Abstract
Free full text

Cardiac fibroblast–derived microRNA passenger strand-enriched exosomes mediate cardiomyocyte hypertrophy
Associated Data
Abstract
In response to stress, the heart undergoes extensive cardiac remodeling that results in cardiac fibrosis and pathological growth of cardiomyocytes (hypertrophy), which contribute to heart failure. Alterations in microRNA (miRNA) levels are associated with dysfunctional gene expression profiles associated with many cardiovascular disease conditions; however, miRNAs have emerged recently as paracrine signaling mediators. Thus, we investigated a potential paracrine miRNA crosstalk between cardiac fibroblasts and cardiomyocytes and found that cardiac fibroblasts secrete miRNA-enriched exosomes. Surprisingly, evaluation of the miRNA content of cardiac fibroblast–derived exosomes revealed a relatively high abundance of many miRNA passenger strands (“star” miRNAs), which normally undergo intracellular degradation. Using confocal imaging and coculture assays, we identified fibroblast exosomal–derived miR-21_3p (miR-21*) as a potent paracrine-acting RNA molecule that induces cardiomyocyte hypertrophy. Proteome profiling identified sorbin and SH3 domain-containing protein 2 (SORBS2) and PDZ and LIM domain 5 (PDLIM5) as miR-21* targets, and silencing SORBS2 or PDLIM5 in cardiomyocytes induced hypertrophy. Pharmacological inhibition of miR-21* in a mouse model of Ang II–induced cardiac hypertrophy attenuated pathology. These findings demonstrate that cardiac fibroblasts secrete star miRNA–enriched exosomes and identify fibroblast-derived miR-21* as a paracrine signaling mediator of cardiomyocyte hypertrophy that has potential as a therapeutic target.
Introduction
Cardiac remodeling is a hallmark in the progression of many cardiovascular diseases and is characterized by cardiomyocyte hypertrophy and cardiac fibrosis that lead often to heart failure, a major and increasing cause of mortality worldwide (1–3). Cardiomyocytes and nonmuscle cells, especially cardiac fibroblasts are dominating cell types in the process of cardiac remodeling. Cardiac stress leads to cardiomyocyte hypertrophy, whereas fibroblasts start to proliferate and to secrete extracellular matrix proteins and proinflammatory cytokines, triggering tissue fibrosis (4). These fibroblast-derived mediators may act in an autocrine or paracrine fashion between fibroblasts and cardiomyocytes to stimulate cardiac remodeling (5, 6). Further, adult murine cardiomyocytes develop hypertrophy when cocultured with cardiac fibroblasts or treated with conditioned fibroblast media (7, 8). However, the underlying mechanisms of fibroblast-cardiomyocyte communication are not yet fully understood.
Recently, microRNAs (miRNAs) have emerged as regulators of cell-cell communication and paracrine signaling mediators during physiological and pathological processes (9–13). miRNAs are short, noncoding nucleotides regulating expression of target genes by mRNA degradation or translational repression (14, 15). Altered expression of miRNAs has been linked to cardiovascular disease conditions, such as heart failure and cardiac hypertrophy (16–20). The discovery of circulating extracellular miRNAs in body fluids indicates a new role of miRNAs to serve as paracrine signaling mediators (9). Circulating miRNAs are actively transported, either by the inclusion in microvesicles/exosomes (9, 11, 12) or by binding to RNA-binding proteins (21, 22), allowing protection from ribonuclease-dependent degradation. Of note, a role for miRNA/vesicle-mediated communication between endothelial cells and other cardiovascular cells was discovered recently (10, 13).
Here, we report data about a novel paracrine miRNA/exosome-mediated crosstalk between cardiac fibroblasts and cardiomyocytes, leading to cardiomyocyte hypertrophy.
Results
Cardiac fibroblasts produce and release exosomes.
Recent reports have shown that miRNAs are actively secreted in microvesicles or exosomes from different cell types, highlighting their potential to serve as paracrine intercellular signaling molecules (9–12). Several assays were performed to determine whether cardiac fibroblasts produce and secrete exosomes; we first used electron microscopy studies and identified the presence of typical multivesicular bodies (MVBs) in the cytoplasm of pure cardiac fibroblast cultures (Figure (Figure1A1A and Supplemental Figure 1; supplemental material available online with this article; 10.1172/JCI70577DS1), which are formed during exosome biogenesis (23). The plasma membrane of the MVBs invaginated inward (Figure (Figure1A,1A, arrows), forming intraluminal vesicles within cardiac fibroblasts (Figure (Figure1A,1A, inset). The MVBs are directed to the plasma membrane of cardiac fibroblasts (Figure (Figure1B),1B), in which they fuse with the plasma membrane and release the intraluminal vesicles into the extracellular fluid or conditioned media, which are then termed exosomes (23). Next, exosomes were isolated from the conditioned media of cardiac fibroblasts and characterized extensively. Electron microscopic analysis revealed a typical size of 50 to 100 nm and a characteristic cup-shaped morphology (Figure (Figure1C),1C), which has been shown for a majority of isolated exosomes (11, 12, 24). Western blotting and FACS analyses confirmed the presence of the exosomal marker protein CD63 in fibroblast-derived exosomes (Figure (Figure1,1, D and E). When we compared isolated RNA from cardiac fibroblasts with that of fibroblast-derived exosomes, we found the RNA profile of fibroblast-derived exosomes to contain only minute amounts of ribosomal RNA (18S and 28S), compared with that of fibroblasts, but large amounts of small RNAs, such as miRNAs (Figure (Figure1,1, F and G). The secretion of miRNAs via exosomes has been shown to be regulated in part by the neutral sphingomyelinase 2 (SMPD3, also known as NSMASE2) (25), which we found highly expressed in rat cardiac fibroblasts (Figure (Figure1H),1H), indicating NSMASE2 to be potentially involved in the secretion of miRNAs in cardiac fibroblasts. The high purity of our fibroblast cell cultures derived from neonatal rat hearts was confirmed by phenotypic immunohistological and molecular protein expression analyses (Supplemental Figure 1).
