Abstract
Free full text

MicroRNA-137 Is a Novel Hypoxia-responsive MicroRNA That Inhibits Mitophagy via Regulation of Two Mitophagy Receptors FUNDC1 and NIX*
Abstract
Mitophagy receptors mediate the selective recognition and targeting of damaged mitochondria by autophagosomes. The mechanism for the regulation of these receptors remains unknown. Here, we demonstrated that a novel hypoxia-responsive microRNA, microRNA-137 (miR-137), markedly inhibits mitochondrial degradation by autophagy without affecting global autophagy. miR-137 targets the expression of two mitophagy receptors NIX and FUNDC1. Impaired mitophagy in response to hypoxia caused by miR-137 is reversed by re-expression of FUNDC1 and NIX expression vectors lacking the miR-137 recognition sites at their 3′ UTR. Conversely, miR-137 also suppresses the mitophagy induced by fundc1 (CDS+3′UTR) but not fundc1 (CDS) overexpression. Finally, we found that miR-137 inhibits mitophagy by reducing the expression of the mitophagy receptor thereby leads to inadequate interaction between mitophagy receptor and LC3. Our results demonstrated the regulatory role of miRNA to mitophagy receptors and revealed a novel link between miR-137 and mitophagy.
Introduction
Mitophagy is a process through which dysfunctional mitochondria are selectively removed by autophagy (1,–4). In this process, damaged mitochondria are incorporated into double-membrane structures known as autophagosomes, which are then delivered to lysosomes for degradation (2, 3, 5, 6). Mitophagy has a crucial function in controlling the quality of mitochondria, and thus, cellular homeostasis. Defects in this process have been implicated in numerous diseases, including neurodegenerative disorders and cancers (7,–10).
Mitophagy is mainly mediated by two signaling pathways, namely, PARKIN and mitophagy receptor (11,–17). E3 ubiquitin ligase PARKIN and phosphatase and tensin homolog-induced putative protein kinase 1 (PINK1) have essential roles in mitochondrial quality control. As mitochondrial membrane potential (Δψ) is lost, PINK1 is stabilized and accumulates on the outer membrane of damaged mitochondria where it selectively recruits PARKIN (11). PARKIN-mediated mitophagy is possible through ubiquitination of a subset of mitochondrial substrates (18,–20). Mitophagy can also be mediated by a family of receptors. Hypoxia induces a marked increase in LC3 (mammalian ATG8 homolog) puncta formation and extensive fragmentation of mitochondria, which is an HIF-1-dependent adaptive metabolic response (13, 21,–24). FUNDC1, an outer mitochondrial membrane protein, promotes mitophagy through interaction with LC3 in response to hypoxia (13). In addition to being a hypoxia-responsive protein, NIX also acts as a mitophagy receptor that is essential for erythroid maturation through autophagic removal of unwanted mitochondria (25,–27).
MicroRNAs, a class of short non-coding RNAs that post-transcriptionally regulate gene expression primarily through partial base pairing with regions of 3′ untranslated regions (3′ UTRs) of target mRNAs, promote translational inhibition or degradation of mRNAs (28,–30). MicroRNAs (miR)4 regulate a wide range of biological processes, including proliferation, stress responses, and metabolism (31). miR-137, the microRNA abundant in the brain, has an important role in controlling embryonic neural stem cell fate determination (32). The down-regulated expression of miR-137 in brain cancer stem cells is necessary for self-renewal in glioma stem cells (33, 34). miR-137 regulates neuronal maturation by targeting ubiquitin ligase Mind Bomb-1 (35). miR-137 is also down-regulated in the blood serum of Alzheimer disease patients (36). A recent study indicated that hematopoietic-restricted deletion of Kap1, the cofactor for KRAB-containing zinc finger proteins, fails to induce mitophagy-associated genes and retains mitochondria due to lack of repression of a subset of microRNAs, which target mitophagy transcripts by stage-specific KRAB-zinc finger proteins (37). This study reveals an emerging role of miRNA in modulating mitophagy.
Here we show that miR-137 significantly inhibits mitophagy induced by hypoxia by targeting two mitophagy receptors, FUNDC1 and NIX. This effect is rescued by re-introduction of FUNDC1 or NIX expression plasmids that lack the miR-137 pairing sites. miR-137 suppresses fundc1 (CDS+3′UTR), but not fundc1 (CDS)-induced mitophagy by lowering the expression of the mitophagy receptor thus decreases the number of mitophagy receptors bound to LC3. Our study thus reveals that microRNA regulates mitophagy by modulating mitophagy receptors.
EXPERIMENTAL PROCEDURES
Animals, Regents, and Antibodies
Mice were maintained and cared for on the basis of animal protocols approved by the Institutional Animal Care Committee. Adult C57BL/6(B6) mice were treated in either normoxic or 8% hypoxic conditions in a hypoxia chamber for 48 or 72 h, and then brain tissues were prepared, miRNAs were extracted, and the expression of miR-137 was examined by real-time quantitative PCR.
The following antibodies: anti-TIM23 (BD Biosciences, 611222), anti-TOM20 (BD Biosciences, 612278), anti-β-ACTIN antibody (Santa Cruz, sc-47778), anti-LC3B polyclonal antibody (Sigma, L7543), anti-FUNDC1 polyclonal antibody (AVIVA, ARP53280_P050), anti-Mitofusin2 (Epitomics, 3272-1), anti-VDAC1 monoclonal antibody (Abcam, ab14734), anti-OPA1 antibody (Thermo, 1E8-1D9), anti-P62/SQSTM1 antibody (MBL, PM045), anti-BNIP3 polyclonal antibody (Abcam, ab8399), anti-Bnip3L antibody (Sigma, HPA015652), anti-β-TUBULIN monoclonal antibody (Earthox, E021040), HRP Affinipure goat anti-mouse IgG (Earthox, E030110), and HRP Affinipure goat anti-rabbit IgG (Earthox, E030120) were used for Western blot. Anti-LC3B (Sigma, L7543) was used for immunoprecipitation assay. Purified mouse anti-CYTOCHROME c (BD Biosciences, 556432), anti-LC3 polyclonal antibody (MBL, PM036), anti-LC3B monoclonal antibody (MBL, M152–3), anti-c-Myc monoclonal antibody (Sigma, C3956), and anti-FLAG (Sigma, F7425) were used for immunofluorescence. The following fluorescent secondary antibodies were used: Alexa Fluor 555-labeled donkey anti-mouse IgG antibodies (Invitrogen, A31570), Alexa Fluor 488-labeled donkey anti-mouse IgG antibodies (Invitrogen, A21202), Alexa Fluor 488-labeled donkey anti-rabbit IgG antibodies (Invitrogen, A21206), Alexa Fluor 647-labeled donkey anti-mouse IgG antibodies (Invitrogen, A31571), and Alexa Fluor 647-labeled donkey anti-rabbit IgG antibodies (Invitrogen, A31573). Protein A/G plus-agarose immunoprecipitation reagent (Santa Cruz, sc-2003) and Lipofectamine 2000 (Invitrogen, 11668027) were also used. Bafilomycin A1 was purchased from Sigma.
