Abstract
Free full text

Reversion of advanced Ebola virus disease in nonhuman primates with ZMapp™
Abstract
Without an approved vaccine or treatment, Ebola outbreak management has been limited to palliative care and barrier methods to prevent transmission. These approaches, however, have yet to end the 2014 outbreak of Ebola after its prolonged presence in West Africa. Here we show that a combination of monoclonal antibodies (ZMapp™), optimized from two previous antibody cocktails, is able to rescue 100% of rhesus macaques when treatment is initiated up to 5 days post-challenge. High fever, viremia, and abnormalities in blood count and chemistry were evident in many animals before ZMapp™ intervention. Advanced disease, as indicated by elevated liver enzymes, mucosal hemorrhages and generalized petechia could be reversed, leading to full recovery. ELISA and neutralizing antibody assays indicate that ZMapp™ is cross-reactive with the Guinean variant of Ebola. ZMapp™ currently exceeds all previous descriptions of efficacy with other therapeutics, and results warrant further development of this cocktail for clinical use.
Introduction
Ebola virus (EBOV) infections cause severe illness in humans, and after an incubation period of 3 to 21 days, patients initially present with general flu-like symptoms before a rapid progression to advanced disease characterized by hemorrhage, multi-organ failure and a shock-like syndrome1. In the spring of 2014, a new EBOV variant emerged in the West African country of Guinea2, an area in which EBOV has not been previously reported. Despite a sustained international response from local and international authorities including the Ministry of Health (MOH), World Health Organization (WHO) and Médecins Sans Frontières (MSF) since March 2014, the outbreak has yet to be brought to an end after five months. As of 15th August 2014, there are 2127 total cases and 1145 deaths spanning Guinea, Sierra Leone, Liberia and Nigeria3. So far, this outbreak has set the record for the largest number of cases and fatalities, in addition to geographical spread4. Controlling an EBOV outbreak of this magnitude has proven to be a challenge and the outbreak is predicted to last for at least several more months5. In the absence of licensed vaccines and therapeutics against EBOV, there is little that can be done for infected patients outside of supportive care, which includes fluid replenishment, administration of antivirals, and management of secondary symptoms6,7. With overburdened personnel, and strained local and international resources, experimental treatment options cannot be considered for compassionate use in an orderly fashion at the moment. However, moving promising strategies forward through the regulatory process of clinical development has never been more urgent.
Over the past decade, several experimental strategies have shown promise in treating EBOV-challenged nonhuman primates (NHPs) after infection. These include recombinant human activated protein C (rhAPC)8, recombinant nematode anticoagulant protein c2 (rNAPc2)9, small interfering RNA (siRNA)10, positively-charged phosphorodiamidate morpholino oligomers (PMOplus)11, the vesicular stomatitis virus vaccine (VSVΔG-EBOVGP)12, as well as the monoclonal antibody (mAb) cocktails MB-003 (consisting of human or human-mouse chimeric mAbs c13C6, h13F6 and c6D8)13 and ZMAb (consisting of murine mAbs m1H3, m2G4 and m4G7)14. Of these, only the antibody-based candidates have demonstrated substantial benefits in NHPs when administered greater than 24 hours past EBOV exposure. Follow-up studies have shown that MB-003 is partially efficacious when administered therapeutically after the detection of two disease “triggers”15, and ZMAb combined with an adenovirus-based adjuvant provides full protection in rhesus macaques when given up to 72 hours after infection16.
The current objective is to develop a therapeutic superior to both MB-003 and ZMAb, which could be utilized for outbreak patients, primary health-care providers, as well as high-containment laboratory workers in the future. This study aims to first identify an optimized antibody combination derived from MB-003 and ZMAb components, before determining the therapeutic limit of this mAb cocktail in a subsequent experiment. In order to extend the antibody half-life in humans and to facilitate clinical acceptance, the individual murine antibodies in ZMAb were first chimerized with human constant regions (cZMAb). The cZMAb components were then produced in Nicotiana benthamiana17, using the large-scale, cGMP-compatible Rapid Antibody Manufacturing Platform (RAMP) and magnICON vectors that currently also manufactures the individual components of cocktail MB-003, before efficacy testing in animals.
Selecting for the best mAb combinations
Our efforts to down-select for an improved mAb cocktail comprising components of MB-003 and ZMAb began with the testing of individual MB-003 antibodies in guinea pigs and NHPs. In guinea pig studies, animals were given one dose of mAb c13C6, h13F6, or c6D8 individually (totaling 5 mg per animal) at 1 day post-infection (dpi) with 1000 × LD50 of guinea pig-adapted EBOV, Mayinga variant (EBOV-M-GPA). Survival and weight loss were monitored over 28 days. Treatment with c13C6 or h13F6 yielded 17% survival (1 of 6 animals) with a mean time to death of 8.4 ± 1.7 and 10.2 ± 1.8 days, respectively. The average weight loss for c13C6 or h13F6-treated animals was 9% and 21% (Table 1). In nonhuman primates, animals were given three doses of mAb c13C6, h13F6, or c6D8, beginning at 24 hours after challenge with the Kikwit variant of EBOV (EBOV-K)18, and survival was monitored over 28 days. Only c13C6 treatment yielded any survivors, with 1 of 3 animals protected from EBOV challenge (Table 1), confirming in two separate animal models that c13C6 is the component that provides the highest level of protection in the MB-003 cocktail.
Table 1
Efficacy of individual and combined monoclonal antibody treatments in guinea pigs and nonhuman primates.
| Treatment groups, time of treatment | Dose (mg) | Mean time to death (days) | # Survivors / # Total | % Survival | % Weight loss | p-value, compared with | |
|---|---|---|---|---|---|---|---|
| cZMAb | MB-003 | ||||||
|
| |||||||
| Guinea pigs | - | - | |||||
| PBS, 3 dpi | N/A | 7.3 ± 0.5 | 0/4 | 0 | 9% | - | - |
| cZMAb, 3 dpi | 5 | 11.6 ± 1.8 | 1/6 | 17 | 7% | - | - |
| MB-003, 3 dpi | 5 | 8.2 ± 1.5 | 0/6 | 0 | 40% | - | - |
| ZMapp1, 3 dpi | 5 | 9.0 ± 0.0 | 4/6 | 67 | <5% | 0.190 | 0.0147 |
| ZMapp2, 3 dpi | 5 | 8.3 ± 0.6 | 3/6 | 50 | 8% | 0.634 | 0.0692 |
| ZMapp3, 3 dpi | 5 | 8.6 ± 1.1 | 1/6 | 17 | 9% | 0.224 | 0.411 |
| c13C6, 1 dpi | 5 | 8.4 ± 1.7 | 1/6 | 17 | 9% | - | - |
| h13F6, 1 dpi | 5 | 10.2 ± 1.8 | 1/6 | 17 | 21% | - | - |
| c6D8, 1 dpi | 5 | 10.5 ± 2.2 | 0/6 | 0 | 38% | - | - |
| Treatment groups, time of treatment | Dose (mg/kg) | Mean time to death (days) | # Survivors / # Total | % Survival |
|---|---|---|---|---|
|
| ||||
| Nonhuman primates | ||||
| PBS, 1 dpi | N/A | 8.4 ± 1.9 | 0/1 | 0 |
| MB-003, 1 dpi | 50 | 14.0 ± 2.8 | 1/3 | 33 |
| c13C6, 1 dpi | 50 | 9.0 ± 1.4 | 1/3 | 33 |
| h13F6, 1 dpi | 50 | 9.0 ± 2.0 | 0/3 | 0 |
| c6D8, 1 dpi | 50 | 9.7 ± 0.6 | 0/3 | 0 |
We then tested mAb c13C6 in combination with two of three mAbs from ZMAb in guinea pigs. The individual antibodies composing ZMAb were originally chosen for protection studies based on their in vivo protection of guinea pigs against EBOV-M-GPA19, and all three possible combinations were tested: ZMapp1 (c13C6+c2G4+c4G7), ZMapp2 (c13C6+c1H3+c2G4) and ZMapp3 (c13C6+c1H3+c4G7), and compared to the originator cocktails ZMAb and MB-003. Three days after challenge with 1000 × LD50 of EBOV-M-GPA, the animals received a single combined dose of 5 mg of antibodies. This dosage is purposely given to elicit a suboptimal level of protection with the cZMAb and MB-003 cocktails, such that potential improvements with the optimized mAb combinations can be identified. Of the tested cocktails, ZMapp1 showed the best protection, with 4 of 6 survivors and less than 5% average weight loss (Table 1). ZMapp2 was next with 3 of 6 survivors and 8% average weight loss, and ZMapp3 protected 1 of 6 animals (Table 1). The level of protection afforded by ZMapp3 was not a statistically significant increase over cZMAb (p = 0.224, log-rank test compared to ZMAb, χ2 = 1.479, df = 1), and showed the same survival rate along with a similar average weight loss (Table 1). As a result, only ZMapp1 and ZMapp2 were carried forward to NHP studies.
