Abstract
Free full text

An engineered Axl ‘decoy receptor’ effectively silences the Gas6/Axl signaling axis
Abstract
Aberrant signaling through the Axl receptor tyrosine kinase has been associated with a myriad of human diseases, most notably metastatic cancer, identifying Axl and its ligand Gas6 as important therapeutic targets. Using rational and combinatorial approaches, we engineered an Axl ‘decoy receptor’ that binds Gas6 with high affinity and inhibits its function, offering an alternative approach from drug discovery efforts that directly target Axl. Four mutations within this high affinity Axl variant caused structural alterations in side chains across the Gas6/Axl binding interface, stabilizing a conformational change on Gas6. When reformatted as an Fc-fusion, the engineered decoy receptor bound to Gas6 with femtomolar affinity, an 80-fold improvement compared to the wild-type Axl receptor, allowing effective sequestration of Gas6 and specific abrogation of Axl signaling. Moreover, increased Gas6 binding affinity was critical and correlative with the ability of decoy receptors to potently inhibit metastasis and disease progression in vivo.
INTRODUCTION
The Axl receptor, a member of the TAM (Tyro3, Axl, Mer) family of receptor tyrosine kinases,1 has attracted significant attention as a promising oncology target. In a number of human cancers, Axl signaling has been shown to drive metastasis2, confer therapeutic resistance3–5, and promote disease progression6. Additionally, Axl overexpression has been observed in multiple solid and hematological malignancies7, with expression levels often correlating with disease stage and poor clinical prognosis8–10. These observations have cemented Axl as an important target for cancer therapy and have created a critical need for molecular entities capable of modulating Axl signaling. Such compounds would be invaluable both as translational therapeutics and, more fundamentally, as research tools that could help unravel the complex mechanisms that govern Axl biology.
Towards this goal, classic drug development approaches have generated Axl antagonists including small molecule tyrosine kinase inhibitors (TKI’s)11,12, antibodies13,14, and nucleic acid aptamers15. However, several features of these molecules have limited their collective utility. For example, the polypharmacology of existing Axl TKIs16 can increase clinical off-target toxicities and prevent their application as biological tools to probe specific functional relationships17. Additionally, the competitive antagonists developed to date all possess nanomolar affinities to Axl13–15, hindering their ability to disrupt the stronger interaction between Axl and its ligand growth arrest-specific 6 (Gas6)18,19. Finally, a commonality amongst these compounds is that they all target Axl, which has raised questions regarding the role of Gas6 in Axl activation20. Thus, there remains a critical need for molecules that can effectively and specifically disrupt the Gas6/Axl signaling axis.
Exploiting natural components of a target system to develop antagonists is an attractive concept21; an elegant derivative is to use soluble fragments of receptors to bind to and neutralize cognate ligands (Fig. 1a)22. The blockbuster success of the TNFα antagonist Enbrel (etanercept) is a prominent example of this effective approach23. However, protein engineering is often required to optimize the biochemical and biophysical properties of such “decoy receptors”24. In the present study, we employ this strategy and engineer high affinity Axl-based antagonists capable of effectively sequestering Gas6. A combination of biochemical and structural studies was used to elucidate the molecular mechanism for high affinity Gas6 binding. Furthermore, we show that the engineered Axl decoy receptor effectively antagonizes the Gas6/Axl signaling axis, and exhibits a remarkable ability to inhibit metastasis and disease progression in vivo.
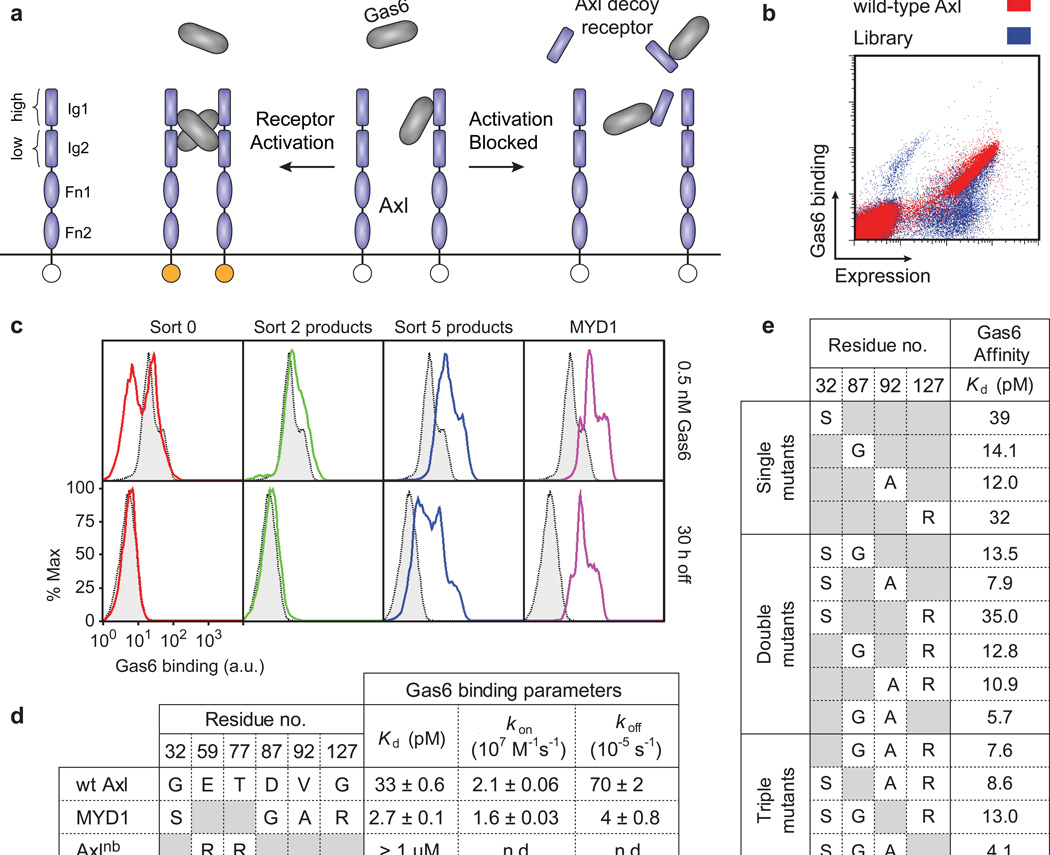
Engineering and characterization of receptor-based Axl antagonists. (a) Axl’s extracellular domain consists of two immunoglobulin-like (Ig) domains containing high and low affinity Gas6 binding sites, followed by two fibronectin type III domains. Binding of Gas6 to Axl leads to receptor dimerization and activation of downstream signaling. Axl decoy receptors sequester Gas6, preventing activation of the Axl signaling cascade. (b) Overlaid flow cytometry dot plots representing binding of yeast-displayed wild-type Axl Ig1 (red) and unsorted Axl Ig1 library (blue) to 10 nM Gas6 (y-axis), and expression levels on the yeast cell surface (x-axis). (c) Flow cytometry histograms of the initial Axl library and intermediate sort products compared to wild-type Axl Ig1 (gray), measuring binding to 0.5 nM Gas6 (top row) and persistent Gas6 binding after 30 h incubation with excess competitor (bottom row). MYD1 is also included for comparison. For clarity, only the gated population of yeast expressing Axl is shown. (d) Binding affinities of wild-type Axl Ig1, MYD1, and Axlnb to Gas6 as determined by KinExA. (e) Binding affinities to Gas6 of every permutation of the four mutations found in MYD1. Raw KinExA data and associated error values can be found in Supplementary Figs. 2 and 3.