Cell Culture and Transfection
SKNSH, SY5Y, HEK293, HeLa, and mouse embryonic fibroblast (MEF) cells were grown in Dulbecco's modified Eagle's medium (Invitrogen) supplemented with 10% fetal calf serum (Hyclone) and 1% penicillin/streptomycin (Beyotime, referred to as complete medium) at 37 °C under 5% CO2. Hypoxic conditions were achieved with a hypoxia chamber (Billups-Rothenberg) flushed with a pre-analyzed gas mixture of 1% O2, 5% CO2, and 95% N2. miR-137 inhibitor was synthesized by GenePharma (Suzhou, China). Control oligo and hsa-miR-137 were chemically synthesized by RiboBio (Guangzhou, China). RNA oligonucleotides were transfected into cells using Lipofectamine 2000 (Invitrogen) according to the manufacturer's protocol.
Plasmids
The 3′ UTR of the human fundc1 gene (NM_173794) and nix gene (NM_004331) were PCR amplified from human cDNA using primers: 5′-GCTCGCTAGCCTCGAGACATGAATATTCTCCCATAACGG-3′ (fundc1, forward); 5′-ATGCCTGCAGGTCGACTCTAAAGCTTTCCACAAAAGGG-3′ (fundc1, reverse); 5′-GCTCGCTAGCCTCGAGTTTCCCTAAGTTTGAGTTGATAGT-3′ (nix, forward); 5′-ATGCCTGCAGGTCGACTTTTGTGAGAGCTTTAATGGCACA-3′ (nix, reverse) and then cloned into the pmirGLO dual-Luciferase miRNA target expression vector (Promega). The corresponding mutant constructs were created by mutating the seed regions of the miR-137 binding sites. The primers used for cloning mutant plasmids were: 5′-GAGAAGAGAAGTGGCACGTTAAAGGCAGTCTCTCAAAAG-3′ (fundc1-mutant1, forward); 5′-CTTTTGAGAGACTGCCTTTAACGTGCCACTTCTCTTCTC-3′ (fundc1-mutant1, reverse); 5′-TTTAAGATATGTTTCGTTAAAGTTAAAAAACTG-3′ (fundc1-mutant2, forward); 5′-CAGTTTTTTAACTTTAACGAAACATATCTTAAA-3′ (fundc1-mutant2, reverse); 5′-CAAAAAAATATCCTGGGGTTATAAAAAAATATTTTAAACC-3′ (nix-mutant, forward); 5′-GGTTTAAAATATTTTTTTATAACCCCAGGATATTTTTTTG-3′ (nix-mutant, reverse). fundc1-myc, nix-FLAG, and gfp-lc3 constructs were described previously (12,–14). The fundc1 (CDS+3′UTR)-myc plasmid was constructed by a PCR-fusion based approach. The fused PCR product was derived from two overlapping PCR fragments. One fragment contained linear Fundc1 (CDS)-myc plasmid. The other fragment contained the 3′ UTR of the human fundc1 gene obtained from reporter plasmid. The PCR primers were, respectively, forward, 5′-AATATGCATACCGCTTGAGACATGAATATTCTCC-3′; reverse, 5′-GTGATGGTGATGATGTCTAAAGCTTTCCAC-3′ and forward, 5′-CACCATCACCATTGAGACATGAATATTCTCC-3′, reverse, 5′-TCAGCGGGTTTAAACTCTAAAGCTTTCCAC-3′. All constructs were verified by direct sequencing.
Luciferase Assay
Cells were seeded in 24-well plates 1 day before transfection. For reporter assays, the cells were transiently cotransfected with 0.8 μg of WT reporter plasmid or mutant plasmid in the presence of 100 nm control miRNA, miR-137, or miR-137 inhibitor using Lipofectamine 2000 (Invitrogen). Firefly and Renilla luciferase activities were measured consecutively by using Dual Luciferase Assay (Promega) according to the manufacturer's instructions. Three independent experiments were performed in triplicate.
qRT-PCR Gene Expression Analysis (Quantification of Mature miRNAs)
Total RNA was isolated with TRIzol (Life Technologies) followed by a DNase treatment to eliminate contaminating genomic DNA and reverse transcription reaction. Relative gene expression was determined using 2-step quantitative RT-PCR. Quantitative PCRs were performed with SYBR PrimeScript miRNA RT-PCR Kit (TaKaRa, RR716) on a Roche LightCycler480II Real-time PCR System. Fold-changes were calculated using the ΔΔCt method with normalization to U6 rRNA endogenous control. Primers used were: 5′-TTATTGCTTAAGAATACGCG-3′ (mature miR-137), 5′-TCGTATCCAGTGCAGGGTC-3′ (miR-Univer), 5′-CTCGCTTCGGCAGCACA-3′ (U6-F), 5′-AACGCTTCACGAATTTGCGT-3′ (U6-R). miR-210, miR-21, and miR-9 qPCR primers were purchased from RiboBio (Guangzhou, China). Three independent experiments were performed in triplicate.
Immunofluorescence Microscopy
Cells were grown to 60% confluence on a coverslip. After treatment, the cells were washed twice with PBS (Shanghai Sangon Biotech) and fixed with freshly prepared 4% formaldehyde at 37 °C for 15 min. Antigen accessibility was increased by treatment with 0.2% Triton X-100 (Shanghai Sangon Biotech). Cells were incubated with primary antibodies for 1 h, and after washing with PBS, stained with a secondary antibody for 50 min. Cell images were captured with a TCS SPF5 II Leica confocal microscope.
