Neu tumors (152). Conversely, activation of TGF-β using TGFBR1CA (151,152) or TGF-β1S223/225 (147) resulted in a subtantially increased number of metastases arising from MMTV-Neu mammary tumors. These observations represent a clear illustration of the TGF-β paradox in vivo. The tumor suppressive effect of TGF-β observed on normal and hyperplastic tissues is obliterated and even reversed in established tumors.
To address the effect of TGF-β during late stages of mammary tumorigenesis, a mouse model was engineered in which expression of TGF-β1 in mammary tumors is temporally controlled by doxycycline (149). This study showed that doxycycline-mediated induction of active TGF-β in late mammary tumors could substantially accelerate metastases, thus confirming that TGF-β behaves as a tumor promoter during metastases.
The skin has also provided very informative mouse models illustrating the tumor promoter role of TGF-β. As expected from its tumor suppressor role, it was shown that respective activation (45,153,154) or inhibition (155,156) of TGF-β can accelerate or inhibit the formation of 7,12-dimethylbenz(α)anthracene-induced/12-O-tetradecanoylphorbol-13-acetate (DMBA/TPA)–induced skin squamous cell carcinoma, with this effect being abrogated in the presence of TGFBR2DN (96). It was observed that TGF-β1S223/225 prevents the onset of benign papillomas induced after DMBA/TPA treatment but unexpectedly enhances progression to invasive spindle carcinomas in transgenic mice (45). Human skin cancers frequently overexpress TGF-β but exhibit decreased expression of the TGFBR2.
Mouse models of pancreatic cancer have also provided crucial information about the role of TGF-β during carcinogenesis. Recurrent genetic alterations were found in human pancreatic ductal adenocarcinoma (PDAC). Common alterations include KRAS-activating mutations in more than 90% of cases and Ink4A/Arf or TP53-inactivating mutations each in approximately 50% of cases [for review, see Hansel et al. (157) and Landi (158)]. LSL-KRASG12D is a conditional allele encoding a constitutively active KRAS mutant (KRASG12D) in the presence of Cre-recombinase. Pancreas-specific activation of the LSL-KRASG12D induces the development of age-dependent precursor lesions in 100% of mice and PDAC after 1 year in approximately 10% of mice [159]. The LSL-KRASG12D allele was combined with genetic modifications that inactivated TGF-β signaling, such as that of SMAD4 (160–162) and TGFBR2 (163). These models validate the tumor suppressive role of TGF-β, reaffirmed clinically by the deletion of SMAD4 in roughly 50% of human PDAC cases. Yet it was also demonstrated that Tgfbr1 haploinsufficiency inhibited KRAS-induced pancreatic tumorigenesis in mice (164). This observation strongly suggests that decreased TGF-β signaling may result in protection against tumorigenesis, representing new in vivo evidence of the tumor-promoting effect of TGF-β.
Combining the KRASG12D allele with inactivation of the tumor suppressor Ink4A/Arfnull led to highly aggressive PDAC with a sarcomatoid histology (165). Interestingly, whereas SMAD4 deficiency promoted progression of KRASG12D lesions to PDAC, SMAD4 deficiency converted KRASG12D/Ink4A-Arfnull tumors into tumors that retain differentiated ductal morphology (160). This effect is potentially attributed to the abrogation of TGF-β–induced EMT.
Additional evidence suggests that migration and EMT can be uncoupled, demonstrated by experiments performed in vitro in human keratinocytes and pancreatic cancer cells (166) and in vivo in mouse models of skin tumors. Findings from these works reported that TGF-β–induced EMT and invasion occur by distinct mechanisms (96). These observations suggest that migration requires other factors than TGF-β, including hepatocyte growth factors (167), epithelial growth factor (168,169), and connective tissue growth factor (170). Indeed, epithelial–stromal interaction during carcinogenesis is of crucial importance, and TGF-β plays a central role in the complex dialogue between these two compartments.
The direct effect of TGF-β is not solely responsible for influencing tumor behavior. This was attested by mouse xenograft models of prostate cancer, in which treatment of primary tumors with TGF-β promotes the accumulation of reactive stromal cells, including fibroblasts, enhancing the growth and progression of these tumors (171–173). Interestingly, it has been shown that fibroblasts devoid of TGFBR2 result in increased mammary gland tumor progression (167,174–176). Genetically engineered mice presenting a conditional pancreatic activation of the KRAS oncogene combined with an inactivation of TGFBR2 secrete high levels of several CXC chemokines, which in turn induces in the pancreatic stromal fibroblasts the expression of connective tissue growth factor, a profibrotic and tumor-promoting factor (170).