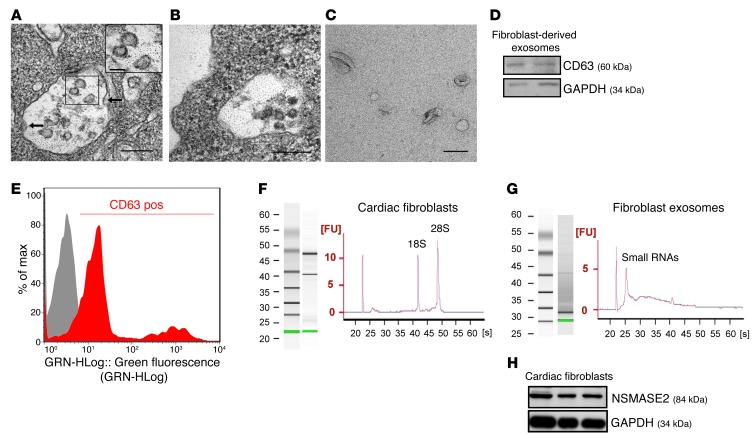
(A and B) Electron microscopy images of rat cardiac fibroblasts. (A) Cytoplasm of rat cardiac fibroblasts with MVB. MVB membrane invaginated inward (arrows), forming intraluminal vesicles (inset: higher magnification of intraluminal vesicles; scale bar: 100 nm). (B) MVB fusing with the cell membrane. (A and B) Scale bar: 200 nm (n = 3). (C) Electron microscopy image of rat cardiac fibroblast–derived exosomes, showing a size of approximately 50 to 100 nm in diameter. Scale bar: 100 nm (n = 4). (D) Western Blot of fibroblast-derived exosomes for CD63 (60 kDa) and GAPDH (34 kDa). (E) Flow cytometry analysis of CD63 of fibroblast-derived exosomes. Fibroblast-derived exosomes were immunostained against CD63 (red curve) and compared with the appropriate isotype control (gray curve). (F and G) Total RNA from (F) rat cardiac fibroblasts and (G) fibroblast-derived exosomes was analyzed by bioanalyzer. Gels and electropherograms are shown. The left gel lane is the ladder standard and the right lane is the total RNA from (F) cardiac fibroblasts or (G) exosomes. Y axis of the electropherogram is the arbitrary fluorescence unit intensity (FU) and x axis is migration time in seconds (s). (H) Western blot analysis of NSMASE2 (84 kDa) and GAPDH (34 kDa) in neonatal rat cardiac fibroblast lysates (n = 3).
Enriched miRNA star strands in fibroblast-derived exosomes.
To analyze the miRNA content of fibroblast-derived exosomes, a miRNA profiling assay (388 rat miRNAs) was performed. Only miRNAs, which were detected in all samples and showed a Ct value <35, were considered for further analysis. When comparing the miRNA ratio derived from exosomes and fibroblast cells, 50 miRNAs were found to be enriched in fibroblast-derived exosomes (Figure (Figure2A2A and Supplemental Table 1). Interestingly, 25.5% of fibroblast-derived exosome-enriched miRNAs were so called “star” miRNAs, representing the passenger (star) strand that often is degraded within a cell. In a previous study, our group showed that miR-21 is predominantly expressed in cardiac fibroblasts and plays a major role in fibroblast biology (17). The present miRNA profiling in exosomes identified miR-21* to be enriched in fibroblast-derived exosomes, suggesting that miR-21* might be transported out of the cardiac fibroblast via exosomal transfer, whereas miR-21 remains inside. Indeed, TaqMan qRT-PCR validation experiments revealed miR-21 levels to be higher in cardiac fibroblasts when compared with those of miR-21* (Figure (Figure2,2, B and C), whereas, in exosomes, miR-21* expression was significantly higher compared with miR-21 expression (Figure (Figure2,2, B and D). In agreement with the qRT-PCR data, RNA sequencing from cardiac fibroblasts and fibroblast-derived exosomes confirmed an enrichment of miR-21 (5.2 log2 fold change) in cardiac fibroblasts, whereas, in cardiac fibroblast–derived exosomes, miR-21* was significantly enriched (–2.1 log2 fold change) (Figure (Figure2E).2E). To clarify whether exosome-generating NSMASE2 is also involved in the secretion of miRNA-enriched fibroblast-derived exosomes, we treated cardiac fibroblasts with a chemical NSMASE2 inhibitor (GW4869). Treatment with this NSMASE2 inhibitor for 48 hours resulted in a strong accumulation of CD63-positive vesicles within cardiac fibroblasts (Figure (Figure2F).2F). Inhibition of NSMASE2 also led to a significant reduction of miR-21* content in fibroblast-derived exosomes (Figure (Figure2G).2G). Reduced levels were also observed for many other but not all miRNAs (see Supplemental Figure 2A). This finding indicates that the secretion of miRNAs in cardiac fibroblasts is regulated at least in part by NSMASE2. In contrast, treatment with the profibrotic and prohypertrophic molecule Ang II increased the levels of miR-21* in fibroblast-derived exosomes, suggesting disease-specific regulation of miRNA-enriched fibroblast-derived exosomes (Figure (Figure2H).2H). Treatment with Ang II also led to higher levels of several other miRNAs (see Supplemental Figure 2B). Expression and secretion of miR-21* in cardiac fibroblasts was not changed by adding a miR-21 inhibitor (data not shown). We next focused on potential functional roles of secreted miR-21*.

(A) miRNA profiling assays were performed with rat cardiac fibroblast–derived exosomes and cardiac fibroblasts (n = 2). Fold change of the ratio exosomes/cells is shown as log10. Only reproducibly detectable miRNAs, having a Ct <35 were considered. miR-21* is shown in red. Other miRNA star strands are shown in green. Data are normalized to U1. The fold changes of miR-9-star, miR-135a, miR-181c, miR-547 are not shown in the graph because their fold change is more than 100. Mean ratios of exosomes/cells are shown in Supplemental Table 1. (B) Ratio of exosomes/cells of miR-21 and miR-21* expression level using TaqMan qRT-PCR. (C) Expression levels of miR-21 and miR-21* using TaqMan qRT-PCR in rat cardiac fibroblasts and (D) in secreted fibroblast-derived exosomes (n = 3–4). Data are normalized to a C. elegans miRNA (cel-miR-54). (E) Differential expression analysis of miR-21 and miR-21* in cardiac fibroblasts and fibroblast-derived exosomes using next-generation deep sequencing. Normalized read counts were used for fold change of the ratio cells/exosomes, which is shown as log2. n = 3 for fibroblast-derived exosomes and cardiac fibroblasts. (F) Immunofluorescence staining of control and NSMASE2 inhibitor–treated rat cardiac fibroblasts. Treatment with 10 μM NSMASE2 inhibitor for 48 hours resulted in accumulation of CD63-positive microvesicles (green) in rat cardiac fibroblasts. Actin cytoskeleton is stained with Phalloidin-TRITC (red), and nuclei are stained with DAPI (blue). Scale bar: 50 μm. (G) Level of miR-21* in fibroblast-derived exosomes treated with NSMASE2 inhibitor (10 μM) for 48 hours and after treatment with (H) Ang II (1 nM) for 24 hours. Data are normalized to U1. (n = 3). Data are mean ± SEM. #P = 0.07, *P < 0.05, **P < 0.01, ***P < 0.005.
Fibroblast-derived miR-21* is transported to cardiomyocytes, leading to cellular hypertrophy.