Electron Microscopy
Electron microscopy was done as previously described (38). Cells were first fixed with 2% paraformaldehyde and 0.2% glutaraldehyde in sodium cacodylate buffer (pH 7.4) at 37 °C for 2 h, and then dehydrated in a graded ethanol series and embedded. Approximately 75-nm ultrathin sections were mounted on nickel grids. The samples were then stained and visualized using a 120 kV Jeol electron microscope (JEM-1400) at 80 kV. Images were captured using a Gatan-832 digital camera.
SDS-PAGE and Western Blot
Cells were lysed in lysis buffer (20 mm Tris, pH 7.4, 2 mm EDTA, 1% Nonidet P-40, and protease inhibitors). Equivalent protein quantities (20 μg) were subjected to SDS-PAGE, and transferred to PVDF membranes (Millipore, Bedford, MA). Membranes were probed with the indicated primary antibodies, followed by appropriate HRP-conjugated secondary antibodies (KPL). Immunoreactive bands were visualized with Pro-light HRP (Tiangen) or Immobilon Western Chemiluminescent HRP Substrate (Milllipore).
Immunoprecipitation
HeLa cells were transiently transfected using Lipofectamine 2000 (Invitrogen) according to the manufacturer's protocol. About 24 h post-transfection, the cells were lysed with 1 ml of lysis buffer containing protease inhibitors (Roche Applied Science) for 30 min on ice. After 12,000 × g centrifugation for 15 min, the lysates were immunoprecipitated with specific antibody and protein A/G plus-agarose immunoprecipitation regent (Santa Cruz) overnight at 4 °C. Thereafter, the precipitants were washed three times with lysis buffer, and the immune complexes were eluted with sample buffer containing 1% SDS for 10 min at 100 °C and analyzed by 15% SDS-PAGE.
Statistical Analysis
Assays for characterizing cell phenotypes were analyzed by Student's test, and correlations between groups were calculated using Pearson's test. p values <0.05 were deemed statistically significant.
RESULTS
miR-137 Is a Novel Hypoxia Down-regulated MicroRNA
Recently, we performed several genome-wide microRNA profiling studies to find autophagy- or mitophagy-relevant microRNAs between normoxia and hypoxia (data not shown), or between hypoxia and hypoxia/reoxygenation (39). We found that the brain-enriched miR-137 is a novel hypoxia-responsive microRNA that has not been reported in previous microRNA profiling data (Fig. 1, A and B) (40,–42). The expression of miR-137 is largely suppressed in mouse brains or in cells subjected to hypoxic conditions (Fig. 1, A and B). As a control, miR-210, a well established hypoxia-responsive microRNA, is significantly up-regulated in either mouse brain or cells subjected to hypoxia treatments (Fig. 1, A and B) (42,–44). The expression of miR-21, another hypoxia-responsive microRNA identified in breast and colon cancer cells, also increases in mouse brain upon 72 h hypoxia treatment (Fig. 1A) (42,–44). The expression of miR-21 augments in SKNSH, SY5Y, or HeLa cells but goes back to the control level in MEFs after 24 h exposure to hypoxia (Fig. 1B). We then examined the expression of miR-9, which was not in the known list of hypoxia-responsive microRNAs (42,–44). The level of miR-9 in mouse brain does not change after 48 or 72 h exposure to hypoxia (Fig. 1A). Nevertheless, the expression of miR-9 increases in SKNSH, SY5Y, MEFs, or HeLa cells in response to hypoxia (Fig. 1B).
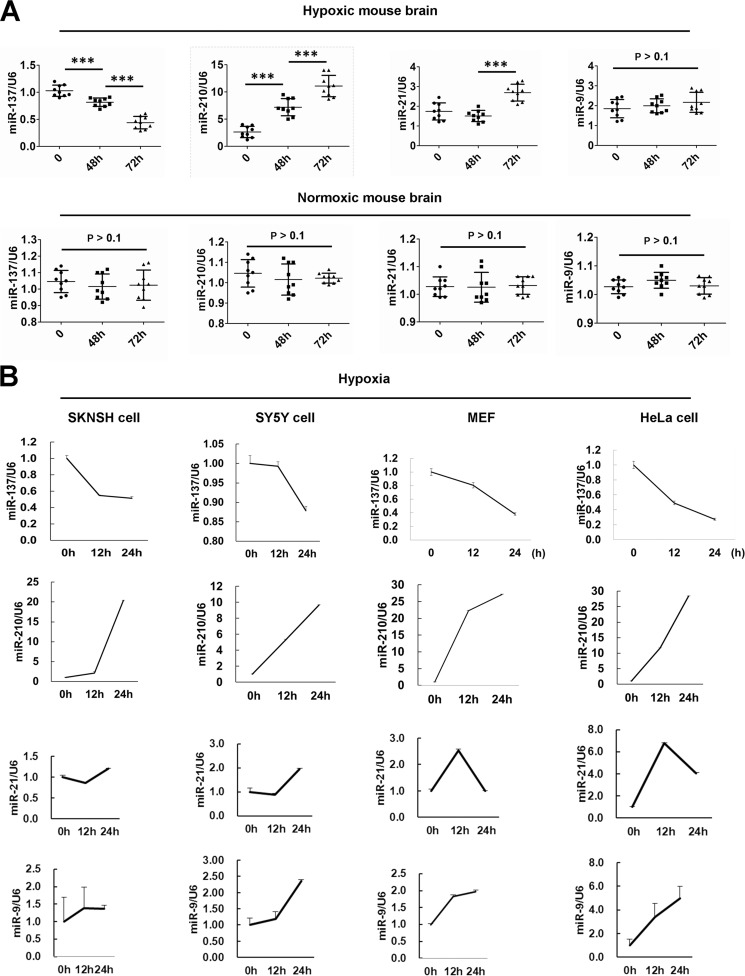
miR-137 is a hypoxia down-regulated microRNA. A, the expression level of miR-137, miR-210, miR-21, and miR-9 in mouse brain in response to hypoxic or normoxic condition. Data are from three independent experiments (n = 3). ***, p < 0.001. B, the expression levels of miR-137, miR-210, miR-21, and miR-9 in neuronal SKNSH and SY5Y cells, or in non-neuronal MEFs or HeLa cells treated by hypoxia. Three independent experiments were performed in triplicate.