RESULTS
Engineering Axl variants with increased Gas6 binding
Axl interacts with Gas6 through two independent epitopes: a high-affinity major binding site on its first immunoglobulin-like domain (Ig1) and a low-affinity minor site on Ig2 (Fig. 1a)25. The major binding site drives Axl’s overall affinity for Gas6 by leveraging an unusual geometry in which anti-parallel β-strands from Gas6 and Axl align on edge, forming a continuous β-sheet that bridges the interface. This architecture creates an elongated binding epitope that involves residues throughout the Ig1 domain. Due to the importance of the major binding site and the extended nature of its interface, we designed a protein engineering strategy in which mutations were randomly introduced into Axl Ig1 through error-prone PCR. The resulting library was displayed as fusion proteins on the yeast cell surface where a subset retained binding to Gas6 (Fig. 1b). Using a combination of equilibrium binding and kinetic off-rate screens, the library was enriched for variants displaying increased Gas6 binding (Fig. 1c, Supplementary Results, Supplementary Fig. 1). After five rounds of fluorescence activated cell sorting (FACS), the library still retained significant diversity; however, a consensus mutation (V92A) had emerged (Supplementary Table 1). Following a sixth and final round of stringent off-rate screening, three unique Axl variants were isolated (Supplementary Table 2) and initially characterized. Qualitative yeast-displayed binding studies showed that MYD1 was the most improved variant, and was thus chosen for further characterization.
Two mutations confer increased Gas6 binding
MYD1 Ig1 was recombinantly expressed and its affinity to Gas6 was measured using the Kinetic Exclusion Assay (KinExA)26 and compared to wild-type Axl Ig1 (Supplementary Fig. 2, see Methods for detailed experimental setup). As a control, we evaluated a non-binding Axl variant containing two mutations that abolish Gas6 binding (Axlnb: E59R/T77R25). Wild-type Axl Ig1 bound to Gas6 with an affinity (Kd) of 33 pM (Fig. 1d), which is significantly tighter than the nanomolar binding values often described in the literature, and the strongest affinity for the native Gas6/Axl interaction reported27. Analysis of the binding kinetics of the Gas6/Axl interaction indicated a fast association rate (kon) of 2.1×107 M−1s−1. In comparison, MYD1 Ig1 bound Gas6 with a Kd of 2.7 pM, a twelve-fold increase over wild-type that was predominantly driven by a slower dissociation rate (koff) (Fig. 1d). Axlnb displayed minimal binding to Gas6 at concentrations up to 1 µM. Additionally, the specificity of the native interaction was retained as MYD1 had negligible binding to protein S (Supplementary Fig. 2), a Gas6 homolog and activating ligand of Tyro3 and Mer28.
To parse the contributions of MYD1’s four amino acid mutations to the observed improvement in binding, all permutations of single, double, and triple mutants were recombinantly expressed and analyzed by KinExA. Thermodynamic mutant cycle analysis demonstrated that two independently acting mutations, D87G and V92A, accounted for nearly the entirety of the increased affinity (Fig. 1e, Supplementary Fig. 3 – 5). The N-terminal G32S mutation had a minor deleterious effect on binding affinity while G127R bound Gas6 at wild-type levels. In studying the double mutants, a beneficial interaction was uncovered between G32S and V92A that further enhanced affinity (Supplementary Table 3). This same interaction was observed in the triple mutants, demonstrating its independence with respect to the mutations at residues 87 and 127. Finally, while G127R did not impact affinity independently, its inclusion with the other three mutations conferred a slight improvement in Gas6 binding. Collectively, these analyses reveal that the majority of MYD1’s increase in Gas6 binding affinity comes from two mutations, D87G and V92A, with the remainder of the improvement derived mostly from a synergistic interaction between G32S, a slightly deleterious mutation on its own, and V92A.
Effects of the single point mutations on protein stability were also explored. Thermal melts showed similar stabilities between wild-type Axl and MYD1 (Tm = 53 ± 0.6 °C versus 54 ± 0.9 °C, respectively), though significant offsetting differences existed between the single point mutants (Supplementary Table 4). In particular, the high-affinity V92A mutation showed a 6 °C decrease in Tm with respect to wild-type, which was largely offset by the stabilizing effects of G127R.
Structural basis for high affinity Gas6/Axl binding
To gain insights into the structural basis for the affinity increase we solved the crystal structure of the Gas6/MYD1 co-complex to a 3.1 Å resolution (Fig. 2a, Supplementary Fig. 6). The resulting structure maintained the overall geometry and 2:2 stoichiometry of the wild-type Gas6/Axl co-complex (Cα r.m.s.d. 1.9 Å, Supplementary Table 5)25. Furthermore, the buried surface area of the major binding site remained unchanged, as the wild-type interaction (1,098.5 Å2) was nearly identical to that of the engineered MYD1 interaction (1,121.5 Å2). A detailed comparison of these structures revealed two mechanisms underlying the enhanced affinity: the alteration of Gas6/Axl contacts, and a MYD1-induced conformational change on Gas6 that stabilized the bound structure of the ligand.
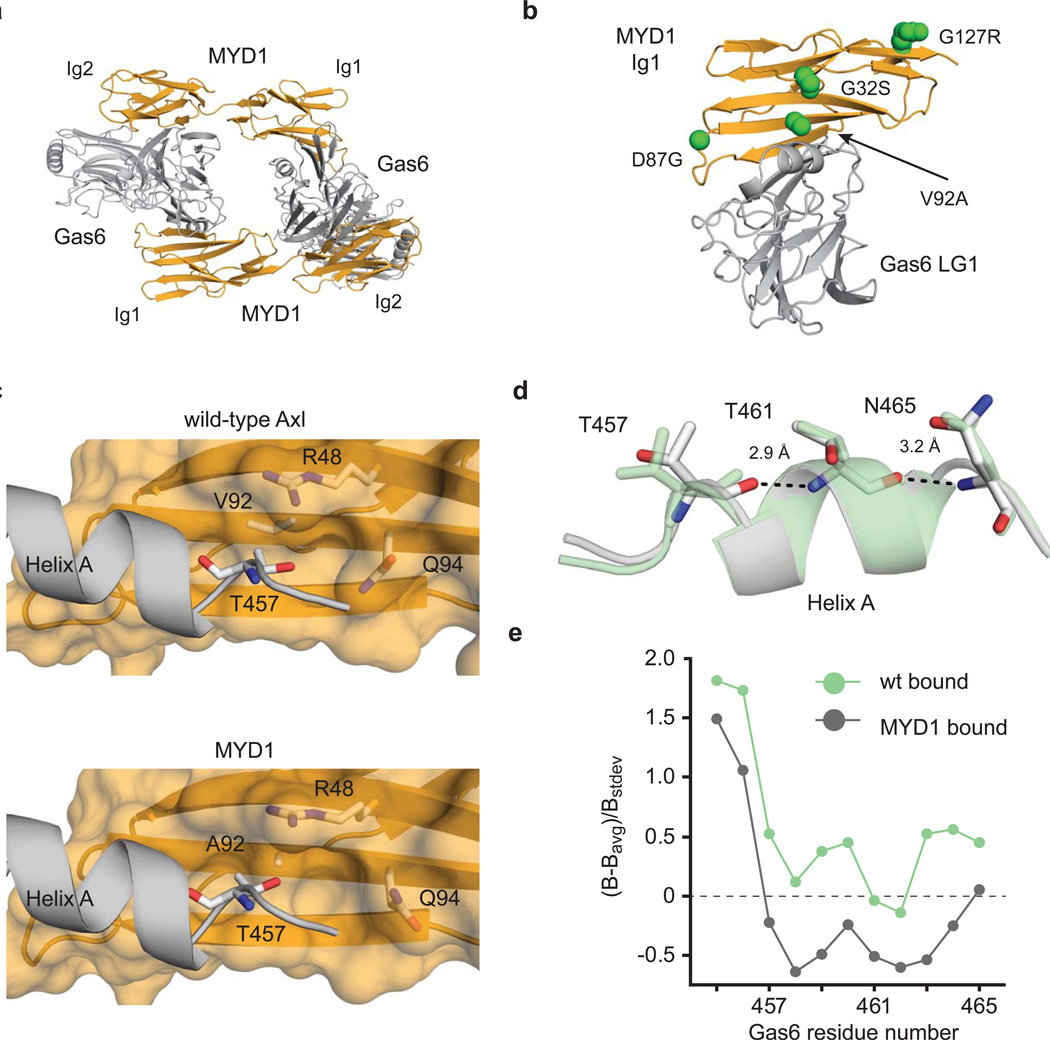
Structural basis for high-affinity binding. (a) Gas6/MYD1 co-complex showing overall architecture and 2:2 stoichiometry. (b) MYD1 Ig1 (orange) and Gas6 LG1 (gray) domains showing the location of the four mutations in MYD1 with respect to the major binding site which lies at the interface of these two domains. (c) Analysis of the wild-type structure (PDB 2C5D) reveals steric crowding between the side chains of T457Gas6 and V92Axl. The V92A mutation alleviates crowding in the MYD1 co-complex and facilitates local reorganization of side chains around V92A, exemplified by R48 and Q94. This in turn creates an elongated groove on MYD1 at the binding interface that allows reorientation of T457 on Gas6. (e) Reorientation of T457 results in capping of the N-terminus of Helix A. The wild-type (green) and MYD1 (gray) structures are overlaid for comparison. (f) Capping stabilizes Helix A, as seen by B-factor analysis (see Methods).
