Abstract
Free full text

White-to-brown metabolic conversion of human adipocytes by JAK inhibition
Abstract
The rising incidence of obesity and related disorders such as diabetes and heart disease has focused considerable attention on the discovery of novel therapeutics. One promising approach has been to increase the number or activity of brown-like adipocytes in white adipose depots, as this has been shown to prevent diet-induced obesity and reduce the incidence and severity of type 2 diabetes. Thus, the conversion of fat-storing cells into metabolically active thermogenic cells has become an appealing therapeutic strategy to combat obesity. Here, we report a screening platform for the identification of small molecules capable of promoting a white-to-brown metabolic conversion in human adipocytes. We identified two inhibitors of Janus Kinase (JAK) activity with no precedent in adipose tissue biology that stably confer brown-like metabolic activity to white adipocytes. Importantly, these metabolically converted adipocytes exhibit elevated UCP1 expression and increased mitochondrial activity. We further found that repression of interferon signalling and activation of hedgehog signalling in JAK-inactivated adipocytes contributes to the metabolic conversion observed in these cells. Our findings highlight a novel role for the JAK/STAT pathway in the control of adipocyte function and establish a platform to identify compounds for the treatment of obesity.
Mammals possess two distinct types of adipose tissue: white and brown fat. White adipose tissue (WAT) stores excess energy and has a number of endocrine functions such as regulating satiety via leptin secretion. In contrast, brown adipose tissue (BAT) maintains body temperature via non-shivering thermogenesis. BAT releases energy in the form of heat by uncoupling the respiratory chain via uncoupling protein 1 (UCP1). In addition to thermogenesis, BAT activation in rodents accelerated plasma clearance of triglycerides, ameliorated insulin resistance and protected against obesity1, 2. Recently, PET/CT imaging revealed adipose tissue with thermogenic activity and UCP1 expression in human adults3. These studies also found that BAT is inversely associated with adiposity, high body mass index and hyperglycemia. Based on these findings, there has been an increased interest in BAT as a therapeutic target to treat metabolic disorders.
Mouse studies have reported the emergence of UCP1-expressing cells in WAT upon cold exposure, β-adrenergic stimulation and peroxisome proliferator-activated receptor gamma (PPARG) activation4-10, a phenomenon referred to as browning. These brown-like cells arise from the recruitment of specific precursor cells11 and/or the conversion of white into brown-like cells12. Two human trials have also demonstrated de novo generation of brown adipocytes upon cold acclimation combined with increased non-shivering thermogenesis and decreased body fat mass13, 14. These studies suggest identifying inducers of browning in humans may ameliorate obesity related diseases. To this end, we established a screening platform to discover small molecules capable of promoting white-to-brown metabolic conversion in human adipocytes and identified Janus kinase (JAK) inhibitors as molecules with browning potential. In addition, we show that human pluripotent stem cell-derived adipocytes provide a scalable, robust and reliable cell model for adipocyte browning studies, compound screening and drug discovery.
RESULTS
A screening platform for adipocyte browning identifies inducers of UCP1
We used a human pluripotent stem cell (PSC)-derived adipocyte model to study the pharmacological conversion of white to brown-like adipocytes. In this approach, adipocytes are obtained via the inducible expression of transcription factors in PSC-derived mesenchymal progenitor cells (MPCs) and the addition of an adipogenic cocktail to the media15. Inducible expression of peroxisome proliferator-activated receptor gamma2 (PPARG2) alone or a combination of PPARG2, CCAAT/enhancer-binding protein beta (CEBPB) and PR domain containing 16 (PRDM16) drives cell differentiation towards the white (PSC-WA) or brown (PSC-BA) lineage respectively15. We reasoned that CEBPB and PRDM16 could be replaced by small molecules to direct PPARG2-expressing MPCs towards a brown-like phenotype (Fig. 1a). We monitored the responsiveness of PSC-WA to known modulators of UCP1 expression16 and observed up-regulation of UCP1 mRNA levels upon treatment with forskolin, 3-isobutyl-1-methylxanthine (IBMX), rosiglitazone and bone morphogenic protein 7 (BMP7), validating the use of PSC-WA for browning assays (Supplementary Figure 1).
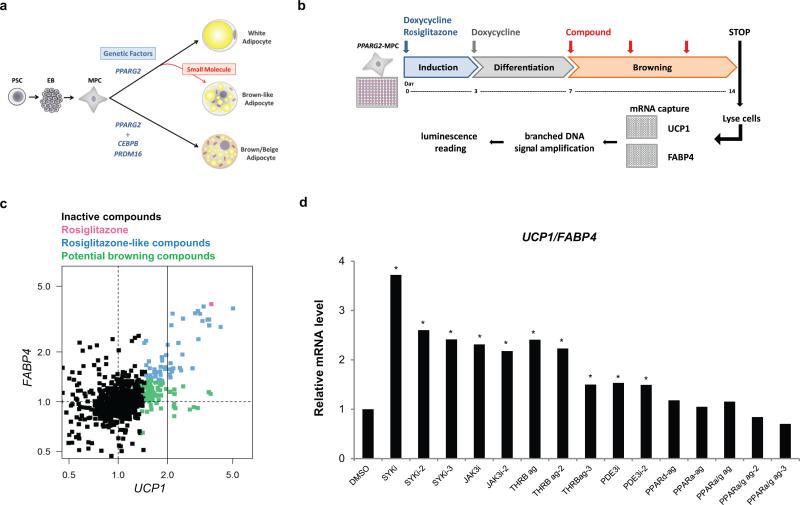
a) Conceptual strategy to identify small molecules with adipocyte browning effect using human stem cells. PSC: pluripotent stem cells, EB: embryoid bodies, MPC: mesenchymal progenitor cells.
b) Adipocyte browning screen, assay workflow. PPARG2 expressing-MPC (PPARG2-MPC) were maintained in adipogenic medium containing doxycycline and rosiglitazone for 3 days in order to induce adipogenesis, and differentiated in the absence of rosiglitazone for 4 days. A library of 867 compounds of known mode of action was applied to PSC-WA at day 7, 10 and 12. Total mRNA was collected at day 14, and UCP1 and FABP4 mRNA levels were quantified using the branched DNA technology. PPARG2-MPC: mesenchymal progenitor cells transduced with rtTA and doxycycline-inducible PPARG2 expression vectors. For more details see the methods section.
c) Scatter plot display of browning screen results. Each data point represents the average of two biological replicates per compound, normalized on DMSO control. X axis: UCP1 mRNA level as an indicator of adipocyte browning, Y axis: FABP4 mRNA as an indicator of general adipogenesis. The color code distinguishes inactive compounds (black) from active ones: Rosiglitazone (red), Rosiglitazone-like compounds that increase UCP1 and FABP4 (blue), and potential browning compounds that induce UCP1 specifically (green). Dashed lines indicate neutral conditions, solid line delineates UCP1 induction above 2 fold.
d) Validation of browning hits by bDNA analysis showing that JAK3 inhibitors, SYK inhibitors and THRB agonists scored as best UCP1/FABP4 inducers. All compounds were added at a 5 μM final concentration. X axis: Compounds are identified by target and mode of action. i= inhibitor, ag = agonist. For chemical nomenclature see the methods section. Values represent the mean of two biological replicates.
Utilizing this model we established a screening assay for assessing the conversion of white to brown-like adipocytes (Figure 1b). A focused library of 867 small molecules that has a large degree of activity annotation, facilitating deconvolution of mechanism of action was applied at day 7, when cells are differentiating and will adopt a white phenotype if no additional stimulus is applied. Adipocytes were exposed to compounds for 7 days and collected at day 14 for analysis. As a browning index, UCP1 expression was monitored by UCP1 mRNA capture plates followed by branched DNA amplification. Expression of fatty acid binding protein 4 (FABP4), an adipocyte specific gene, served as internal control to eliminate anti and pro-adipogenic compounds not specific to UCP1, such as the PPARG agonist rosiglitazone (Fig. 1c, red dot). Out of 135 UCP1 inducers (see online methods for statistical analysis), 52 compounds induced both UCP1 and FABP4 levels (Fig. 1c, blue dots). Among the 83 compounds that induced UCP1 specifically (Fig. 1c, green dots), compounds that induced UCP1 at least 2 fold were selected as browning hits and compared to rosiglitazone-like compounds in an independent experiment (Fig. 1d and Supplementary Table 1). UCP1 induction by thyroid hormone receptor beta (THRB) agonists and phosphodiesterase enzyme 3 (PDE3) inhibitors is in accordance with previously reported up-regulation of UCP1 promoter activity by thyroid hormone and cAMP17-20. Of particular interest, three annotated inhibitors of Spleen Tyrosine Kinase (SYK) and two inhibitors of Janus Kinase 3 (JAK3) showed the highest UCP1/FABP4 ratio. JAK3 and SYK are best characterized for their role in immune cells development and physiology and as mediators of pro-inflammatory pathways21. Recently, the JAK-STAT pathway was found to modulate early adipogenesis upstream of PPARG but little evidence supported a role in adipose tissue remodeling and thermogenicity22.