ZMapp1 or ZMapp2-treated NHPs
Rhesus macaques were used to determine whether administration of ZMapp1 or ZMapp2 was superior to ZMAb and MB-003 in terms of extending the treatment window. Due to mAb availability constraints, m4G7 was utilized in place of c4G7 for this NHP experiment. The experiment consisted of six NHPs per group receiving three doses of ZMapp1 (Group A) or ZMapp2 (Group B) at 50 mg/kg intravenously (IV) at 3-day intervals, beginning 3 days after a lethal intramuscular (IM) challenge with 4000 × TCID50 (or 2512 PFU) of EBOV-K. Control animals were given phosphate-buffered saline (PBS) or mAb 4E10 (C1 and C2, respectively). Mock-treated animals succumbed to disease between 6–7 dpi with symptoms typical of EBOV (Figure 1a), characterized by high clinical scores but no fever (Figures 1b and 1c), in addition to viral titers up to ~108 and ~109 TCID50 by the time of death (Figure 1d). Analysis of blood counts and serum biochemistry revealed leukocytopenia, thrombocytopenia, severe rash, decreased levels of GLU, as well as increased levels of ALP, ALT, BUN and CRE at end-stage EBOV disease (Figures 1e to 1o, Table 2).
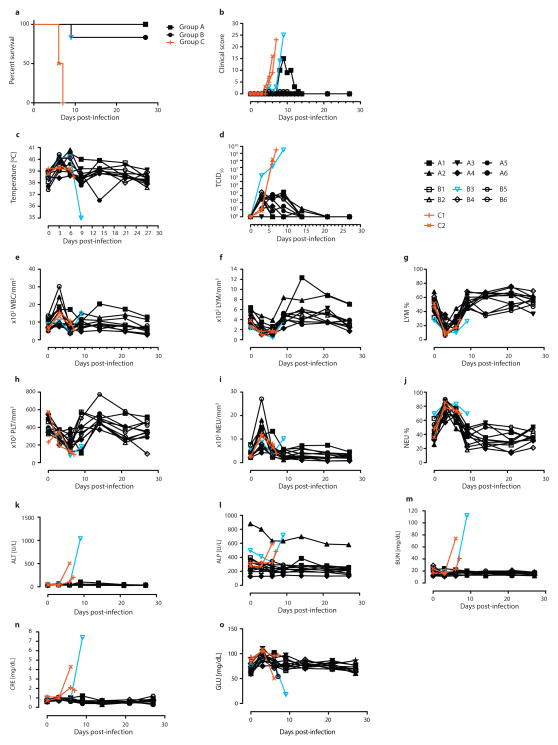
Rhesus macaques were challenged with EBOV-K, and 50 mg/kg of ZMapp1 (Group A) or ZMapp2 (Group B) were administered on days 3, 6, and 9 (n=6 per treatment group, n=2 for controls). Non-specific IgG mAb or PBS was administered as a control (Group C). (a) Kaplan-Meier survival curves. (b) Clinical score. (c) Rectal temperature. (d) EBOV viremia by TCID50. Blood parameters: (e) white blood cell count; (f) lymphocyte count; (g) lymphocyte percentage; (h) platelet count; (i) neutrophil count; (j) neutrophil percentage; (k) ALT; (l) ALP; (m) BUN; (n) CRE; (o) GLU.
Table 2
Clinical findings of EBOV-infected NHPs from 1 to 27 dpi.
| Animal ID | Treatment group | Clinical findings | Outcome | ||||
|---|---|---|---|---|---|---|---|
| Body temperature | Rash | White blood cells | Platelets | Biochemistry | |||
| A1 | 50 mg/kg c13C6+c2G4+m4G7, 3 dpi | Fever (6, 9, 14 dpi) | Thrombocytopenia (6, 9 dpi) | ALT↑ (9, 14 dpi), TBIL↑ (9 dpi), PHOS↓ (6 dpi) | Survived | ||
| A2 | 50 mg/kg c13C6+c2G4+m4G7, 3 dpi | Fever (3 dpi) | Leukocytosis (3 dpi) | CRE↓ (14 dpi) | Survived | ||
| A3 | 50 mg/kg c13C6+c2G4+m4G7, 3 dpi | Fever (3 dpi) | Leukocytosis (3 dpi) | Thrombocytopenia (6 dpi) | Survived | ||
| A4 | 50 mg/kg c13C6+c2G4+m4G7, 3 dpi | Leukocytopenia (9 dpi) | Thrombocytopenia (3, 6, 14, 21, 27 dpi) | Survived | |||
| A5 | 50 mg/kg c13C6+c2G4+m4G7, 3 dpi | Fever (3, 6, 9 dpi) | Leukocytopenia (9 dpi) | Thrombocytopenia (3, 21 dpi) | Survived | ||
| A6 | 50 mg/kg c13C6+c2G4+m4G7, 3 dpi | Fever (3 dpi) | Survived | ||||
| B1 | 50 mg/kg ZMapp2, 3 dpi | Fever (3, 14, 21 dpi) | Leukocytopenia (6, 14, 21, 27 dpi) | Thrombocytopenia (6 dpi) | Survived | ||
| B2 | 50 mg/kg ZMapp2, 3 dpi | Fever (3, 6 dpi) | Thrombocytopenia (6, 9 dpi) | Survived | |||
| B3 | 50 mg/kg ZMapp2, 3 dpi | Fever (3, 6 dpi), Hypothermia (9 dpi) | Severe rash (9 dpi) | Thrombocytopenia (6, 9 dpi) | ALT↑↑↑ (9 dpi), TBIL↑↑ (9 dpi), BUN↑↑↑ (9 dpi), CRE↑↑↑ (9 dpi), GLU↓↓ (9 dpi) | Died, 9 dpi | |
| B4 | 50 mg/kg ZMapp2, 3 dpi | Fever (3, 6 dpi) | Leukocytopenia (6 dpi) | Thrombocytopenia (6, 27 dpi) | Survived | ||
| B5 | 50 mg/kg ZMapp2, 3 dpi | Fever (3, 6, 14, 21 dpi) | Leukocytosis (3 dpi) | Thrombocytopenia (3, 6 dpi) | Survived | ||
| B6 | 50 mg/kg ZMapp2, 3 dpi | Fever (3 dpi) | Leukocytosis (3 dpi), Leukocytopenia (6, 9, 14, 21, 27 dpi) | Thrombocytopenia (6 dpi) | PHOS↓ (3 dpi), CRE↓ (27 dpi) | Survived | |
| C1 | PBS, 3 dpi | Moderate rash (6 dpi), Severe rash (7 dpi) | Leukocytosis (3 dpi) | Thrombocytopenia (6, 7 dpi) | ALB↓ (7 dpi), ALT↑ (7 dpi), BUN↑ (7 dpi) | Died, 7 dpi | |
| C2 | Control mAb, 3 dpi | Severe rash (6 dpi) | Leukocytopenia (6, 7 dpi) | Thrombocytopenia (6, 7 dpi) | ALP↑ (3 dpi), ALT↑↑↑ (6 dpi), BUN↑ (6 dpi), CRE↑↑↑ (6 dpi) | Died, 6 dpi | |
Hypothermia was defined as below 35°C. Fever was defined as >1.0°C higher than baseline. Mild rash was defined as focal areas of petechiae covering <10% of the skin, moderate rash as areas of petechiae covering 10 to 40% of the skin, and severe rash as areas of petechiae and/or ecchymosis covering >40% of the skin. Leukocytopenia and thrombocytopenia were defined as a >30% decrease in numbers of WBCs and platelets, respectively. Leukocytosis and thrombocytosis were defined as a twofold or greater increase in numbers of WBCs and platelets over baseline, where WBC count >11.000. ↑, two- to threefold increase; ↑↑, four- to fivefold increase; ↑↑↑, greater than fivefold increase; ↓, two- to threefold decrease; ↓↓, four- to fivefold decrease; ↓↓↓, greater than fivefold decrease. ALB, albumin; AMY, amylase; TBIL, total bilirubin; BUN, blood urea nitrogen; PHOS, phosphate; CRE, creatinine; GLU, glucose; GLOB, globulin.