Of the four mutations in MYD1, only V92A lies within the binding epitope (Fig. 2b); however, amino acid side chains throughout the engineered interface underwent significant reorganization (Supplementary Fig. 7), including the emergence of several additional contacts with respect to the wild-type structure (Supplementary Fig. 8, Supplementary Table 6). The structural data supports the conclusion from the biochemical experiments that the V92A mutation is critical for increased binding affinity (Fig. 2c). Specifically, the V92A mutation alleviates steric crowding and permits local structural reorganization, as exemplified by the reordering of Axl residues R48 and Q94. As a result, a groove is created within MYD1 at the engineered binding interface, which in turns likely induces the observed restructuring of an α-helix (Helix A) within the bound state of Gas6 (Fig. 2c, d). In the wild-type co-complex structure, Helix A, which is disordered in the unbound state, is loosely formed and makes minimal contact with Axl (Supplementary Fig. 7)29. In the MYD1 co-complex structure, two complimentary interactions stabilize the helix. Adjacent to position 92, the R48MYD1 side chain rotates, forming a more favorable interaction with E460Gas6 (Supplementary Fig. 8). More importantly, the additional space provided by the new groove permits the reorientation of residues in Helix A, allowing a new backbone hydrogen bond to form between T457Gas6 and T461Gas6 (Fig. 2d). This hydrogen bond is consistent with the commonly observed mechanism of helix stabilization through N-terminal capping30, and together these changes stabilize the bound conformation of Gas6 as reflected by the lower crystallographic B-factors in Helix A relative to the rest of the MYD1 structure, compared to wild-type Axl (Fig. 2e). Moreover, this receptor-induced conformational change represents a unique mechanism for affinity enhancement, as engineered interactions often achieve improved shape complementarity through restructuring of the mutated binding partner31. Such indirect and far reaching structural effects, mediated by a small number of distant mutations, would have been difficult to predict using rational engineering or computational design methods.
Axl Fc fusions exhibit increased Gas6 binding affinity
Increasing the size of a small protein to avoid glomerular filtration can substantially prolong its serum half-life32. Therefore, to facilitate in vivo studies, the Ig1 fragments of wild-type Axl, Axlnb, and MYD1 were cloned back into the full-length receptor and fused to the Fc domain of a human IgG1 (Fig. 3a, Supplementary Fig. 9). Significant improvements in the apparent affinities of the Fc fusions were observed relative to the affinities of the Ig1 constructs, as measured by KinExA (Fig. 3b, Supplementary Fig. 10). This is illustrated by MYD1 Fc, which bound Gas6 with an apparent affinity of 420 fM, a 6-fold increase over MYD1 Ig1, and an 80- fold increase over wild-type Axl Ig1. To establish the mechanism underlying this affinity improvement, a panel of MYD1 Fc variants was created and studied (Fig. 3a). Compared to Axl Ig1, the full-length Fc fusions include the minor Gas6 binding site on Axl Ig2. To interrogate the role of this site, we removed it both through mutagenesis (MYD1−minor Fc; K204E/T208E25) and by truncating the Fc fusion to contain only Axl Ig1 (MYD1 Ig1 Fc). In each case, loss of the minor binding site resulted in Fc fusions that displayed little affinity improvement over MYD1 Ig1 (Fig. 3b). Furthermore, reintroducing an intact minor site was alone insufficient as an Fc fusion containing the Ig1 and Ig2 domains (MYD1 Ig1–2 Fc) similarly failed to attain improved binding. Collectively, these data suggest a heterobivalent binding mechanism in which a molecule of Gas6 interacts with both Axl molecules in the Fc fusion: one binds the major site on Gas6, while the other binds the minor site (Fig. 3c). These interactions only occur when the minor site is functional as well as spatially accessible, and affords an avidity effect that increases the apparent affinity of the overall interaction.

Design and characterization of Axl Fc fusions. (a) Schematic representation of the panel of MYD1 Fc fusions generated. The major and minor Gas6 binding sites are located on Axl’s Ig1 and Ig2 domains, respectively. (b) Apparent binding affinities of Axl Fc fusions to human and mouse Gas6. Raw KinExA data and associated error values can be found in Supplementary Fig. 8. (c) Proposed model of multivalent binding, where both arms of the fulllength Axl Fc fusion contact Gas6. While a 1:1 complex is shown, the same multivalent binding may occur in a 2:1 Gas6:Axl Fc ratio reminiscent of the physiologic active complex.
MYD1 Fc inhibits Axl signaling and sequesters Gas6
To determine whether the Axl decoy receptors could effectively neutralize Gas6 and antagonize Axl signaling, we tested their activity in a cellular context. Skov3.ip human ovarian cancer cells were stimulated with Gas6 in the presence and absence of the decoy receptors, and Axl phosphorylation was measured. Both wild-type Axl Fc and MYD1 Fc efficiently reduced Gas6-mediated Axl phosphorylation, while Axlnb Fc displayed negligible effects, demonstrating the ligand dependent activity of the decoy receptors (Fig. 4a, Supplementary Fig. 11, Supplementary Fig. 12). Inhibition of Axl activation led to a decrease in phosphorylated Akt and Erk1/2 (Fig. 4b), two critical downstream effectors of Axl signaling7. Additionally, modulation of Axl signaling by MYD1 Fc increased expression of the epithelial marker e-cadherin (Fig. 4b), further illustrating the link between Axl and the epithelial-to-mesenchymal transition (EMT)33.