A subset of UCP1-inducers modulate adipocyte lipid droplet morphology
White adipocytes have a single or a few large lipid droplets, whereas the brown adipocytes contain multiple small lipid droplets. We exploited this morphological difference to evaluate the browning effects of newly identified UCP1-inducer compounds. Following treatment of PSC-WA with selected compounds (Fig. 1b), we used fluorescent dyes to mark nuclei and lipid droplets, and performed high content imaging to quantify the number and size of lipid droplets per cell. After 7 days of treatment with selected JAK3 and SYK inhibitors, we observed changes in lipid droplet morphology that are typical of brown-like adipocytes (Fig. 2a). Contrastingly, a THRB agonist was inert in this assay despite elevation of UCP1 expression (Fig. 2a and and1d).1d). We determined the lipid area of small (<1070μm2) and large (>1070μm2) droplets per well (Fig. 2b, curve graphs) and used the ratio small/large droplet area as a ‘brown-like lipid index’ for each compound, as exemplified in Figure 2c for DMSO, JAK3 inhibitor, SYK inhibitor and THRB agonist. A total of 39 compounds that induced UCP1 or displayed visible effects on lipid morphology during the browning screen were thereby quantified and ascribed a brown-like lipid index (Fig. 2d, Y-axis). When the two browning indices were taken into account, i.e. UCP1/FABP4 and lipid droplet morphology, JAK3 and SYK inhibitors were the most potent browning compounds from our screen (Fig. 2d). In addition, this analysis revealed that brown-like lipid morphology was poorly correlated with elevated UCP1 expression, indicating that quantification oflipid droplet size is not a suitable phenotypic readout for adipocyte browning, at least in our system.
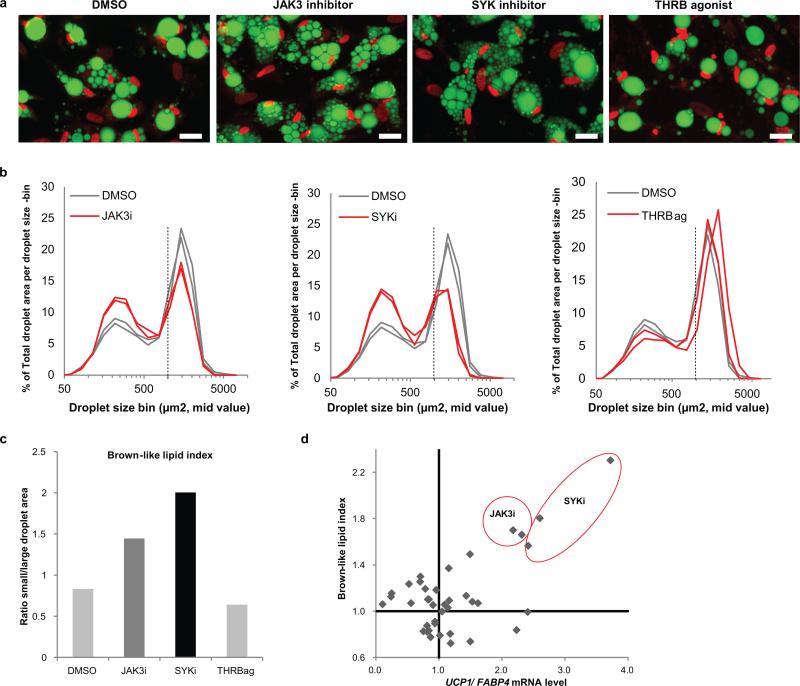
a) PSC-WA were differentiated and treated as described in Fig1B. At day 14, cells were fixed, stained and imaged by confocal microscopy. Green: lipids, Red: nuclei. Scale bars, 50μm. Images are representative of n =3 biological replicates.
b) Quantification of changes in lipid morphology is depicted as fraction of total lipid area (Y axis) per lipid droplet size (X axis). JAK3 inhibitor, SYK inhibitor and THRB agonist-treated cells are shown in red and DMSO in gray. Values represent the mean of two biological replicates.
c) Bar graph illustrating the quantification of a brown-like lipid index, determined by calculating the ratio of total area for small (<1070μm2) versus large (>1070μm2) lipid droplets for the graphs in b), normalized to the DMSO control sample. Values represent the mean of two biological replicates.
d) Scatter plot showing the relation between UCP1/FABP4 mRNA (X axis) and brown-like lipid morphology (Y axis) upon treatment with 39 selected compounds. Brown-like lipid index refers to the ratio of small/large lipid droplets normalized on DMSO as determined in b). Values represent the mean of two biological replicates.
Validation of tofacitinib and R406 browning compounds in primary adipocytes
As model browning compounds, we selected the JAK3 inhibitor tofacitinib and the SYK inhibitor R406 for their potency in our browning assay and for their clinical relevance (Supplementary Figure 3). We examined the dose-response of tofacitinib and R406 in PSC-WA and found that the UCP1/FABP4 ratio reaches a plateau at 2 μM for tofacitinib and 1 μM for R406 (Fig. 3a). Graphical representation of UCP1 and FABP4 normalized on housekeeping gene revealed a UCP1-specific effect of tofacitinib, while R406 induced both UCP1 and FABP4 expression when dosed above 1 μM (Fig. 3a). We confirmed that tofacitinib and R406-mediated increases in UCP1 mRNA levels translated into accumulation of UCP1 protein (Fig. 3b) and observed concomitant elevation in PRDM16 protein (Fig. 3b).
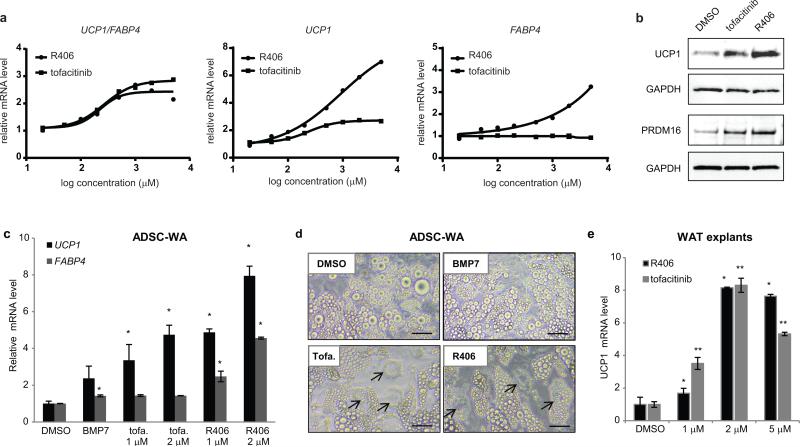
a) bDNA analysis of dose response with tofacitinib and R406. At high doses, R406 increases both UCP1 and FABP4 expression but UCP1/FABP4 remains above 2. Values represent the mean of two biological replicates.
b) Western blot analysis showing that up-regulation of UCP1 and PRDM16 protein levels correlates with up-regulation of UCP1 mRNA by tofacitinib and R406.
c) bDNA analysis showing that tofacitinib (tofa.) and R406 increase UCP1 expression in human primary adipocytes. ADSC: Adipose tissue-derived stromal cells. Values are mean ± s.d. of n = three biological replicates and differences from DMSO are significant for * P < 0.05. P values were calculated using the two-tailed paired Student's t-test.
d) Bright field images showing that tofacitinib (tofa.) and R406 induce brown-like lipid morphology (arrows) in human primary adipocytes more prominently than BMP7. ADSC: Adipose tissue-derived stromal cells. Scale bars, 20μm. Data representative of 3 independent experiments. For uncropped images see Supplementary Figure 3.
e) RT-PCR analysis of UCP1 gene expression in mouse subcutaneous white adipose tissue (WAT) explants following 7 days of treatment with the indicated compound. Values are mean ± S.E.M. of n = three biological replicates of pooled tissue from 5 mice and differences from DMSO are significant for * P < 0.05 and ** P < 0.005. P values were calculated using the two-tailed paired Student's t-test.
Importantly, we validated the effects of tofacitinib and R406 on UCP1 and FABP4 expression in lentiviral-free, human primary adipose tissue-derived stem cells (ADSCs) differentiated according to standard procedures (Fig. 3c). Tofacitinib and R406 also caused the emergence of brown-like lipid droplet morphologies in ADSC-derived adipocyte cultures and appeared superior to BMP7 in this assay (Fig. 3d). Finally, we subjected mouse white adipose tissue explants to increasing doses of tofacitinib and R406 and observed up-regulation of UCP1 expression in explants of sub-cutaneous, but not visceral, origin (Fig. 3e and Supplementary Figure 4).
