Abstract
Free full text

Epigenetic programming during monocyte to macrophage differentiation and trained innate immunity
Structured Abstract
Introduction
Monocytes circulate in the bloodstream for up to 3–5 days. Concomitantly, immunological imprinting of either tolerance (immunosuppression) or trained immunity (innate immune memory) determines the functional fate of monocytes and monocyte-derived macrophages, as observed after infection or vaccination.
Methods
Purified circulating monocytes from healthy volunteers were differentiated under the homeostatic M-CSF concentrations present in human serum. During the first 24 hours, trained immunity was induced by β-glucan (BG) priming, while post-sepsis immunoparalysis was mimicked by exposure to LPS, generating endotoxin-induced tolerance. Epigenomic profiling of the histone marks H3K4me1, H3K4me3 and H3K27ac, DNase I accessibility and RNA sequencing were performed at both the start of the experiment (ex vivo monocytes) and at the end of the six days of in vitro culture (macrophages).
Results
Compared to monocytes (Mo), naïve macrophages (Mf) display a remodeled metabolic enzyme repertoire and attenuated innate inflammatory pathways; most likely necessary to generate functional tissue macrophages. Epigenetic profiling uncovered ~8000 dynamic regions associated with ~11000 DNase I hypersensitive sites. Changes in histone acetylation identified most dynamic events. Furthermore, these regions of differential histone marks displayed some degree of DNase I accessibility that was already present in monocytes. H3K4me1 mark increased in parallel with de novo H3K27ac deposition at distal regulatory regions; H3K4me1 mark remained even after the loss of H3K27ac, marking decommissioned regulatory elements. β-glucan priming specifically induced ~3000 distal regulatory elements, whereas LPS-tolerization uniquely induced H3K27ac at ~500 distal regulatory regions.
At the transcriptional level, we identified co-regulated gene modules during monocyte to macrophage differentiation, as well as discordant modules between trained and tolerized cells. These indicate that training likely involves an increased expression of modules expressed in naïve macrophages, including genes that code for metabolic enzymes. On the other hand, endotoxin tolerance involves gene modules that are more active in monocytes than in naïve macrophages. About 12% of known human transcription factors display variation in expression during macrophage differentiation, training and tolerance. We also observed transcription factor motifs in DNase I hypersensitive sites at condition-specific dynamic epigenomic regions, implying that specific transcription factors are required for trained and tolerized macrophage epigenetic and transcriptional programs. Finally, our analyses and functional validation indicate that the inhibition of cAMP generation blocked trained immunity in vitro and during an in vivo model of lethal C. albicans infection, abolishing the protective effects of trained immunity.
Discussion
We documented the importance of epigenetic regulation of the immunological pathways underlying monocyte-to-macrophage differentiation and trained immunity. These dynamic epigenetic elements may inform on potential pharmacological targets that modulate innate immunity. Altogether, we uncovered the epigenetic and transcriptional programs of monocyte differentiation to macrophages that distinguish tolerant and trained macrophage phenotypes, providing a resource to further understand and manipulate immune-mediated responses.
Introduction
Monocytes are generally considered an intermediate developmental stage between bone marrow precursors and tissue macrophages (1). However, circulating monocytes have important effector functions, both during homeostasis through patrol and repair functions (2), as well as during infections by exerting inflammatory effects (3). Although several populations of tissue resident macrophage originate from yolk sack embryonic precursors (4), tissues such as dermis and the intestine contain adult monocyte-derived macrophages (5–7), while during infections, adult blood inflammatory monocytes migrate to inflamed tissues and differentiate into monocyte-derived macrophage populations that can eliminate the pathogen and restore tissue integrity (8).
In homeostasis or during infections, monocytes can follow different functional programs. During the process of ‘tolerance/immunoparalysis’, a common complication of severe sepsis, monocytes enter a refractory functional state, characterized by the incapacity to produce proinflammatory cytokines and decreased HLA-DR expression (9). Tolerance can be mimicked in vitro and in vivo by challenging the cells with endotoxins such as LPS: after an initial stimulation phase, cells enter a state of long-term immunotolerance. In contrast, other infections or vaccinations (e.g. Candida albicans infection, Bacille Calmette-Guerin (BCG) or measles vaccination) increase the long-term responsiveness of monocytes to microbial stimuli, a state termed ‘trained immunity’ which confers resistance to secondary infections (10–14). In the case of Candida infection, training requires the β-glucan receptor Dectin-1 and the non-canonical Raf-1 pathway, and is associated with stable changes in histone trimethylation at H3K4 at a small subset of promoters (10). Tolerance and training represent clinically relevant functional states such as the immune paralysis encountered during bacterial sepsis or endotoxin shock (9) or the non-specific protective effects of live microorganism vaccination (e.g. BCG, measles, yellow fever, (15)) and strongly influence susceptibility to secondary infections.
Histone and DNA modifications have been hypothesized to play a role in regulating monocyte and macrophage phenotypes (16), but data are limited to a few genes and histone modifications (17). Moreover, the epigenetic modifications after LPS stimulation were studied for relatively short periods of up to one day (18). In addition, the two major events that determine the functional fate of monocytes and macrophages, immune tolerance (19) and innate immune training (10), have not been thoroughly characterized at the transcriptional and epigenomic level. The BLUEPRINT consortium aims at defining the epigenomic maps and transcription profiles of a wide variety of blood cells from healthy primary human cells as well as blood-based diseases (20, 21) (www.blueprint-epigenome.eu). Here we report on the epigenome and transcriptome of monocyte to macrophage differentiation and the response to tolerance and training.
Monocyte differentiation in response to external stimuli
Upon migration into the tissues, monocytes differentiate into tissue macrophages. In addition, under differential inflammatory conditions, monocytes can be directed into tolerance or trained immunity functional programs (Fig. 1A). To analyze the different functional programs of monocytes and macrophages, human circulating monocytes (Mo) were purified by triple depletion of T- and B-lymphocytes, NK-cells from the peripheral blood mononuclear cells of healthy human volunteers (Fig. S1). FACS and transcriptome analyses revealed high purity monocytes similar to cell surface markers of CD14+ positively selected monocytes (Fig. S1).