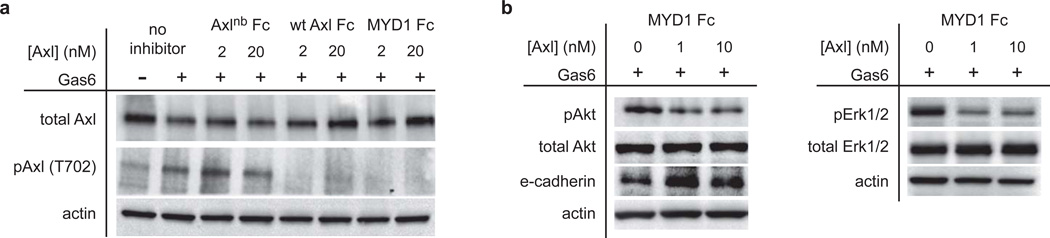
MYD1 Fc inhibits Axl activation and downstream signaling in skov3.ip cells. (a) Wild-type Axl Fc and MYD1 Fc, but not Axlnb Fc, can inhibit Gas6-mediated Axl activation in vitro. (b) Inhibition of Axl activation leads to reduced levels of phosphorylated Akt and Erk1/2, and an increase in the epithelial marker e-cadherin. For full (uncut) blots, see Supplementary Fig. 11.
We next asked whether the decoy receptor’s ability to sequester Gas6 would translate in vivo. Non-tumor bearing mice were given a single dose of MYD1 Fc ranging from 0.1 – 1 mg/kg, and the amount of unbound (i.e. free) serum Gas6 was measured after 12 hours (Fig. 5a). At a dose of 1 mg/kg, a negligible amount of free Gas6 remained in the serum, and a linear dose response was observed with decreasing concentrations of MYD1 Fc. To characterize the kinetics of Gas6 sequestration, mice were administered a 1 mg/kg dose of MYD1 Fc and free Gas6 levels were monitored over time. Rapid and complete elimination of free Gas6 was observed, and after forty-eight hours Gas6 levels approached the untreated baseline (Fig. 5b, black). The pharmacokinetic profile of MYD1 Fc supports these results, as a single dose cleared within 48 hours (Fig. 5b, red). To further study biodistribution, mice were given a single dose of fluorescently labeled MYD1 Fc and monitored using whole body in vivo optical imaging. The imaging experiment showed elimination of MYD1 Fc after 48 hours, qualitatively confirming the pharmacokinetic profile (Supplementary Fig. 13). Predosing mice with unlabeled MYD1 Fc did not significantly alter clearance, indicating the absence of target-mediated disposition.

Sequestration of Gas6 by MYD1 Fc inhibits metastasis (a) Amount of free Gas6 in serum of mice 12 h after administration of a single dose of MYD1 Fc. (b) Kinetics of Gas6 sequestration (black) and MYD1 Fc clearance (red) following a 1 mg/kg dose of MYD1 Fc. (c) Two mice were analyzed for each data point in (b) and (c). Using the off-rates of the Gas6/Axl Fc interactions (Fig. 3b), dissociation of Gas6 bound to either wild-type Axl Fc (red) or MYD1 Fc (blue) is plotted over time. The in vivo clearance of the Axl decoy receptors as measured in (c) is overlaid in black. (d – f) Tumor burden in in vivo models of metastatic human ovarian cancer. The number of visible metastases in animals treated with Axlnb Fc, wild-type Axl Fc, or MYD1 Fc was counted in the skov3.ip (d) and OVCAR (f) tumor models. Representative images of mice from each treatment group in the skov3.ip model are shown, arrows indicate disease (e). In both models, animals were administered 10 mg/kg of the indicated protein twice weekly. (g) Lung metastases in the 4T1 luciferase breast cancer model as quantified by ex vivo bioluminescent imaging. Mice received IV injections of the indicated treatment twice weekly. (h) Representative bioluminescent images of lungs and spleens from each treatment group. Error bars represent ± s.d., n = 6 – 12 mice per group, *P < 0.05; **P < 0.01, ***P < 0.001; ****P < 0.0001.
Taken together, the PK profile and binding kinetics of the Axl Fc’s highlight the functional gains obtained through protein engineering. The half-life of the Gas6/wild-type Axl Fc binding interaction is significantly shorter than the serum half-life of the decoy receptor (Fig. 5c). This can paradoxically lead to increased ligand concentrations as the decoy receptor protects Gas6 from degradation while allowing dissociation prior to complex clearance. In contrast, the half-life of the engineered Gas6/MYD1 Fc interaction is significantly longer than the serum half-life of the Axl Fc, effectively allowing irreversible binding to Gas6 as the complex is cleared from the blood well before dissociation occurs.
MYD1 Fc inhibits metastasis and disease progression
Most Axl antagonists developed to date aim to disrupt the Gas6/Axl signaling axis by interacting with the receptor, leaving sequestration of the ligand an underrepresented approach for therapeutic intervention. MYD1 Fc was used to treat several advanced models of ovarian and breast cancer to test whether systemically neutralizing Gas6 could disrupt pathologic Axl signaling and lead to reductions in metastasis and disease progression. In these models, we compared the activity of wild-type Axl Fc, MYD1 Fc, and Axlnb Fc to determine the relationship between the Gas6 binding affinity of the decoy receptors and their therapeutic efficacy.
In the first model, mice were injected intraperitoneally with skov3.ip human ovarian cancer cells resulting in diffuse, metastatic disease throughout the peritoneal cavity. The model was allowed to establish for one week prior to treatment, at which time the presence of macroscopic metastatic disease was confirmed by visual inspection in a subset of mice. Treatment with MYD1 Fc was highly effective, as metastasis was inhibited by 90% compared to the Axlnb Fc treated controls (Fig. 5d – e). These results correlated with a significant reduction in phosphorylated Axl in tumor tissue (Supplementary Fig. 14). In comparison, treatment with wild-type Axl Fc only modestly reduced metastasis, highlighting a correlation between increased Gas6 binding affinity and improved therapeutic efficacy.
Similar results were observed when the Axl decoy receptors were used to treat a second in vivo model of human ovarian cancer. Using human OVCAR8 cells, the amount of pre-existing disease was substantially increased by implanting five times as many cells as in the skov3.ip model and waiting two weeks prior to treatment. In this advanced model, MYD1 Fc treatment reduced metastases over an order of magnitude compared to control animals (Fig. 5f), while wild-type Axl Fc again demonstrated minimal therapeutic benefit.
An important caveat to the human ovarian cancer models is the presence of two independent pools of Gas6: endogenous mouse Gas6 and autocrine human Gas6 produced by the tumor cells (Supplementary Fig. 12). We show that mouse Gas6 binds wild-type human Axl Fc with higher affinity compared to human Gas6 (Fig. 3b, 1.5 pM vs 9.2 pM, respectively), allowing it to act as a potent stimulatory factor to the human cancer cells. As MYD1 Fc does not possess improved affinity to murine Gas6, the increase in efficacy observed between MYD1 Fc and wild-type Axl Fc is derived from differential binding to the pool of human Gas6, and thus efficacy is likely underestimated in these studies.