Tofacitinib and R406 block the JAK-STAT1/3 pathway during adipocyte browning
Tofacitinib is a potent inhibitor of signaling through JAK1 and JAK3 with 5-100 fold selectivity over JAK2 in cell-based assays23, 24. R406 is described as a potent and specific SYK inhibitor, but also targets JAKs, c-kit, Lck, and FLT3 when profiled at 2 μM25. We sought to determine the pharmacological signature of tofacitinib and R406 in human adipocytes where target abundance and signaling activity might differ from immune cells. First we used RNA sequencing analysis of PSC-WA to evaluate the abundance of known targets of tofacitinib and R406. JAK1 was the most abundantly expressed member of the Janus kinase family, followed by Tyk2, JAK2 and JAK3 (Figure 4a). The abundance of SYK, c-kit, Lck and FLT3 was below statistical significance suggesting that R406 may exert its browning effect via JAK inhibition.
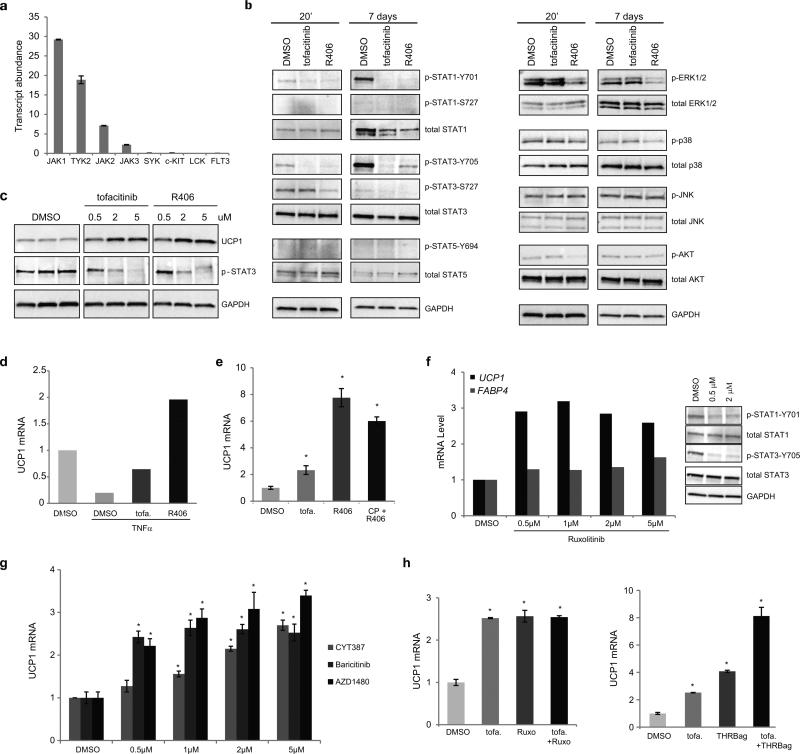
a) Transcript abundance in RPKM (reads per kilobase transcript per million reads) of known targets of tofacitinib and R406 indicating that the JAK kinases are predominantly represented in PSC-WA. Values are mean ± s.d. of n = three biological replicates.
b) Western blot analyses of STATs, AKT and MAPKs in PSC-WA previously treated with DMSO, tofacitinib and R406 for 20 minutes or 7 days showing a pronounced inhibition of STAT phosphorylation by tofacitinib and R406 at both time points. R406 also significantly decreased phosphorylation levels of AKT and ERK1/2. The data shown is representative of two independent experiments.
c) Western blot analysis of a dose-response with tofacitinib and R406 showing the correlation between UCP1 accumulation and inhibition of STAT3 phosphorylation. The data shown is representative of two independent experiments.
e) bDNA analysis of PSC-WA differentiated as in figure 1b) and treated with TNFα at day 12, indicating that the negative effect of TNFα on UCP1 expression is rescued by tofacitinib and R406. Values represent the mean of two biological replicates.
f) Tofacitinib (tofa.) and R406 do not synergize during adipocyte browning. PSC-WA were treated with tofacitinib, R406 or a combination of both and analyzed for UCP1 expression by bDNA. Values are mean ± s.d. of n = three biological replicates and differences from DMSO are significant for * P < 0.05. P values were calculated using the two-tailed paired Student's t-test.
d) The JAK 1/2 inhibitor Ruxolitinib positively modulates UCP1 expression (graph) and inhibits STAT1/3 phosphorylation (right panels) in PSC-WA. Graph values of bDNA analysis represent the mean of two biological replicates. Images of western blot analysis are representative of two independent experiments.
g) bDNA analysis showing that the JAK1/2 inhibitors CYT387, AZD1480 and Baricitinib positively regulate UCP1 expression in PSC-WA. Values are mean ± s.d. of n = three biological replicates and differences from DMSO are significant for * P < 0.005. P values were calculated using the two-tailed paired Student's t-test.
h) Tofacitinib (tofa.) synergizes with THRB agonist (right panel) but not with Ruxolitinib (Ruxo., left panel) during adipocyte browning. Values are mean ± s.d. of n = three biological replicates and differences from DMSO are significant for * P < 0.005. P values were calculated using the two-tailed paired Student's t-test.
To profile possible downstream targets, PSC-WA were treated with tofacitinib and R406 at the determined ‘browning dose’ (2 and 1 μM respectively) for 20 minutes and screened by reverse phase protein array (RPPA) covering 51 different phospho-isoforms. RPPA analysis revealed STAT3 as the main downstream target for both tofacitinib and R406 (Supplementary Figure 5). While the effect of tofacitinib was restricted to STAT3 in the RPPA panel, R406 had additional downstream targets including AKT, EGFR and ERK1/2 (Supplementary Figure 5). We further investigated the effect of tofacitinib and R406 on JAK/STAT, AKT and ERK1/2 pathways by immunoblot analysis. As shown in Figure 4b, tofacitinib and R406 blocked tyrosine phosphorylation of STAT1 and STAT3 after 20 minutes, an effect that is exacerbated upon 7 days of treatment. In a dose response experiment, the gradual increase in UCP1 levels negatively correlated with STAT3 phosphorylation (Fig. 4c). Noticeably, STAT1 protein level decreased after treatment with tofacitinib and R406. Tofacitinib had no effect on AKT and MAPKs, whereas R406 significantly decreased the phosphorylation level of AKT, ERK1 and ERK2 (Fig. 4b). AKT phosphorylation was restored at day 7, suggesting an adaptive response to maintain adipocyte homeostasis and insulin signaling. Phospho-STAT5 was undetectable in our assay and p38 and JNKs were unaltered at both times. Thus, JAK inhibitors likely change UCP1 expression via a different route than the cAMP/p38 pathway downstream of β3-adrenergic stimulation26, 27.
Tumor necrosis factor alpha (TNF-α) represses UCP1 expression in brown adipocytes28, an effect also observed in PSC-WA (Fig.4d). Since TNF-α can signal through JAK/STAT and ERK pathways, we tested CP-690550 and R406 ability to antagonize TNF-α repression of UCP1 expression. CP-690550 and R406 partially and fully restored UCP1 levels respectively, in agreement with the JAK/STAT-specific and JAK/STAT-ERK1/2 effects of each compound (Fig.4b). In addition, the combination of CP-690550 and R406 was not superior to R406 alone in increasing UCP1 expression (Fig.4e), supporting evidence that CP-690550 and R406 functionally overlap.
Finally, we tested Ruxolitinib, a JAK1/2 inhibitor, and observed up-regulation of UCP1 expression without significant regulation of FABP4 with concomitant inhibition of STAT1/3 phosphorylation (Fig. 4f). Three other JAK1/2 inhibitors, AZD1480, CYT387 and Baricitinib, also showed up-regulation of UCP1 expression (Fig. 4g). A combination of tofacitinib and Ruxolitinib did not further increase UCP1 expression (Fig. 4h, left panel), unlike a combination of tofacitinib with a THRB agonist (Fig. 4h, right panel). Altogether, the data highlight a central role for the JAK1/2-STAT1/3 pathway in UCP1 expression in adipocytes.