miR-137 Represses Hypoxia-induced Mitophagy
Hypoxia potentially promotes mitophagy (13, 21, 22, 45, 46). So, we investigated the role of the brain-enriched miR-137 in hypoxia-induced mitophagy. Consistent with our previous report, hypoxia induces a marked increase and enlargement of the endogenous LC3 puncta and extensively fragmented mitochondria (Fig. 2, A–C) (13, 21). The number of cells that contains fragmented mitochondria induced by hypoxia is dramatically reduced upon expression of miR-137 compared with control microRNA (NC) (Fig. 2, B and C). miR-137 restores the fragmented mitochondria to normal network structures and causes a significant decrease in co-localization of endogenous LC3 puncta with mitochondria without affecting global LC3 puncta formation under hypoxia or starvation conditions, compared with NC (Fig. 2). Electron micrographs further show that the mitochondrion-containing autophagosome can be observed in NC-transfected cells in response to hypoxia, compared with normoxic or miR-137-transfected hypoxic samples (Fig. 2D). Biochemical analysis demonstrates that miR-137 represses the loss of outer and inner mitochondrial membrane proteins (Tom20 and Tim23) without apparently affecting the LC3-I to LC3-II transition. The degradation of LC3 and p62 proteins is dramatic in response to hypoxic conditions, which is inhibited by the lysosome inhibitor bafilomycin A1, suggesting that hypoxia promotes a strong autophagy flux (Fig. 2E).
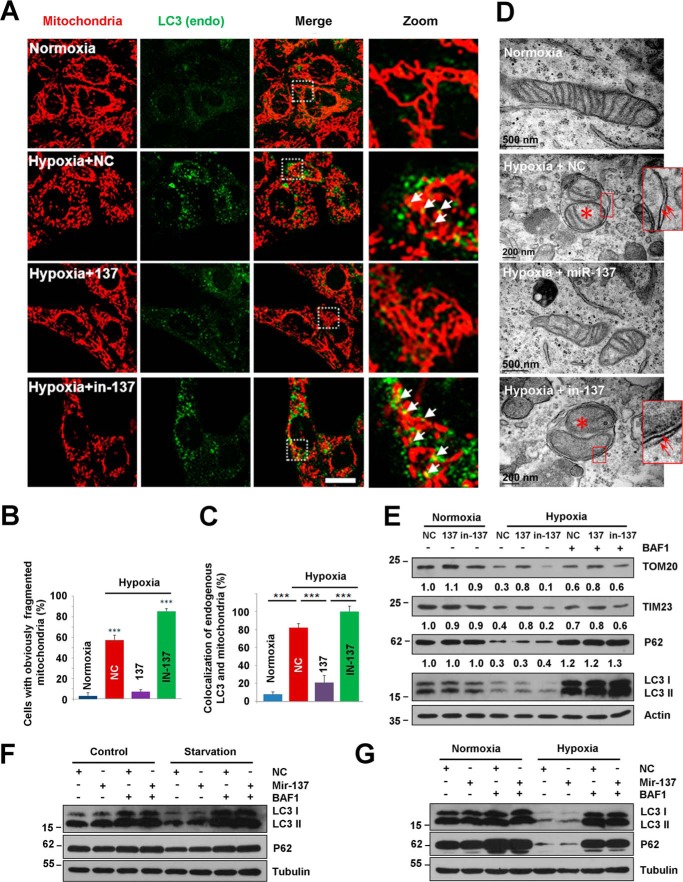
miR-137 suppresses hypoxia-induced mitophagy. A, MEFs were cultured in normal growth conditions (normoxia) or treated with hypoxia (1% O2) after being transfected with control microRNA (NC) and miR-137 or inhibitor of miR-137 (in-137) for 24 h. Samples were stained with anti-cytochrome c (red) and anti-LC3 (green) antibodies. White arrows indicate colocalized mitochondria with LC3. Scale bar, 10 μm. B and C, quantitative analysis of cells that contain fragmented mitochondria or co-localization of LC3 dots with mitochondria per cell in the experiments (A) (mean ± S.D.; n = 100 cells from three independent experiments; ***, p < 0.001). Co-localization is performed by ImageJ software. D, after transfection with NC, miR-137, or in-137, MEFs were maintained in normoxic or hypoxic conditions for 24 h. Samples were then processed for electron microscopy to assess the inhibitory role of miR-137 in hypoxia-induced mitophagy. Red arrows indicate double-membrane autophagic structures. Red asterisk shows engulfed mitochondrion in autophagosome. E, HeLa cells were transfected with NC, miR-137, or in-137, then exposed to hypoxic or normoxic conditions for 12 h accompanied with bafilomycin A1 (BFA) for another 12 h or not in the same condition. Samples were collected for Western blot to analyze the expression of LC3, P62, and mitochondrial proteins TOM20 and TIM23. Densitometric ratios of P62 and mitochondrial marker proteins from samples are quantified by ImageJ software (data are from three independent experiments; *, p < 0.05; **, p < 0.01). F, HeLa cells were transfected with NC or miR-137 for 24 h, then left starved or in complete medium for another 2 h with or without bafilomycin A1 (BFA). Samples were then analyzed by Western blot analysis probed by the indicated antibodies. G, HeLa cells were transfected with NC or miR-137 for 24 h, then maintained in normoxic or hypoxic conditions for another 12 h with or without bafilomycin A1. Samples were then analyzed by Western blot analysis.
miRNA inhibitors counteract miRNA effects by specifically pairing mature endogenous miRNAs. To test the effects of endogenous miR-137 inhibition on mitophagy, we used a miR-137-specific inhibitor (in-137) to analyze mitophagy under hypoxic conditions. The mitochondrial fragmentation is exacerbated by 34 ± 4% through the inhibition of endogenous miR-137 by the miR-137 inhibitor (in-137) compared with control microRNA (NC) (Fig. 2, A and B). The co-localization of endogenous LC3 puncta with damaged mitochondria becomes more prominent upon inhibition of endogenous miR-137 under hypoxic conditions compared with NC (Fig. 2, A and C). The autophagic degradation of mitochondria (Tom20 and Tim23) is accelerated by the inhibition of endogenous miR-137, indicating that endogenous miR-137 contributes to the limitation of hypoxia-activated mitophagic responses (Fig. 2, A–E).