Building on these results, we evaluated the ability of MYD1 Fc to inhibit metastatic disease in a murine 4T1 breast cancer model. This model, in which tumors orthotopically implanted in the mammary fat pad spontaneously metastasize to the lungs, faithfully recapitulates metastasis patterns of human disease34. After three weeks of treatment, mice receiving twice weekly doses of 1 or 10 mg/kg of MYD1 Fc had on average 60% and 78% fewer lung metastases, respectively, compared to those treated with Axlnb Fc (Fig. 5g, h). These studies collectively demonstrate that MYD1 Fc antagonizes Axl signaling by specifically neutralizing Gas6, affording significant therapeutic benefits across several models of aggressive metastatic disease. Importantly, these benefits come without significant toxicity as chronic administration of MYD1 Fc was found to be well-tolerated. Non-tumor bearing mice dosed with MYD1 Fc at 1 or 10 mg/kg twice a week for four weeks displayed no adverse events or signs of toxicity (Supplemental Fig. 14, Supplementary Table 7).
Discussion
Advancements in early detection and the emergence of targeted therapies have significantly improved the management of localized cancers; however, effective treatment options for the most insidious forms of the disease, those characterized by therapeutic resistance and metastasis, remain elusive35. Recently, the signaling axis formed by the Axl receptor tyrosine kinase and its ligand, Gas6, has been shown to play a causal role in the development of advanced disease, highlighting new opportunities for therapeutic intervention. Interest in Axl as a therapeutic target has been further fueled by transgenic animal studies, as both Gas6−/− and Axl−/− knockout mice are viable, fertile, and display no significant debilitating pathologies, suggesting that specific modulation of the Gas6/Axl signaling pathway carries a low risk of toxicity to complement its therapeutic potential36,37.
A number of strategies have been pursued to develop Axl antagonists, including using antibodies, aptamers, and small molecules to block signaling through the receptor. Despite these efforts, current anti-Axl therapeutics either exhibit modest anti-tumor efficacy or induce significant off-target effects11–15,38. As our current study shows, the unusually high picomolar binding affinity of the native Gas6/Axl interaction has made the development of competitive antagonists particularly challenging. Moreover, homology between the kinase domains of Axl and other receptors confers promiscuity among small molecule inhibitors and associated dose limiting toxicity.
To address these limitations, we engineered receptor-based antagonists that leverage the innate affinity and specificity of the native interaction to effectively sequester Gas6. Using combinatorial protein engineering, an Axl-based decoy receptor was developed that bound to Gas6 with an apparent affinity of 420 fM, as measured by KinExA. The KinExA is uniquely suited for quantifying high-affinity interactions, as it measures the concentration of free (i.e. unbound) ligand in a binding reaction equilibrated prior to instrument analysis26. This circumvents artifacts derived from ligand depletion, binding partner immobilization, and mass transport effects that can be significant sources of error in other techniques such as surface plasmon resonance. KinExA also allows fractional ligand activity to be calculated thus correcting for errors associated with protein quantification. This latter point is significant as we found Gas6 to be highly unstable, and preparations of commercial Gas6 to be largely inactive. Compared to our measurement of 33 pM, literature values of the native Gas6/Axl interaction measured by alternative methods are consistently reported to be in the nanomolar range. This underestimation of the tight binding affinity between Gas6 and Axl provides an explanation for the poor therapeutic efficacy observed with previously developed competitive antagonists with nanomolar binding affinities.
The increased Gas6 binding affinity of our most improved Axl variant, MYD1, is the result of altered molecular interactions mediated by its four mutations, with D87G and V92A largely driving the affinity increase. D87G and V92A appeared multiple times in clones sequenced from earlier library screening rounds, including several times as single point mutants, further demonstrating that they are independent and beneficial. G32S and G127R only appear in MYD1, a feature that highlights the fact that their contributions to the affinity improvement are highly context dependent. Further improvements in Gas6 affinity were obtained when MYD1 was reformatted as a full-length Fc fusion, by leveraging a mechanism analogous to cytokine “traps”, which rely on heterobivalent binding to achieve ultra-high affinity22.
In an interesting parallel to convergent evolution, Nature appears to mediate orthologous high affinity Gas6/Axl interactions through a similar molecular mechanism that instead relies on alterations to Gas6. In particular, we showed that MYD1 Fc bound human Gas6 significantly better than wild-type Axl Fc; however, both bound mouse Gas6 at equivalent levels (Fig. 3b). Sequence analysis reveals that the murine and human forms of Gas6 have alanine and threonine at residue 457, respectively, while V92Axl is conserved. As previously noted, the side chains of T457Gas6 and V92Axl cause steric crowding in the human complex, and the V92A mutation in MYD1 alleviates this issue to improve the affinity. In the murine interaction a similar mechanism seems to have evolved, but in this case through the removal of the offending side chain on Gas6 (A457Gas6). It is likely that the mutation to A457Gas6 is functionally equivalent to the V92AAxl mutation found in our study, and while definitive conclusions regarding the mouse interaction cannot be made without directly measuring it, this offers an explanation for why MYD1’s improved binding affinity is not translated across species.
The affinity improvements gained from rational and combinatorial protein engineering efforts enabled efficient disruption of Axl signaling in a cellular context. While both wild-type Axl Fc and MYD1 Fc inhibited Axl activation in vitro, the non-binding Axlnb Fc had no affect with respect to controls. These results demonstrate that the decoy receptors specifically act through Gas6 sequestration, and that ligand-mediated Axl signaling drives receptor activation in our experiments. This latter point has been the focus of some ambiguity, as recent reports suggest that pathologic Axl signaling is, in part, a consequence of ligand-independent mechanisms20. As our decoy receptors are the first molecules that effectively disrupt the signaling axis by specifically targeting the ligand, this hypothesis can now be directly tested. Upon administration, systemic levels of free Gas6 are reduced to near zero, and these levels stay depressed for up to 48 hours. While it cannot be ruled out that in some cell lines Axl signaling is decoupled from Gas6, in the three mouse models tested the decoy receptors significantly reduce tumor burden and metastasis, reaffirming that ligand-dependent Axl signaling drives disease progression in these systems. Furthermore, these effects were directly correlated with affinity, establishing that the efficacy of a decoy receptor is directly tied to its affinity for the target ligand.
In conclusion, we demonstrate that Gas6 sequestration is a viable method for effective disruption of Axl signaling. This strategy, which moves beyond directly targeting the Axl receptor, offers a more judicious approach for therapeutic intervention. Importantly, the high affinity engineered decoy receptor described here further validates Axl as a therapeutic target and creates a path for its potential to be realized.
ONLINE METHODS
Synthesis of yeast-displayed Axl library
DNA encoding human Axl Ig1, amino acids Ala19 – Pro131, was cloned into the pCT yeast display plasmid using NheI and BamHI restriction sites39. Sequence numbering was done to match that used in Sasaki et al25 to facilitate comparisons to their work with the wild-type proteins. An error-prone library was created using the Axl Ig1 DNA as a template and mutations were introduced by using low-fidelity Taq polymerase (Invitrogen) and the nucleotide analogs 8-oxo-dGTP and dPTP (TriLink Biotech). Six separate PCR reactions were performed in which the concentration of analogs and the number of cycles were varied to obtain a range of mutation frequencies; five cycles (200 µM), ten cycles (2, 20, or 200 µM), and 20 cycles (2 or 20 µM). Products from these reactions were amplified using forward and reverse primers each with 50 bp homology to the pCT plasmid in the absence of nucleotide analogs. Amplified DNA was purified using gel electrophoresis and pCT plasmid was digested with NheI and BamHI. Purified mutant cDNA and linearized plasmid were electroporated in a 5:1 ratio by weight into EBY100 yeast where they were assembled in vivo through homologous recombination40. Library size was estimated to be 7.4×107 by dilution plating.