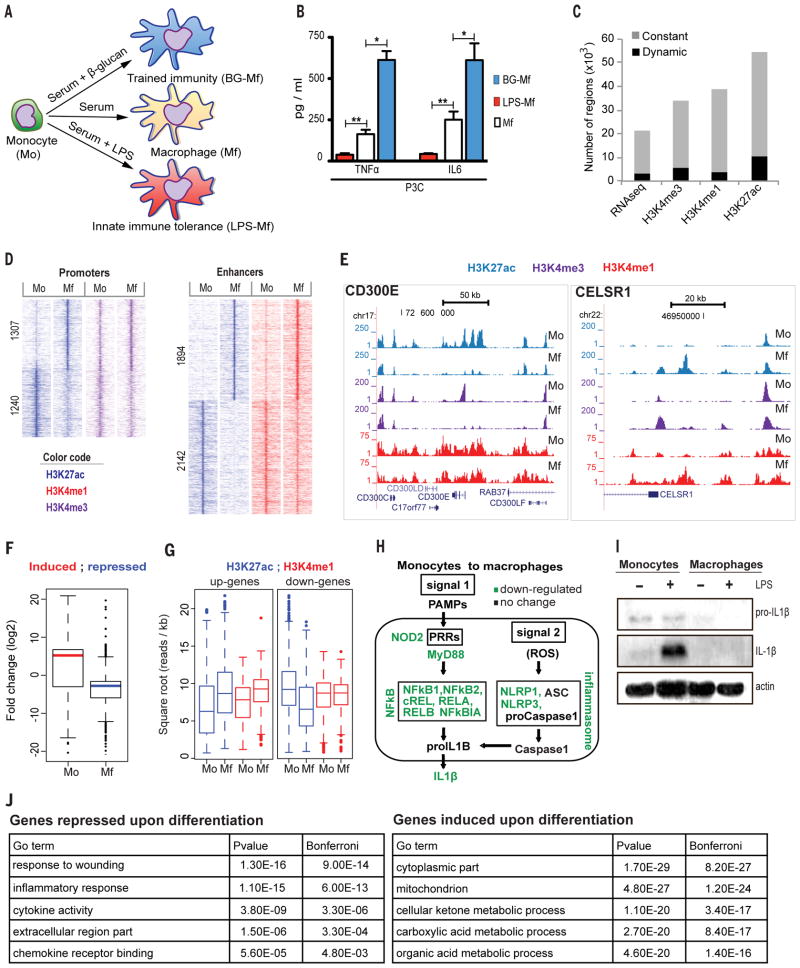
(A) Upon migration into tissues under homeostatic conditions, monocytes (Mo) differentiate into macrophages (Mf). In addition, monocytes will enter into either a refractory state described as endotoxin ‘tolerance’ that is mimicked in vitro with a short LPS stimulation (LPS-Mf) or a state of increased responsiveness described as ‘trained immunity’ that is mimicked in vitro with a short β-glucan incubation (BG-Mf) (See also Fig. S4E). (B) Cytokine production determined by ELISA in supernatants of monocytes primed for 24 h with: cell culture medium (Mf), β-glucan (BG-Mf) or LPS (LPS-Mf), and re-stimulated on day 6 for an additional 24 hours. * p<0.05 and ** p<0.005 (Wilcoxon signed rank test). Data are presented as mean ± SEM (pooled data from n ≥10 individuals in 4 independent experiments). (C) The proportion of dynamic transcripts and epigenetic regions that differ by at least two median absolute deviations for the H3K27ac, H3K4me3 and H3K4me1 epigenetic marks as a function of monocyte differentiation and/or priming regimens. (D) Pileup heat map of the epigenetic marks from ‘C’ at promoters and enhancers in monocytes (Mo) and macrophages (Mf). Rows are genomic regions from −12 to +12 kb around the center of the peaks; the signal intensity was determined in windows of 400 bp. (E) Screen shots of the epigenetic landscape at the CELSR1 (right) or CD300E (left) loci, two representative examples of loci that display important changes during macrophage differentiation. (F) Boxplot presentation of changes in transcript levels during differentiation (Mo to Mf) for the closest differentially expressed genes (within 100 kb) from the dynamic H3K27ac marked regions. (G) Boxplot presentation of changes in the H3K27ac and H3K4me1 signal at distal H3K27ac binding sites, in the vicinity (closest within 100 kb) of up-(left) and down-regulated (right) genes during Mo to Mf differentiation. (H) Modulation of the inflammasome and NF-κB pathways during monocyte to macrophages differentiation. (I) Monocytes (Mo) and macrophages (Mf) were left unstimulated (−) or stimulated with LPS (10 ng/mL) for 24 hours (+). Inflammasome activation was determined by western blot for IL-1β and pro-interleukin-1 β (proIL-1β). The results are representative of at least three independent experiments. (J) GO analysis. Enriched GO categories in a set of significantly (4-fold change, RPKM > 2) down- and up-regulated genes when comparing monocytes (Mo) to macrophages (Mf).
Using an in vitro approach, we differentiated monocytes (Mo) into macrophages (Mf) by long-term incubation in medium supplemented with human serum (22). Alternatively, monocytes were exposed for the first 24 hours to lipopolysaccharide (LPS) to induce endotoxin tolerance (immune paralysis) (Fig. 1A, B) (9). The LPS exposure of monocytes induced long-term tolerant cells (LPS-Mf) that produce less pro-inflammatory mediators, such as TNFα and IL-6, upon an immune challenge with tripalmitoyl glyceryl cysteine (Pam3Cys, a ligand of TLR2) than unprimed naïve macrophages (Mf). In contrast, a short priming of monocytes with β-glucan induces trained immune cells (BG-Mf) (Fig. 1A, B) (10) that are characterized by an enhanced inflammatory status (10, Fig. 1B). On the basis of expression of cell surface markers we concluded that LPS and β-glucan priming followed by continued culture for 5 days yields macrophage-like cells, but with distinct functional programs (Fig. S1). This was supported by our epigenomic and transcriptomic analyses (Fig. S3, see below).
Epigenomic profiles were generated for monocytes (Mo), naïve macrophages (Mf), tolerized (LPS-Mf) and trained cells (BG-Mf) (Fig. 1C) for three histone marks positively associated with gene expression: H3K4me3 which marks promoters, H3K4me1 which marks distal regulatory elements (enhancers) and H3K27ac which marks active promoters and enhancers (23, 24) following the guidelines of the International Human Epigenome Consortium (www.ihec-epigenome.org). We also examined the DNA accessibility through DNaseI-seq and RNA-seq (25). In total, 17% of the H3K4me3 peaks, 10% of the H3K4me1 blocks and 19% of the H3K27ac peaks are dynamic, changing during differentiation (Mo relative to Mf, LPS-Mf and BG-Mf) by at least two median absolute deviations (Fig. 1C). H3K27ac was by far the most dynamic mark, more often fluctuating distally (7611, termed ACetylated elements–ACe) compared to the regions within 2.5 kb of an annotated transcription start site (TSS), 3063, termed ACetylated promoters–ACp). Importantly, all regions examined displayed some degree of DNase I cleavage sensitivity, indicating that the vast majority of locations, including those with the de novo deposition of H3K27ac, are likely already accessible to some extent in monocytes, i.e. before stimulation (see also Fig. 2).
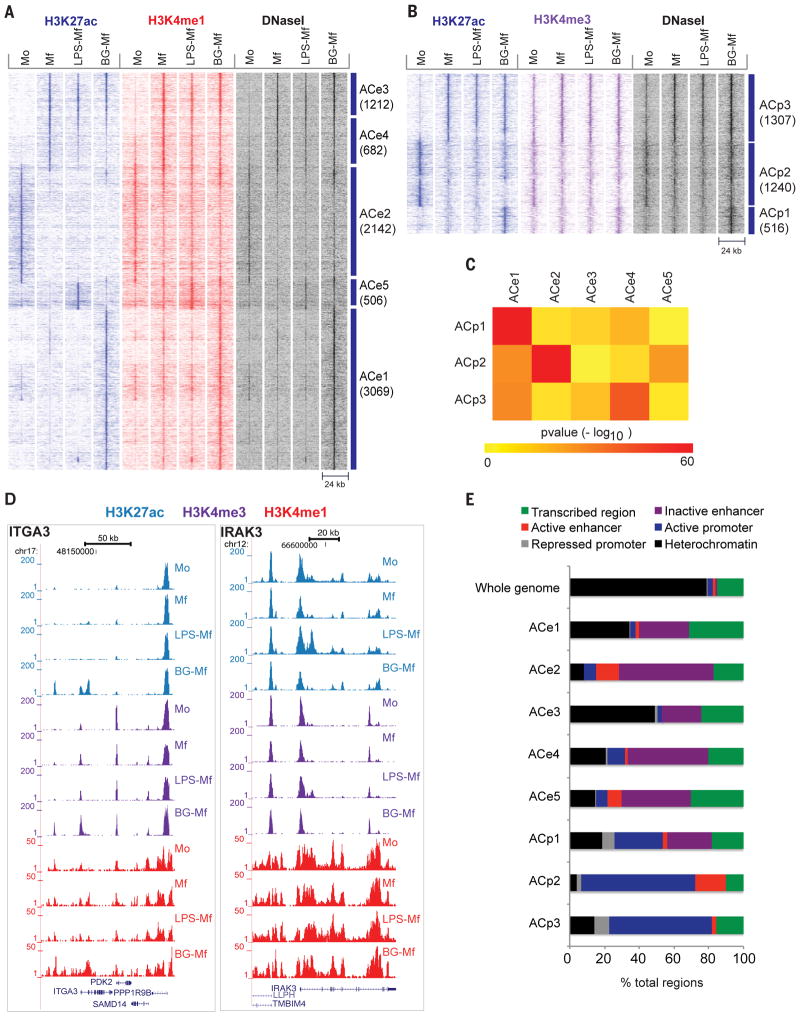
(A, B) Heat map showing dynamic acetylation marks in enhancers (A) and promoters (B). Dynamic H3K4me regions devoid of H3K27ac are displayed in fig. S2. (C) Heat map of pair-wise overlaps of genes associated with dynamic promoter (ACp) and distal regulatory element (ACe) cluster. P-values (−log10) were obtained by the hypergeometric test, reflecting the probability that the obtained number of shared target genes would be shared by any two equivalent random gene sets. (D) Two representative gene loci corresponding to ITGA3 and IRAK3 that gain H3 lysine modifications in BG-Mf and LPS-Mf, respectively. (E) Epigenetic clusters as assigned by ChromHMM analysis (37) for primary human monocytes. For each cluster, the percentage of elements with the designations heterochromatic (H3K9me3, H3K27me3 or empty), active promoter (H3K4me3), inactive promoter (H3K4me3 and H3K27me3), active regulatory element (H3K4me1 and H3K27ac), inactive regulatory element (H3K4me1) and transcribed segment (H3K36me3), was calculated.