All six Group A NHPs survived the challenge with mild signs of disease (Figure 1a, Table 2) (p = 0.0039, log-rank test, χ2 = 8.333, df = 1, comparing to Group C), with the exception of A1 which showed an elevated clinical score (Figure 1b), increased levels of ALT, TBIL, and decreased PHOS (Figure 1, Table 2). However, this animal recovered after the third ZMapp1 dose and the clinical score dropped to zero by 15 dpi (Figure 1b). A fever was detected in all but one of the NHPs (A4) at 3 dpi, the start of the first ZMapp1 dose (Figure 1c). Viremia was also detected beginning at 3 dpi by TCID50 in all but one animal from blood sampled just before the administration of the treatment (A3) (Figure 1d), and similar results were observed by RT-qPCR (Extended Data Table 1). The viremia decreased to undetectable levels by 21 dpi. EBOV shedding was not detected from oral, nasal and rectal swabs by RT-qPCR in any of the Group A animals (Extended Data Tables 2–4).
Extended Data Table 1
Blood viremia measured by RT-qPCR for the ZMapp1- and ZMapp2-treated NHPs.
| DAY | A1 | A2 | A3 | A4 | A5 | A6 | B1 | B2 | B3 | B4 | B5 | B6 | C1 | C2 |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 0 | UD | UD | UD | UD | UD | UD | UD | UD | UD | UD | UD | UD | UD | UD |
| 3 | UD | 3.98E+02 | UD | UD | 9.99E+02 | 1.27E+03 | 8.05E+03 | 1.65E+04 | 9.36E+03 | 9.77E+03 | 9.27E+02 | 9.48E+02 | UD | 4.34E+02 |
| 6 | 3.10E+03 | 4.49E+02 | UD | 8.34E+02 | 5.81E+03 | 2.09E+03 | UD | 1.22E+04 | 1.04E+05 | 4.26E+03 | 3.14E+02 | 4.49E+03 | 5.57E+06 | 2.05E+07 |
| 7 | 5.50E+05 | |||||||||||||
| 9 | UD | UD | UD | UD | 5.24E+02 | UD | 1.74E+05 | 5.03E+05 | 1.87E+03 | 5.16E+02 | UD | |||
| 14 | 3.62E+03 | UD | UD | UD | UD | UD | UD | UD | UD | UD | UD | |||
| 21 | UD | UD | UD | UD | UD | UD | UD | UD | UD | UD | UD | |||
| 27 | UD | UD | UD | UD | UD | UD | UD | UD | UD | UD | UD |
Footnote: UD = Undetectable.
For Group B, 5 of 6 NHPs survived with B3 succumbing to disease at 9 dpi (Figure 1a) (p = 0.0039, log-rank test, χ2 = 8.333, df = 1, comparing to Group C). Surviving animals showed only mild signs of disease (Table 2). The moribund animal showed increased clinical scores (Figure 1b), in addition to a drastic drop in body temperature shortly before death (Figure 1c). At the time of death, animal B3 had elevated levels of ALT, TBIL, BUN, CRE, in addition to decreased levels of GLU, suggesting multi-organ failure (Figure 1). All six Group B animals showed fever in addition to viremia at 3 dpi by TCID50 and RT-qPCR (Figure 1D, Extended Data Table 1). It was interesting to note that in B3, the viremia reached ~106 TCID50 after 3 dpi (Figure 1d), suggesting that this NHP was particularly susceptible to EBOV infection. No escape mutants were detected with this animal. The administration of ZMapp2 at the reported concentrations was unable to effectively control viremia at this level. Virus shedding was also detected from the oral and rectal swabs by RT-qPCR in the moribund NHP B3 (Extended Data Tables 2–4). Since ZMapp1 demonstrated superior protection to ZMapp2 in this survival study, ZMapp1 (now trademarked as ZMapp™ by MappBio Pharmaceuticals) was carried forward to test the limits of protection conferred by this mAb cocktail in a subsequent investigation.
ZMapp™-treated NHPs
In this experiment, rhesus macaques were assigned into three treatment groups of six and a control group of three animals, with all treatment NHPs receiving three doses of ZMapp™ (c13C6+c2G4+c4G7, 50 mg/kg per dose) spaced three days apart. After a lethal IM challenge with 1000 × TCID50 (or 628 PFU) of EBOV-K18, we treated the animals with ZMapp™ at 3, 6 and 9 dpi (Group D); 4, 7, and 10 dpi (Group E); or 5, 8 and 11 dpi (Group F). The control animals (Group G) were given mAb 4E10 as an IgG isotype control (n = 1) or PBS (n = 2) in place of ZMapp™ starting at 4 dpi (Figure 2a). All animals treated with ZMapp™ survived the infection, whereas the three control NHPs (G1, G2 and G3) succumbed to EBOV-K infection at 4, 8 and 8 dpi, respectively (p = 3.58E-5, log-rank test, χ2 = 23.25, df = 3, comparing all groups) (Figure 2b). At the time ZMapp™ treatment was initiated, fever, leukocytosis, thrombocytopenia and viremia could be detected in the majority of the animals (Figure 2c-f, Table 3). All animals presented with detectable abnormalities in blood counts and serum biochemistry during the course of the experiment (Figure 2g-l, Table 3).
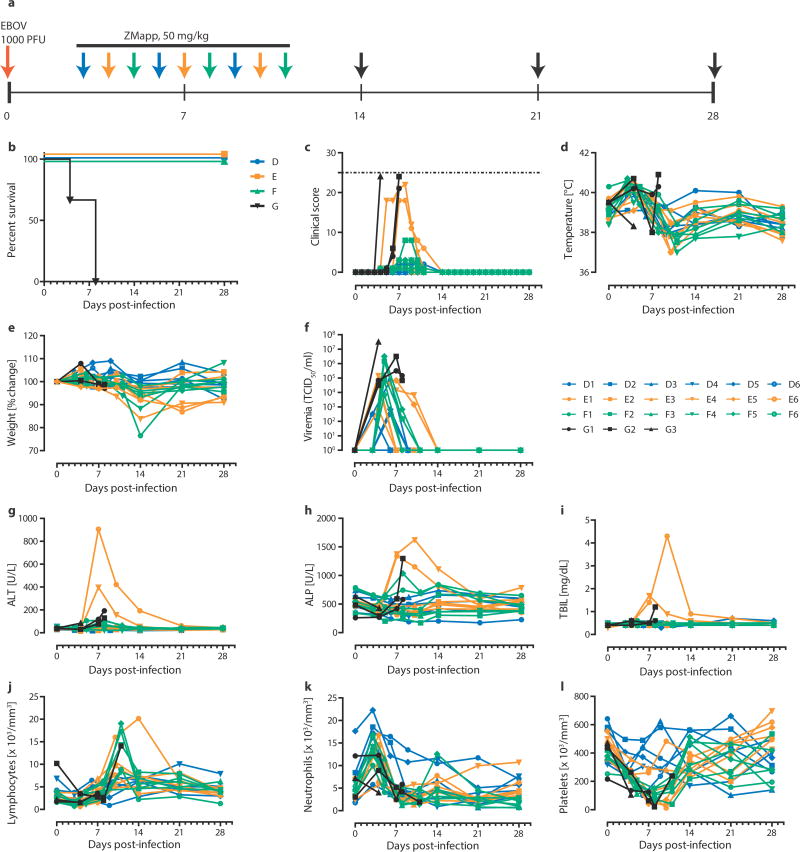
™.