Conditioned supernatant of cardiac fibroblasts is known to induce cellular hypertrophy in cardiomyocytes (7, 8). To investigate whether fibroblast-derived exosomes are crucially involved in this paracrine action, we first treated cardiomyocytes with conditioned medium of cardiac fibroblasts; surprisingly, depletion of exosomes by sequential ultracentrifugation completely abolished the prohypertrophic effects (Supplemental Figure 3). Further studies were performed to test whether miRNAs are transported via exosome transfer between cardiac fibroblasts and cardiomyocytes. To study exosome uptake mechanisms by cardiomyocytes, we labeled secreted fibroblast-derived exosomes with a green fluorescent marker, PKH67, and incubated cultured cardiomyocytes with the labeled exosomes (Figure (Figure3A).3A). Analysis of exosome uptake performed in the cardiomyocyte cell line HL-1 by confocal microscopy revealed an increasing uptake of labeled exosomes over time (Figure (Figure3B).3B). This was further confirmed in cultures of primary cardiomyocytes (Supplemental Figure 4A). To confirm that exosomes are taken up into cardiomyocytes and that they do not simply attach to the cell surface, we performed 3D reconstructions of the confocal image z-stacks, which confirmed the cytoplasmatic localization of internalized exosomes (Supplemental Video 1). Incubation of labeled exosomes with cardiomyocytes either at 4°C for 2 hours or in the presence of cytochalasin D (an inhibitor of actin polymerization) at 37°C at 2 hours resulted in decreased exosome uptake by cardiomyocytes (Figure (Figure3,3, C–E), indicating a temperature- and actin-dependent uptake mechanism.
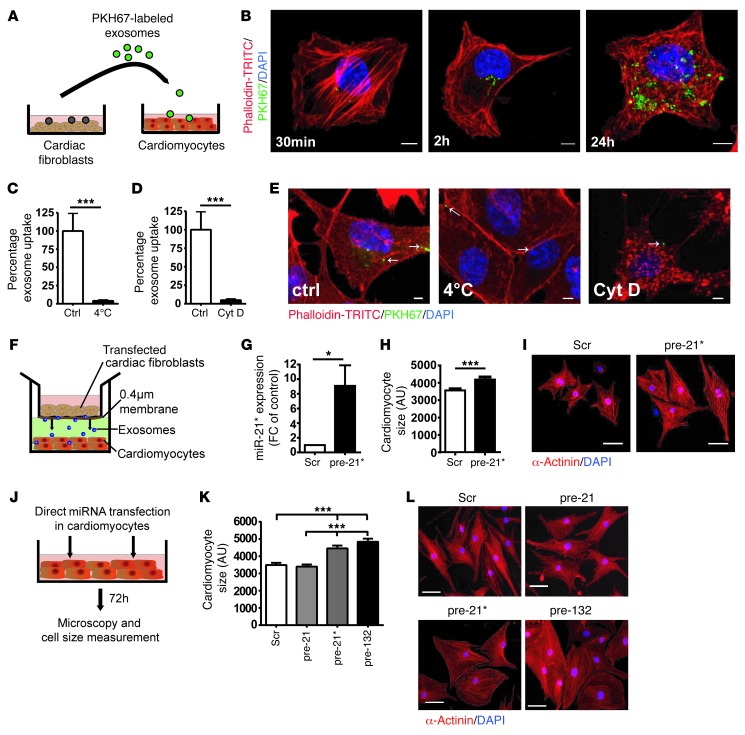
(A) Exosome uptake experiment. Purified fibroblast-derived exosomes were labeled with green fluorescent dye and incubated with cardiomyocytes. (B) Murine cardiomyocytes were incubated with PKH67-labeled (green) exosomes from mouse fibroblasts and fixed for confocal imaging. Cardiomyocytes were incubated with PKH67-labeled exosomes for 30 minutes, 2 hours, and 24 hours (3 independent experiments; n = 3–7). Scale bar: 5 μm. (C and E) Cardiomyocytes were incubated with PKH67-labeled exosomes for 2 hours (4°C) or (D and E) treated with cytochalasin D (Cyt D, 0.5 μM) for 30 minutes and then incubated with PKH67-labeled fibroblast-derived exosomes for 2 hours (37°C). (C and D) Exosome uptake was quantified as percentage of fluorescence intensity. Control is exosome uptake for 2 hours (37°C). HL-1 cardiomyocytes were stained with Phalloidin-TRITC (red) and DAPI (blue). Scale bar: 5 μm (n = 3). (F) A transwell coculture assay was used, with cardiac fibroblasts in the top well, cardiomyocytes in the bottom well, and a 0.4-μm porous membrane between the 2 wells inhibiting cell-cell contact. (G) Cardiac fibroblasts were transfected with a precursor of miR-21* (pre-21*) or a control precursor miRNA (scr), cocultured with cardiomyocytes for 72 hours, and expression of miR-21* was measured in cardiomyocytes (n = 3). (H and I) Cardiomyocyte size was measured after incubation with scrambled (scr) or pre-miR-21*–transfected cardiac fibroblast–derived exosomes (n = 63–87). (J) Experimental setup of cardiomyocyte transfection. (K and L) Cardiomyocytes were transfected with a precursor of miR-21, miR-21*, and miR-132 (pre-132) or a control miRNA (scr) for 72 hours. Cardiomyocytes were stained, and cell size was measured (n = 62–82). (I and L) Cardiomyocytes were stained with α-actinin (red) and nuclei with DAPI (blue). Scale bar: 50 μm. *P < 0.05, ***P < 0.005.
Next, a coculture assay system was used to study the miRNA crosstalk between cardiac fibroblasts and cardiomyocytes (Figure (Figure3F).3F). To visualize whether miRNAs released from cardiac fibroblasts are transported to cardiomyocytes, we transfected cardiac fibroblasts with a Cy3-labeled precursor miRNA and cocultured the transfected fibroblasts with cardiomyocytes for 24 hours. Confocal imaging of cardiomyocytes demonstrated that Cy3-labeled miRNAs derived from fibroblasts were detectable in separated cardiomyocytes during the coculture period (Supplemental Figure 4B). As cardiac fibroblasts are not able to migrate through the 0.4-μm porous membrane filter, the Cy3-labeled miRNA must have been transported and taken up into cardiomyocytes. We next transfected cardiac fibroblasts with a precursor of miR-21* and measured miR-21* expression in cardiomyocytes 72 hours after coculturing. Indeed, fibroblast-derived miR-21* was enriched in cardiomyocytes after the coculture period (Figure (Figure3G).3G). In contrast, transfecting fibroblasts with an inhibitor of miR-21* resulted in a decrease in miR-21* concentration in cardiomyocytes (Supplemental Figure 4C). In line, transfection of fibroblasts with a miRNA that is only present in Caenorhabditis elegans confirmed a miRNA transport mechanism from fibroblasts to cardiomyocytes (Supplemental Figure 4D).