Epigenetic and transcriptional changes during monocyte to macrophage differentiation
Epigenetic alterations during differentiation of monocytes (Mo) into naïve macrophages (Mf) were considered separately at promoters and distal regulatory elements (Fig. 1D). Promoters were operationally defined as regions within 2.5 kb of a TSS that also bear a diagnostic H3K4me3 peak. After six days of culture, as monocytes develop into macrophages, we observed a decrease in H3K27 acetylation in 1240 promoters and increase in 1307 promoters (z-test, p-value < 0.05). Hence, at the epigenetic level, approximately equal numbers of promoters are turned on or off during macrophage differentiation (Fig. 1D). In general, H3K4me3 mark is largely constant at promoters that display dynamic H3K27 acetylation (Fig. 1D), suggesting that H3K27ac appears to function more as a mark of changes in promoter activity than H3K4me3.
In addition, as monocytes develop into macrophages, dynamic distal regulatory elements were operationally defined by H3K27ac at regions that are not known TSSs marked with H3K4me3 signal, and that significantly (z-test, p-value < 0.05) lose (2142) or gain (1894) acetylation (Fig. 1D). We observed that distal regulatory elements that gain H3K27ac generally gain H3K4me1, starting from a low level of H3K4me1. However, elements that lose H3K27ac generally do not lose their H3K4me1 mark, supporting the notion that H3K4me1 provides an epigenetic memory function in macrophages (Fig. 1D, E) (18). Furthermore, DNase I cleavage frequencies reveal that the majority of distal regulatory DNA elements that lose H3K27ac remain accessible in macrophages (see also Fig. 2A).
In order to evaluate the relation between H3K27ac-bearing elements and gene expression, we plotted the fold change in RNA level of the closest gene against H3K27ac distal elements revealing a positive correlation between epigenomic marking and transcription from the closest dynamic gene (Fig. 1F). In contrast, the H3K4me1 mark does not correlate well with adjacent gene expression changes (Fig. 1G). This observation is in line with the hypothesis that H3K4me1 marks active (23, 24) as well as latent regulatory elements (18). Altogether, our analysis indicates that in the course of monocyte to macrophage differentiation, histone H3 marks correlate positively with the activity of promoters (H3K4me3/H3K27ac) and distal regulatory elements that are presumed enhancers (H3K4me1/H3K27ac).
Biological pathways correlated with epigenetic marks
In order to identify the biological processes affected during differentiation of monocytes into macrophages, we performed a Gene Ontology (GO) analysis on differentially expressed genes (851 down, 1292 up, Table S1). We observed previously described differences in gene expression between monocytes and monocyte-derived macrophages including the response to wounding (GO:0009611), inflammatory response (GO:0006954), and defense response (GO:0006952) (Fig. 1J, Table S2) (26–29). As expected, pathogen-associated molecular patterns (PAMP)-mediated signal transduction through pattern recognition receptors (PRRs) is remodeled as monocytes differentiate into macrophages, through changes in the epigenetic and transcription state of PRR genes. Furthermore, the NF-κB transcription factor subunits REL, RELA, RELB, NFκB1 and NFκB2, which are key regulators of the cellular response to inflammation (30), are all down-regulated (> 2-fold lower, Fig. 1H). Among the modulated inflammatory pathways during monocyte to macrophages differentiation, we detected components of the inflammasome ((31), Fig. 1H). In line with the expectation that tissue macrophages are more tolerant to challenges of the immune system, in vitro-differentiated-macrophages, unlike monocytes, showed no activation of the inflammasome component caspase-1and they lacked the capacity to secrete the active form of the pro-inflammatory IL-1β obtained after maturation of pro- IL-1β by caspase-1 ((32), Fig. 1I, S4).
The most significant GO terms associated with induced RNA expression during monocyte to macrophage differentiation (Table 2) were monocarboxylic acid and cellular ketone metabolic processes. Up-regulated metabolic enzymes include dehydrogeneases HSD17B1, -B4 and -B7, of which HSDB4 has been implicated in the peroxisomal beta-oxidation pathway and PPAR signaling, which play essential roles in the regulation of cellular development and metabolism, as well as enzymes involved in glycine, serine and threonine metabolism (33), and the TCA cycle. The TCA cycle, which has recently been shown together with glycolysis to enhance ER and golgi membrane synthesis and induce innate activation of dendritic cells upon TLR-mediated stimulation (34), appears to be remodeled such that the cytoplasmic versions of isocitrate dehydrogenase (IDH1) and malate dehydrogenase (MDH1) are induced > 50 and > 10-fold respectively, while their mitochondrial counterparts are induced about 2-fold reaching ~20% of their cytoplasmic equivalents. These data support that re-orchestration of cellular metabolism during macrophage differentiation is an important components of the phenotypic switch between inflammatory and tolerant cells, with resting or tolerant macrophages exhibiting TCA cycle and oxidative metabolism (35). The balance between glycolysis and oxidative metabolism also plays a role in trained immunity and tolerance (accompanying manuscript of Cheng et al. (36)).
The epigenetic profiles of tolerant versus trained cells
After an infectious episode, monocytes can retain an “immunological scar” from a previous encounter, entering either in a refractory state (tolerance) or a state of increased responsiveness (trained). These processes that result in non-specific innate immune memory, have been suggested to be mediated through epigenetic mechanisms (10, 19) and describe very important in vivo phenomena such as post-sepsis immunoparalysis (tolerance), or BCG-induced non-specific (or heterologous) protective effects (training). Tolerance and training can be recapitulated in vitro using monocytes (Mo) exposed for 24 hours with LPS or β-glucan, respectively, followed by culture without additional stimuli for five days. These procedures yield differentiated cells that are either tolerant or trained macrophages-like cells (10) (Fig 1A, S4E).
At the epigenomic level, 17% of all dynamic promoters (ACp1) and 40.3 % of all dynamic distal elements (ACe1) gain H3K27ac de novo exclusively in β-glucan pre-treated cells (BG-Mf). Furthermore, a small subset of the dynamic distal regulatory elements (6%; ACe5) gain de novo H3K27ac marks exclusively in LPS stimulated cells (LPS-Mf). These changes likely underlie the long-term effects of cell tolerization or training (Fig. 2A, B, Tables S3), mirroring post-sepsis immunoparalysis and post-vaccination non-specific (heterologous) protection, respectively.