Progressive and stable conversion of adipocytes by JAK inhibition
Molecules that increase UCP1 expression by directly activating the UCP1 promoter26 may not be suitable for in vivo induction of browning or therapeutically useful to combat obesity. Alternative mechanisms of browning such as adipocyte metabolic remodeling are needed, and our screening platform was designed for this purpose. We examined the kinetics of tofacitinib and R406-mediated browning in comparison to THRB agonist, which also ranked in the top-10 best UCP1 inducers but failed to modify lipid droplet morphology (Fig. 1d and and2).2). The UCP1 promoter contains a thyroid hormone responsive element (TRE)20 and as expected, a THRB agonist rapidly increased UCP1 levels (Fig. 5a). Contrastingly, UCP1 mRNA levels increased more slowly and progressively in tofacitinib and R406-treated adipocytes (Fig. 5a), consistent with remodeling of cellular metabolism rather than direct modulation of UCP1 promoter activity.
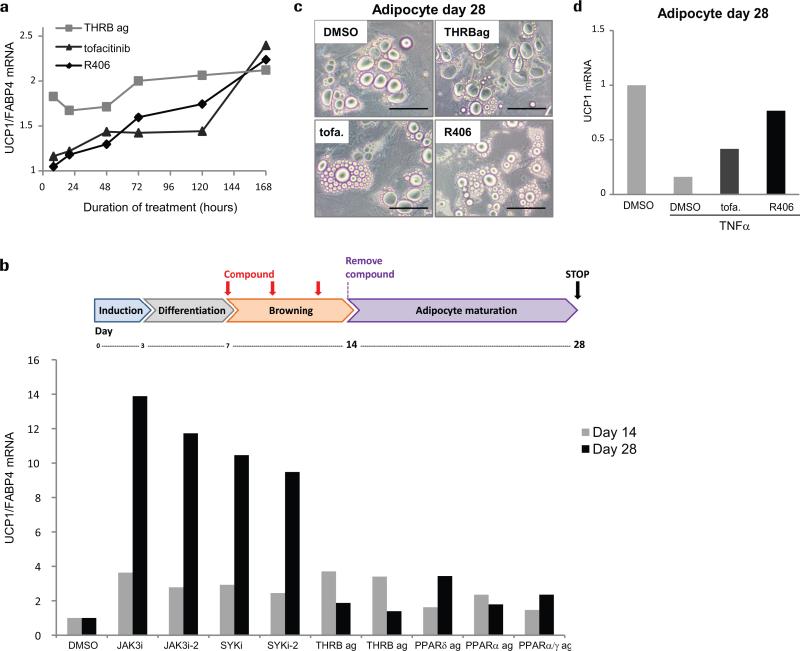
a) bDNA quantification of UCP1 mRNA levels over time, showing that the progressive accumulation of UCP1 induced by tofacitinib and R406 contrasts with the acute effect of THRB agonists. Values represent the mean of two biological replicates.
b) Schematic illustration of experimental design for b), c), and d): PSC-WA were treated with compounds for 7 days, washed 3 times with compound-free medium, and maintained in compound-free medium for an additional 14 day-period. Images were captured at day 28 prior to addition of lysis buffer and bDNA analysis. Only JAK3i and SYKi-pre-treated cells displayed high levels of UCP1 mRNA relative to DMSO. Values represent the mean of two biological replicates.
c) Bright field images showing a reduced lipid vacuole size at day 28 for tofacitinib (tofa.) and R406-pre-treated adipocytes. Scale bars, 50μm. Images are representative of two independent experiments.
d) DMSO, tofacitinib (tofa.) and R406-pre-treated adipocytes were exposed to TNFα from day 26 to 28. bDNA analysis shows that pre-treatment with tofacitinib and R406 protects adipocytes from TNFα-mediated down-regulation of UCP1 expression. Values represent the mean of two biological replicates.
If a stable white to brown-like conversion is achieved via JAK inhibition, the brown-like phenotype should persist upon removal of CP-690550 and R406. To test this, we selected 9 inducers of UCP1 mRNA levels from the original screen and treated PSC-WA with each compound for 7 days, washed with compound-free medium, and maintained in compound-free medium for an additional 14 day-period (Fig. 5b). Remarkably, all cells pre-treated with JAK inhibitors displayed high levels of UCP1 mRNA and reduced lipid droplet size at day 28 compared to DMSO-treated cells while all other compounds lost their browning effect after washout (Fig. 5b-c). Finally, we repeated the washout experiment with tofacitinib and R406 and added TNF-α at day 26, 48 hours prior to cells collection. Even in the absence of compounds, adipocytes pre-treated with tofacitinib and R406 maintained higher level of UCP1 when challenged with TNF-α (Fig. 5d). Altogether, these results indicate that JAK inhibition leads to the stable acquisition of brown-like metabolic properties in human adipocytes.
JAK-inactivated adipocytes acquire a brown-like metabolic program
Brown adipocytes are characterized by high mitochondrial content as well as high metabolic activity1. Mitochondrial content was assessed by determining the ratio of subunit I of Complex IV (COX-I), which is mitochondrial DNA-encoded, on the 70kDa subunit of Complex II (SDH-A), which is nuclear DNA-encoded. Quantification of COX-1 and SDH-A immunoblots showed a significant up-regulation of mitochondrial content in R406 and tofacitinib-treated PSC-WA and ADSC adipocytes (Fig. 6a). To determine the impact of these changes on metabolic activity, oxygen consumption rate (OCR) was measured. We found that basal, uncoupled and maximal respiration were increased when PSC-WA and ADSC adipocytes were pre-treated for 7 days with tofacitinib or R406 as compare to a DMSO control (Fig. 6b). Thermogenesis in brown adipocytes is fuelled by lipid catabolism, which can be measured by release of glycerol from fatty acid. We quantified the level of free glycerol from adipocyte culture medium and observed a ~2.5 fold increase of basal lipolysis in tofacitinib and R406-treated adipocytes (Fig. 6c), but no significant increase in forskolin (FSK) stimulated lipolysis (Fig. 6c). Altogether, these data suggest the engagement of a brown-like metabolic program upon inhibition of JAK/STAT in human adipocytes.

a) Mitochondrial content was assessed by determining the ratio of COX-I on SDH-A protein levels by immunoblots as shown in upper panel for PSC-WA. Quantification of immunoblots revealed up-regulation of mitochondrial content in tofacitinib and R406-treated PSC-WA and ADSC adipocytes. Values are mean ± s.e.m. of n = three biological replicates and differences from DMSO are significant for * P < 0.05 and ** P < 0.01. P values were calculated using the two-tailed paired Student's t-test.
b) Tofacitinib and R406-treated PSC-WA and ADSC adipocytes have higher oxygen consumption rate (OCR) compare to DMSO-treated adipocytes. Values are mean ± s.e.m. of n = four (DMSO and tofacitinib) and n = three (R406) biological replicates.
c) The degree of lipolytic activity was assessed by quantification of glycerol release. tofacitinib (tofa.) and R406-treated ADSC adipocytes showed increased lipolysis in the basal state (upper graph), but no in Forskolin (FSK)-stimulated cells (lower graph. Values are mean ± s.e.m. of n = three biological replicates and differences from DMSO are significant for * P < 0.05. P values were calculated using the two-tailed paired Student's t-test.
JAK-inactivated adipocytes maintain a white adipocyte transcriptional identity
To determine whether tofacitinib and R406 treatment had converted the transcriptional identity of white adipocytes, we performed RNA sequencing and analyzed the whole transcriptome of MPC, PSC-WA, PSC-BA and compound-treated PSC-WA. We used multidimensional scaling to project expression data, where distances between samples reflect the differences between global gene expression profiles. As expected, PSC-WA and PSC-BA moved away from undifferentiated PSC-MPC, and away from each other (Fig. 7a). Despite the acquisition of a brown-like metabolic profile, tofacitinib and R406-treated PSC-WA global expression profiles clustered with untreated PSC-WA samples and not with PSC-BA (Fig. 7a). Consistent with our findings, RNAseq analyses revealed a strong induction of UCP1 expression by tofacitinib and R406 (Fig. 7b). Among known positive regulators of brown adipogenesis, R406 promoted mRNA expression of PGC1α, PGC1β and PPARG (Supplementary Figure 6). Intriguingly, PRDM16 mRNA expression was decreased in treated-PSC-WA despite an increase at the protein level (Supplementary Figure 6 and Fig. 3). These data indicate that adipocyte browning by JAK inhibition occurs by functionally remodelling white adipocytes and through the acquisition of brown-like metabolic activities rather than via cell fate conversion.