We previously showed, by in vitro and in vivo experiments, that miR-21 has no significant effect in cardiomyocyte hypertrophy (17). However, the role of the passenger strand miR-21* in cardiomyocytes is unknown so far. To investigate this, we first confirmed that miR-21* binds to Ago2 in cardiac fibroblast cells by applying RNA immunoprecipitation, which revealed that miR-21* may have functional effects (Supplemental Figure 5). We next analyzed the role of exosome-derived miR-21* on cardiomyocytes; exosomes were isolated from the conditioned medium of miR-21*–transfected fibroblasts and then incubated with cardiomyocytes, followed by analyses of cardiomyocyte cell size changes. Fibroblast exosomal–derived miR-21* led to a significant increase in cardiomyocyte cell size (Figure (Figure3,3, H and I). In line with this, transfection of a miR-21* precursor directly to cardiomyocytes (Figure (Figure3J)3J) similarly resulted in cardiomyocyte hypertrophy, which was not seen after transfection of miR-21 (Figure (Figure3,3, K and L). Transfection of miR-132 served as a positive control (26).
In contrast, inhibiting miR-21* in cardiomyocytes led to a decrease in cardiomyocyte size (Supplemental Figure 6A). Inhibition of miR-21* in cardiomyocytes treated with Ang II to induce cellular hypertrophy rescued the Ang II–induced prohypertrophic effect, suggesting a potential therapeutic use (Supplemental Figure 6B). Ang II treatment of cardiomyocyte did not have direct effects on miR-21* levels (data not shown).
SORBS2 and PDLIM5 are targets of miR-21* in cardiomyocytes.
We next applied a proteome profiling approach to identify miR-21*–regulated targets in cardiomyocytes. Proteome profiling in cardiomyocytes transfected either with miR-21 or miR-21* identified differently regulated protein candidates (Figure (Figure4A4A and Supplemental Table 2), many of them involved in important pathways in cardiomyocyte biology (Figure (Figure4B).4B). Because miRNAs normally bind to the 3′ untranslated region (3′UTR) of their target mRNAs and lead to their repression, we were interested in targets that were downregulated after transfection of miR-21*. Among them, sorbin and SH3 domain-containing protein 2 (SORBS2) was strongly silenced and a very interesting candidate (Table (Table1),1), as SORBS2 was predicted to have 2 miRNA recognition sites in the 3′UTR for miR-21*. Indeed, SORBS2, also known as Arg-binding protein 2 (ARGBP2), is expressed at high levels in the heart and is repressed during cardiac pathologies (27, 28). We revealed that transfection of miR-21* in cardiomyocytes led to repression of SORBS2 protein expression, which was not observed after transfection of miR-21 (Figure (Figure4,4, C and D). Consistent with this finding, incubation of cardiomyocytes with fibroblast-derived exosomes transfected with miR-21* resulted in reduced SORBS2 expression in cardiomyocytes (Supplemental Figure 7A). Mechanistically, we found that miR-21* directly targeted SORBS2 3′UTR, as shown by luciferase gene reporter assays (Figure (Figure44F).

(A) Principal component analysis of protein expression patterns obtained by a proteomics approach from rat cardiomyocytes 72 hours after transfection with a scrambled control (blue dots), a miR-21 precursor (red dots), or a miR-21* precursor (green dots). (B) Pathway analysis of regulated protein patterns. GO term enrichment graph of regulated proteins in cardiomyocytes using functional annotation cluster tool DAVID 6.7. The top 11 gene groups that are regulated in cardiomyocytes after transfection of miR-21* are shown. (C) Protein expression of SORBS2, PDLIM5, and GAPDH in cardiomyocytes transfected with control miRNA (Scr) and precursor of miR-21 and miR-21*. Quantification of (D) SORBS2 protein expression and (E) PDLIM5 protein expression (n = 3 per transfection group). (F) Activity of luciferase reporter construct containing the 3′UTR of Sorbs2 mRNA relative to β-gal control plasmid after transfection of scrambled miRNA (Scr) or precursor of miR-21*. (G) Measurement of cardiomyocyte cell size and (H) immunofluorescence staining after transfection of control, SORBS2, and PDLIM5 siRNA in cardiomyocytes. Scale bar: 25 μm. *P < 0.05, ***P < 0.005.
Table 1
Deregulated proteins in cardiomyocytes after transfection of miR-21/miR-21*
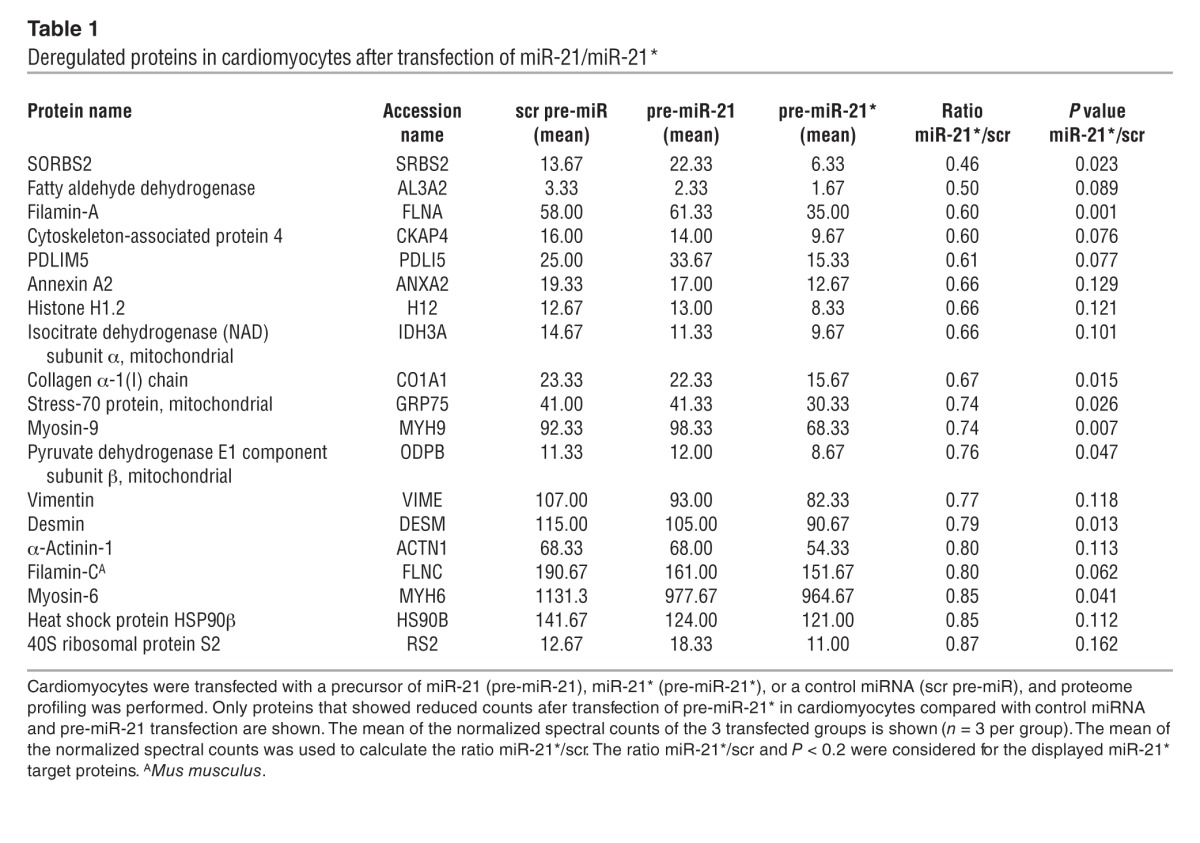
Based on the results from the proteome profiling, we also identified PDZ and LIM domain 5 (PDLIM5, also known as Enigma homolog [Enh]) as a further silenced target of miR-21* (Table (Table1).1). This is of interest, as cardiomyocyte-specific PDLIM5 knockout mice develop cardiomyopathy, suggesting PDLIM5 to play a major role in cardiac muscle structure and function (29). Validating Western blot experiments showed transfection of miR-21* in cardiomyocytes to reduce PDLIM5 protein expression (Figure (Figure4,4, C and E). Consistently, PDLIM5 was repressed in cardiomyocytes after incubation with miR-21*–transfected fibroblast-derived exosomes (Supplemental Figure 7B). However, using bioinformatic prediction tools, we did not find a predicted miR-21* recognition element in the 3′UTR, indicating PDLIM5 to be an indirect miR-21* target or to have putative binding sites in the 5′UTR or coding sequence.