Finally, tumor-derived TGF-β inhibits the immune system, allowing the cancer cells to escape the immune surveillance program. The most striking in vivo evidence is provided by TGF-β1null mice that develop autoimmune disorders, culminating in their death by 3 weeks of age (128,177). Deprivation of TGF-β signaling selectively in T cells is associated with a phenotype resembling that observed in TGF-β1null animals (129,178). Mouse models have shown that abrogation of TGF-β in T cells could result in a potentiation of the immune system, which becomes more efficient to eradicate melanoma cells (179) and prostate cancer cells (142). SMAD4-deficient intestinal tumors recruit a specific subset of myeloid cells able to promote invasion mediated by TGF-β (180). It was demonstrated that Snail-induced EMT (Snail being a transcriptional target of TGF-β) accelerates melanoma metastasis through enhanced invasion but also through induction of immunosuppression (181).
The described animal models succeed in underlining the complexity of the cooperative interactions between cancer and surrounding cells to facilitate invasion. This is particularly relevant when examining the close relationship between matrix remodeling, EMT, and immune suppression. These models represent valuable tools to test anti-TGF-β therapeutic approaches aimed at abrogating positive feedback loops that fuel these different compartments to facilitate tumor invasion.
Roles of TGFBR1 in Cancer Risk and Progression
Although in vivo models have done much to uncover the contributions of TGF-β signaling, they fail to determine whether these phenomena are a cause or consequence of carcinogenesis. The first clinical TGFBR1 mutation, a 9-basepair in-frame deletion of TGFBR1 exon 1 named TGFBR1*6A (182), has been identified. Molecular characterization of this mutation revealed that the 9-basepair deletion was located in the signal sequence and did not affect generation of the mature receptor (183). However, functional assays showed that TGFBR1*6A was less effective than wild-type TGFBR1 with respect to TGF-β signaling (184,185). The hypomorphic nature of TGFBR1*6A and the discovery that its allelic frequency was higher among patients with cancer compared with normal control subjects led us to hypothesize that TGFBR1*6A may act as a tumor susceptibility allele (185).
It was further hypothesized that constitutively decreased TGFBR1-mediated TGF-β signaling might predispose to the development of cancer (46). Numerous follow-up studies have assessed the association between TGFBR1*6A and risk for various forms of cancer, and several recent meta-analyses have demonstrated that TGFBR1*6A is associated with risk for breast (186), ovarian (186), and colorectal cancer (187). It was also shown that somatically acquired TGFBR1*6A may switch TGF-β growth inhibitory signals into growth stimulatory signals (188). Additional evidence suggests TGFBR1*6A is acquired by both epithelia and stromal cells in colorectal as well as head and neck tumors (189). Somatic acquisition of TGFBR1*6A appears to be an early genetic event, which is not accompanied by acquisition of other tumor-specific mutations. Although TGFBR1*6A is not an oncogene, it enhances the migration and invasion of cancer cells (190). These prometastatic properties were confirmed by the discovery that 30% of colorectal cancer metastases somatically acquired TGFBR1*6A (188).
It has been demonstrated that constitutively decreased TGFBR1-mediated signaling through Tgfbr1 haploinsufficiency leads to cancer development (191), as evident in the two-fold increase in the incidence of premalignant polyps in wild-type Tgfbr1 mice when crossed with Apc Min/+ mice. Also, adenocarcinoma was only observed in Tgfbr1 haploinsufficient mice (191). These findings were further corroborated in humans, with constitutively decreased TGFBR1-mediated TGF-β signaling substantially more common among patients with a diagnosis of colorectal cancer than among healthy control subjects (192).
Summary and Clinical Relevance
The traditional terms “oncogene” and “tumor suppressor gene” fail to describe the role of TGF-β in tumorigenesis and progression. TGF-β can have potent tumor suppressive properties in early-stage disease (4,43,44) while harboring a tumor-promoting effect in more advanced stages. Thus, therapies targeting TGF-β too early in the disease process may be detrimental, and the timing of these therapies needs to be carefully considered.