For the distal regulatory elements, consistent loss of H3K27ac is observed in Mf, LPS-Mf and BG-Mf at 2142 distal elements (ACe2, Fig. 2A) relative to the starting monocytes (Mo). Distal elements that are marked de novo with H3K27ac, following differentiation from monocytes (Mo) to macrophages (Mf, LPS-Mf and BG-Mf), fall into two clusters; ACe3 which shows a consistent gain of H3K27ac and ACe4 which are dampened relative to macrophages concordantly in both LPS and β-glucan pre-treated cells (Fig. 2A). The most extensive pre-treatment specific epigenetic response is obtained with β-glucan, whereby 3069 distal elements become marked by H3K27ac (ACe1, Fig. 2A, Fig. 2D left panel). Some diversity in marking of distal elements with H3K27ac in ACe1 can be observed in other states: there are sub-clusters with low marking in macrophages (Mf), or in monocytes (Mo), and there is also a very small sub-cluster of ACe1 distal elements that is specifically marked upon LPS treatment (Fig. 2A). This indicates a modular, treatment dependent activation of distinct pathways. Altogether, β-glucan appears to induce an epigenetic program that involves many promoters and distal elements. Some parts of this program are shared with untreated, naïve macrophages (Mf) and others with LPS-tolerized cells (LPS-Mf), while neither LPS-tolerized nor naïve macrophages show such a strong exclusive epigenomic signature (Fig. 2A, B).
We counted the genes targeted by more than one epigenetically marked cluster and computed the probability of randomly obtaining such overlaps (Fig. 2C). The most significant overlaps (hypergeometric test, p-value <0.01) are between elements that show similar dynamics (ACp1/ACe1, ACp2/ACe2, ACp3/ACe4), in keeping with the idea that epigenetically marked promoters and enhancers cooperate to drive gene expression. Notably, β-glucan induced ACe1 distal elements also appear to be associated with ACp2 and ACp3 promoters more often than expected, suggesting modulation of differentiation-sensitive promoters by β-glucan induced enhancers. Finally, the ACe5 distal elements, which are marked only upon LPS-tolerization are associated with ACp2 promoters which are down-regulated upon monocyte differentiation to macrophages. This suggests that LPS-Mf stimulate the activity of a subset of monocyte specific genes in macrophages via ACe5 elements (Fig. 2C).
To determine the provenance of these epigenetic elements in more detail, we used the Blueprint ChromHMM data for primary human monocytes. ChromHMM indexes chromatin into functional states such as active or repressed promoters, distal regulatory element (enhancer), and heterochromatic (repressed) chromatin (37). The majority of dynamic acetylated promoters and distal regulatory elements were already marked with low but appreciable levels of H3K4me3 (promoters) or with H3K4me1 at distal elements in monocytes (Mo), indicating changes in activity rather than in chromatin state during differentiation to Mf, LPS-induced tolerance (LPS-Mf) or β-glucan induced training (BG-Mf) (Fig. 2E).
Polytomous analysis of the transcriptome of Mf, LPS-Mf and BG-Mf
To gain insight into the effects of LPS and β-glucan on non-specific innate immune memory, we grouped genes according to a Bayesian methodology (MMSEQ and MMDIFF) (38, 39) and applied a polytomous model selection approach that allows for classifying genes according to their expression pattern by considering multiple conditions simultaneously instead of generating multiple pairwise comparisons. This approach thus allowed us to identify statistically distinct expression patterns with biological relevance at once and define cell state-specific as well as co-regulated gene modules during monocyte to macrophage differentiation and in response to training/tolerization. A total of fourteen models were compared to the base model of no differential expression and polytomous model selection was applied by computing the posterior probability of each model for each gene. Out of 11698 genes expressed > 2 RPKM, 7514 showed more than two-fold changes, and 5847 fit is one of the models with a posterior probability >0.35. The models were condensed into expression modules (M1-6) to bring together genes as a function of their differential expression in LPS-Mf and BG-Mf relative to Mf cells. The genes of the modules that change four-fold are plotted in Fig. 3A and the top associated GO terms are shown (Fig. 3B, Tables S2, S4). Interestingly, GO analysis on these modules (Fig. 3B) resulted in obtaining similar terms from our comparison of monocytes (Mo) and macrophages (Mf) in the absence of LPS or β-glucan (Fig. 3B, ,1J).1J). This suggests that both LPS and β-glucan pre-treatments modulate the monocyte to macrophage differentiation program.
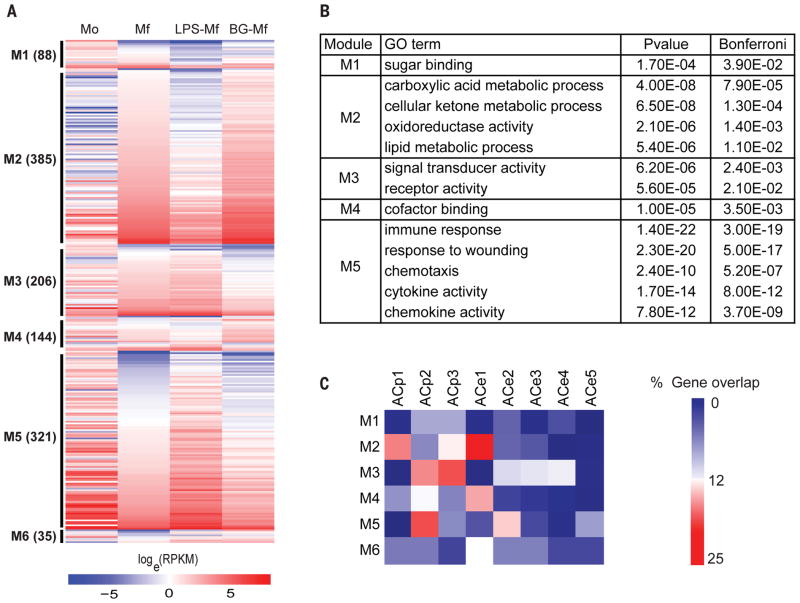
(A) Gene expression patterns were analyzed using a polytomous analysis of gene expression profiles that contrasts LPS-Mf and BG-Mf relative to Mf, as explained in detail in the text. Genes plotted are those with a fold change > 4 and a polytomous post-hoc probability > 0.35. (B) Enriched GO categories in the major expression modules represented in Fig. 3A. (C) Heat map presentation of the percentage of genes in a module that overlap with a dynamic ACp or ACe cluster.
Modules are condition and cell-specific
In order to link the expression modules to the epigenome, we determined the percentage of the genes of each module associated with an epigenetically dynamic promoter or distal element cluster (Fig. 3C, Table S3, S4). Surprisingly, LPS-Mf do not down-regulate a large set of genes that are expressed in monocytes (Mo) to the same extent as Mf and BG-Mf (Fig. 3A, M5) suggesting the LPS-Mf retains a monocyte-like expression level for these genes. Indeed, module M5 is enriched in genes from the GO categories ‘immune response’, ‘response to wounding’ and ‘chemotaxis’ (Fig. 3B) and shows up-regulation of monocyte-specific genes (Fig. 1J), as well as a ‘response to LPS’ (Table S2). An example of a gene with a monocyte-like expression level in LPS-Mf is IRAK3 kinase, an essential regulator of innate immune homeostasis that functions as a negative-feedback regulator of Toll-like receptor signaling (40) ((41) Fig. 2D right panel). This finding is expected from the perspective of the low inflammatory profile of tolerant macrophages. Module M5 harbors relatively many ACp2, ACe2 and ACe5 elements (Fig. 3C), suggesting that some parts of the monocyte specific gene expression program are retained in LPS-Mf cells, possibly via induction of a new set of distal elements (ACe5). Furthermore, some of these elements may sustain the expression of target genes that are otherwise down-regulated in Mf and BG-Mf cells. Similarly, module M3 is generally better expressed in LPS-Mf than BG-Mf. M3 includes factors involved in signal transduction, such as IL10RA involved in controlling intestinal inflammation (42), the aryl hydrocarbon receptor repressor AHRR that has been shown to be essential for endotoxin tolerance (43), and the adenosine receptor ADORA3. Module M3 genes often harbor ACp2, ACp3, ACe2, ACe3 and ACe4 elements, and calls the GO terms ‘signal transducer activity’ and ‘receptor activity’ (Fig. 3B) likely reflecting functional linkage between the execution of the naïve macrophage differentiation program and the LPS- and β-glucan induced epigenetic programs.