a-f) Adipocytes were differentiated according to scheme 1B, treated with tofacitinib and R406 at day 7, and collected at day 8 (24h time point) or day 14 (d7 time point). N= 3 biological replicates. Each independent biological replicate was pooled from two individual wells.
a) Multi-dimensional scaling of RNA sequencing data revealing the white lineage identity of tofacitinib (tofa.) and R406 treated PSC-WA.
b) Levels of UCP1 transcripts served as experimental control for adipocyte browning. UCP1 transcripts are higher in BA versus WA and higher in tofacitinib (tofa.) and R406-treated PSC-WA compare to DMSO-treated PSC-WA.
c) Interferon targets and pro-inflammatory pathways are significantly down-regulated by both compounds at 7d in PSC-WA. Enrichment scores of 9116 gene sets are compared between two time points (24h and 7d) for both compounds. Each circle represents one gene set that is coherently regulated by an upstream pathway. Black lines indicate the change of average scores, and blue lines the change of individual pathways that are significantly reduced (|ΔES|>=2).
d) Differential expression profiles of interferon targets induced by tofacitinib and R406. A substantial subset of target genes are negatively regulated in both cases, making the density curves of logFC shifts toward left and thereby forming a “red shoulder”. Compared with 24h, the expression of interferon pathway targets are repressed by both compounds at 7d (P =2.94E-6 and 2.06E-5, respectively; one-sided Kolmogorov-Smirnov test).
e) Differential expression profiles of selected interferon target genes in heatmap.
f) Whole transcriptome analysis revealed that the sonic hedgehog responsive genes GLI1, SFRP5, KLHL31 and SHH were up-regulated in tofacitinib (tofa.) and R406-treated adipocytes at day 7 compare to DMSO control. Values are mean ± s.d. of n = three biological replicates and differences from DMSO are significant for * P < 0.05 and ** P < 0.01. P values were calculated using the two-tailed paired Student's t-test.
Downregulation of IFN and activation of hedgehog signaling contribute to metabolic browning downstream of JAK inhibition
To decipher the molecular mechanisms underlying metabolic browning of adipocytes via JAK inhibition, gene set analyses were performed and enrichment scores calculated for 9116 gene sets. Strikingly, all gene sets altered by tofacitinib were down-regulated, most prominently interferon (IFN) response (IFNα/β/γ), STAT1/3/5 pathway and pro-inflammatory pathways (OSM and IRF) (Fig. 7c). Similarly, R406 largely attenuated IFN response and mediators of inflammation (OSM, TNF, chemokine) (Fig. 7c). Tofacitinib and R406 treated adipocytes closely resembled one another in the differential expression profiles of IFN targets, of which 25 genes where nearly equally regulated by the two inhibitors (Fig. 7d and e). IFNα/β/γ bind to JAKs and activate STAT1/2/329. Thus the gene set enrichment score confirm our findings that tofacitinib and R406 inhibit the JAK-STAT pathway in adipocytes. The transcriptional changes downstream of R406 displayed a broader spectrum of regulated gene sets in comparison to tofacitinib including up-regulation of PPARG, SREBF and BMP target genes (Fig. 7c and Supplementary Figure 6), an observation in accordance with its positive effect on FABP4 expression (Fig. 3a).
We further interrogated RNAseq data for individual genes up-regulated in both tofacitinib and R406-treated PSC-WA. 54 genes were up-regulated by both compounds, 17 of which were BAT-specific (Supplementary Figure 6). Notably, we observed that GLI1 was up-regulated in tofacitinib and R406-treated PSC-WA and in PSC-BA (Fig. 7f). Moreover, three GLI1-target genes, SFRP5, KLHL31 and SHH, were up-regulated, suggesting activation of sonic hedgehog (SHH) signalling downstream of JAK/STAT inhibition during adipocyte browning.
The common gene signature of tofacitinib and R406-treated PSC-WA suggests that inhibition of JAK/STAT signaling short-circuits the IFN/JAK/STAT positive feedback loop and thereby progressively alleviates anti-browning and/or activates browning signals. We verified this hypothesis by inducing a white phenotype after addition of IFNγ to PSC-WA, observable by the formation of unilocular lipid droplets, an effect that was reversed by addition of tofacitinib or R406 (Fig. 8a). Moreover, IFNγ treatment of PSC-WA and PSC-BA decreased the UCP1/FABP4 ratio and increased expression of the white adipocyte marker HSL (Fig. 8b).
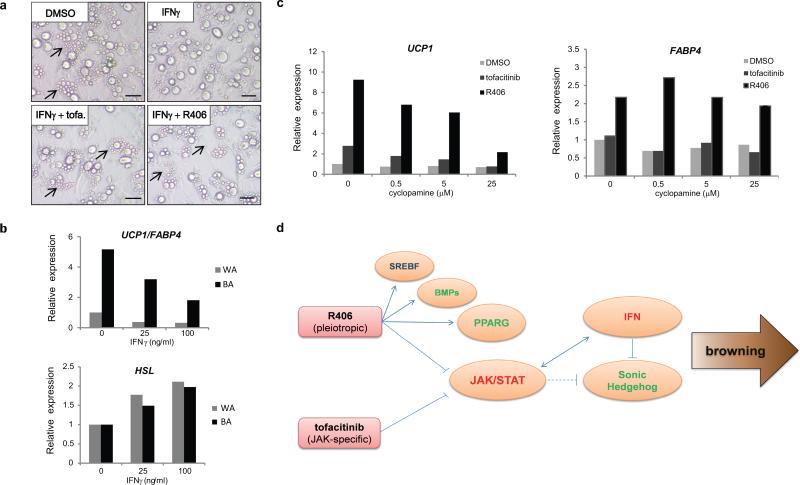
a) Whitening of IFNγ-treated adipocytes is visible as lipid accumulation forms a single, large vacuole. Treatment with tofacitinib (tofa.) and R406 restores the formation of small lipid droplets (arrows). Bright field images representative of two independent experiments. Scale bars, 20μm.
b) IFNγ treatment decreases the UCP1/FABP4 ratio (upper graph) and increases the expression of HSL, a marker of white adipocyte (lower graph) both in WA and BA. Values represent the mean of two biological replicates.
c) The SHH pathway antagonist cyclopamine fully blocks CP-6990550-mediated browning as judged by UCP1 level and partially blocks SYKi-mediated browning (left graph). Cyclopamine didn't restore R406-induced FABP4 levels, thereby decoupling the regulation of UCP1 and FABP4 by R406 (right graph). Values represent the mean of two biological replicates.
d) Model of adipocyte browning by pharmacological inhibition of JAK. Tofacitinib and R406 inhibit the JAK-STAT pathway in human adipocytes, leading to down-regulation of the interferon alpha, beta and gamma responses. Sustained shut down of IFN signaling relieves inhibition of the sonic hedgehog (SHH) pathway and thereby contributes to accumulation of UCP1. R406 acts as a pleiotropic drug with broad effects on adipocytes through activation of PPARG, BMPs and SREBF target genes. Red fonts: negative regulator of browning; Green fonts: positive regulator of browning; Arrows: activation; Flat lines: inhibition; Dash lines: hypothetical.
We observed that repression of IFN response was followed by up-regulation of BA-specific transcripts in tofacitinib and R406-treated PSC-WA including the GLI1 transcription factor (Fig. 7f). The previously reported antagonistic link between IFN and SHH30 prompted us to evaluate the functional relevance of up-regulation of GLI and GLI-target genes in tofacitinib and R406-mediated metabolic browning. To examine the contribution of SHH signalling downstream of JAK/STAT inhibition during metabolic browning of adipocytes, PSC-WA were subjected to increasing doses of cyclopamine, a SHH pathway inhibitor, in combination with DMSO, tofacitinib or R406 for 7 days. Addition of cyclopamine did not significantly alter gene expression in DMSO-treated PSC-WA but completely blocked tofacitinib-mediated induction of UCP1 expression (Fig. 8c). Interestingly, high doses of cyclopamine antagonized the effect of R406 on UCP1 expression but did not block the induction of FABP4, thereby decoupling the regulation of UCP1 and FABP4 by R406 (Fig. 8c). The up-regulation of PPARG activity by R406 (Supplementary Figure 6) likely accounts for this observation. UCP1 expression was also down-regulated in PSC-BA upon treatment with cyclopamine (Supplementary Figure 7). These results support a role for SHH signaling in the acquisition of brown-like properties in human adipocytes.
Overall, we provide evidence that inhibition of JAK down-regulates interferon signaling in human adipocytes. The persistent repression of IFN signaling relieves inhibition of the SHH pathway30 and thereby contributes to the upregulation of UCP1 and promotes the metabolic browning of adipocytes (Fig. 8d).
DISCUSSION
The remarkable phenomenon of adipocyte browning –or the emergence of brown-like adipocytes in white adipose depots– and its associated metabolic benefits has spurred interest in the discovery of pharmacotherapies targeting adipose tissue to treat metabolic diseases such as obesity and type 2 diabetes. Here, we established a small molecule screen to unravel pharmacologically accessible pathways capable of inducing a brown-like thermogenic program in human adipocytes. We identified tofacitinib and R406 as potent inducers of UCP1 and via secondary screening established their role in remodeling lipid droplet morphology in human adipocytes. We found that both compounds target the JAK-STAT1/3 pathway in adipocytes and have a distinct progressive and long-lasting effect that confers brown-like metabolic properties to white adipocytes. Our results further highlight important antagonistic interplays between interferon and sonic hedgehog signaling in adipocyte function.