A number of different approaches have been used to target TGF-β signaling. Monoclonal neutralizing antibodies targeting TGF-β proteins bind to ligand(s) and prevent access to TGF-β receptors. GC1008 is a neutralizing antibody that targets all three TGF-β molecules (193) and is being tested in phase I/II clinical trials for the treatment of metastatic breast cancer and malignant pleural mesothelioma (http://ClinicalTrials.gov). Soluble forms of TGFBR2 and TGFBR3 have also been used to sequester TGF-β from the cellular receptors (194,195). Another approach involves inhibiting the kinase activity of the TGF-β receptors. For example, LY2157299 is a kinase inhibitor that targets the ATP-binding site of TGFBR1 (196) and is currently in clinical trials for the treatment of a variety of cancers, including hepatocellular, glioma, and pancreatic cancer (http://ClinicalTrials.gov). Other kinase inhibitors are being developed that inhibit both TGFBR1 and TGFBR2. For example, LY2109761 is a dual TGFBR1/2 inhibitor that is currently undergoing preclinical studies (197). There are also attempts to decrease expression of TGF-β levels through the use of antisense technology. Antisense oligonucleotides that target TGF-β1 (AP 11014) and TGF-β2 (AP 12009) effectively reduced secretion of TGF-β from tumor cells (198,199). AP 12009 dramatically decreased secretion of TGF-β2 from cancer cell lines and suppressed tumor growth in vivo. Early-stage clinical trials in patients with advanced melanoma, colorectal carcinoma, or pancreatic carcinoma demonstrated promising results (200). A phase III clinical trial with AP12009 is currently underway in patients with high-grade gliomas (http://ClinicalTrials.gov).
Although results from current trials suggest potential promise for targeting the TGF-β pathway in humans, given the dual nature of TGF-β signaling, the challenge lies in targeting only the tumor-promoting aspect of TGF-β and minimizing the effect on the tumor-suppressive arms of TGF-β signaling. Also, it will be important to identify only those patients who will most likely respond to anti-TGF-β therapies and to exclude those patients who may be harmed by anti-TGF-β therapies.
Other potential targets are downstream of TGF-β. A novel target downstream of TGF-β is vimentin, a relative endpoint for several pathways involved in disease progression and that, apart from its role in the contractile phase of wound healing, is not entirely necessary for normal cell function (201). As discussed, another potential target is ADAM17, also a promising target for rheumatologic and autoimmune disease because of its promotion of TNF signaling. Some suggest caution regarding ADAM17 inhibition in cancer because the resultant surplus of SMAD signaling can have detrimental effects (59). Another example is the use of fibroblast activation protein-activated promelittin protoxin to target cancer-associated stromal cells for the treatment of prostate cancer (202). Additionally, the strategy of adoptive transfer of the tumor-reactive TGF-β–insensitive CD8+ T cells should be pursued with caution as another potential approach for late-stage cancer treatment (141–143,203).
Because perturbation of the TGF-β signaling network has a variety of tumorigenic effects (Figure 5), its mechanisms must be studied further to identify novel points of convergence with other pathways and maximize both the clinical efficacy and tumor specificity of future therapies. Through investigation of the TGF-β pathway and its relationship with other oncogenic factors in the tumor microenvironment, additional strategic points of convergence can be identified and exploited as a means to prevent or reverse tumor progression.
Aberrant transforming growth factor β (TGF-β) signaling in cancer. TGF-β has several tumor suppressive functions in benign cells. However, these effects are commonly lost in advanced cancers, and TGF-β begins to actively contribute to disease progression. The figure describes a series of partial mechanisms underlying the uncontrolled response to TGF-β in cancer cells and the dysregulation of several associated oncogenic factors leading to enhanced cell proliferation and migration.
Funding
This review was supported in part by grants from the Department of Defense (W81XWH-09-1-0311); American Cancer Society, Illinois (08-22); Portes Center/Institute of Medicine of Chicago; American Cancer Society Institutional Research Grant (ACS-IRG 93-037-12); Illinois Department of Public Health (contract 83284034); American Urological Association Foundation; National Cancer Institute (P50CA090386;, U01CA152738;, R01 CA137000;, CA126888;, and CA112520); the Institut National de la Santé Et de la Recherche Médicale (INSERM–Avenir Program); the Ligue Nationale Contre le Cancer; the Association pour la Recherche sur le Cancer; the Institut National du Cancer (“PANSTRESS” grant 2010–203); and by the Barnum and Zell Family Foundations at Northwestern University.