Strikingly, module M2 represents a large set of genes that tend to be induced in Mf and are markedly less expressed in LPS-Mf, but equally or even better induced in BG-Mf (Fig. 3A, M2). M2 invokes the GO metabolic terms ‘cellular ketone metabolic process’ and ‘carboxylic acid metabolic process’ that are significant macrophage differentiation markers (Fig. 1J, Table S2). Notably, M2 harbors ATF3, a transcription repressor known to target cytokine genes in mouse macrophages ((44), see also Fig. 4B). Module M4 is also most up-regulated in β-glucan pre-treated cells, and like M2 it is enriched in ACe1 distal elements (Fig. 3C). This suggests that sustained expression of module M4 in BG-Mf relative to the starting monocytes may depend on the presence of β-glucan-induced distal elements, explaining the apparently paradoxical overlap between the ACe1 elements and ACp2 promoters (Fig. 2C). M4 is enriched in the GO category ‘co-factor binding’, including the pentose phosphate pathway-linked enzymes pyridoxine kinase PDXK and glucose-6-phosphate dehydrogenase G6PD, the glycolysis enzyme pyruvate dehydrogenase subunit DLAT and the cytoplasmic version of the TCA cycle enzyme malate dehydrogenase MDH1 (Table S2); (also see (36)). Intriguingly, a small subset of mitosis-specific genes that were previously reported in the context of monocyte-derived macrophages (27, 29) are also present in module M4 (Table S4). However, we assayed DNA replication directly and found no indication that Mf nor BG-Mf cells are replicating (Fig. S5).
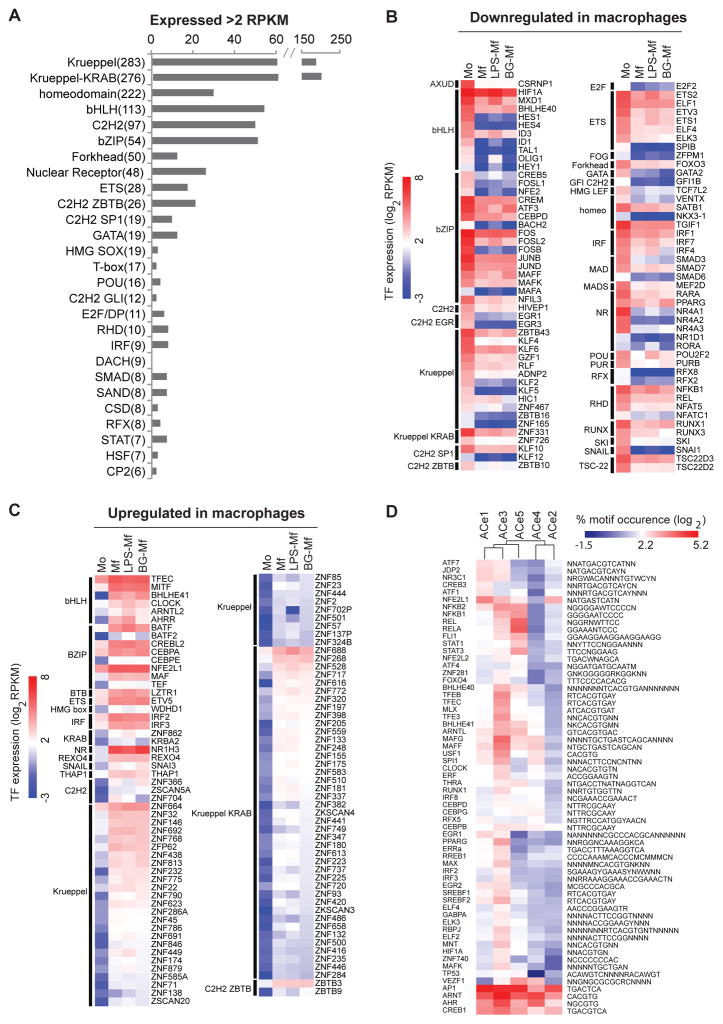
(A) Bar representation of the transcription factor family members that are expressed in at least one of the four cell samples (Mo, Mf, LPS-Mf, BG-Mf). The number of TFs with RPKM values > 2 is plotted and the total number of TF family members in humans is indicated between brackets. (B–C) Heat map of the expression levels (log2 RPKM) of TFs that change expression at least fourfold in at least one of the four conditions examined. TFs are grouped according to families and whether they are up- or down-regulated upon differentiation. Scale (left) indicates the level of expression (log2 RPKM). (D) Hierarchical clustering of the TF motif occurrence frequency (log2 percentage, scale at top) in each ACe cluster-associated DHSs from all TFs motifs that have more than 5% occurrence in at least one instance and that are over-represented in at least one of the ACe clusters compared to all non-dynamic acetylated regions (hypergeometric test, p-value <0.01).
Multiple modules contain G-protein coupled receptors (GPCRs), protein kinases and epigenetic enzymes (Fig. S6, Table S4), indicating extensive remodeling of the trained cell’s repertoire of signal transduction molecules. Indeed, the beta-adrenergic receptor ADRA2B is specifically induced in β-glucan trained, rather than in LPS-tolerized cells. Furthermore, monocytes and the three Mf states could be distinguished by differential expression of human adenosine receptor and SRC kinase family members (27, Fig. S6). ADORA2B and the FGR kinase are most strongly expressed in trained BG-Mf cells, while HCK kinase transcript levels are highest in LPS-Mf, providing potential pharmacological entry points to modulate immunoparalysis and trained innate immunity-related health conditions.
In summary, the epigenetic effects observed in BG-Mf and LPS-Mf appear to modulate the well-documented M-CSF-induced macrophage differentiation program (26) and polytomous transcriptome analysis reveals that LPS-Mf cells sustain expression of certain monocyte pro-inflammatory pathways that are dampened in Mf and BG-Mf cells (M3, M5). On the other hand, β-glucan pre-treatment appears to reinforce specific gene expression programs including metabolic pathway remodeling (M2, M4) through epigenetic modification of both ACp1 promoters and ACe1 distal regulatory elements.
The transcription factor repertoires of differentiation, tolerance and training
Transcription factors (TFs) bind to specific DNA sequences and are the effectors of cell phenotype and function, including numerous signal transduction pathways. As they recruit chromatin-modifying enzymes (Fig. S6), which modulate gene expression, they are cornerstones of epigenetic signaling. Hence, we focused on the expression patterns of the human TFs to identify those whose expression patterns may determine monocyte (Mo) and the monocyte-derived macrophage fates (Mf, LPS-Mf and BG-Mf). There are about 1600 bona fide human sequence-specific TFs (Table S5). Of these, 74% are expressed at a detectable level (RPKM > 0.1), and 41% are expressed above 2 RPKM (Fig. 4A). Of the highly expressed TFs, 197 TFs change transcript levels over four-fold between at least two states (Mo, Mf, LPS-Mf and BG-Mf) and are henceforth referred to as ‘dynamic’ TFs. Altogether, 92 TFs are 4-fold down- and 105 TFs are 4-fold up-regulated (Fig. 4B, C).
From a differentiation program perspective, it is informative to study dynamic changes within TF families (Fig. 4A). The largest TF super-family are the C2H2 zinc finger Krueppel factors with and without KRAB domains, encompassing 559 factors (Fig. 4A). Of these 15% are dynamic; 14 are down- regulated and 75 are up-regulated, albeit to low levels (Fig. 4B, C).