(a to f) Rhesus macaques (n=6 per ZMapp™ treatment group, n=3 for controls) were challenged with EBOV-K, and 50 mg/kg of ZMapp™ were administered beginning at 3 (Group A), 4 (Group B) or 5 (Group C) days after challenge. Non-specific IgG mAb or PBS was administered as a control (Group D). (a) Timeline of infection, treatment and sample days. (b) Kaplan-Meier survival curves. (c) Clinical scores; the dashed line indicates the minimum score requiring mandatory euthanasia. (d) Rectal temperature. (e) Percentage body weight change. (f) EBOV viremia by TCID50. (g to l) Selected clinical parameters of Group A to D animals. (g) ALT. (h) ALP. (i) TBIL. Counts for (j) Lymphocytes, (k) Neutrophils and (l) Platelets over the course of the experiment.
Table 3
Clinical findings of EBOV-infected NHPs from 1 to 28 dpi.
| Animal ID | Treatment group | Clinical findings | Outcome | ||||
|---|---|---|---|---|---|---|---|
| Body temperature | Rash | White blood cells | Platelets | Biochemistry | |||
| D1 | 50 mg/kg ZMapp™, 3 dpi | Fever (3, 6, 14, 21 dpi) | Leukocytosis (3, 6, 21 dpi) | Thrombocytopenia (3, 6, 9, 14, 21 dpi) | ALB↓ (14, 21 dpi), ALP↓ (9, 14, 21, 28 dpi), AMY↓ (9 dpi), GLOB↑ (21, 28 dpi) | Survived | |
| D2 | 50 mg/kg ZMapp™, 3 dpi | Leukocytopenia (21, 28 dpi) | Thrombocytopenia (28 dpi) | PHOS↓ (9 dpi) | Survived | ||
| D3 | 50 mg/kg ZMapp™, 3 dpi | Fever (3 dpi) | Leukocytosis (3, 14 dpi) | Thrombocytopenia (3, 21, 28 dpi) | ALT↓ (6 dpi) | Survived | |
| D4 | 50 mg/kg ZMapp™, 3 dpi | Leukocytopenia (14 dpi) | Thrombocytopenia (14, 21 dpi) | ALT↓ (9 dpi), CRE↑ (14 dpi) | Survived | ||
| D5 | 50 mg/kg ZMapp™, 3 dpi | Fever (3 dpi) | Leukocytopenia (21, 28 dpi) | Thrombocytopenia (6, 9 dpi) | ALB↓ (9 dpi), BUN↓ (3, 6, 14, 21, 28 dpi) | Survived | |
| D6 | 50 mg/kg ZMapp™, 3 dpi | Thrombocytopenia (6 dpi) | Survived | ||||
| E1 | 50 mg/kg ZMapp™, 4 dpi | Thrombocytopenia (4, 7, 21 dpi) | AMY↓↓ (4, 21 dpi), AMY↓ (7, 10, 14 dpi), CRE↓ (21, 28 dpi) | Survived | |||
| E2 | 50 mg/kg ZMapp™, 4 dpi | Fever (4 dpi) | Leukocytosis (4, 10 dpi) | Thrombocytopenia (4, 7, 10, 21 dpi) | ALT ↓↓ (4 dpi), GLU↑ (4 dpi) | Survived | |
| E3 | 50 mg/kg ZMapp™, 4 dpi | Fever (4 dpi) | Leukocytosis (4, 10 dpi) | Thrombocytopenia (7, 10, 14 dpi) | CRE↓ (14 dpi) | Survived | |
| E4 | 50 mg/kg ZMapp™, 4 dpi | Severe rash (5, 6, 7, 8 dpi), Mild rash (9 dpi) | Leukocytosis (10, 14, 21, 28 dpi) | Thrombocytopenia (4, 7, 10, 14 dpi) | ALP↑ (7, 10, 14 dpi), ALT ↑↑↑ (7 dpi), ALT ↑↑ (10 dpi), AMY↓ (4, 7, 10 dpi), TBIL↑↑↑ (7 dpi), TBIL↑ (10, 14 dpi), PHOS↓ (7, 10 dpi), K+↓ (4 dpi) | Survived | |
| E5 | 50 mg/kg ZMapp™, 4 dpi | Fever (7 dpi) | Leukocytosis (4 dpi) | Thrombocytopenia (4, 7, 10, 14 dpi) | ALT↑ (7 dpi), AMY↓ (4, 7 dpi), PHOS↓ (10 dpi) | Survived | |
| E6 | 50 mg/kg ZMapp™, 4 dpi | Fever (4 dpi) | Mild rash (7, 8, 9 dpi) | Leukocytosis (4, 10, 14 dpi) | Thrombocytopenia (4, 7, 10, 14 dpi) | ALP↑ (7, 10 dpi), ALT ↑↑↑ (7, 10, 14 dpi), AMY↓ (7, 10 dpi), TBIL↑↑ (7 dpi), TBIL↑↑↑ (10 dpi), TBIL↑ (14 dpi), PHOS↓ (7 dpi), GLOB↑ (21 dpi) | Survived |
| F1 | 50 mg/kg ZMapp™, 5 dpi | Leukocytosis (11 dpi) | Thrombocytopenia (3, 5, 8, 11 dpi) | AMY↓ (5 dpi), PHOS↓ (11 dpi), CRE↓ (28 dpi) | Survived | ||
| F2 | 50 mg/kg ZMapp™, 5 dpi | Fever (3, 5 dpi) | Mild rash (8 dpi) | Leukocytosis (3, 5, 11 dpi) | Thrombocytopenia (3, 5, 8, 11, 14, 21 dpi) | PHOS↓ (11 dpi), CRE↓↓ (11 dpi) | Survived |
| F3 | 50 mg/kg ZMapp™, 5 dpi | Leukocytopenia (8 dpi), Leukocytosis (3 dpi) | Thrombocytopenia (5, 8, 11, 21 dpi | ALT↑ (8 dpi), CRE↓↓ (14 dpi) | Survived | ||
| F4 | 50 mg/kg ZMapp™, 5 dpi | Fever (3, 5 dpi) | Leukocytopenia (8 dpi) | Thrombocytopenia (5, 8, 11, 28 dpi) | PHOS↓ (8 dpi) | Survived | |
| F5 | 50 mg/kg ZMapp™, 5 dpi | Fever (3 dpi) | Leukocytosis (3, 11, 14 dpi) | Thrombocytopenia (5, 8, 11 dpi) | PHOS↓ (5, 8 dpi), CRE↓ (8, 11, 21, 28 dpi) | Survived | |
| F6 | 50 mg/kg ZMapp™, 5 dpi | Fever (3 dpi) | Leukocytopenia (8, 21, 28 dpi) | Thrombocytopenia (8, 11, 21 dpi) | PHOS↓ (5, 8, 11 dpi), GLU↑ (5 dpi) | Survived | |
| G1 | PBS, 4 dpi | Severe rash (4 dpi) | Leukocytopenia (4 dpi) | Thrombocytopenia (4 dpi) | AMY↓ (4 dpi) | Died, 4 dpi | |
| G2 | Control mAb, 4 dpi | Severe rash (8 dpi) | Leukocytopenia (7, 8 dpi) | Thrombocytopenia (4, 7, 8 dpi) | ALP↑ (8 dpi), ALT↑ (7 dpi), ALT ↑↑↑ (8 dpi), CRE↑ (8 dpi) | Died, 8 dpi | |
| G3 | PBS, 4 dpi | Fever (4, 8 dpi) | Severe rash (8 dpi) | Leukocytopenia (7, 8 dpi) | Thrombocytopenia (4, 7, 8 dpi) | ALP↑ (8 dpi), ALT↑ (7, 8 dpi), AMY↓ (7 dpi), AMY ↓↓ (8 dpi), TBIL↑ (8 dpi), PHOS↓ (7 dpi) | Died, 8 dpi |
Hypothermia was defined as below 35°C. Fever was defined as >1.0°C higher than baseline. Mild rash was defined as focal areas of petechiae covering <10% of the skin, moderate rash was defined as areas of petechiae covering 10 to 40% of the skin, and severe rash was defined as areas of petechiae and/or ecchymosis covering >40% of the skin. Leukocytopenia and thrombocytopenia were defined as a >30% decrease in the numbers of WBCs and platelets, respectively. Leukocytosis and thrombocytosis were defined as a twofold or greater increase in numbers of WBCs and platelets above baseline, where WBC counts are greater than 11.0. ↑, two- to threefold increase; ↑↑, four- to fivefold increase; ↑↑↑, greater than fivefold increase; ↓, two- to threefold decrease; ↓↓, four- to fivefold decrease; ↓↓↓, greater than fivefold decrease. ALB, albumin; ALP, alkaline phosphatase; ALT, alanine aminotransferase; AMY, amylase; TBIL, total bilirubin; BUN, blood urea nitrogen; PHOS, phosphate; CRE, creatinine; GLU, glucose; K+, potassium; GLOB, globulin.