Mitophagy Receptors FUNDC1 and NIX Are Direct Targets of miR-137
To unravel the direct molecular mechanism of mitophagy inhibition by miR-137, we searched for genes containing potential miR-137 recognition sites in their 3′ UTRs using bioinformatics tools Target scan, miRBase, and DIANA LAB. Interestingly, mitophagy receptors fundc1 and nix are identified as two potential miR-137 targets (Fig. 3A). The predicted interaction between miR-137 and the fundc1 or nix 3′ UTR is shown in Fig. 2A. fundc1 holds two sites within the 3′ UTR at nucleotides 51–58 and 464–470, respectively, whereas nix contains a single 7-mer seed match to miR-137 at nucleotide 556–562 (Fig. 3A).
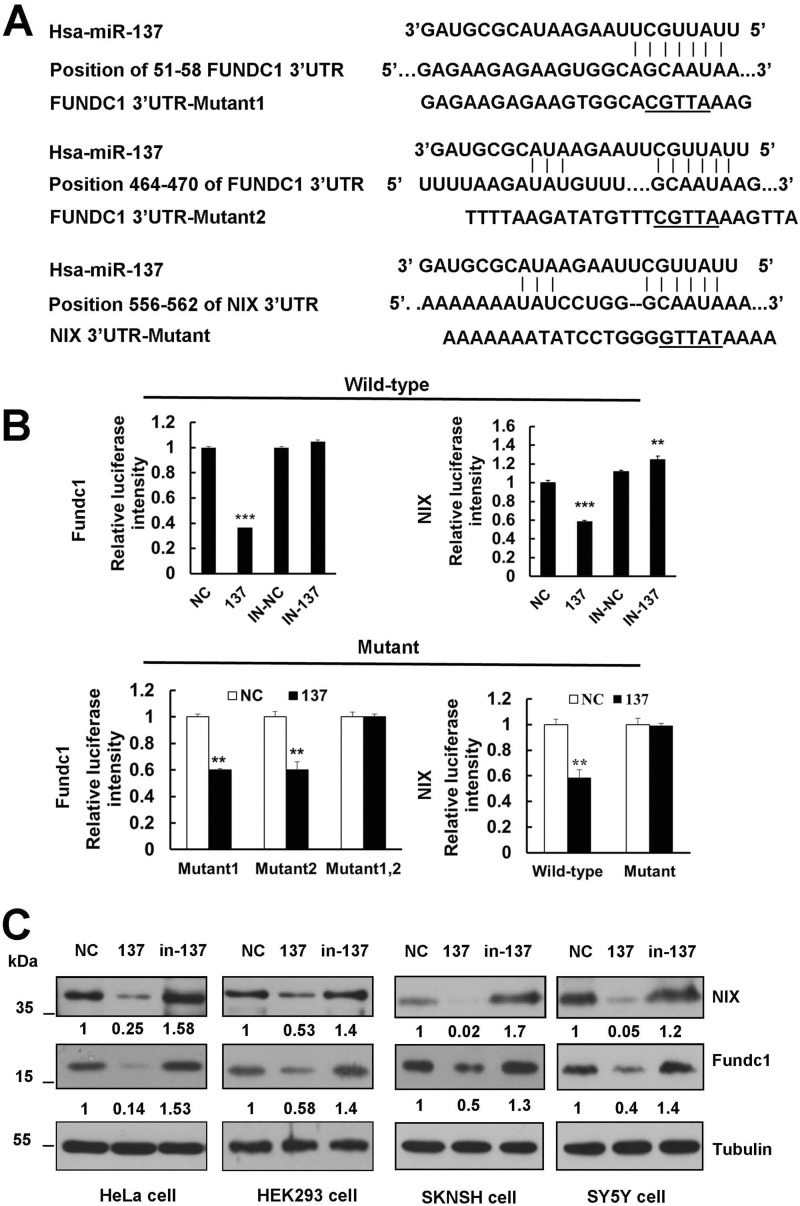
Mitophagy receptors FUNDC1 and NIX are direct targets of miR-137. A, diagram of WT and mutant luciferase report constructs. The matched base pairs are connected by a vertical line, and the mutated nucleotides in the target sites are underlined. fundc1 and nix 3′ UTR relevant fragments were inserted downstream of the firefly luciferase gene of the pmirGLO vector. B, luciferase reporter assay by the interaction between miR-137 and the predicted microRNA response elements in HEK293 cells. Each WT luciferase construct was co-transfected with control, in-control, miR-137, or in-137. Meanwhile, the mutant luciferase construct was co-transfected with control and miR-137. About 24 h after transfection, luciferase activity was detected. The firefly luciferase activity was normalized to Renilla. Data are shown as the mean ± S.D. from three independent experiments. *, p < 0.05; **, p < 0.01; ***, p < 0.001. C, miR-137 represses endogenous FUNDC1 and NIX expression. NC, miR-137 mimics, and in-137 were transfected into HeLa, HEK293, SKNSH, and SY5Y cells, respectively. At 48 h after transfection, cell lysates were prepared and subjected to Western blot analysis by using anti-FUNDC1 and NIX antibodies.