Library screening
Yeast displaying high affinity Axl variants were isolated from the library using fluorescence-activated cell sorting (FACS). For FACS rounds 1 – 3, equilibrium binding sorts were performed in which yeast were incubated at room temperature in phosphate buffered saline with 1 mg/ml BSA (PBSA) with the following nominal concentrations of Gas6 (R&D Systems): sort 1) 10 nM Gas6 for 3 h; sort 2) 2 nM Gas6 for 3 h; sort 3) 0.2 nM Gas6 for 24 h. After incubation with Gas6, yeast were pelleted, washed, and resuspended in PBSA with 1:250 of chicken anti-c-Myc (Invitrogen) for 1 h at 4 °C. Yeast were then washed, pelleted and secondary labeling was performed for on ice for 30 min using PBSA with a 1:100 dilution of goat anti-chicken Alexa Fluor 555 (Invitrogen) and mouse anti-HIS Hilyte Fluor 488 (Anaspec).
For FACS rounds 4 – 6, kinetic off-rate sorts were conducted in which yeast were incubated with 2 nM Gas6 for 3 hours at room temperature, after which cells were washed twice to remove excess unbound Gas6, and resuspended in PBSA containing a ~50 fold molar excess of Axl Fc (R&D Systems) to render unbinding events irreversible. The length of the unbinding step was as follows: sort 4) 4 h; sort 5) 4 h; sort 6) 24 h, with all unbinding reactions performed at room temperature. During the last hour of the dissociation reaction, chicken anti-c-Myc was added to a final dilution of 1:250. Yeast were pelleted, washed, and secondary labeling was performed as previously described. Labeled yeast were sorted by FACS using a Vantage SE flow cytometer (Stanford FACS Core Facility) and CellQuest software (Becton Dickinson). Sorts were conducted such that the 1–3% of clones with the highest Gas6 binding/c-Myc expression ratio were selected, enriching the library for clones with the highest binding affinity to Gas6. In sort 1, 108 cells were screened and subsequent rounds analyzed a minimum of ten-fold the number of clones collected in the prior sort round to ensure adequate sampling of the library diversity. Selected clones were propagated and subjected to further rounds of FACS. Following sorts 5 and 6, plasmid DNA was recovered using a Zymoprep kit (Zymo Research Corp.), transformed into XL-1 blue supercompetent cells, and isolated using plasmid miniprep kit (Qiagen). Sequencing was performed by Sequetech Corp.
Analysis of yeast-displayed sort products was performed using the same reagents and protocols and described for the library sorts. Samples were analyzed on a FACSCalibur (BD Biosciences) and data was analyzed using FlowJo software (Treestar Inc.).
Recombinant protein production
Axl Ig1 variants were cloned into the pPIC9K plasmid (Invitrogen) with N- and C-terminal FLAG and 6xHIS tags, respectively. Plasmid DNA was transformed into the yeast P.pastoris GS115 strain according to the manufacturer’s protocol and proteins were purified from culture supernatant using nickel affinity chromatography. N-linked glycosylations were removed using endoglycosidase H (EndoHf, New England Biolabs) and monomeric Axl Ig1 was further purified using size exclusion chromatography.
The Gas6 and Axl constructs used for crystallography studies were produced as previously described25,29. Axl Fc fusions were obtained by cloning the cDNA encoding the signal peptide and extracellular domain (amino acids 1 – 440) of Axl into the cytomegalovirus-driven pAdd2 adenoviral shuttle vector directly upstream of the gene encoding a human IgG1 Fc domain. Human embryonic kidney (HEK) cells were transiently transfected with pAdd2 plasmid DNA using the FreeStyle Max 293 Expression System (Invitrogen). Axl Fc fusions were purified from culture supernatant using protein A affinity chromatography and disulfide-linked dimers were isolated by size exclusion chromatography.
Kinetic Exclusion Assay (KinExA)
A KinExA 3200 instrument (Sapidyne Instruments Inc.) was used to measure equilibrium binding and association rate constants for the Gas6/Axl interactions. For all experiments, MYD1 Fc coated polymethyl methacrylate (PMMA) beads were used as a capture reagent. Adsorption coating of 200 mg of beads was performed using 45 µg of protein at room temperature for 1 h. The protein solution was then removed and beads were blocked using PBS with 1% BSA for 2 h at room temperature. Beads were stored in blocking buffer at 4 °C and used within two weeks. All binding reactions were carried out in PBS with 1 mg/ml BSA and 0.02% sodium azide.
For equilibrium binding studies, three independent titrations were completed for each Axl Ig1 or Axl Fc fusion. Depending on the affinity of interaction, three of the following four titrations were performed: 1) 1 ml reactions of 5 nM Gas6 (nominal), Axl serially diluted 1:2 twelve times starting at 30 nM, 3 h RT incubation; 2) 2 ml reactions of 500 pM Gas6, Axl serially diluted 1:2.5 twelve times starting at 10 nM, 18 h RT incubation; 3) 12 ml reactions of 50 pM Gas6, Axl serially diluted 1:3 twelve times starting at 9 nM, 1 day RT incubation; 4) 12 ml reactions of 15 pM Gas6, Axl serially diluted 1:3 twelve times starting at 1 nM, 4 day RT incubation. After the appropriate incubation time, reactions were flowed over MYD1 Fc coated beads and captured free Gas6 was probed using an anti-6xHIS Dylight 649 antibody (Rockland Immunochemicals Inc.). Each sample was measured twice and data from the three independent equilibrium binding experiments were globally analyzed using n-curve analysis in the KinExA Pro 3.6.2 software (Sapidyne Instruments Inc.) to obtain the Kd value.
To analyze the association rate of the interactions, the direct inject method was used. For these experiments, 1 µM Axl was serially diluted 1:2.5, and equal volumes of each Axl sample and 500 nM Gas6 (nominal) were briefly mixed and flowed over the capture beads. Free Gas6 was detected as described and the data was fit using the KinExA Pro 3.6.2 software to obtain the association rate (kon) of the interaction. The dissociation rate (koff) was calculated as the product of the Kd and the kon.
To test binding of Protein S (OriGene) to MYD1, 700 µl of a 10 nM solution was flowed over the capture beads. Binding of Protein S to the beads was probed using an anti-c-Myc Dylight 649 antibody (Rockland Immunochemicals Inc, 200-343-382).
Circular dichroism spectroscopy
To measure the thermal stabilities of the Axl Ig1 variants, temperature melts were performed using a Jasco J-815 circular dichroism spectropolarimeter. Recombinant protein was diluted to 10 µM in PBS and ellipticity was monitored at 206 nm as temperature was increased from 20 – 80 °C at a rate of 1 °C/min. The resulting temperature melt was fit to a two-state unfolding curve in order to calculate the Tm.