Of the 48 human nuclear receptors, 24 are expressed at least in one of our four cell types and eight are dynamic. Monocytes (Mo) express relatively high levels (RPKM > 100) of the three pro-inflammatory NR4A1-3 nuclear receptor subfamily members NUR77, NURR1 and NOR1 (45), while derived macrophages show a dramatic reduction in expression of these nuclear receptors (Fig. 4B). The retinoic acid receptor RARA (NR1B1) also follows this pattern (Fig. 4B). On the other hand, LXRα (NR1H3), a nuclear receptor implicated amongst others in liver cell and macrophage cholesterol metabolism (46, 47), increases in Mf, LPS-Mf and BG-Mf cells (85-, 50- and 117-fold), attaining 300 RPKM in BG-Mf (Fig. 4C). Accordingly, the cytochrome (CYP27A1), an enzyme involved in the formation and breakdown of various molecules such as the 25-hydroxy-7-dehydrocholesterol which activates the LXRα (48), is itself induced ~25-fold in macrophages. Interestingly, the peroxisome proliferator-activated receptor PPARG (NR1C3) is reduced in Mf and LPS-Mf, but it is maintained in BG-Mf relative to purified monocytes (Mo), in line with its proposed role in inflammation resolution ((49), Fig. 4B). Thus, nuclear receptors appear as major regulators of monocyte and macrophage biology, with LXRα levels qualitatively reflecting the immunological phenotypes of Mf, LPS-Mf and BG-Mf (Fig. 1A).
Aside from nuclear receptors, multiple TF families display coordinated family member switches. For instance, SNAI1, a major mesoderm lineage marker (50, 51) is reduced in expression about 100-fold during macrophage differentiation, but its paralog SNAI3 is 5- to 10-fold higher in macrophages, most likely outcompeting SNAI1 for binding to its cognate DNA binding sites in Mf’s (Fig. 4B, C). Interferons (IFNs) and IFN responses are one essential arm of the host immune responses. Upon infection, PAMPs recognition by PRRs elicit antimicrobial responses by inducing target genes that include those encoding type I IFNs, proinflammatory cytokines, and chemokines. The interferon regulatory factor (IRF) family are transcriptional regulators of type I IFNs and IFN-inducible genes, and help regulate facets of innate and adaptive immune responses (52). Five of the nine human interferon regulatory factors are dynamic, as IRF1, IRF4 and IRF7 significantly decrease while IRF2 and IRF3 increase, in a background where IRF8 and IRF9 remain the most abundantly expressed IRF family members. This may reflect the decrease in IRF1 levels, which activates type I interferon IFN-β production, whereas levels of its antagonistic family member IRF2 increases ((53–55). Thus, specific remodeling within TF families occurs in the course of monocyte to macrophage differentiation.
The bZIP TFs appear as an influential family in the course of monocyte differentiation, since 94% are expressed above 2 RPKM and 44% are dynamic (Fig. 4A, B, C). AP1 complexes composed of heterodimers of the bZIP-FOS and -JUN subfamilies are the most abundantly expressed (RPKM > 200) in monocytes and are reduced upon monocyte-to-macrophage differentiation (Fig. 4B), suggesting a down-modulation of AP1 signaling pathways. The CCAAT/enhancer-binding (C/EBP) bZIP subfamily also shows strong dynamics; CEBPA is induced 7-fold whilst the paralog CEBPD is down-regulated 4-fold, indicating that there may be a coordinated switch in C/EBP heterodimerization which occurs in a complex along with the non-dynamic, highly expressed CEBPB (Fig. 4B, C). The 10-fold down regulation of CREM, a bZIP subfamily member, also likely reflects a biological impact (Fig. 4B) because CREM codes for a dominant negative heterodimerization partner of most cAMP response element (CRE) binding proteins (56). CREM down-regulation is therefore likely to capacitate cAMP response element binding factors (see below).
Integration of epigenetic regions with transcription factor binding sites
We used 1406 sequence motifs for 658 human TFs (57, 58), to scan DNase I hypersensitive sites (DHS) for TF-binding motifs. We determined the overlap between DHSs and dynamic distal elements in our dataset. We performed hypergeometric tests using the motif frequency in all distal (non-dynamic) H3K27ac regions as the background, to yield TF motifs that are likely candidates to regulate the activity of the distal regulatory regions. As an example, Fig. 4D plots the proportion of dynamically acetylated regions within one cluster that bear a given motif in their associated DHSs. To be included, a motif had to occur in at least 5% of any one epigenetically marked cluster (Fig. 4D, Table S6). This reveals enriched TF motifs and permits a comparative analysis of putative TF repertoires in epigenetically marked regions.
The motifs for the bZIP AP1 and CREB1, and bHLH factors ARNT and AHR are the most abundant occurring in 10 to 36% of ACe1 to -5 associated DHSs, in keeping with a broad role for the pathways that control these transcription factors in monocytes and macrophages, namely Map kinases, cAMP and metabolic signaling (59). Motifs for bZIP (sub-) family members, most prominently ATF1, -7, CREB3 and JDP2, are enriched in the β-glucan induced ACe1 cluster. Furthermore, motif for the glucocorticoid receptor (NR3C1) is also relatively enriched in ACe1. This suggests that cAMP and cortisol-mediated signaling pathways intersect with the epigenetic response instigated by β-glucan training. By contrast, the immunoparalyzed LPS-Mf cell-specific ACe5 cluster is characterized by NF-κB and REL motifs (Fig. 4D), which are also present in the monocyte-specific ACe2 cluster (Fig. 1H). This is in keeping with the RNA-seq analysis which points to a large transcriptome module (M5) where LPS-Mf cells resemble monocytes (Mo) more than naïve Mf or BG-Mf (Fig. 3A). In fact, module M5 harbors the NF-κB complex subunit genes that are known to be involved in inflammatory pathways.
Interestingly, the TF motif frequencies in the monocyte-specific ACe2 dynamic distal regions resemble those of ACe4 regions, which are induced in Mf but suppressed when monocytes are challenged with LPS or with β-glucan. However, ACe2 and ACe4 are distinguished by the enrichment of bZIP macrophage (MAFF, MAFG (60, 61) subfamily motifs in ACe4, which are also relatively more frequent in the differentiation-linked epigenetic clusters ACe1, ACe3 and ACe5 and thus likely drive ACe4 element activity upon differentiation.
The bHLH family, including the cAMP hypoxia inducible factor HIF1A as well as the circadian factors BHLHE40, -E41, and CLOCK (Fig. S7) (62, 63), show related patterns, and are most frequent in the monocyte-to-macrophage differentiation cluster ACe3 and the least in the monocyte-specific cluster ACe2. This suggests that bHLH TFs play important roles in macrophage differentiation. Moreover, IRF (interferon-regulatory factors) motifs are also enriched in the Mf differentiation cluster ACe3, as are smaller TF families such as EGR and ETS factors (Fig. 4D).
In summary, the immune tolerant phenotype elicited by LPS exposure appears to depend on epigenetically marked putative distal regulatory regions that frequently harbor NF-κB motifs, while the motifs that are enriched in the trained BG-Mf-marked epigenetic elements are most frequently also in epigenetic regions turned on in Mf macrophages, indicating reinforcement of specific Mf (sub) programs by monocyte exposure to β-glucan.