To investigate whether SORBS2 and PDLIM5 are involved in the hypertrophic phenotype mediated by miR-21* in cardiomyocytes, we silenced either SORBS2 or PDLIM5 in cardiomyocytes using siRNA approaches. Silencing of the miR-21* targets SORBS2 and PDLIM5 in cardiomyocytes significantly induced cellular hypertrophy (Figure (Figure4,4, G and H).
Several other proteins were also regulated by miR-21*, as depicted by proteome profiling analysis (Figure (Figure4A,4A, Table Table1,1, and Supplemental Table 2), some of which may additionally contribute to the observed phenotype. In conclusion, we revealed that SORBS2 and PDLIM5 are regulated by miR-21*, which are involved in the development of cardiomyocyte hypertrophy.
In vivo relevance of miR-21* for cardiac hypertrophy.
To study a potential cardiac-derived secretion mechanism in vivo, we detected miR-21* levels in pericardial fluid of mice with cardiac hypertrophy. We collected pericardial fluid of sham-operated mice and mice with left ventricular pressure overload–induced hypertrophy after aortic constriction and determined miR-21* levels (Figure (Figure5A).5A). miR-21* was significantly increased in pericardial fluid of mice with cardiac hypertrophy compared with that in sham-operated mice (Figure (Figure5B).5B). Consistent with that, in situ PCRs of heart tissue sections of mice revealed strong miR-21* detection signals in cardiomyocytes after pressure overload (Supplemental Figure 8). As in vitro miR-21* inhibition was able to block Ang II–mediated cardiomyocyte hypertrophy, we next implanted mice with osmotic Ang II minipumps for 2 weeks to mimic a prohypertrophic condition in vivo and injected specific chemically cholesterol-modified single-strand RNA antagomirs (Figure (Figure5C).5C). Mice implanted with Ang II–containing minipumps or sham-operated mice that served as controls were either injected with scrambled antagomir (control) or miR-21* antagomir (80 mg/kg) at day 0 and day 2 (Figure (Figure5C),5C), and cardiac function and dimensions were measured after 2 weeks. The expression level of miR-21* was repressed successfully in hearts of mice and cultured neonatal cardiomyocytes (Supplemental Figure 9, A and B) treated with miR-21* antagomir but not when treated with a scrambled antagomir. Ang II treatment led to increased intraventricular left ventricular pressure (Supplemental Table 3). The heart/body weight ratio (mg/g) was increased in mice implanted with Ang II minipumps compared with that in sham-operated mice (Figure (Figure5D).5D). Treatment with miR-21* antagomir resulted in a reduced heart/body weight ratio in mice implanted with Ang II minipumps compared with that in scrambled antagomir-injected mice (Figure (Figure5D).5D). Consistently, the average cardiomyocyte diameter was increased in mice implanted with Ang II minipumps, whereas, in mice treated with miR-21* antagomir, the cardiomyocyte diameter was decreased compared with that in scrambled antagomir-injected mice (Figure (Figure5,5, E and F). The findings revealed that antagonism of miR-21* in mice with Ang II–induced cardiac hypertrophy leads to the attenuation of cardiomyocyte hypertrophy. A schematic summary of our findings is shown in Figure Figure66.
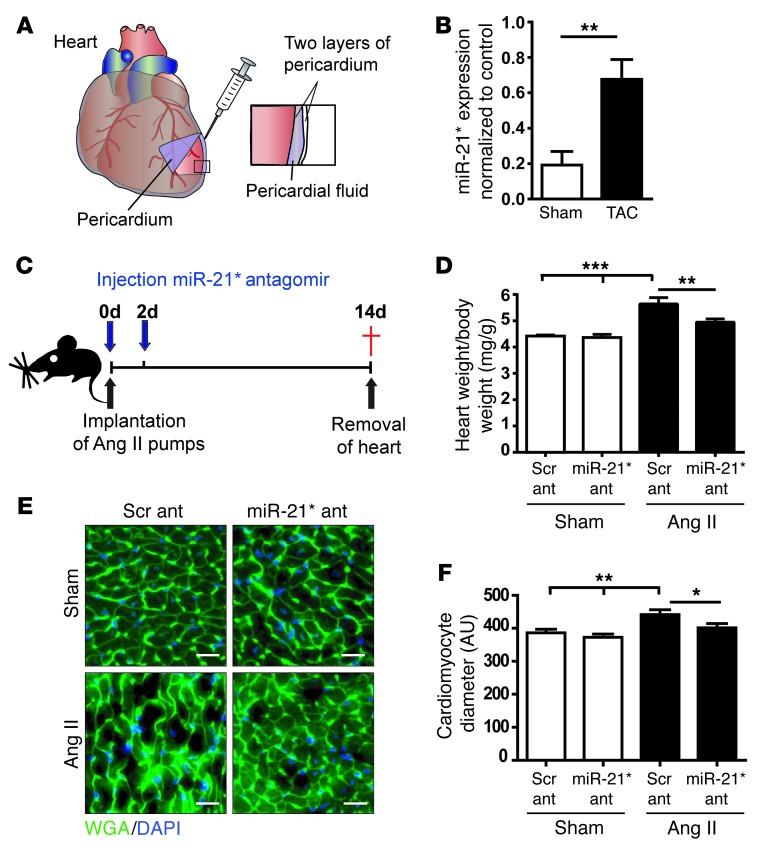
(A) Overview of the experimental setup. (B) Pericardial fluid was collected from 4-week-old TAC and sham mice, and miR-21* expression level was measured by qRT-PCR (n = 5 for sham group, n = 6 for TAC group). (C) Overview of the experimental in vivo procedure. (D) Heart weight/body weight ratio of sham mice and Ang II minipump–implanted mice treated either with scrambled (Scr ant) or miR-21* antagomir (miR-21* ant). (E) Immunofluorescence staining and (F) measurement of cardiomyocyte diameter in sham mice or mice with Ang II minipumps treated either with scrambled or miR-21* antagomir. Three different heart section areas from each animal were selected, and at least 50 cells were measured per area. Scale bar: 25 μm. Data are mean ± SEM. *P < 0.05, **P < 0.01, ***P < 0.005. WGA, wheat germ agglutinin (membrane stain).