To confirm the in silico-based predictions, we next examined the ability of miR-137 to regulate fundc1 and nix. We individually cloned 559-base pair 3′ UTR fragments from fundc1 and 2721-base pair 3′ UTR fragments from nix to a dual luciferase reporter system, and tested the ability of miR-137 to regulate the reporters. As shown in Fig. 3B, the 3′ UTRs of two putative target genes responded markedly to miR-137 overexpression relative to a scrambled control. Notably, the luciferase vectors are susceptible to repression of endogenous miR-137 as the inhibition of miR-137 specifically relieves the repression of the reporters (Fig. 3B). To further validate the specificity of miR-137, we constructed several luciferase vectors that carry mutants of fundc1 or nix (Fig. 3A). Disruption of either of the pairing sites did not prevent the susceptibility of the fundc1-reporter to miR-137, whereas double mutants did, because fundc1 has two miR-137 pairing sites at the 3′ UTR (Fig. 3B). Substitution of several nucleotides at the nix-reporter blocks the repressive effect by miR-137 (Fig. 3B). Finally, we performed immunoblot analysis in the negative control (NC) and in miR-137 or in-137-transfected cell extracts using anti-FUNDC1 or NIX specific antibody. Overexpression of miR-137 results in a potent down-regulation of both FUNDC1 and NIX protein levels in HeLa, HEK293, SKNSH, or SY5Y cells (Fig. 3C). Conversely, introduction of the antagomir in-137 results in an increase in FUNDC1 or NIX protein levels in these cells, as compared with NC (Fig. 3C).
Re-expression of NIX and FUNDC1 without miR-137 Response Elements Restores Mitophagy Inhibited by miR-137
To further validate that the repression of FUNDC1 and NIX is due to the mitophagy-related effects of miR-137, we determined whether the expression of fundc1 and nix plasmids lacking the miR-137 recognition elements is resistant to miR-137-mediated suppression. In agreement with previous reports, under normoxic conditions, overexpression of fundc1 and nix could induce potent mitochondria fragmentation and mitochondria-LC3 colocalization (Fig. 4, A–C). We also confirmed the critical function of FUNDC1 or NIX in hypoxia-induced mitophagy by separately knocking down FUNDC1 or NIX by siRNAs (Fig. 4, D and E). Hypoxia induces mitochondrial fragmentation and mitophagy with or without overexpression of either fundc1 or nix (Fig. 4, A–C). Under hypoxia, miR-137 overexpression significantly decreases endogenous FUNDC1 and NIX protein levels so that the autophagic degradation of mitochondrial proteins VDAC1, MFN2, and TOM20 are reduced compared with NC (Fig. 4C). However, re-introduction of either Fundc1 or Nix that lacks the miR-137 recognition sites is sufficient to induce more mitophagy even in the presence of miR-137 (Fig. 4C). The autophagic degradation of mitochondria is markedly prevented by the lysosome inhibitor bafilomycin A1 (Fig. 4C). Therefore, under these conditions, miR-137-mediated suppression of mitophagy and mitochondrial fission is reversed upon re-expression of the FUNDC1 or NIX protein (Fig. 4).
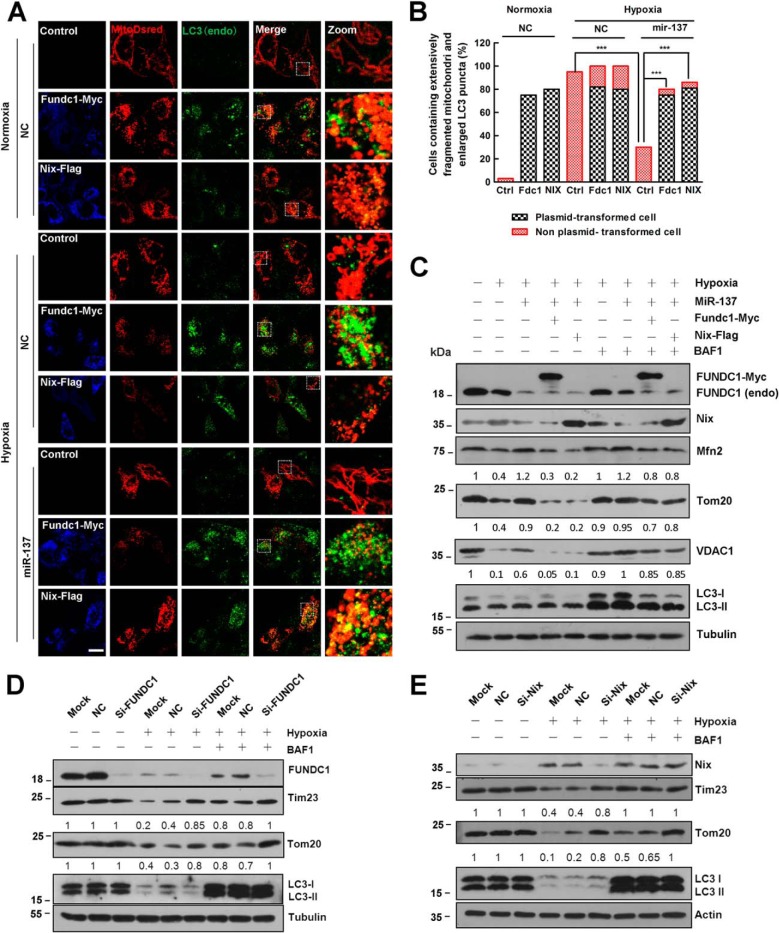
Re-expression of FUNDC1 and NIX without miR-137 response elements rescue mitophagy inhibited by miR-137. A, in HeLa cells, control vector, fundc1-myc, and nix-FLAG were individually co-transfected with Mito-dsRed in the presence of miR-137 or NC. HeLa cells were then treated with either hypoxia or normoxia for 24 h. Samples were processed for immunofluorescence with anti-LC3 (green) and anti-MYC (blue) or anti-FLAG (blue) antibodies. Scale bar, 20 μm. B, cells that contain extensively fragmented mitochondria and enlarged LC3 dots are quantified (data are shown as the mean ± S.D. from three independent experiments. ***, p < 0.001). C, HeLa cells were co-transfected with microRNA (NC or miR-137) and plasmids (fundc1-myc or nix-flag) or not as indicated. Cells were then treated with either hypoxia or normoxia for 12 h accompanied with bafilomycin A1 (BFA1) for another 12 h or not in the same condition. Samples were collected and analyzed for the expression of endogenous or exogenous NIX, FUNDC1, LC3, and mitochondrial proteins VDAC1, TOM20, and MFN2 by Western blot analysis. D, HeLa cells were transfected with siRNA (scrambled or si-fundc1) or not (Mock) as indicated. Cells were then treated with either hypoxia or normoxia for 24 h in the presence of BFA for another 12 h or not in the same condition. Samples were collected and analyzed by Western blot. E, HeLa cells were transfected with siRNA (scrambled or si-nix) or not (Mock) as indicated. Cells were then treated with either hypoxia or normoxia for 24 h in the presence of BFA for another 12 h or not in the same condition. Samples were collected and analyzed by Western blot. Tubulin or actin were used as loading controls. Representative mitochondrial proteins/tubulin densitometric ratios were marked (n = 3). The samples were derived from the same experiment and densitometric ratios of blots were quantified by ImageJ software and processed in parallel.