Based on clinical scores (Figure 2c, Extended Data Figure 1), the Group F animals did not appear to be as sick as animals E4 and E6, both of whom were near the clinical limit for IACUC mandated euthanasia at 5 and 7 dpi, respectively. Animal E4 had a flushed face and severe rash on more than 40% of its body surface between 5 to 8 dpi in addition to nasal hemorrhage at 7 dpi, whereas animal E6 had a flushed face and petechiae on its arms and legs between 7 to 9 dpi, in addition to jaundice between 10 to 14 dpi. This indicates that host genetic factors may play a role in the differential susceptibility of individual NHPs to EBOV-K infections. Fever, leukocytosis, thrombocytopenia, and a severe rash symptomatic of EBOV disease progression was detected in both E4 and E6 (Table 3). Increases in the level of liver enzymes ALT (10- to 30-fold increase), ALP (2- to 3-fold), and total bilirubin (TBIL, 3- to 11-fold) indicate significant liver damage (Figure 2g-l), a hallmark of filovirus infections. However, ZMapp™ was successful in reversing observed disease symptoms and physiological abnormalities after 12 dpi, 2 days after the last ZMapp™ administration (Table 3). Furthermore, ZMapp™ treatment was able to lower the high virus loads observed in animals F2 and F5 (up to 106 TCID50/ml) to undetectable levels by 14 dpi (Figure 2f, Extended Data Figure 2).
ZMapp™ cross-reacts with Guinea EBOV
While the results were very promising with EBOV-K infected NHPs, it was unknown whether therapy with ZMapp™ would be similarly effective against the Guinean variant of EBOV (EBOV-G), the virus responsible for the West African outbreak. Direct comparison of published amino acid sequences between EBOV-G and EBOV-K showed that the epitopes targeted by ZMapp™ 20,21 were not mutated between the two virus variants (Figure 3a), suggesting that the antibodies should retain their specificity for the viral glycoprotein. To confirm this, in vitro assays were carried out to compare the binding affinity of c13C6, c2G4 and c4G7 to sucrose-purified EBOV-G and EBOV-K. As measured by ELISA, the ZMapp™ componentsshowed slightly better binding kinetics for EBOV-G than for EBOV-K (Figure 3b). Additionally, the neutralizing activity of individual mAbs was evaluated in the absence of complement for c2G4 and c4G7, and in the presence of complement for c13C6, as they have previously been shown to neutralize EBOV only under this condition13 (Figure 3c). The results supported the ELISA binding data, with comparable neutralizing activities between the two viruses.

(a) Sequence alignment of the EBOV glycoprotein from the Kikwit (EBOV-K) and Guinea (EBOV-G) variants, with the binding epitopes of ZMapp™ pointed with an arrow, and (b) an ELISA (for each antibody, the EC50 are different (p < 0.05, regression analysis) between the two antigens) as well as (c) a neutralizing antibody assay showing the activity of the individual mAbs composing ZMapp™ against EBOV-K (black) and EBOV-G (purple), and the samples were run in triplicate.
Discussion
The West African outbreak of 2014 has highlighted the troubling absence of available vaccine or therapeutic options to save thousands of lives and stop the spread of EBOV. The lack of a clinically acceptable treatment offer limited incentive for people who suspect they might be infected to report themselves for medical help. Several previous studies have showed that antibodies are crucial for host survival from EBOV22,23,24. Prior NHP studies have also demonstrated the ZMAb cocktail could protect 100% or 50% of animals when dosing was initiated 1 or 2 dpi, while the MB-003 cocktail protected 67% of animals with the same dosing regimen. Before the success with mAb-based therapies, other candidate therapeutics had only demonstrated efficacy when given within 60 minutes of EBOV exposure.
Our results with ZMapp™, a cocktail comprising of individual mAbs selected from MB-003 and ZMAb, demonstrate for the first time the successful protection of NHPs from EBOV disease when intervention was initiated as late as 5 dpi. In the preceding ZMapp1/ZMapp2 experiment, 11 of 12 treated animals had detectable fever (with the exception of A4), and live virus could be detected in the blood of 11 of 12 animals (with the exception of A3) by 3 dpi. Therefore, for the majority of these animals, treatment was therapeutic (as opposed to post-exposure prophylaxis), initiated after two detectable triggers of disease. ZMapp2 was able to protect 5 of 6 animals when administered at 3 dpi. For reasons currently unknown, the lone non-survivor (B3) experienced a viremia of 106 TCID50 at 3 dpi, which is 100-fold greater than all other NHPs and approximately 10-fold higher than what ZMAb has been reported to suppress in a previous study16. This indicates enhanced EBOV replication in this animal, possibly due to host factors. It was important to note that despite the high levels of live circulating virus detected in B3, ZMapp2 administration was still able to prolong the life of this animal to 9 dpi, and suggests that in cases of high viremia such as this, the dosage of mAbs should be increased.
The highlight of these experimental results is undoubtedly ZMapp™, which was able to reverse severe EBOV disease as indicated by the elevated liver enzymes, mucosal hemorrhages and rash in animals E4 and E6. The high viremia (up to 106 TCID50/ml of blood in some animals at the time of intervention) could also be effectively controlled without the presence of escape mutants, leading to full recovery of all treated NHPs by 28 dpi. In the absence of direct evidence demonstrating ZMapp™ efficacy against lethal EBOV-G infection in NHPs, results from ELISA and neutralizing antibody assays show that binding specificity is not abrogated between EBOV-K and EBOV-G, and therefore the levels of protection should not be affected. The compassionate use of ZMapp™ in two infected American healthcare workers with apparently positive results pertaining to survival and reversion of EBOV disease25, supports this assertion. Rhesus macaques have approximately 55–80 ml of blood per kg of body weight26; at a dose of 50 mg/kg of antibodies, the estimated starting concentration is approximately 625–909 μg/ml of blood (total; ~200–300 μg/ml for each antibody). Therefore, the low EC50 values for EBOV-G (0.004 – 0.02 μg/ml) bode well for treating EBOV-G infections with ZMapp™.
Since the host antibody response is known to correlate with and is required for protection from EBOV infections23,24, mAb-based treatments are likely to form the centerpiece of any future therapeutic strategies for fighting EBOV outbreaks. However, whether ZMapp™-treated survivors can be susceptible to re-infection is unknown. In a previous study of murine ZMAb-treated, EBOV-challenged NHP survivors, a re-challenge of these animals with the same virus at 10 and 13 weeks after initial challenge yielded 6 of 6 survivors and 4 of 6 survivors, respectively27. While specific CD4+ and CD8+ T-cell responses could be detected in all animals, the circulating levels of glycoprotein (GP)-specific IgG were shown to be 10-fold lower in non-survivors compared to survivors, suggesting that antibody levels may be indicative of protective immunity27. Sustained immunity with experimental EBOV vaccines in NHPs remain unknown, however in a recent study, a decrease in GP-specific IgG levels due to old age or a suboptimal reaction to the VSVΔG/EBOVGP vaccine in rodents also appear to be indicative of non-survival28.