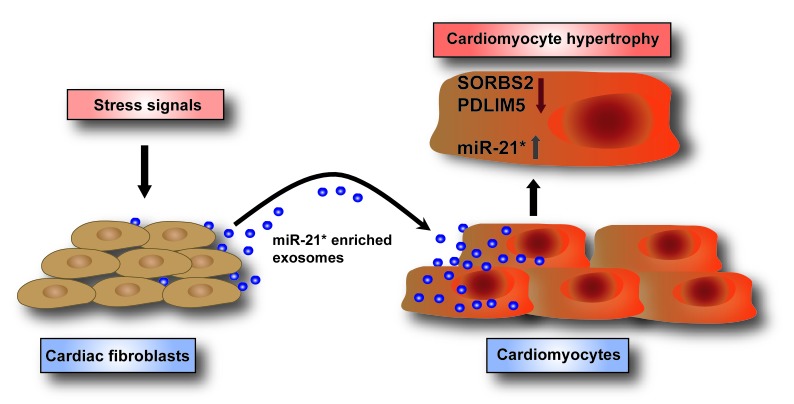
During cardiac stress, cardiac fibroblasts secrete exosomes enriched with miR-21*, which are transported to cardiomyocytes. Fibroblast-derived miR-21* regulates the expression of the targets SORBS2 and PDLIM5 in cardiomyocytes, leading to cardiomyocyte hypertrophy.
Discussion
In the current study, we identified a novel miRNA/exosome-mediated communication mechanism between cardiac fibroblasts and cardiomyocytes, contributing to the development of fibroblast-derived cardiomyocyte hypertrophy.
miRNA secretion.
Cardiac fibroblasts produced and secreted exosomes enriched with miRNA passenger strands (star miRNAs), which are usually degraded during miRNA biogenesis (14). We identified miR-21* to be enriched in fibroblast-derived exosomes compared with the intracellular compartment of these cells. In contrast, the mature strand miR-21 was abundant in fibroblast cells compared with exosomes, indicating that there might be a selective packaging process of miRNAs into exosomes. Our findings further indicate that miR-21* is exported from the cardiac fibroblast via exosomes and is transported to neighboring cells. Consistent with our results, several studies showed that the miRNA content can vary in exosomes and their parental cells, as depicted in mast cell exosomes as well as B cell– and T cell–derived exosomes (12, 30). Interestingly, miR-21* was found to be abundant in T cell– and B cell–derived exosomes, but miR-21* was more abundant in cells than in exosomes (30). Our finding that about one-quarter of the miRNAs found in fibroblast-derived exosomes represent miRNAs derived from the passenger strand suggests the existence of a general transport mechanism of star miRNAs. This notion is further supported by the presence of natural antisense RNAs in exosomes from human colorectal cancer cell lines that were transported to donor cells (31).
With regard to the mechanism underlying the secretion of miRNAs, it was shown that the secretion of miRNAs via exosomes in HEK293 cells is ceramide-dependent and is regulated by NSMASE2, the rate-limiting enzyme of ceramide biosynthesis (25). In this study, inhibition of NSMASE2 in HEK293 cells resulted in a dose-dependent reduction of extracellular miR-146a and miR-16 and subsequent reduction of exosome secretion. In accordance, inhibition of NSMASE2 in cardiac fibroblasts resulted in a significant decrease of miR-21* levels in fibroblast-derived exosomes and in accumulation of CD63-positive vesicles. NSMASE2 inhibition led to the reduction of some other but not all miRNAs, suggesting that miRNA secretion via exosomes in cardiac fibroblasts is in part but not totally regulated by NSMASE2. miR-21* secretion from cardiac fibroblasts was increased after stimulation with the profibrotic and prohypertrophic molecule Ang II, indicating a disease-specific regulation of miRNA-enriched fibroblast-derived exosomes. However, other molecules, such as components of the endosomal sorting complex required for transport (ESCRT) complex and RAB and SNARE proteins, which have been shown to be involved in exosome secretion (32, 33), may also play a role in regulating miRNA-exosome release in cardiac fibroblasts but have not been investigated.
miRNA uptake.
Although several studies showed that exosomes can interact with their recipient cells, the underlying mechanism of how exosomes are taken up and exert their function in the recipient cells is still unclear. Possible mechanisms of exosome uptake by target cells include specific binding to surface receptors (34, 35), fusion to the plasma membrane (36), or internalization (37). In our study, we demonstrated that fibroblast-derived exosomes are taken up by cardiomyocytes in an actin- and temperature-dependent manner. In accordance to our findings, the involvement of temperature and actin during exosome uptake was also shown in PC12 cells (38). We also provide evidence that exosome uptake by cardiomyocytes is mediated either via internalization, such as endocytosis, or via a receptor-mediated mechanism, because extensive PBS washing steps could not remove fibroblast-derived exosomes from cardiomyocytes, indicating the effective internalization. However, we can exclude macropinocytosis (distinct pathway of endocytosis) as a possible uptake mechanism, because incubation of fibroblast-derived exosomes with the macropinocytosis marker FITC-dextran showed no colocalization (data not shown). Our present study shows that exosomes released by cardiac fibroblasts are major components triggering cardiomyocyte hypertrophy.
miRNA function.
We showed previously that miR-21 has an essential role in fibroblast biology and is increased in human and murine failing hearts (17). However, neither increase nor decrease of miR-21 expression levels in cardiomyocytes influenced morphology or size, suggesting that miR-21 plays no functional role in cardiomyocytes (17). In the current study, we provide the first evidence that miR-21* acts as a paracrine signaling mediator and is involved in the development of cardiomyocyte hypertrophy. Specifically, fibroblast-derived miR-21* induced cellular hypertrophy in cardiomyocytes but not miR-21, which indicates that miRNA passenger strands can act as mature functional miRNAs. Consistent with this, miR-21* has been shown recently to have a pivotal immunopathological function in controlling apoptosis in granulocyte eosinophils in vitro (39). Another study found that miR-21* expression was differentially expressed in the aging mouse heart (40). Of note, extensive deep RNA sequencing in failing human hearts identified miR-21* to be expressed in the human heart and increased during heart failure (41). Others have shown functional roles for star miRNAs also in other pathologies, such as colorectal and prostate cancer or obesity/metabolic syndrome (42–44). RNA deep sequencing studies in Drosophila melanogaster identified that a large number of miRNA* sequences are not degraded but are able to bind to Ago2-RISC complexes (45–47). Moreover, the expression of miRNA guide and miRNA* strand is controlled either by pri-miRNA transcription or Ago1/Ago2 proteins during adult aging (40). In accordance to these studies, we found that miR-21* binds to Ago2 in cardiac fibroblasts, revealing that miR-21* is not degraded in cardiac fibroblasts and might act as a functional miRNA. However, the exact mechanism of miRNA loading into exosomes was not further addressed in this study and remains to be investigated. It is also possible that other currently unknown factors/proteins are involved in the miRNA-loading process.