Our previous results show that FUNDC1 could potentially induce mitophagy (13). To further examine the specificity of miR-137 on mitophagy receptor inhibition, we constructed a fundc1 (CDS+3′UTR) plasmid containing the recognition sequences of miR-137 and compared it with fundc1 (CDS)-induced effects. We found that transfection of both plasmids can effectively induce mitochondrial fragmentation and mitophagy in the presence of control microRNA (NC) (Fig. 5, A–C). By contrast, fundc1 (CDS+3′UTR)-induced mitochondrial fragmentation or mitophagosome formation in the presence of miR-137 could be strongly inhibited (Fig. 5, A–D). However, inhibition of fundc1 (CDS)-induced mitophagy by miR-137 is not observed (Fig. 5, A–D). Biochemical experiments are in agreement with the immunofluorescence results. fundc1 (CDS+3′UTR)-induced autophagic degradation of mitochondrial proteins (Tim23, Tom20, and VDAC1) is suppressed by miR-137, whereas miR-137 does not prevent mitophagy induced by fundc1 (CDS) (Fig. 5D).
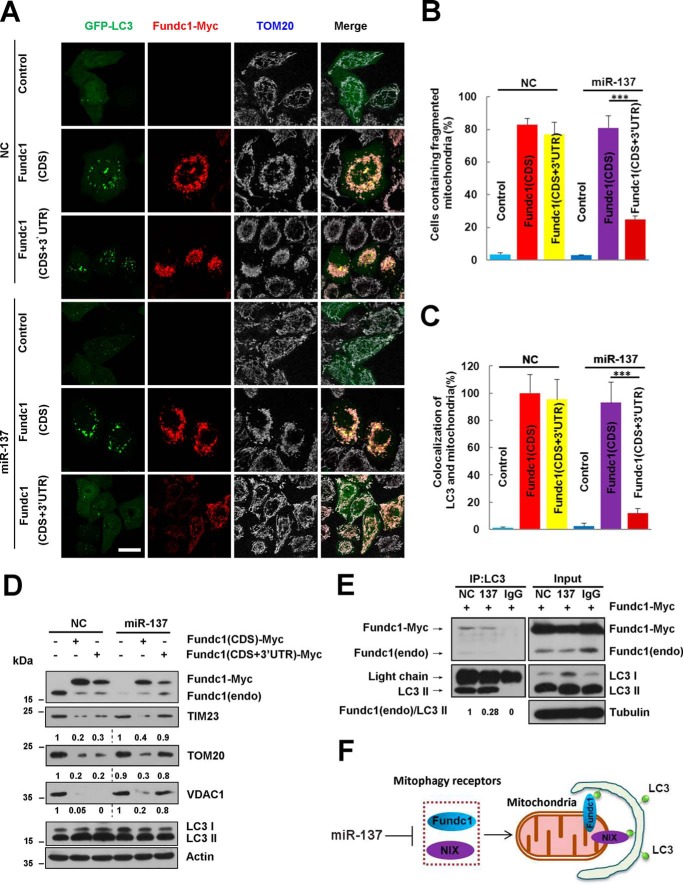
miR-137 inhibits mitophagy by reducing the number of mitophagy receptors bound to LC3. A, GFP-LC3 (green) positive HeLa cells were co-transfected with the indicated constructs (control vector, fundc1 (CDS), or fundc1 (CDS+3′ UTR)) and microRNA (miR-137 or NC) for 24 h. Cells were processed for immunofluorescence with anti-MYC (red) and anti-TOM20 (gray) antibodies. Scale bar, 20 μm. B and C, the number of cells containing fragmented mitochondria and the co-localization of GFP-LC3 and mitochondria quantified (data are shown as the mean ± S.D. from three independent experiments. ***, p < 0.001). D, the experiments are the same as in A without introduction of GFP-LC3 constructs. Samples were collected and analyzed for the expression of endogenous or exogenous FUNDC1, LC3, and mitochondrial proteins TOM20, TIM23, and VDAC1 by Western blot analysis. ACTIN was used as a loading control. Mitochondrial proteins/Actin and LC3-II/LC3-I densitometric ratios were marked. E, fundc1-myc was co-transfected with either miR-137 or NC. Exogenous and endogenous FUNDC1 were immunoprecipitated with anti-LC3 antibody or IgG. Samples were then analyzed by Western blot analysis. Endogenous FUNDC1/precipitated LC3-II densitometric ratios were marked (n = 3). The samples were derived from the same experiment and densitometric ratios of blots were quantified by ImageJ software and processed in parallel. F, provisional working model of miR-137 in regulation of mitophagy.
miR-137 Inhibits Mitophagy by Reducing the Number of Mitophagy Receptors Bound to LC3
To further explore the mechanism of the inhibitory role of miR-137 in mitophagy, we used FUNDC1-induced mitophagy as a cellular model. In HeLa cells transfected with NC or miR-137, co-immunoprecipitation analysis showed that miR-137 markedly reduces the binding of endogenous LC3 and FUNDC1 compared with NC (Fig. 5E). Thus, miR-137 represses mitophagy by reducing the protein level of the mitophagy receptor, thereby affecting the number of mitophagy receptors bound to LC3, which in turn causes mitophagy inhibition by miR-137.