ZMapp™ consists of a cocktail of highly purified mAbs; which constitutes a less controversial alternative than whole blood transfusions from convalescent survivors, as was performed during the 1995 EBOV outbreak in Kikwit29. The safety of mAb therapy is well-documented, with generally low rates of adverse reactions, the capacity to confer rapid and specific immunity in all populations, including the young, the elderly and the immunocompromised, and if necessary, the ability to provide higher-than-natural levels of immunity compared to vaccinations30. The evidence presented here suggests that ZMapp™ currently offers the best option of the experimental therapeutics currently in development for treating EBOV-infected patients. We hope that initial safety testing in humans will be undertaken soon, preferably within the next few months, in order to enable the compassionate use of ZMapp™ as soon as possible.
Materials and Methods
Ethics statement
The guinea pig experiment, in addition to the second and third NHP study (ZMapp1, ZMapp2 and ZMapp™) were performed at the National Microbiology Laboratory (NML) as described on Animal use document (AUD) #H-13-003, and has been approved by the Animal Care Committee (ACC) at the Canadian Science Center for Human and Animal Health (CSCHAH), in accordance with the guidelines outlined by the Canadian Council on Animal Care (CCAC). The first study with MB-003 in NHPs was performed at United States Army Medical Research Institute of Infectious Diseases (USAMRIID) under an Institutional Animal Care and Use Committee (IACUC) approved protocol in compliance with the Animal Welfare Act, Public Health Service Policy, and other federal statutes and regulations relating to animals and experiments involving animals. The facility where this research was conducted in accredited by The Association for Assessment and Accreditation of Laboratory Animal Care International and adheres to principles stated in the 8th edition of the Guide for the Care and Use of Laboratory Animals, National Research Council 2011.
mAb production
The large-scale production of mAb cocktails cZMAb, MB-003, ZMapp1, ZMapp2 and ZMapp™ in addition to control mAb 4E10 (anti-HIV) from N. benthamiana under GMP conditions was done by Kentucky BioProcessing (Owensboro, KY) as described previously13,15,31. The large-scale production of m4G7 was performed by the Biotechnology Research Institute (Montreal, QC) using a previously described protocol16.
Viruses
The challenge virus used in NHPs was Ebola virus H.sapiens-tc/COD/1995/Kikwit-9510621 (EBOV-K) (order Mononegavirales, family Filoviridae, species Zaire ebolavirus; GenBank accession #AY354458)18. Passage three from the original stock was used for the studies at the NML and passage four was used for the study performed at USAMRIID (the NHP study with the individual MB-003 mAbs). Sequencing of 112 clones from the passage three stock virus revealed that the population ratio of 7U:8U in the EBOV GP editing site was 80:20; sequencing for the passage four stock virus was not performed, and therefore the ratio of 7U:8U in the editing site was unknown. The virus used in guinea pig studies was guinea pig-adapted EBOV, Ebola virus VECTOR/C.porcellus-lab/COD/1976/Mayinga-GPA (EBOV-M-GPA) (order Mononegavirales, family Filoviridae, species Zaire ebolavirus; Genbank accession number AF272001.1)32. The Guinean variant used in IgG ELISA and neutralizing antibody assays was Ebola virus H.sapiens-tc/GIN/2014/Gueckedou-C05 (EBOV-G) (order Mononegavirales, family Filoviridae, species Zaire ebolavirus; GenBank accession #KJ660348.1)2.
Animals
Outbred 6–8 week old female Hartley strain guinea pigs (Charles River) were used for these studies. Animals were infected IP with 1000 × LD50 of EBOV-M-GPA. The animals were then treated with one dose of ZMAb, MB-003, ZMapp1, ZMapp2, c13C6, h13F6 or c6D8 totaling 5 mg per guinea pig, and monitored every day for 28 days for survival, weight and clinical symptoms. This study was not blinded, and no animals were excluded from the analysis.
For the MB-003 study performed at USAMRIID, thirteen rhesus macaques (Macaca mulatta) were obtained from the USAMRIID primate holding facility, ranging from 5.1 to 10 kg. This study was not blinded, and no animals were excluded from the analysis. Animals were given standard monkey chow, primate treats, fruits, and vegetables for the duration of the study. All animals were challenged IM with a target dose of 1000 PFU. Treatment with either monoclonal antibody, MB-003 cocktail, or PBS was administered on 1, 4, and 7 dpi via saphenous intravenous infusion. Animals were monitored at least once daily for changes in health, diet, behavior, and appearance. Animals were sampled for chemical analysis, complete bloods counts and viremia on 0, 3, 5, 7, 10, 14, 21, and 28 dpi.
For the ZMapp1 and ZMapp2 study, fourteen male and female rhesus macaques (Macaca mulatta), ranging from 4.1 to 9.6 kg (4–8 years old) were purchased from Primgen (USA). This study was not blinded, and no animals were excluded from the analysis. Animals were assigned groups based on gender and weight. Animals were fed standard monkey chow, fruits, vegetables, and treats. Husbandry enrichment consisted of visual stimulation and commercial toys. All animals were challenged IM with a high dose of EBOV [backtiter: 4000 × TCID50 or 2512 PFU] at 0 dpi. Administration of the first treatment dose was initiated at 3 dpi, with identical doses at 6 and 9 dpi. Animals were scored daily for signs of disease, in addition to changes in food and water consumption. On designated treatment days in addition to 14, 21, and 27 dpi, the rectal temperature and clinical score were measured, and the following were sampled: blood for serum biochemistry and cell counts and viremia. This study was not blinded, and no animals were excluded from the analysis.
For the ZMapp™ study, twenty-one male rhesus macaques, ranging from 2.5 to 3.5 kg (2 years-old) were purchased from Primgen (USA). This study was not blinded, and no animals were excluded from the analysis. Animals were assigned groups based on gender and weight. Animals were fed standard monkey chow, fruits, vegetables, and treats. Husbandry enrichment consisted of visual stimulation and commercial toys. All animals were challenged IM with EBOV [backtiter: 1000 × TCID50 or 628 PFU] at 0 dpi. Administration of the first treatment dose was initiated at 3, 4 or 5 dpi, with two additional identical doses spaced three days apart. Animals were scored daily for signs of disease, in addition to changes in food and water consumption. On designated treatment days in addition to 14, 21, and 28 dpi, the rectal temperature and clinical score were measured, and the following were sampled: blood for serum biochemistry and cell counts and viremia.
Blood counts and blood biochemistry
Complete blood counts were performed with the VetScan HM5 (Abaxis Veterinary Diagnostics). The following parameters were shown in the figures: levels of white blood cells (WBC), lymphocytes (LYM), percentage of lymphocytes (LYM%), levels of platelets (PLT), neutrophils (NEU) and percentage of neutrophils (NEU%). Blood biochemistry was performed with the VetScan VS2 (Abaxis Veterinary Diagnostics). The following parameters were shown in the figures: levels of alkaline phosphatase (ALP), alanine aminotransferase (ALT), blood urea nitrogen (BUN), creatinine (CRE), and total bilirubin (TBIL).
Enzyme-linked immunosorbent assays (ELISAs)
IgG ELISA with c13C6, c2G4 or c1H3 was performed as described previously16 using gamma-irradiated EBOV-G and EBOV-K virions purified on a 20% sucrose cushion as the capture antigen in the ELISA. Each mAb was assayed in triplicate.
Neutralizing antibody assays
Two-fold dilutions of c13C6, c2G4 or c1H3 ranging from 0.0156 to 2 μg/ml were first incubated with 100 PFU of EBOV-G at room temperature for 1 hour with or without complement, transferred to Vero E6 cells and incubated at 37°C for 1 hour, and then replaced with DMEM supplemented with 2% fetal bovine serum and scored for the presence of cytopathic effect (CPE) at 14 dpi. The lowest concentrations of mAbs demonstrating the absence of CPE were averaged and reported.
EBOV titration by TCID50 and RT-qPCR
Titration of live EBOV was determined by adding 10-fold serial dilutions of whole blood to VeroE6 cells, with three replicates per dilution. The plates were scored for cytopathic effect at 14 dpi, and titers were calculated with the Reed and Muench method33. Results were shown as median tissue culture infectious dose (TCID50).