miRNA targets.
miRNAs can regulate dozens to hundreds of targets simultaneously. In our study, we thus performed proteome profiling in cardiomyocytes to identify miR-21*–regulated protein candidates. We demonstrated that miR-21* regulates the expression of SORBS2 and PDLIM5 in cardiomyocytes, which were further shown to be the main mediators of the prohypertrophic phenotype of miR-21*. We identified SORBS2 as a major direct target of miR-21*. In line, SORBS2 was detected recently in human heart tissue and was found to be repressed in cardiac pathologies (28). SORBS2 is localized in the Z-bands of mature myofibrils and regulates important processes in cardiomyocytes, including the assembly of myofibrils or regulation of signaling at the Z-band level (27). SORBS2 can interact with a number of binding partners, including signaling molecules such as tyrosine kinases and cytoskeletal molecules (27). Importantly, we found SORBS2 silencing to induce cardiomyocyte hypertrophy. Our proteome results showed that PDLIM5 is also silenced by miR-21* in cardiomyocytes and PDLIM5 knockout mice develop dilated cardiomyopathy (29), suggesting PDLIM5 to play a major role in cardiac muscle structure and function.
In vivo results.
We found that the miR-21* expression level was upregulated in pericardial fluid of mice with cardiac hypertrophy compared with that in sham-operated mice. Changes in miRNA expression in pericardial fluid may provide information about the pathological status of the heart. However, no comprehensive analysis was made to identify the origin of the circulating miR-21* in pericardial fluid. Further studies are needed to find out the origin of miR-21*, which was secreted during cardiomyocyte hypertrophy, and how circulating miR-21* is protected from RNAse in pericardial fluid.
Despite permanent progress in therapeutic applications, disorders of the cardiovascular systems are still a major cause of human morbidity and mortality in the industrialized nations (1–3). Notably, several miRNA-based therapeutics are used in clinical trials, including antagomir (inhibitor) against miR-122 for hepatitis treatment, which is in phase 2 clinical trials (48), or miRNA mimic for miR-34 to treat cancer, which is in preclinical development (49). Pharmacological inhibition of miR-21* by injection of an antagomir against miR-21* in mice with Ang II–induced cardiac hypertrophy attenuated the development of cardiac hypertrophy. This finding demonstrated a potential therapeutic use of miR-21* antagonism in cardiac hypertrophy.
In conclusion, this is the first report showing that miRNAs are involved in the crosstalk between cardiac fibroblasts and cardiomyocytes. We demonstrated that miR-21* is enriched in exosomes, which are shuttled to cardiomyocytes, leading to cellular hypertrophy by affecting target genes. Antagonism of miR-21* in mice with Ang II–induced cardiac hypertrophy prevented the development of cardiac hypertrophy. Our findings reveal that fibroblast-derived miR-21* acts as a crucial paracrine signaling mediator during fibroblast-derived cardiomyocyte hypertrophy and illustrate a novel role for miR-21* as therapeutic target in cardiac failure.
Methods
Cell culture and reagents.
Neonatal rat cardiac fibroblasts and cardiomyocytes were isolated from newborn rats by enzymatic digestion as described previously (17). Neonatal cardiomyocytes were cultivated in minimal essential medium (Animed) containing vitamin B12, NaHCO3, l-glutamine, BrdU, penicillin/streptomycin, and 5% FBS (Invitrogen) at 37°C in 1% CO2. Primary rat cardiac fibroblasts were cultured in DMEM (PAA) supplemented with 10% FBS and 1% penicillin/streptomycin at 37°C in 5% CO2. HL-1 cells were cultured in Claycomb Medium (Sigma-Aldrich, catalog no. 51800C) with 10% FBS (Sigma-Aldrich, catalog no. 12103C), penicillin/streptomycin (100 U/ml:100 μg/ml; Sigma-Aldrich, catalog no. P4333), 0.1 mM Norepinephrine (Sigma-Aldrich, catalog no. A0937), 2 mM l-glutamine (Sigma-Aldrich, catalog no. G7513) at 37°C in 5% CO2. Mouse fibroblast NIH3T3 cells were cultured in DMEM supplemented with 10% FBS and 1% penicillin/streptomycin at 37°C in 5% CO2.
Exosome purification, analysis, and labeling.
Exosomes were purified from conditioned medium of neonatal rat cardiac fibroblasts cultured in DMEM supplemented with 1% exosome-depleted FBS (FBS is depleted of contaminating bovine exosomes by ultracentrifugation for at least 6 hours at 100,000 g). Conditioned medium of rat cardiac fibroblasts was collected for 48 hours, and exosomes were purified by several centrifugation and filtration steps as described previously (50). Briefly, the supernatant was centrifuged at 300 g for 10 minutes, 2,000 g for 10 minutes, and 10,000 g for 30 minutes, followed by filtration through a 0.22-μm filter to eliminate cells, dead cells, and cellular debris. For exosome purification, the supernatant was ultracentrifuged at 100,000 g for 70 minutes, followed by an additional washing step of the exosome pellet with PBS at 100,000 g for 70 minutes (Ultracentrifuge, Beckman Coulter, L8-70M). The exosome pellet was resuspended in 100 μl PBS and stored at –80°C. The protein content was measured using MicroBCA protein assay (Thermo Scientific). Fibroblast-derived exosomes were analyzed for the presence of the exosomal marker protein CD63 by Western blot and flow cytometry as previously described (50). For exosome uptake experiments, fibroblast-derived exosomes were labeled with the PKH26 Red Fluorescent Cell Linker Kit or the PKH67 Green Fluorescent Cell Linker Kit (Sigma-Aldrich) according to the manufacturer’s protocol with minor modifications. Exosomes diluted in PBS were added to 1 ml Diluent C (Sigma-Aldrich). In parallel, 4 μl PKH26/PKH67 dye was added to 1 ml Diluent C and incubated with the exosome solution for 4 minutes. To bind excess dye, 2 ml 0.5% BSA/PBS was added. The labeled exosomes were washed at 100,000 g for 1 hour, and the exosome pellet was diluted in 100 μl PBS and used for uptake experiments.
RNA sequencing.