Our findings are in line with previously proposed functions of Hedgehog in determining white versus brown adipose cell fate31. By using the Hedgehog activator Smoothened Agonist (SAG) in various mouse cell lines, Pospisilik et al. showed that activation of Hedgehog blocks white, but not brown, adipocyte differentiation and solely in the absence of cAMP and glucocorticoid signaling. In our study, activation of Hedgehog signaling by JAK inhibition occurred in PPARG2 expressing cells in the presence of cAMP and glucocorticoid signaling and promoted the acquisition of a brown-like metabolic phenotype. It is thus conceivable that activation of Hedgehog signaling after adipogenesis is initiated and during the maturation of white adipocytes may promote the acquisition of brown-like properties. We further demonstrated that pharmacological inhibition of Hedgehog decreased UCP1 expression in genetically programmed BA and brown-like adipocytes. Interestingly, Pospisilik et al. reported the up-regulation of UCP1 mRNA levels in WAT and BAT tissue upon genetic activation of Hedgehog in the aP2-SufuKO mouse, reminiscent of the data presented here upon treatment with tofacitinib and R406. More recently, an antagonistic crosstalk between Hedgehog and IFNγ in white adipose tissue has been described, wherein inhibition of Hedgehog signaling by IFNγ rescues white adipocyte differentiation30. Our findings further reinforce and refine the role of an IFN-JAK/STAT-SHH axis in human adipocyte biology. A recent study by Derecka et al. revealed a positive role for Tyk2 and Stat3 in the regulation of Myf5+ pre-adipocyte differentiation and BAT development in mice32. In light of our findings, it will be interesting to investigate the role of TYK2/STAT3 and other JAK/STATs in committed, functional brown adipocytes. A fine tuning of the JAK/STAT pathway at various stages of differentiation and maturation might be key for optimum BAT expansion and metabolic function.
The utility of JAK inhibition as a therapeutic for obesity is complicated by the well-described role of this signalling pathway in the immune system. In fact, tofacitinib is currently approved in the United States to treat rheumatoid arthritis. Thus, if one were to envision targeting adipose tissue by in vivo administration of an IFN/JAK/STAT inhibitor or similar compound it would almost certainly need to be delivered locally and prevented from spreading systemically or alternatively targeted selectively to white adipocytes. One could also conceive of a cell-based therapy wherein JAK inhibition of patient-derived adipocytes ex vivo is followed by transplantation to treat obesity, but this therapeutic modality would need to overcome numerous and significant obstacles before becoming a reality. A further limitation of the current study is the lack of evidence that JAK inhibition would promote metabolic browning in vivo, in particular in humans were evidence supporting this phenomenon is scant. Thus, additional research is required before inhibition of IFN/JAK/STAT signalling could be utilized therapeutically for the treatment of metabolic disorders.
Recent progress in the development of human embryonic stem cells and induced pluripotent stem cell -based models of disease have delivered unprecedented tools for basic research and translational medicine. However, their applicability for drug discovery remains to be fully realized. Here, we tested the suitability of a human PSC-derived adipocyte model15 for screening and functional browning assays. We found this model provided an inexhaustible and rapidly expandable source of human adipocytes for screening and downstream assays. Moreover, it proved easily scalable and highly reliable in our screening assay. The responsiveness of human PSC-derived adipocytes to known and newly identified browning molecules closely mirrored that of primary adipose derived stromal cells (ADSC) further validating this model for adipocyte cell fate and browning studies.
To conclude, we successfully performed a proof of concept screen utilizing an innovative stem cell-based model to identify compounds that convert human adipocytes to metabolically active brown-like cells. Given that this platform is amenable to very high-throughput screening it has considerable potential to contribute to the discovery of novel biological modulators of adipocyte cell fate and function and possibly to the development of new drugs for patients with metabolic disorders.
METHODS
Maintenance of human MPCs and ADSCs and differentiation into adipocytes
MPCs, white adipocytes (PSC-WA) and brown adipocytes (PSC-BA) were generated and differentiated as described by Ahfeldt et al.15 with the following modifications: MPCs were derived from human embryonic stem cell line SA001 (Cellartis), MPCs were seeded at 75% confluency in 15 cm dishes, incubated overnight with lentiviral particles at an MOI of 50 for each virus and maintained in growth medium without virus for 24 hours. Infected MPCs (herein PPARG2-MPCs) were counted and seeded in 96 well plates at a density of 37 500 cells/cm2 in growth medium. The next day (Day 0), growth medium was changed for induction medium consisting of adipocyte medium (DMEM, Knockout Replacement Serum, non-essential amino acids, insulin, dexamethasone) supplemented with 700 ng/ml doxycycline and 0.5 μM rosiglitazone. 72 hours later (Day 3), induction medium was replaced by differentiation medium (adipocyte medium supplemented with 700 ng/ml doxycycline) for 96 hours. From day 7, white (PSC-WA) and brown (PSC-BA) adipocytes were maintained in adipocyte medium and refreshed every two or three days.
Adipose tissue-derived stem cells (ADSCs) were obtained from PromoCell (human mesenchymal stem cell from adipose tissue or hMSC-AT, catalog number C-12977) and maintained in mesenchymal stem cell growth medium (PromoCell C-28010) according to manufacturer's instructions. For differentiation studies, ADSCs were seeded in 96 well plates at a density of 37 500 cells/ cm2. At 90% confluency, growth medium was changed for adipogenic differentiation medium (PromoCell C-28011) and refreshed every third day.
Compound library for adipocyte browning screen
A library of 867 small molecules of known mode of action was assembled by combining Roche internal reference compounds and commercially available screening libraries (Tocriscreen Collections #3514 Kinase inhibitor toolbox, Selleckchem Stem cell #L2100, Selleckchem Epigenetic #L1900 and Selleckchem GPCR #L2200). All compounds were pre-dissolved in DMSO, diluted to 5mM stock solutions and stored at -80°C. A list of compounds ID, mode of action and CAS numbers is available upon request.
Browning screen, statistical analysis and hit validation
PPARG2-MPCs were seeded in 96-well plates, induced and differentiated into PSC-WA as described above. A library of small molecules was applied at a 5 μM final concentration in biological duplicates at day 7, 10 and 12 in fresh adipogenic medium. DMSO and rosiglitazone (0.5 μM final concentration) were included on each plate as negative and positive controls respectively. Adipocytes were collected at day 14 and processed for UCP1 and FABP4 mRNA analysis by bDNA.
For statistical analysis, the raw mRNA signal was log-transformed and normalized to fold-change of the per plate median of the negative controls for all calculations, xnorm = log(sraw)/Mneg, where Mneg = medianplage{log(sneg)}. The data quality was assessed by calculating the Z’ factor based on positive and negative controls for each plate (Supplementary Figure 1b). For UCP1, <Z’> = 0.50, and Z’ > 0.50 for 11 out of 24 plates; for FABP4, <Z’> = 0.64, and Z’ > 0.50 for 18 out of 24 plates. This is characteristic for a good quality HCS34. To evaluate the replicates performance, biological duplicates were aggregated to their mean; 5% of the compounds were excluded from further analysis since their UCP1 difference was larger than 0.07; Supplementary Figure 1c shows that the replicate difference distribution is constant over the entire range, suggesting a constant replicate variance.
The remaining 832 compounds were used for hit selection as described before35. Knowing the constant variance from the controls, a p-value could be assigned to each compound following the test statistics T = (xnorm − 1)/σneg, where σneg = MADplate{log(sneg)}, xnorm(Mneg) = 1, which is normal distributed for inactive compounds (i.e., under the null hypothesis H0). We next estimated the false discovery rate (FDR) by p-value distribution analysis36 and applied a selection threshold of <0.01. Supplementary Figure 1d, left panel, illustrates the selected primary actives among the non-selected compounds in a quantile-quantile plot and the p-value distribution (inset). Next, a second hit selection process for FABP4 was included, again at a FDR cutoff <0.01 (Supplementary Figure 1d, right panel). We thereby found 135 UCP1 hits, of which 52 were assigned as “Rosiglitazone-like” compounds that induced both UCP1 and FABP4. In the validation experiment, we selected browning hits that induced UCP1 by at least two fold and UCP1/FABP4 by at least 1.5 fold. PPAR agonists were also included for a total of 33 re-tested compounds, of which 26 were confirmed (Supplementary Figure 1d, left panel). 15 validated hits and PPAR agonists are displayed in figure 1d and chemical names are provided in Supplementary Table 1.