For titers measured by RT-qPCR, total RNA was extracted from whole blood with the QIAmp Viral RNA Mini Kit (Qiagen). EBOV was detected with the LightCycler 480 RNA Master Hydrolysis Probes (Roche) kit, with the RNA polymerase (nucleotides 16472 to 16538, AF086833) as the target gene. The reaction conditions were as follows: 63°C for 3 min, 95°C for 30 s, and cycling of 95°C for 15 s, 60°C for 30 s for 45 cycles on the ABI StepOnePlus. The lower detection limit for this assay is 86 genome equivalents/ml. The sequences of primers used were as follows: EBOVLF2 (CAGCCAGCAATTTCTTCCAT), EBOVLR2 (TTTCGGTTGCTGTTTCTGTG), and EBOVLP2FAM (FAM-ATCATTGGCGTACTGGAGGAGCAG-BHQ1).
Sequence alignment
Protein sequences for EBOV-K and EBOV-G surface glycoproteins were obtained from GenBank, accession numbers AGB56794.1 and AHX24667.1 respectively. The sequences were aligned using DNASTAR Lasergene 10 MEGAlign using the Clustal W algorithm.
Statistical analysis
For the guinea pig and nonhuman primate studies, each treatment group consisted of six animals. Assuming a significance threshold of 0.05, a sample size of six per group will give >80% power to detect a difference in survival proportions between the treatment (83% survival or higher) and the control group using a one-tailed Fisher’s exact test.
Survival was compared using the log-rank test in GraphPad PRISM 5, differences in survival were considered significant when the p-value was less than 0.05. Antibody binding to EBOV-G and EBOV-K was compared by fitting the data to a 4-parameter logistic regression using GraphPad PRISM 5. The EC50 were considered different if the 95% Confidence Intervals excluded each other. For all statistical analyses, the data conformed to the assumptions of the test used.
Extended Data
Extended Data Figure 1
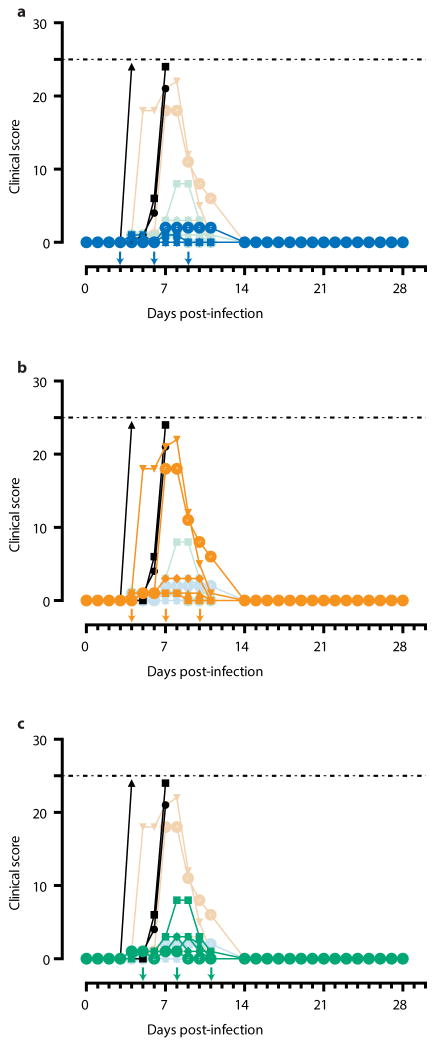
Arrows indicate treatment days. Dashed line represents humane endpoint threshold. Faded symbols/lines are the other two treatment groups, for comparison. Control group (Group G) is shown in black on all three panels. (a) Clinical score of Group D (blue); (b) Clinical score of Group E (orange); (c) Clinical score of Group F (green).
Extended Data Figure 2
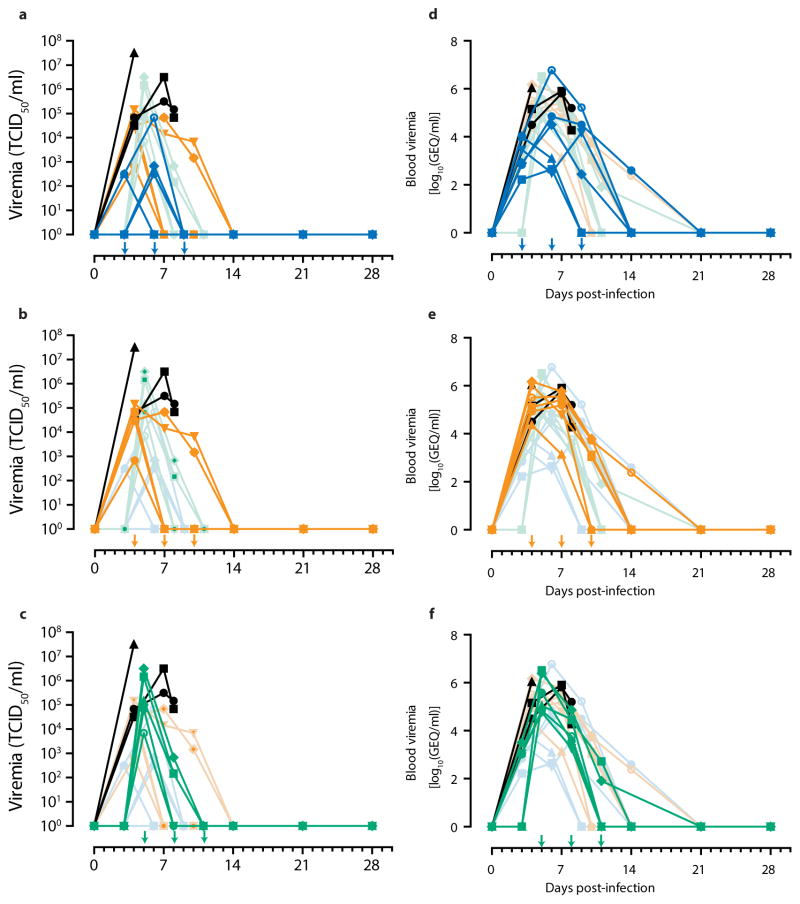
Arrows indicate treatment days. Faded symbols/lines are the other two treatment groups, for comparison. Control group (Group G) is shown in black on all three panels. (a) TCID50 of Group D (blue); (b) TCID50 of Group E (orange); (c) TCID50 of Group F (green). (d) Viremia by RT-qPCR of Group D (blue); (e) Viremia by RT-qPCR of Group E (orange); (f) Viremia by RT-qPCR of Group F (green).
Extended Data Table 2
Oral swab viremia measured by RT-qPCR for the ZMapp1- and ZMapp2-treated NHPs.
| DAYS | A1 | A2 | A3 | A4 | A5 | A6 | B1 | B2 | B3 | B4 | B5 | B6 | C1 | C2 |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 0 | UD | UD | UD | UD | UD | UD | UD | UD | UD | UD | UD | UD | UD | UD |
| 3 | UD | UD | UD | UD | UD | UD | UD | UD | UD | UD | UD | UD | UD | UD |
| 6 | UD | UD | UD | UD | UD | UD | UD | UD | UD | UD | UD | UD | UD | UD |
| 7 | 5.05E+03 | |||||||||||||
| 9 | UD | UD | UD | UD | UD | UD | UD | 4.81E+04 | UD | UD | UD | |||
| 14 | UD | UD | UD | UD | UD | UD | UD | UD | UD | UD | UD | |||
| 21 | UD | UD | UD | UD | UD | UD | UD | UD | UD | UD | UD | |||
| 27 | UD | UD | UD | UD | UD | UD | UD | UD | UD | UD | UD |
Footnote: UD = Undetectable.
Extended Data Table 3
Nasal swab viremia measured by RT-qPCR for the ZMapp1- and ZMapp2-treated NHPs.
| DAYS | A1 | A2 | A3 | A4 | A5 | A6 | B1 | B2 | B3 | B4 | B5 | B6 | C1 | C2 |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 0 | UD | UD | UD | UD | UD | UD | UD | UD | UD | UD | UD | UD | UD | UD |
| 3 | UD | UD | UD | UD | UD | UD | UD | UD | UD | UD | UD | UD | UD | UD |
| 6 | UD | UD | UD | UD | UD | UD | UD | UD | UD | UD | UD | UD | UD | 3.75E+02 |
| 7 | 1.98E+04 | 2.16E+03 | ||||||||||||
| 9 | UD | UD | UD | UD | UD | UD | UD | UD | UD | UD | UD | |||
| 14 | UD | UD | UD | UD | UD | UD | UD | UD | UD | UD | UD | |||
| 21 | UD | UD | UD | UD | UD | UD | UD | UD | UD | UD | UD | |||
| 27 | UD | UD | UD | UD | UD | UD | UD | UD | UD | UD | UD |
Footnote: UD = Undetectable.