DISCUSSION
Recent microRNA profiling data have revealed a group of hypoxia-relevant microRNAs from several cancer cells (40, 42). But their biological functions are less clear. Among them, miR-210 is consistently up-regulated in response to hypoxia in several independent studies (41). In this study, we chose hypoxia-responsive microRNAs including miR-210 and miR-21, or non-responsive microRNA, miR-9 as controls. miR-210 and miR-21 are indeed induced by hypoxia in vivo and in vitro, except that in MEFs the expression of miR-21 goes back to the basal level upon 24 h hypoxic treatment. Consistent with previous results in breast and colon cancer cells, miR-9 shows no responsiveness to hypoxia in vivo. However, it is up-regulated in response to hypoxia in SKNSH cells, SY5Y cells, HeLa cells, and MEFs. Except for miR-210, the limited overlap in microRNAs regulated by hypoxia from different studies is not surprising, given the differences in cells examined, oxygen concentrations, as well as time investigated (42, 44). Interestingly, in this study we identified that miR-137 is a novel down-regulated microRNA in response to hypoxia not only in different cell lines but also in vivo. miR-137 is involved in mitophagy because overexpression of miR-137 suppresses mitophagy induced by hypoxia by inhibiting the autophagic degradation of overall mitochondrial proteins, including inner and outer mitochondrial membrane proteins Tim23, Tom20, and VDAC1 without affecting global autophagy. In addition, the mitophagic effect is accelerated by antagomir-mediated repression of cellular endogenous miR-137. We further demonstrated that the inhibitory effect of miR-137 on mitophagy consists of targeting mitophagy receptors FUNDC1 and NIX, which impairs the binding of the mitophagy receptor to LC3, thereby inhibiting the targeting of mitochondria to autophagosomes. Our study established a novel connection between microRNA and mitophagy receptors.
Several E3 ubiquitin ligases participate in mitochondria turnover, including cytosolic Parkin and mitochondrial E3 ubiquitin ligase MARCH-5 and MUL1 (12, 47,–50). They ubiquitinate a series of mitochondrial substrates and may contribute to mitochondrial clearance through mitophagy. Among them, the mechanism of PARKIN/carbonyl cyanide p-trifluoromethoxyphenylhydrazone-mediated mitophagy has been most intensively studied (51, 52). Despite the well established mechanism of the PARKIN/FCCP-mediated mitophagy pathway, little is known about the role of mitophagy receptors, including FUNDC1 and NIX (52). The ubiquitinated proteins are possibly recognized by P62 family proteins, whereas the latter interacts with autophagosome components such as LC3, which may mediate the full engulfment of mitochondria by autophagosomes (12, 53). Notwithstanding the proposal of P62-independent mitophagy mediated by PARKIN/carbonyl cyanide p-trifluoromethoxyphenylhydrazone, more studies are required to understand this complicated cargo-autophagosome recognition process (54, 55). Our observations that miR-137 represses mitophagy by down-regulating the expression of mitophagy receptors, and thus, reducing mitophagy receptors bound to LC3 suggests another parallel cargo-autophagosome recognition framework in which mitophagy receptors could be directly involved in mitophagy (Fig. 5F).
*This work was supported by grants from the Scientific Research Start-up from Guangdong Medical College (B2012043, B2012044, and 701B01206), the National Natural Science Foundation of China (81171244 and 31301104), the Guangdong Natural Science Foundation of China (S2011010004095), the Startup fund of the Ministry of Education for returning-back scholars of China ([2012]940), and the Foundation for Distinguished Young Talents in Higher Education of Guangdong, China (2013LYM_0035).
4The abbreviations used are:
- miR
- microRNA
- MEF
- mouse embryonic fibroblast.
REFERENCES
Articles from The Journal of Biological Chemistry are provided here courtesy of American Society for Biochemistry and Molecular Biology
Full text links
Read article at publisher's site: https://doi.org/10.1074/jbc.m113.537050
Read article for free, from open access legal sources, via Unpaywall:
http://www.jbc.org/article/S0021925820403333/pdf
Citations & impact
Impact metrics
Citations of article over time
Alternative metrics
Article citations
The Role of Non-Coding RNAs in Mitochondrial Dysfunction of Alzheimer's Disease.
J Mol Neurosci, 74(4):100, 28 Oct 2024
Cited by: 0 articles | PMID: 39466447
Review
The Role of miR-137 in Neurodegenerative Disorders.
Int J Mol Sci, 25(13):7229, 30 Jun 2024
Cited by: 0 articles | PMID: 39000336
Review
Potential application of traditional Chinese medicine in age-related macular degeneration-focusing on mitophagy.
Front Pharmacol, 15:1410998, 17 May 2024
Cited by: 0 articles | PMID: 38828456 | PMCID: PMC11140084
Review Free full text in Europe PMC
The structure and function of FUN14 domain-containing protein 1 and its contribution to cardioprotection by mediating mitophagy.
Front Pharmacol, 15:1389953, 17 May 2024
Cited by: 0 articles | PMID: 38828457 | PMCID: PMC11140143
Review Free full text in Europe PMC
miRNA-137-5p improves spatial memory and cognition in Alzheimer's mice by targeting ubiquitin-specific peptidase 30.
Animal Model Exp Med, 6(6):526-536, 18 Dec 2023
Cited by: 3 articles | PMID: 38111333 | PMCID: PMC10757218
Go to all (79) article citations
Data
Data behind the article
This data has been text mined from the article, or deposited into data resources.
BioStudies: supplemental material and supporting data
HPA - The Human Protein Atlas
- (1 citation) HPA - HPA015652
Similar Articles
To arrive at the top five similar articles we use a word-weighted algorithm to compare words from the Title and Abstract of each citation.
Mitochondrial outer-membrane protein FUNDC1 mediates hypoxia-induced mitophagy in mammalian cells.
Nat Cell Biol, 14(2):177-185, 22 Jan 2012
Cited by: 901 articles | PMID: 22267086
BNIP3L/NIX and FUNDC1-mediated mitophagy is required for mitochondrial network remodeling during cardiac progenitor cell differentiation.
Autophagy, 15(7):1182-1198, 22 Feb 2019
Cited by: 150 articles | PMID: 30741592 | PMCID: PMC6613840
ULK1 translocates to mitochondria and phosphorylates FUNDC1 to regulate mitophagy.
EMBO Rep, 15(5):566-575, 26 Mar 2014
Cited by: 305 articles | PMID: 24671035 | PMCID: PMC4210082
Receptor-mediated mitophagy in yeast and mammalian systems.
Cell Res, 24(7):787-795, 06 Jun 2014
Cited by: 212 articles | PMID: 24903109 | PMCID: PMC4085769
Review Free full text in Europe PMC