High content imaging
At day 14 of browning assay, cells were fixed with 4% Paraformaldehyde in PBS for 15 minutes at room temperature, incubated with blocking buffer (5% Donkey Serum, 0.5% triton X-100 and 0.02% Azid NaN3 in PBS) for 30 minutes at RT and stained with 10 μM Hoechst [B2261, Sigma] and 20ng/ml of Bodipy ® 493/503 (D-3922, Life technologies) in PBS for 30 minutes at room temperature. Samples were kept in PBS. Fluorescence Images where acquired using the Opera QEHS reader (Perkin-Elmer, Hamburg, Germany) equipped with a Nipkow Spinning disc for confocality using a 20× objective. Hoechst stain was imaged with laser excitation at 405nm laser and a 455/70nm band path filter for emission. The lipid stain was imaged using laser excitation at 488nm, and a 535/60nm / BP filter for emission. The image segmentation was done with the instrument integrated software Acapella. A custom made algorithm was used to identify the morphologically heterologous nuclei in the mixed cell population: Larger, round and relatively dark (brown adipocytes, non-differentiated cells); smaller, bright, more irregular shaped nuclei (white). The lipid droplets were segmented independently as either larger round bright objects or darker spots and assigned to individual nuclei based on an equidistant ring around the nuclei. The ratio in the total area of cell related droplets above and below 1070 μm2 was quantified as shift between a more white and brown phenotype in the whole cell population. Cell classification; Adipocyte: total droplet area above 750 μm2 and/or >10 spots <50 μm2. White subtype: Minimum one droplet above 830 μm2, brown subtype: No larger droplets, minimum 10 spots.
Browning assay with tofacitinib, R406, JAK inhibitors and BMP7
Following screening and hit validation, browning assays were performed with commercially available JAK3 inhibitor (tofacitinib, Selleckchem S5001) and SYK inhibitor (R406, Selleckchem S1533). PSC-WA and ADSCs were differentiated and treated following the assay design of the browning screen. Tofacitinib was added at 2 μM and R406 at 1 μM final concentration unless otherwise indicated on figures and legends. THRB agonist (CAS 219691-94-8) was added at 5 μM final concentration. Ruxolitinib (Selleckchem S1378), Baricitinib (Selleckchem S2851), CYT387 (Selleckchem S2219) and AZD1480 (Selleckchem S2162) were added at 0.1-5 μM final concentration as indicated on figures.
Recombinant human BMP7 (R&D systems, 354-BP) was added at 10 ng/ml final concentration in adipogenic medium supplemented with 250 μM IBMX (3-Isobutyl-1-methylxanthine, Sigma-Aldrich I7018) at day 0, 3 and 5. In ADSCs, BMP7 was added at 10 ng/ml at day 7, 10 and 12. Tofacitinib and R406 were added following the same conditions as BMP7 for proper comparison.
Browning of mouse adipose tissue explants
Subcutaneous and visceral adipose depots were dissected from 8 week old male C57/Bl6 mice (n=5) and minced in standard media (DMEM containing 10 % FBS and 1% penicillin/streptomycin). Minced pieces were pooled, placed into a 100 μM nylon strainer and rinsed 3 times with standard media to remove red blood cells and excess lipid. Explants were carefully removed from the top of the strainer and plated into 24-well plates. Following 48 hours of acclimatization in adipogenic medium (DMEM, 10% FBS, 1% penicillin/streptomycin, 17 nM insulin, 1 μM dexamethasone) cells were treated with adipogenic medium containing DMSO, tofacitinib or R406 at 1, 2 and 5 μM final concentrations. Each treatment was performed in triplicate. Media with fresh compound was replaced every other day. Following 7 days of treatments, RNA was extracted in Trizol (Life Technologies) following standard protocol. RT-PCR was performed using Superscript III (Life Technologies) followed by analysis on a ViiA7 Real-Time PCR System (Life Technologies). Expression was analyzed relative to 18s RNA levels. Taqman probes were purchased from Life Technologies (UCP1: catalog number Mm01244861_m1, 18S: catalog number Mm03928990_g1).
Treatment with TNFα, INFγ and cyclopamine
TNFα (human recombinant Tumor necrosis factor alpha, Sigma-Aldrich T0157) was added directly to cell medium 48 hours or 24 hours before end of assay, at a final concentration of 0.25 ng/ml or 1 ng/ml respectively. IFNγ (human recombinant Interferon-gamma, Sigma-Aldrich I3265) and cyclopamine (Tocris ,1623) were added to fresh cell medium containing DMSO, tofacitinib or R406, at day 7, 10 and 12. For all treatments, corresponding volume of solvent (PBS, DMSO or ethanol, according to supplier's recommendation) was added to control wells and used for normalization.
Gene expression analysis by branched DNA (bDNA)
Adipocytes grown in 96 well plates were collected by replacing adipocyte medium with 100 μl/well of 1× RNA lysis mixture (Panomics QuantiGene2.0 Assay kit). Lysates were transferred to mRNA capture plates containing a specific capture probe (Supplementary Table 2), incubated overnight, processed for branched DNA amplification and analyzed according to the manufacturer's instructions (Panomics QuantiGene2.0 Assay). ACTB and PPIB probes were used as housekeeping genes for normalization. Data are presented as average and standard deviation of 2 or 3 biological replicates and normalized on DMSO control.
Western blot analysis
For protein analysis, browning assays were performed in 6 well plates initially seeded with 5×105 PPARG2-MPCs. At differentiation day 7, PSC-WA were treated with DMSO, tofacitinib (2 μΜ) or R406 (1 μM) for 20 minutes and lysed in 75 ul/well of cold RIPA buffer (60 mM Tris–HCl at pH 7.4, 150 mM NaCl, 0.25% SDS, 1% NP40) containing Complete protease inhibitor (Roche) and phosphoSTOP (Roche). For browning assays over 7 days, cells were lysed in cold RIPA buffer at day 14. Western blot analysis was carried out using the following antibodies: STAT1 (Cell Signaling 9172, 1:1000), phospho-STAT1-S727 (Cell Signaling 9177, 1:1000), phospho-STAT1-Y701 (Cell Signaling 9167, 1:1000), STAT3 (Cell Signaling 4904, 1:1000), phospho-STAT3-S727 (Cell Signaling 9134, 1:1000), phospho-STAT3-Y705 (Cell Signaling 9131, 1:1000), STAT5 (Cell Signaling 9363, 1:1000), phospho-STAT5-Y694 (Cell Signaling 9351, 1:1000), Akt (Cell Signaling 4691, 1:1000), phospho-Akt-Ser473 (Cell Signaling 4060, 1:1000), Erk1/2 (Cell Signaling 9102, 1:1000), phospho-Erk1/2-Thr202/Tyr204 (Cell Signaling 4370, 1:2000), p38 MAPK (Cell Signaling 8690, 1:1000), phospho-p38-Thr180/Tyr182 (Cell Signaling 4511, 1:1000), JNK (R&D AF1387, 1:1000), phospho-JNK-T183/Y185 (R&D AF1205, 1:400), UCP1 (R&D MAB6158, 1:2000), PRDM16 (R&D AF6295, 1:200), GAPDH (SIGMA, G8795, 1:2000), COX-I (abcam 14705, 1:1000) and SDH-A (abcam 14715, 1:1000). The CHEMIDOC MP Imaging System and ImageLab Analysis Tool (BIO-RAD) or the Licor odyssey system was used to scan & quantitate the band intensity.
Reverse phase protein array (RPPA) analysis
For RPPA analysis, browning assays were performed in 10 cm dishes initially seeded with 1×106 PPARG2-MPCs. At differentiation day 7, adipocytes were treated with DMSO, tofacitinib (2 μM) or R406 (1 μM) for 20 minutes and lysed in 100ul of CLB1 lysis buffer. Lysates were snap-frozen in liquid nitrogen, stored at −80°C and shipped to NMI Technology Transfer GmbH (Reutlingen, Germany) for RPP analysis as follow: Lysates were printed at 4 dilutions in duplicates onto ZEPTOMARK chips (Bayer Technology Services GmbH, Leverkusen, Germany) and analyzed for 51 selected phospho-analytes. Array images were analyzed using the ZeptoVIEW 3.1 software.