Importance of cAMP-dependent intracellular signaling for trained immunity
cAMP signals start with a G-protein coupled receptor (GPCR) and culminate in PKA kinase-mediated cAMP response element binding protein activation. cAMP-dependent signaling has been associated with monocyte function, differentiation, and response to infection (64, 65). In Mf, BG-trained and LPS-induced tolerant cells, the heterotrimeric G-protein repertoire is remodeled: adenylate cyclase 3 (ADCY3) and the cAMP-degrading phosphodiesterase (PDE3B) are specifically induced in trained cells (Fig. 5A, S8). Strengthening the hypothesis that β-glucan training and LPS-tolerization differ with respect to cAMP metabolism, we found that LPS-Mf express significantly more of the monocyte-specific cAMP phosphodiesterase (PDE4B) (Fig. 5A, S8). The cAMP-dependent protein kinase (PKA) regulatory subunit anchoring factor AKAP11 transcript is induced (two tailed t-test, p< 0.05) in Mf, BG-Mf and LPS-Mf cells compared to Mo, and so is the type 1 alpha PKA regulatory subunit PRKAR1A. Furthermore, one of the five human PKA isoforms (PRKACB) is more highly induced (two tailed t-test, p< 0.05) in BG-Mf relative to LPS-Mf cells. In addition, the transcripts for cAMP response element binding protein ATF3 and the endoplasmic reticulum stress response element binding CREB3L2 are higher in BG-Mf than in LPS-Mf (Fig. 5A, S8). Importantly, we identified a known cAMP inducible bHLH factor (66, 67), BHLHE41, as a likely key TF motif enriched in ACe1, -3, -4 and -5 (Fig. 4D), strongly suggesting that cAMP-mediated molecular signal transduction is important for Mf, LPS-Mf and BG-Mf cells.
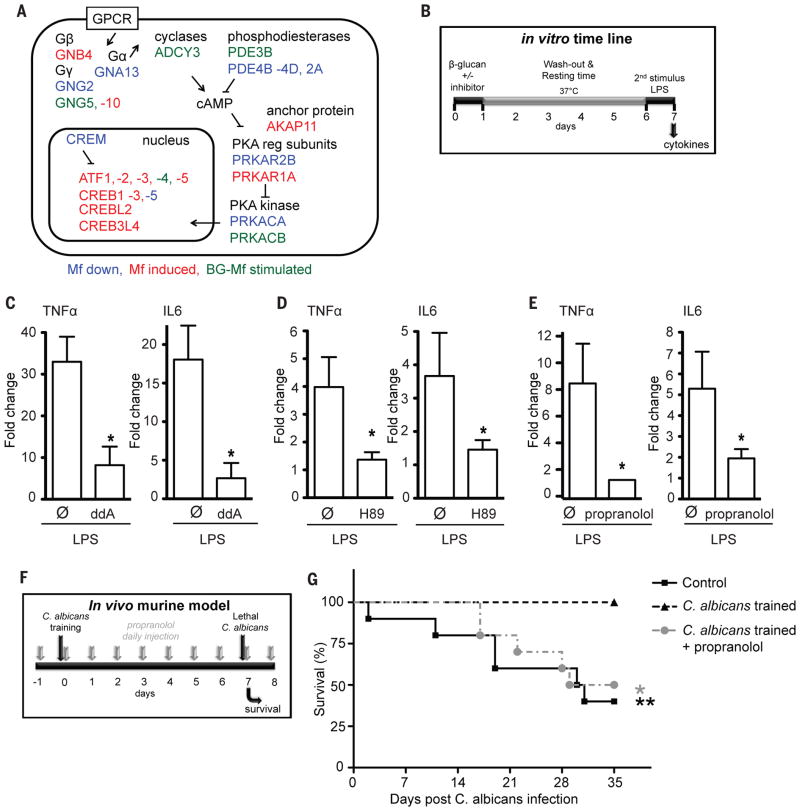
(A) cAMP signaling pathway remodeling. Significant (two-tailed t-test, p<0.05) transcript level decreases (blue) and increases (red) in all macrophage states relative to monocytes (Mo), as well as β-glucan treatment-specific increases are indicated, see also Fig. S8. (B) Diagram of the timeline and experimental setup of the in vitro cAMP inhibition experiment. cAMP inhibitors or vehicle were applied to human primary monocytes during the first 24 hours of “training” (see Fig. S4E and (70) for details). After 6 days, secondary stimulation of cells was performed with LPS to induce cytokine production. (C–E) The inhibitor of adenylate cyclase 2′,5′-Dideoxyadenosine (ddA), the cAMP dependent protein kinase (PKA) inhibitor H89 and the β-adrenergic receptor blocker propranolol inhibited the induction of training by β-glucan, assayed as the response to LPS stimulus on day six. * p<0.05 (Wilcoxon signed rank test). Data show the fold increase in cytokine production upon training as compared to untrained Mf-cells and are presented as mean ± SEM, n > 6 in 3 separate ELISA experiments. (F) Timeline of the in vivo training experiment in mice. Mice were either injected intravenously with PBS (control) or a non-lethal dose of C. albicans (2×104 CFU/mouse) seven days prior to intravenous inoculation of a lethal C. albicans dose (2×106 CFU/mouse). In a third group of mice, propranolol was administered before the inoculation of the non-lethal dose of C. albicans (70). (G) Survival rate of wild-type C57BL/6 mice to a systemic infection with C. albicans (n 8 per group).
We investigated the role of cAMP-dependent signaling in vitro and in an in vivo experimental model (Fig. 5B). Inhibition of adenylate cyclase and subsequent cAMP synthesis by 2′,5′-dideoxyadenosine significantly (Wilcoxon signed rank test, p<0,05) impairs training-induced increased cytokine production (Fig. 5C), and in line with this, the PKA inhibitor H89 also blocks training of monocytes (Fig. 5D). Of note, the PKA inhibitor H89 had no effect on the LPS-triggered immune response (3% increased IL-6 production and 9% decreased TNFα production compared to control stimulation, p>0.05, Fig. S9). Similar effects were observed with propranolol, an inhibitor of β-adrenergic signaling-dependent increase in cAMP (Fig. 5E). Importantly, even though the amplitude of responsiveness to secondary stimulation of each individual varies, the effect of the cAMP-pathway-inhibitors on the trained immunity response is consistent and significant among the 18 tested individuals. A non-lethal infection with C. albicans protects mice against lethal candidiasis in a T/B-cell independent fashion, due to monocyte-dependent trained immunity (10). The protection conferred by the non-lethal dose of C. albicans was completely abolished by the use of propranolol (Fig. 5F, G), validating that cAMP-dependent signaling is important for the protective effects of trained immunity.
Discussion
Monocytes and macrophages are prototype mononuclear phagocytes that mediate fundamental innate immune processes such as pathogen clearance, inflammatory cytokine production and tissue repair. We studied macrophages differentiated from monocytes exclusively in the presence of human serum. This is probably the in vitro model closest to the development of tissue resident macrophages in vivo, as the differentiation is taking place under the influence of endogenous homeostatic levels of M-CSF, rather than stimulated by artificially high concentrations of recombinant colony-stimulating factors (68). In contrast, differentiation with additional recombinant growth factors such as GM-CSF, IL-4 or IFNγ will induce activated M1 or M2 macrophages most often encountered during infectious processes (68). Thus, these observations provide clues as to which pathways to tackle in clinical attempt to reverse immune tolerance to turn on training/activation pathways during immunoparalysis/sepsis.
The present work shows that differentiating monocytes undergo significant epigenetic changes affecting ~19 Mbp, equivalent to 0.6%, of the human genome. Using the guidelines of the International Human Epigenome Consortium (www.ihec-epigenomes.org) we generated genome-wide maps of histone modifications (H3K4me3, H3K4me1, H3K27ac and DNase I accessibility). We delineated eight epigenetically marked clusters reflecting putative transcription regulatory regions, of which four are differentially modulated when monocytes are exposed to LPS or β-glucan. H3K27 acetylation levels changed at thousands of promoters and distal regions, and correlate well with observed changes in gene expression.