Crystallization and data collection for Gas6/MYD1 co-complex
Purified wild-type Gas6 and MYD1 Ig1–2 were mixed in a 1:1 molar ratio and allowed to complex at room temperature for 24 h. The co-complexes were purified using size exclusion chromatography to remove any unreacted components, and were buffer exchanged into 25 mM Na-HEPES, 150 mM NaCl, and 1 mM calcium acetate to a final concentration of 10 mg/ml. Crystals were grown at room temperature by the hanging-drop vapor-diffusion method with a 1:1 mixture (1.2 µl each) of the complex solution (6.2 mg/ml) and the well solution containing 0.4 M Li2SO4, 0.1 M Tris-HCl (pH 8.5), 5% glycerol, and 2 mM Ni2SO4. For cryocooling, the crystals were dipped in a solution containing 38 parts of 1M Li2SO4, 0.1 M Tris-HCl (pH 8.5), 0.1 M NaCl and 12 parts of 100% glycerol. Diffraction data sets for the Gas6/MYD1 co-complex (3.1Å) were collected at 100 K using Stanford Synchrotron Radiation Lightsource (SSRL) beamline 7-1 at a wavelength of 1.03 Å. Data were indexed and integrated using the XDS package41. The crystals belong to space group P3221 and contain one 2:2 complex per asymmetric unit. The crystallographic data are summarized in Supplementary Table 5. Several attempts to improve the resolution by using various additives, crystal dehydration experiments, and changes in the cryocooling procedure were unsuccessful.
Structure determination and refinement
Initial phases were obtained by molecular replacement by using the program MOLREP42 and the coordinates of the wild type Gas6/Axl Ig1–2 crystal structure (PDB ID: 2C5D) as the search model. Density corresponding to the loop on Axl from residue 541 to 550 was visible for chain A. This loop is missing in the wild-type structure. After several cycles of manual model building using COOT43 and refinement using REFMAC44, the final R(working) and Rfree values were 20.2% and 24.3%, respectively. The Ramachandran statistics were as follows: 93.19% (favored) and 0.97% (outlier). The refinement statistics are provided in Supplementary Table 5. All crystal structure figures were created using PyMOL45.
Analysis of intermolecular contacts
The binding interface in the Gas6/MYD1 co-complex was analyzed using the PDBePISA server46 v1.48 [24/10/2013] (http://www.ebi.ac.uk/pdbe/prot_int/pistart.html) utilizing a cutoff of 3.5 Å and 4.0 Å for hydrogen bonds and electrostatic interactions, respectively. Only those contacts that were present in both chains were considered in the analysis. The results are shown in Supplementary Table 6. Normalized B-factors shown in Fig. 2f were calculated as described previously47. For both contact and B-factor analysis the wild-type Gas6/Axl complex (PDB ID: 2C5D) was refined in a manner identical to the Gas6/MYD1 complex to facilitate comparison.
In vitro cell growth analysis
Skov3.ip and 4T1 cells were grown in RPMI media supplemented with 10% FBS. Cells were trypsonized and seeded at an initial density of 200 or 500 cells per well in a 96-well microtiter plate. Cells were grown either in complete media, or complete media supplemented with 100 nM wild-type Axl Fc or MYD1 Fc. Media was changed every other day for the duration of the experiment. Cell number was assessed every other day using the XTT Cell Proliferation Assay Kit (Trevigen).
Analysis of in vivo Gas6 sequestration by Axl Fc fusions
For all in vivo studies, six week old, female nude (nu/nu) mice (Jackson Laboratory) were used. All procedures involving animals and their care and use were approved by the Institutional Animal Care and Usage Committee of Stanford University.
For the dose response studies, mice were administered a single dose of MYD1 Fc ranging from 0.1 – 1 mg/kg via tail vein injection. All doses were formulated in a 200 µl volume. Two mice were analyzed per condition and untreated mice were used to determine baseline Gas6 levels. Twelve hours post-injection, retro-orbital bleeds were performed to obtain blood samples from which serum was isolated. The amount of free Gas6 in each sample was determined using a sandwich ELISA. In this assay, MYD1 Fc was used as a capture reagent in order to ensure the detection of free, unbound Gas6, and not Gas6/Axl Fc complexes. Detection of Gas6 was carried out using a biotinylated polyclonal anti-mouse Gas6 antibody (R&D Systems, BAF986) and streptavidin HRP (Trevigen Inc, 4800-30-06). All ELISAs were developed using the 1-Step Ultra TMB ELISA (Pierce).
For time course studies, the protocols were performed as described above except that all mice received a single dose (1 mg/kg) of Axl Fc. Blood collection was done at 2, 12, 24, 36 and 48 h post-administration and free Gas6 was measured as described.
To measure the pharmacokinetics of the Axl Fc’s, the amount of MYD1 Fc was quantified in serum samples from the time course studies. An anti-human IgG (Abcam, Ab7500) was used as the capture, and MYD1 Fc was detected using biotinylated anti-human Axl (R&D Systems, BAF154) and streptavidin HRP (Trevigen Inc.).
Alexa Fluor 680 labeling of Axl Fc fusions
Fluorescent labeling of MYD1 Fc fusions was done using the NHS ester Alexa Fluor 680 (Invitrogen). 10 nmoles of MYD1 Fc was diluted into 400 µl of 100 mM sodium bicarbonate buffer pH 8.1. 100 nmoles of Alexa Fluor 680 NHS ester was added dropwise to the protein solution while mixing. The reaction was left stirring for 1 h at room temperature after which it was allowed to proceed overnight at 4 °C. Free unreacted dye was removed using a PD10 column (GE Healthcare) and eluted conjugated protein was buffer exchanged into 1×PBS. Labeling resulted in 1.7 dye molecules per protein.
Near infra-red whole body optical imaging
200 µg of Alexa Fluor 680 conjugated MYD1 Fc was administered to mice via tail vein injection and whole body near-infrared optical imaging was performed using an IVIS 200 In vivo imaging system (PerkinElmer Inc). The Alexa 680 was excited at 615 – 665 nm and emission was collected at 695 – 770 nm. Resulting images were processed using the Living Image software v4.3.1 (Caliper Life Sciences). Animals predosed with unlabeled MYD1 Fc received a 10 mg/kg dose on each of the two days preceding the imaging study.
In vivo tumor models
To generate diffuse metastatic disease in the skov3.ip ovarian cancer model10, 1×106 cells (kindly provided by Dr. Gordon Mills, M.D. Anderson Cancer Center) were injected IP and allowed to establish for one week prior to dosing. Five days after tumor inoculation, two animals were sacrificed at random and necropsies were performed to confirm the presence of macroscopic metastatic disease. Seven days post-tumor inoculation, Axl Fc fusions were administered at 10 mg/kg twice a week via IP injections for three weeks. Animals were sacrificed on day 28 and metastatic burden was assessed by counting the number of visible metastatic lesions in the peritoneal cavity. The OVCAR810 study was conducted in a similar manner with two notable exceptions: 5×106 cells were used to seed the model and two weeks were waited to allow establishment prior to dosing.
To establish orthotopic mammary tumors, 5×104 4T1 luciferase cells34 (kindly provided by Dr. Marta Vilalta, Stanford University) were injected in 50 µl of DMEM into the 4th inguinal mammary fat pad. Tumor establishment was confirmed using an IVIS 200 3 days post-tumor inoculation. Treatment began 4 days post-inoculation and consisted 1 or 10 mg/kg dose of Axl Fc administered intravenously (IV) via tail vein injection in a 200 µl volume. Treatment was administered twice a week for 3 weeks, for a total of 6 doses. On day 24, mice were sacrificed 10 minutes after receiving a single intraperitoneal (IP) injection of D-luciferin. Ex-vivo bioluminescent imaging of the lungs and spleen was performed using an IVIS 200 and images were analyzed in the Living Image software v4.3.1 to quantify lung metastases.