Notes
This manuscript is dedicated to the memory of Dr William J. Wasserman (Loyola University Chicago), a wonderful teacher, brilliant scientist, and exemplary human being.
The authors would like to thank Dr Qiang Zhang and Dr John M. Rosen (Children’s Memorial Hospital) and Dr Charles N. Rudick (Northwestern University) for their invaluable reviews of the manuscript, as well as Ryan E. Yaggie and Emman Mascariñas (Northwestern University) for their technical support.
References
Articles from JNCI Journal of the National Cancer Institute are provided here courtesy of Oxford University Press
Full text links
Read article at publisher's site: https://doi.org/10.1093/jnci/djt369
Read article for free, from open access legal sources, via Unpaywall:
https://academic.oup.com/jnci/article-pdf/106/2/djt369/7677178/djt369.pdf
Citations & impact
Impact metrics
Citations of article over time
Alternative metrics
Smart citations by scite.ai
Explore citation contexts and check if this article has been
supported or disputed.
https://scite.ai/reports/10.1093/jnci/djt369
Article citations
Unveiling the therapeutic potential: KBU2046 halts triple-negative breast cancer cell migration by constricting TGF-β1 activation <i>in vitro</i>.
Oncol Res, 32(11):1791-1802, 16 Oct 2024
Cited by: 0 articles | PMID: 39449805 | PMCID: PMC11497199
TEAD transcription factor family emerges as a promising therapeutic target for oral squamous cell carcinoma.
Front Immunol, 15:1480701, 04 Oct 2024
Cited by: 0 articles | PMID: 39430767 | PMCID: PMC11486717
Review Free full text in Europe PMC
GPR56 facilitates hepatocellular carcinoma metastasis by promoting the TGF-β signaling pathway.
Cell Death Dis, 15(10):715, 01 Oct 2024
Cited by: 0 articles | PMID: 39353900 | PMCID: PMC11445230
Prognostic value of tumour-associated regulatory T-cells as a biomarker in non-small cell lung cancer: a systematic review and meta-analysis.
Syst Rev, 13(1):233, 14 Sep 2024
Cited by: 1 article | PMID: 39272135 | PMCID: PMC11401299
Review Free full text in Europe PMC
Targeted therapies in hepatocellular carcinoma: past, present, and future.
Front Oncol, 14:1432423, 29 Aug 2024
Cited by: 0 articles | PMID: 39267840 | PMCID: PMC11390354
Review Free full text in Europe PMC
Go to all (304) article citations
Data
Similar Articles
To arrive at the top five similar articles we use a word-weighted algorithm to compare words from the Title and Abstract of each citation.
Lack of transforming growth factor-β signaling promotes collective cancer cell invasion through tumor-stromal crosstalk.
Breast Cancer Res, 14(4):R98, 02 Jul 2012
Cited by: 47 articles | PMID: 22748014 | PMCID: PMC3680921
The miR-106b-25 cluster targets Smad7, activates TGF-β signaling, and induces EMT and tumor initiating cell characteristics downstream of Six1 in human breast cancer.
Oncogene, 31(50):5162-5171, 30 Jan 2012
Cited by: 200 articles | PMID: 22286770 | PMCID: PMC3342483
Proteinase-Activated Receptor 2 May Drive Cancer Progression by Facilitating TGF-β Signaling.
Int J Mol Sci, 18(11):E2494, 22 Nov 2017
Cited by: 15 articles | PMID: 29165389 | PMCID: PMC5713460
Review Free full text in Europe PMC
Vasohibin-2 is required for epithelial-mesenchymal transition of ovarian cancer cells by modulating transforming growth factor-β signaling.
Cancer Sci, 108(3):419-426, 01 Mar 2017
Cited by: 20 articles | PMID: 28064471 | PMCID: PMC5378260
Funding
Funders who supported this work.
NCI NIH HHS (11)
Grant ID: R01 CA126888
Grant ID: U01 CA152738
Grant ID: U01CA152738
Grant ID: P50CA090386
Grant ID: R01 CA112520
Grant ID: CA112520
Grant ID: CA126888
Grant ID: P30 CA012197
Grant ID: R01 CA137000
Grant ID: P30 CA060553
Grant ID: P50 CA090386