Although we cannot ascertain what fraction of the purified monocytes we cultured participated in any one observed epigenomic or transcriptome change, nor to what extent the laboratory in vitro conditions affect differentiating monocytes relative to the in vivo condition, we can state that monocytes are able to deploy a long-term epigenetic program upon β-glucan-induced training in vitro in the absence of DNA replication. This epigenetic program persists for at least five days, is highly reproducible across monocytes from unrelated donors, and represents β-glucan induced changes in the epigenetic states of promoter and distal elements, indicative of a specific cell differentiation program. Although more modest, a similar epigenomic response of monocytes to LPS is also uncovered, that induces a state akin to post-sepsis immune paralysis. Identification of core signatures of training and tolerance is thus important, as it allows the better characterization of these functional states. Since many epigenetic clusters that differentiate training and tolerance yield significant GO scores, the clusters we identified appear to report physiologically relevant effects. Indeed, we linked cAMP signaling to the trained phenotype in purified human monocytes as well as in a mouse model of disseminated candidiasis.
Our transcription factor analysis suggests that monocyte differentiation relies on quantitative remodeling of the transcription factor repertoire, since coordinated changes affect multiple members of many transcription factor families. DNA sequence motifs detected in DHSs adjacent to epigenetically dynamic elements link to cell state-specific epigenetic clusters that fit RNA expression modules. The present data sets, in particular the genomic coordinates of the dynamic epigenomic sites, therefore represent a rich molecular resource to understand and modulate the functionality of monocyte-derived medically important cell types such as macrophages, dendritic cells, foam cells and osteoclasts (69).
Methodology
Monocyte and monocyte-derived cells
To enable analysis of the different functional programs of monocytes, human primary monocytes were purified from three to six healthy human donors (depending on experiment) by first isolating peripheral blood mononuclear cells by differential centrifugation over Ficoll-Paque, followed by depletion of T-cells, B-cells and NK-cells with anti-CD3, CD19 and CD56-coated beads, respectively (Fig. S1). Macrophages (Mf) were obtained by in vitro culture of the purified monocytes (Mo) for 6 days in cell culture medium enriched with human serum (10%). Monocyte tolerance, a state akin to immune paralysis during bacterial sepsis or endotoxin shock, was induced in vitro by exposing the purified monocytes to lipopolysaccharide (LPS) for 24 h (9). Monocyte training, a functional state in which cells respond more strongly to microbial stimuli that mirrors the non-specific protective effects of live microorganism vaccination (e.g. BCG, measles, yellow fever, (15)) (Fig. 1A, B) was induced in vitro by exposing the purified monocytes to β-glucan for 24 h (Fig. 1B) (10). The LPS or β-glucan was then washed out from the system and the cells were incubated in enriched cell cultured medium for five days.
Genome-wide epigenetic profiling
For all the four types of cells described above: monocytes (Mo), macrophages (Mf), LPS-tolerant (LPS-Mf) and β-glucan-trained (BG-Mf) cells, the epigenomic profiles were generated for three histone H3-borne marks known to be associated with gene expression. They respectively mark promoters (H3K4me3), distal regulatory elements (H3K4me1) and the active forms of both promoters and enhancers (H3K27ac) (23, 24). Additionally, RNA and DNase I accessibility were quantified (25). Raw data from the healthy volunteers is available through EGA. Data can be accessed via GEO accession GSE58310.
Supplementary Material
Suppl Table 1
Suppl Table 2
Suppl Table 3
Suppl Table 4
Suppl Table 5
Suppl Table 6
Supplementary Figures S1-S9, Supplementary Methods
Acknowledgments
The authors are grateful to J. Stamatoyannopoulos for help with DNaseI-seq. We thank S. van Heeringen for access to his databases and expertise. The research leading to the results described in this manuscript has received funding from the European Union’s Seventh Framework Programme (FP7/2007-2013) under grant agreement n° 282510 – BLUEPRINT. M.G.N. was supported by a Vici grant of the Netherlands Organization for Scientific Research and an ERC Consolidator Grant (nr. 310372). J.H.A.M by a VIDI grant of the Netherlands Organization for Scientific Research. All the genome-wide data generated and analyzed in this study as well as the sequencing details can be accessed via GEO accession GSE58310.
References and Notes
Full text links
Read article at publisher's site: https://doi.org/10.1126/science.1251086
Read article for free, from open access legal sources, via Unpaywall:
https://europepmc.org/articles/pmc4242194?pdf=render
Citations & impact
Impact metrics
Citations of article over time
Alternative metrics
Smart citations by scite.ai
Explore citation contexts and check if this article has been
supported or disputed.
https://scite.ai/reports/10.1126/science.1251086
Article citations
Knowledge mapping of trained immunity/innate immune memory: Insights from two decades of studies.
Hum Vaccin Immunother, 20(1):2415823, 21 Oct 2024
Cited by: 0 articles | PMID: 39434217 | PMCID: PMC11497974
Review Free full text in Europe PMC
Multifaceted roles of trained immunity in diverse pathological contexts.
BMB Rep, 57(10):431-440, 01 Oct 2024
Cited by: 0 articles | PMID: 38835118 | PMCID: PMC11524827
Review Free full text in Europe PMC
Single high-fat challenge and trained innate immunity: A randomized controlled cross-over trial.
iScience, 27(11):111103, 05 Oct 2024
Cited by: 0 articles | PMID: 39493874 | PMCID: PMC11530819
Inflammatory Memory in Epidermal Stem Cells - A New Strategy for Recurrent Inflammatory Skin Diseases.
J Inflamm Res, 17:6635-6643, 21 Sep 2024
Cited by: 0 articles | PMID: 39323610 | PMCID: PMC11423832
Review Free full text in Europe PMC
TCA metabolism regulates DNA hypermethylation in LPS and <i>Mycobacterium tuberculosis</i>-induced immune tolerance.
Proc Natl Acad Sci U S A, 121(41):e2404841121, 30 Sep 2024
Cited by: 1 article | PMID: 39348545
Go to all (852) article citations
Data
Data behind the article
This data has been text mined from the article, or deposited into data resources.
BioStudies: supplemental material and supporting data
GEO - Gene Expression Omnibus
- (2 citations) GEO - GSE58310
Quick GO (2)
- (1 citation) GO - GO0006954
- (1 citation) GO - GO0006952
Similar Articles
To arrive at the top five similar articles we use a word-weighted algorithm to compare words from the Title and Abstract of each citation.
β-Glucan Reverses the Epigenetic State of LPS-Induced Immunological Tolerance.
Cell, 167(5):1354-1368.e14, 01 Nov 2016
Cited by: 311 articles | PMID: 27863248 | PMCID: PMC5927328
Candida albicans β-Glucan Differentiates Human Monocytes Into a Specific Subset of Macrophages.
Front Immunol, 9:2818, 30 Nov 2018
Cited by: 28 articles | PMID: 30555483 | PMCID: PMC6284042
Macrophages: innate memory training.
Nat Rev Immunol, 14(11):713, 10 Oct 2014
Cited by: 12 articles | PMID: 25301255
Infectious Agents as Stimuli of Trained Innate Immunity.
Int J Mol Sci, 19(2):E456, 03 Feb 2018
Cited by: 43 articles | PMID: 29401667 | PMCID: PMC5855678
Review Free full text in Europe PMC
Funding
Funders who supported this work.
British Heart Foundation (1)
Premature cardiovascular disease and platelet quantitative traits: identification and functional characterisation of novel high-penetrance mutations and rare alleles
Professor Willem Hendrik Ouwehand, University of Cambridge
Grant ID: RG/09/012/28096
NIDDK NIH HHS (1)
Grant ID: P30 DK043351
National Institute for Health Research (NIHR) (3)
Megakarocyte and Platelet Biology and Genomics
Professor Willem Hendrik Ouwehand, NHS Blood and Transplant
Grant ID: RP-PG-0310-1002
Grant ID: NF-SI-0510-10214
Grant ID: NF-SI-0513-10151