Toxicology study of MYD1 Fc
Six week old non-diseased mice were injected IV via tail vein with a 10 mg/kg dose of MYD1 Fc twice a week for four weeks. Animal weight was measured over the course of the study. After 28 days, mice were sacrificed and complete blood counts and chemistry panels were performed (Stanford Veterinary Service Center). Tissue samples from the lungs, liver, and kidneys were harvested and stained with hematoxylin and eosin (H&E) for histological analysis of toxicity.
Immunoblotting
Cell lysates subjected to sodium dodecyl sulfate polyacrylamide gel electrophoresis followed by transfer to nitrocellulose membrane. The membranes were then probed with primary antibodies against total Axl (R&D system, AF154), phospho-Axl (T702; Cell Signaling #5724), total Akt (Cell Signaling #4691), phosphor-Akt (Cell Signaling #4060), e-cadherin (BD #610404), total Erk1/2 (Cell Signaling #9102), or phosphor-Erk1/2 (Cell Signaling #9100) at 4 °C overnight. The blots were then washed and probed with HRP conjugated anti-goat (Santa Cruz Biotechnology) or HRP conjugated anti-rabbit (Pierce, Rockford, IL) as appropriate. The blots were developed with Bio-Rad western C developing reagent (Bio-Rad, Hercules, CA) and visualized with Chemidoc digital imager (Bio-Rad, Hercules, CA).
To detect expression of Gas6 in vitro, both conditioned serum free media and cell lysate were collected from skov3.ip and OVCAR8 cultures. Samples of each were analyzed as described and blots were probed with a biotinylated polyclonal anti-human Gas6 primary antibody (R&D Systems, BAF885) for two hours at room temperature. Blots were washed and secondary labeling was performed using a streptavidin-HRP conjugate (Trevigen Inc, 4800-30-06) for one hour at room temperature. Blots were developed and imaged as described.
Immunohistochemistry
Immunohistochemical staining of Axl and phosphorylated Axl was performed on tumor tissue samples. Briefly, tumor sections were subjected to antigen retrieval using 10 mM citric acid and were probed with primary anti-Axl antibody (R&D Systems) or anti-pAxl antibody (R&D Systems) overnight at 4 °C followed by secondary detection using streptavidin-HRP.
Statistical analysis
Differences between groups in all in vivo experiments were examined for statistical significance using a two-tailed Student’s t-test. A P value < 0.05 was considered significant.
ACKNOWLEDGEMENTS
The authors thank Dr. Erhard Hohenester for his generous gift of the Gas6 plasmid and helpful discussions regarding protein production, Luo Luo Zheng for technical assistance with protein production and purification, and the Stanford FACS Core Facility for assistance with the flow cytometric sorting. All in vivo and ex vivo imaging was conducted in the Stanford Small Animal Imaging Facility. Portions of this research were performed at the Stanford Synchrotron Radiation Laboratory, a national user facility operated by Stanford University on behalf of the U.S. Department of Energy Office of Basic Energy Sciences. This work was supported by the Wallace H. Coulter Translational Research Grant Program (J.R.C and A.J.G.), Stanford Bio-X and ARCS Graduate Fellowships (M.S.K.), NIH CA-088480 and NIH CA-67166 (A.J.G and Y.M.), and NIH training grant T32 GM008412-15S1 and Siebel Graduate Fellowship (D.S.J.).
Footnotes
AUTHOR CONTRIBUTIONS: M.S.K., Y.R.M., D.S.J., S.K., I.I.M., A.J.G., and J.R.C. designed the research; M.S.K. and D.S.J. performed the protein engineering and characterization; M.S.K., S.K., and I.I.M. did the crystallography; M.S.K. and Y.R.M. conducted the in vivo experiments; all authors analyzed the data; M.S.K. prepared the manuscript with input from all co-authors.
Accession Codes
Protein Data Bank (PDB). Atomic coordinates and structure factors for the Gas6/MYD1 co-complex have been deposited under accession codes XXXXX.
COMPETING FINANCIAL INTERESTS
Stanford University holds a patent "Biologic inhibitors for therapeutic targeting the receptor tyrosine kinase AXL" US8618254 B2, which is related to the work described in this paper, with M.S.K, Y.R.M, D.S.J., A.J.G., and J.R.C. named as inventors. A.J.G. is a founder of Ruga, a company that discovers and develops targeted therapeutics in oncology that has licensed this patent.
REFERENCES
Full text links
Read article at publisher's site: https://doi.org/10.1038/nchembio.1636
Read article for free, from open access legal sources, via Unpaywall:
https://europepmc.org/articles/pmc4372605?pdf=render
Citations & impact
Impact metrics
Citations of article over time
Alternative metrics
Smart citations by scite.ai
Explore citation contexts and check if this article has been
supported or disputed.
https://scite.ai/reports/10.1038/nchembio.1636
Article citations
Clinical functional proteomics of intercellular signalling in pancreatic cancer.
Nature, 13 Nov 2024
Cited by: 0 articles | PMID: 39537929
AXL as immune regulator and therapeutic target in Acute Myeloid Leukemia: from current progress to novel strategies.
Exp Hematol Oncol, 13(1):99, 04 Oct 2024
Cited by: 0 articles | PMID: 39367387 | PMCID: PMC11453060
Review Free full text in Europe PMC
MAGPIE: An interactive tool for visualizing and analyzing protein-ligand interactions.
Protein Sci, 33(8):e5027, 01 Aug 2024
Cited by: 1 article | PMID: 38989559
MERTK Inhibition as a Targeted Novel Cancer Therapy.
Int J Mol Sci, 25(14):7660, 12 Jul 2024
Cited by: 0 articles | PMID: 39062902 | PMCID: PMC11277220
Review Free full text in Europe PMC
Role and Mechanisms of Tyro3 in Podocyte Biology and Glomerular Disease.
Kidney Dis (Basel), 10(5):398-406, 24 Jul 2024
Cited by: 0 articles | PMID: 39430290 | PMCID: PMC11488836
Review Free full text in Europe PMC
Go to all (88) article citations
Other citations
Data
Data behind the article
This data has been text mined from the article, or deposited into data resources.
BioStudies: supplemental material and supporting data
Protein structures in PDBe
-
(3 citations)
PDBe - 2C5DView structure
Similar Articles
To arrive at the top five similar articles we use a word-weighted algorithm to compare words from the Title and Abstract of each citation.
Inhibition of the GAS6/AXL pathway augments the efficacy of chemotherapies.
J Clin Invest, 127(1):183-198, 28 Nov 2016
Cited by: 62 articles | PMID: 27893463 | PMCID: PMC5199716
Axl/Gas6 pathway positively regulates FLT3 activation in human natural killer cell development.
Eur J Immunol, 43(10):2750-2755, 08 Jul 2013
Cited by: 17 articles | PMID: 23722894 | PMCID: PMC3829002
In Silico Approach to Identify Potential Inhibitors for Axl-Gas6 Signaling.
Methods Mol Biol, 1549:221-229, 01 Jan 2017
Cited by: 0 articles | PMID: 27975295
Therapeutic Targeting of the Gas6/Axl Signaling Pathway in Cancer.
Int J Mol Sci, 22(18):9953, 15 Sep 2021
Cited by: 28 articles | PMID: 34576116 | PMCID: PMC8469858
Review Free full text in Europe PMC
Funding
Funders who supported this work.
NCI NIH HHS (6)
Grant ID: P30 CA124435
Grant ID: CA-67166
Grant ID: P01 CA067166
Grant ID: R01 CA088480
Grant ID: CA-088480
Grant ID: R37 CA088480
NIGMS NIH HHS (1)
Grant ID: T32 GM008412