Extended Data Table 4
Rectal swab viremia measured by RT-qPCR for the ZMapp1- and ZMapp2-treated NHPs.
| DAYS | A1 | A2 | A3 | A4 | A5 | A6 | B1 | B2 | B3 | B4 | B5 | B6 | C1 | C2 |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 0 | UD | UD | UD | UD | UD | UD | UD | UD | UD | UD | UD | UD | UD | UD |
| 3 | UD | UD | UD | UD | UD | UD | UD | UD | UD | UD | UD | UD | UD | UD |
| 6 | UD | UD | UD | UD | UD | UD | UD | UD | UD | UD | UD | UD | 4.16E+02 | 8.17E+03 |
| 7 | 4.38E+04 | |||||||||||||
| 9 | UD | UD | UD | UD | UD | UD | UD | 3.90E+02 | UD | UD | UD | |||
| 14 | UD | UD | UD | UD | UD | UD | UD | UD | UD | UD | UD | |||
| 21 | UD | UD | UD | UD | UD | UD | UD | UD | UD | UD | UD | |||
| 27 | UD | UD | UD | UD | UD | UD | UD | UD | UD | UD | UD |
Footnote: UD = Undetectable.
Extended Data Table 5
Blood viremia measured by RT-qPCR for the ZMapp™-treated NHPs.
| DAYS | A1 | A2 | A3 | A4 | A5 | A6 | B1 | B2 | B3 | B4 | B5 | B6 | C1 | C2 | C3 | C4 | C5 | C6 | D1 | D2 | D3 |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 0 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 |
| 3 | 676.08 | 165.96 | 10233 | 2884 | 812.83 | 10965 | 1047.1 | 1122 | 3235.9 | 1148.2 | |||||||||||
| 4 | 85114 | 128825 | 23442 | 1E+06 | 1E+06 | 323594 | 31623 | 144544 | 1E+06 | ||||||||||||
| 5 | 380189 | 3E+06 | 109648 | 58884 | 2E+06 | 69183 | |||||||||||||||
| 6 | 70795 | 446.68 | 1230.3 | 316.23 | 32359 | 6E+06 | |||||||||||||||
| 7 | 154882 | 257040 | 1380.4 | 63096 | 588844 | 363078 | 645654 | 812831 | |||||||||||||
| 8 | 3715.4 | 28184 | 29512 | 1862.1 | 72444 | 5888.4 | 158489 | 18621 | |||||||||||||
| 9 | 31623 | 1 | 1 | 15136 | 275.42 | 165959 | |||||||||||||||
| 10 | 1 | 1071.5 | 1 | 1318.3 | 6166 | 5248.1 | |||||||||||||||
| 11 | 1 | 524.81 | 1 | 1 | 81.283 | 1 | |||||||||||||||
| 14 | 398.11 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 239.88 | 1 | 1 | 1 | 1 | 1 | 1 | |||
| 21 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | |||
| 28 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 |
Supplementary Material
1
2
3
4
5
6
7
Acknowledgments
The authors thank Kevin Tierney, Allen Grolla, Shane Jones, Jessica Dong, and Dr. Darwyn Kobasa for their excellent technical assistance, Dr. Victor Klimyuk and Dr. Yuri Gleba for access to the magnICON expression system, and Dr. Herta Steinkellner for access to transgenic N. benthamiana. This work was supported by the Defense Threat Reduction Agency (DTRA contract HDTRA1-13-C-0018), the National Institutes of Health (U19AI109762), the Public Health Agency of Canada (PHAC), and a Canadian Safety and Security Program (CSSP) grant to G.P.K. and X.Q. G.W. is the recipient of a Doctoral Research Award from the Canadian Institute for Health Research (CIHR).
Footnotes
Author contributions
X.Q., G.K. and L.Z. designed the experiments. X.Q., G.W., J.A., A.B., L.F., J.B.A., H.F., H.W., J.A., J. P., G.G.O. and G.K. performed the experiments. X.Q., G.W., J.A., K.W., B.X., J. E. S., L.Z. and G.K. wrote the manuscript. E.H., A.J., J.M., K.S., O.B., N.B., C.G., D.K., M.H.P., J.V., K.W. and L.Z. contributed reagents for this study.Financial disclosures
Her Majesty the Queen in right of Canada holds a patent on mAbs 2G4, and 4G7, PCT/CA2009/000070, “Monoclonal antibodies for Ebola and Marburg viruses.” K.W. and L.Z. are the owners of Mapp Biopharmaceutical Inc. The authors declare no other competing interests.
References
Full text links
Read article at publisher's site: https://doi.org/10.1038/nature13777
Read article for free, from open access legal sources, via Unpaywall:
https://www.ncbi.nlm.nih.gov/pmc/articles/PMC4214273
Citations & impact
Impact metrics
Article citations
Longitudinal proteome-wide antibody profiling in Marburg virus survivors identifies wing domain immunogen for vaccine design.
Nat Commun, 15(1):8133, 17 Sep 2024
Cited by: 0 articles | PMID: 39285186 | PMCID: PMC11405854
Emerging and reemerging infectious diseases: global trends and new strategies for their prevention and control.
Signal Transduct Target Ther, 9(1):223, 11 Sep 2024
Cited by: 0 articles | PMID: 39256346 | PMCID: PMC11412324
Review Free full text in Europe PMC
Evaluation of human antibodies from vaccinated volunteers for protection against <i>Yersinia pestis</i> infection.
Microbiol Spectr, 12(10):e0105424, 27 Aug 2024
Cited by: 0 articles | PMID: 39189763 | PMCID: PMC11448073
Therapeutic glycan-specific antibody binding mediates protection during primary amoebic meningoencephalitis.
Infect Immun, 92(10):e0018324, 05 Sep 2024
Cited by: 0 articles | PMID: 39235225
Fully human monoclonal antibodies against Ebola virus possess complete protection in a hamster model.
Emerg Microbes Infect, 13(1):2392651, 26 Aug 2024
Cited by: 0 articles | PMID: 39155772 | PMCID: PMC11348817
Go to all (585) article citations
Other citations
Wikipedia
Data
Data behind the article
This data has been text mined from the article, or deposited into data resources.
BioStudies: supplemental material and supporting data
Nucleotide Sequences (Showing 6 of 6)
- (1 citation) ENA - AHX24667
- (1 citation) ENA - AF272001
- (1 citation) ENA - AF086833
- (1 citation) ENA - AY354458
- (1 citation) ENA - KJ660348
- (1 citation) ENA - AGB56794
Show less
Similar Articles
To arrive at the top five similar articles we use a word-weighted algorithm to compare words from the Title and Abstract of each citation.
Ebola-specific therapeutic antibodies from lab to clinic: The example of ZMapp.
Antiviral Res, 226:105873, 03 Apr 2024
Cited by: 1 article | PMID: 38580170
Review
Development of a Human Antibody Cocktail that Deploys Multiple Functions to Confer Pan-Ebolavirus Protection.
Cell Host Microbe, 25(1):39-48.e5, 01 Jan 2019
Cited by: 63 articles | PMID: 30629917 | PMCID: PMC6396299
Delayed treatment of cynomolgus macaques with a FVM04/CA45 monoclonal antibody cocktail provides complete protection against lethal Sudan virus infection.
J Virol, 98(8):e0124223, 16 Jul 2024
Cited by: 0 articles | PMID: 39012096 | PMCID: PMC11334508
Macaque Monoclonal Antibodies Targeting Novel Conserved Epitopes within Filovirus Glycoprotein.
J Virol, 90(1):279-291, 14 Oct 2015
Cited by: 60 articles | PMID: 26468532 | PMCID: PMC4702572
Funding
Funders who supported this work.
CIHR
NIAID NIH HHS (2)
Grant ID: U19 AI109762
Grant ID: U19AI109762