Sample preparation for sequencing was performed as described in the Illumina TruSeq small RNA library preparation protocol. Briefly, approximately 200 ng total RNA was used to enrich for the small RNA fraction using native polyacrylamide gel electrophoresis. Both 3′ and 5′ adaptors were ligated along with the unique index sequence in each sample. The adaptor-ligated RNA fragments were then reverse transcribed and pooled together. The sequencing (for 50-base read length) was performed on a HiSeq 2000 (Illumina Inc.). Demultiplexing (generating individual sample groups) of the reads after sequencing was performed by sorting the different index sequences. For data analysis, the 3′ adapter sequence was removed from the reads using cutadapt ( http://code.google.com/p/cutadapt/). Truncated reads less than 10 bases were removed from the analysis data set. Reads were first mapped to all known miRNA sequences (miRBase; http://www.mirbase.org). For this purpose, a suffix tree approach for string mapping was used (51). Briefly, a minimum of 7 bases is required to be mapped to the miRNA sequence in order for the read to belong to the miRNA, with maximum of 2 mismatches allowed. For the reads that could be mapped to multiple miRNAs using the above criteria, the read was dedicated to the miRNA where it was mapped with maximum length and with least mismatches. The count table for each miRNA was prepared using a custom-made perl script. For differential miRNA expression analysis, the DeSeq package (52) from Bioconductor ( http://bioconductor.org) was applied with default settings, except in the case of dispersion estimation, for which the blind option was used.
Animal experiments.
Animal studies were performed in accordance to the relevant guidelines and regulations of the responsible authorities. The model of left ventricular pressure overload–induced cardiac hypertrophy (transverse aortic constriction [TAC]) in C57BL/6N mice is described in the Supplemental Methods. Pericardial fluid was collected 4 weeks after TAC or sham operation. In the Ang II model of cardiac stress, C57BL/6N mice were implanted with osmotic minipumps according to the manufacturers’ instructions (Alzet). Briefly, an incision was made in the midscapular region, and an osmotic minipump (Alzet, model 2002; 0.5 μl/h for 14 days) was implanted subcutaneously. The implanted minipumps contained Ang II (Sigma Aldrich) dissolved in water, and the infusion rate was 1.5 mg/kg/d. Sham-operated mice underwent an identical surgical procedure. sham-operated mice and Ang II minipump–implanted mice were either treated with a scrambled antagomir or a miR-21* antagomir (Integrated DNA Technology) at day 0 and day 2 (each dose 80 mg/kg) via retroorbital injection. After 2 weeks, cardiac function and dimension were measured and the hearts were removed for biochemical analysis.
Statistics.
Data are presented as mean ± SEM. Statistical analysis was carried out using Prism software (Graph Pad). For statistical analysis of 2 groups, unpaired, 2-tailed Student’s t test was used; for comparison of 3 or more groups, ANOVA followed by Bonferroni’s post-test was applied.
Study approval.
All animal studies were performed in accordance with the relevant guidelines and regulations and with the approval of the Niedersächsisches Landesamt für Verbraucherschutz und Lebensmittelsicherheit (approval no. 33.9-42502-04-09/1793).
Further detailed information can be found in the Supplemental Methods.
Supplementary Material
Acknowledgments
This work was supported by the Fondation Leducq (MIRVAD to T. Thum and M. Mayr), the IFB-Tx (01EO1302 to T. Thum), the DFG (TH903/10-1 to T. Thum), REBIRTH Excellence Cluster, the internal Hannover Biomedical Research School program Molecular Medicine (to C. Bang and T. Thum), EU grant FP7-CIG-294278, FIBROTARGET (7th Framework Program of the European Commission), and the German Federal Ministry of Education and Research (PtJ: 0315690D to A. Zeug),
Footnotes
Conflict of interest: Claudia Bang and Thomas Thum filed a patent about the diagnostic and therapeutic use of miR-21*.
Citation for this article:J Clin Invest. 2014;124(5):2136–2146. 10.1172/JCI70577.
See the related Commentary beginning on page 1896.
References
Articles from The Journal of Clinical Investigation are provided here courtesy of American Society for Clinical Investigation
Full text links
Read article at publisher's site: https://doi.org/10.1172/jci70577
Read article for free, from open access legal sources, via Unpaywall:
http://www.jci.org/articles/view/70577/files/pdf
Citations & impact
Impact metrics
Citations of article over time
Alternative metrics
Article citations
Exosomes Induce Crosstalk Between Multiple Types of Cells and Cardiac Fibroblasts: Therapeutic Potential for Remodeling After Myocardial Infarction.
Int J Nanomedicine, 19:10605-10621, 19 Oct 2024
Cited by: 0 articles | PMID: 39445157 | PMCID: PMC11498042
Review Free full text in Europe PMC
Potential cardiac-derived exosomal miRNAs involved in cardiac healing and remodeling after myocardial ischemia-reperfusion injury.
Sci Rep, 14(1):24275, 16 Oct 2024
Cited by: 0 articles | PMID: 39414956 | PMCID: PMC11484883
GHRH-stimulated pituitary small extracellular vesicles inhibit hepatocyte proliferation and IGF-1 expression by its cargo miR-375-3p.
J Nanobiotechnology, 22(1):649, 22 Oct 2024
Cited by: 0 articles | PMID: 39438882 | PMCID: PMC11494759
Unveiling exosomes: Cutting-edge isolation techniques and their therapeutic potential.
J Cell Mol Med, 28(20):e70139, 01 Oct 2024
Cited by: 1 article | PMID: 39431552 | PMCID: PMC11492151
Review Free full text in Europe PMC
Cardiomyocyte-derived small extracellular vesicle: a new mechanism driving diabetic cardiac fibrosis and cardiomyopathy.
Theranostics, 14(15):5926-5944, 09 Sep 2024
Cited by: 0 articles | PMID: 39346544 | PMCID: PMC11426245
Go to all (609) article citations
Other citations
Wikipedia
Data
Data behind the article
This data has been text mined from the article, or deposited into data resources.
BioStudies: supplemental material and supporting data
Similar Articles
To arrive at the top five similar articles we use a word-weighted algorithm to compare words from the Title and Abstract of each citation.
Extracellular vesicular MicroRNA-27a* contributes to cardiac hypertrophy in chronic heart failure.
J Mol Cell Cardiol, 143:120-131, 01 May 2020
Cited by: 40 articles | PMID: 32370947 | PMCID: PMC7338251
Stargazing microRNA maps a new miR-21 star for cardiac hypertrophy.
J Clin Invest, 124(5):1896-1898, 17 Apr 2014
Cited by: 20 articles | PMID: 24743143 | PMCID: PMC4001563
A critical role of cardiac fibroblast-derived exosomes in activating renin angiotensin system in cardiomyocytes.
J Mol Cell Cardiol, 89(pt b):268-279, 20 Oct 2015
Cited by: 118 articles | PMID: 26497614 | PMCID: PMC4988239
Exosomes as New Intercellular Mediators in Development and Therapeutics of Cardiomyocyte Hypertrophy.
Adv Exp Med Biol, 998:91-100, 01 Jan 2017
Cited by: 5 articles | PMID: 28936734
Review
Funding
Funders who supported this work.
British Heart Foundation (2)
Circulating MicroRNAs as novel biomarkers for cardiovascular risk
Manuel Mayr, King's College London
Grant ID: SP/12/5/29574
Proteomics to Explore Mechanistic Links between Circulating MiRNAs and Response to Vascular Injury
Manuel Mayr, King's College London
Grant ID: FS/13/2/29892