Measurement of cellular Oxygen Consumption Rate (OCR)
PPARG2-MPCs and ADSCs were plated at 7×104 cells/well in gelatin-coated XF 24-well cell culture microplates (Seahorse Bioscience), differentiated into adipocytes at confluence and treated with DMSO, tofacitinib and R406 as described above (browning assay). Day 14 adipocytes were incubated in pre-warmed unbuffered DMEM medium (DMEM containing 2 mM GlutaMAX, 1 mM sodium pyruvate, 1.85 g l−1 NaCl and 25 mM glucose) for 1 h. The oxygen consumption rate was measured by the XF24 extracellular flux analyser (Seahorse Biosciences). Mitochondrial biogenesis was profiled by injecting perturbation drugs, 2 μM oligomycin, 0.5 μM CCCP (carbonyl cyanide-p-trifluoromethoxyphenylhydrazone) and 5 μM antimycin A, in succession. OCR were determined by plotting the oxygen tension and acidification of the medium in the chamber as a function of time and normalized by protein concentration (picomoles per minute per milligram), respectively.
Lipolysis assay
To measure lipolysis activity, DMSO, tofacitinib and R406-treated ADSC adipocytes were starved in DMEM containing 1% FBS for 1 h. Cells were then incubated in HBSS (Hank's balanced salt solution) with 2% fatty-acid-free BSA alone or with 10 μM forskolin. The culture medium was collected for glycerol measurement using the free glycerol reagent (Sigma#F6428). Protein concentrations used to normalize glycerol content were measured using the Bradford protein assay (BioRad). Glycerol release was expressed in micrograms of glycerol per milligram of total protein or the lowest value is set to 1.
RNA sequencing of adipocyte browning
A total of six, 6-well plates were prepared as follow: In each plate, three wells were seeded with PPARG2-MPC, one well with PPARG2/CEBPB/PRDM16-MPC and one well with rtTA-MPC (5×105 cells/well). All cells were induced and differentiated as described above. At day 7, PSC-WA received either DMSO, tofacitinib (2 μM) or R406 (1 μM). PSC-BA and MPC received DMSO only. At day 8, three cell plates were collected by adding 350 ul/well of RLT lysis buffer (Qiagen), constituting the 24h-triplicates. In the remaining three plates, treatment with DMSO, tofacitinib and R406 was repeated at day 10 and 12. At day 14, cells were collected by adding 350 ul/well of RLT lysis buffer (Qiagen), constituting the 7days-triplicates. Total RNA from adipocyte lysate was extracted and purified using the RNeasy mini kit (Qiagen) according to the manufacturer's instructions. Genomic DNA was removed from the extracted RNA using the RNase Free DNase Set (Qiagen) and purified with RNeasy Minelute Clean up Columns (Qiagen). For all samples high quality RNA was obtained (RIN between 8.1 and 9.8). RNA Sequencing libraries were prepared from 1μg of total RNA using the Illumina TruSeq RNA Sample preparation Kit v2 (Illumina) following the manufacturer's instructions. Sequencing libraries were quantified using the Kapa Library Quantification kit (Kapa Biosystems) and quality controlled by capillary electrophoresis on a Bioanalyzer using High Sensitivity chips (Agilent Technologies). Libraries were sequenced in randomized pools of three or four libraries per lane on a HiSeq2000 sequencer (Illumina) for 2× 50 cycles using version 3 cluster generation kits and version 3 sequencing reagents (Illumina). One % of PhiX control library (Illumina) was spiked into each lane as a sequencing control.
Bioinformatics and statistical analysis of RNAseq data
RNA-seq reads were first aligned to human transcriptome, and unmapped reads were then aligned to human genome. Reads that map to annotated genes were modelled by negative binomial distribution, and differential expression was quantified by generalized linear models (GLMs) implemented in the edgeR package. For gene-level analysis, the filter for significantly differentially expressed genes was set as |logFC|>=1 (logFC=log fold-change) and FDR<0.05 (Likelihood-ratio test, adjusted with the Benjamini-Hochberg method for multiple testing). The filtering was not necessary for gene-set-level analysis. Gene set analysis was performed with both Gene Set Enrichment Analysis (GSEA) and the CAMERA method. Gene set enrichment score (ES) was defined as ES=sign(NESGSEA)log10(FDRGSEA), namely the sign of normalised enrichment score reported by GSEA (+1 if NESGSEA>0, and -1 otherwise), multiplied by log10-transformed false-discovery rate (the q-value). The enrichment score ranges between -4 and 4, corresponding to negatively or positively enriched gene sets that are statistically highly significant (FDR q<=1E-4). Gene sets in use included both pathway information from public repositories, including Reactome, StringDB and NCI-Nature pathway database, and manually curated sets of genes whose expression are regulated by upstream pathways.
General statistical analyses
Biological replicates in figure legends refer to independent adipocyte differentiation and treatment, i.e. batch-infected PPARG2-MPCs were seeded in separate wells or dishes, differentiated for 7 days in the absence of compound and matured for another 7 days in the presence of compound. Biological replicates therefore capture the biological variation of stem cell differentiation into mature adipocytes and the cellular response to compound. Repeat experiments performed with different batches of PPARG2 lentiviral particles and MPC infections are shown in Supplementary Table 3. Technical replicates were replicate measurements of a given sample in readout assays (bDNA, RT-PCR, lipid droplet imaging, glycerol release). Statistical analyses were carried using a two-tailed unpaired Student's t-test when n ≥ 3 biological replicates.
Supplementary Material
1
9
10
2
3
4
5
6
7
8
Acknowledgements
The authors thank Inga Clausen, Martine Kapps, Roland Schmucki and Angelika Schuler for technical support, Klaus Christensen and Martin Graf for stem cell support, Laura Badi for preliminary data analysis, Corinne Solier, Annette Schell-Steven and Tobias Bergauer for experimental planning and Michael Pawlak (Natural and Medical Sciences Institute at the University of Tübingen) for RPPA analyses. A.M. was supported by the Roche Postdoctoral Fellowship (RPF) program (2011-2013). This research was supported in part by F.Hoffmann-La Roche; grant R01DK095384 (C.A.C. and Y.K.L.) and R01DK097768 (C.A.C.) from the United States Institutes of Health (NIH); and Harvard University.
Footnotes
Author contributions: A.M. designed and performed experiments, analyzed data and wrote the manuscript; Y-K.L. performed experiments, analyzed data and edited the manuscript; R.G., C.S.H. and F.X. performed experiments; J.D.Z. and M.E. performed bioinformatics analyses and contributed to main text related to figure 7; H.H.T., S.Z. and M.P. performed high content imaging analysis; A.K. performed RNA sequencing; C.A.M. and R.T.S. supervised stem cell activities; K.E.A. supervised the project and C.A.C. supervised the project and wrote the manuscript. A.M., Y-K.L., R.G., C.H., M.P., J.D.Z., H.H.T., S.Z. and A.K. contributed to description of online methods.
AUTHOR INFORMATION
Primary accession number
RNA sequencing data set generated in this study is available online at NCBI GEO, Accession Number GSE57896.
Competing financial interests
The authors declare no competing financial interests.
Primary Accession Numbers
RNA sequencing data set generated in this study is available online at NCBI GEO, Accession Number GSE57896.
REFERENCES
Full text links
Read article at publisher's site: https://doi.org/10.1038/ncb3075
Read article for free, from open access legal sources, via Unpaywall:
https://europepmc.org/articles/pmc4276482?pdf=render
Citations & impact
Impact metrics
Citations of article over time
Alternative metrics
Smart citations by scite.ai
Explore citation contexts and check if this article has been
supported or disputed.
https://scite.ai/reports/10.1038/ncb3075
Article citations
Single-cell transcriptomic analysis reveals dynamic activation of cellular signaling pathways regulating beige adipogenesis.
Exp Mol Med, 56(10):2309-2322, 28 Oct 2024
Cited by: 0 articles | PMID: 39465352 | PMCID: PMC11541910
Identification of Bioactive Substances Derived from the Probiotic-Induced Bioconversion of Lagerstroemia speciosa Pers. Leaf Extract That Have Beneficial Effects on Diabetes and Obesity.
Microorganisms, 12(9):1848, 06 Sep 2024
Cited by: 0 articles | PMID: 39338524 | PMCID: PMC11434581
Head and neck cancer: pathogenesis and targeted therapy.
MedComm (2020), 5(9):e702, 21 Aug 2024
Cited by: 1 article | PMID: 39170944 | PMCID: PMC11338281
Review Free full text in Europe PMC
Non-invasive mapping of brown adipose tissue activity with magnetic resonance imaging.
Nat Metab, 6(7):1367-1379, 25 Jul 2024
Cited by: 0 articles | PMID: 39054361 | PMCID: PMC11272596
Thermogenic Fat as a New Obesity Management Tool: From Pharmaceutical Reagents to Cell Therapies.
Biomedicines, 12(7):1474, 04 Jul 2024
Cited by: 0 articles | PMID: 39062047 | PMCID: PMC11275133
Review Free full text in Europe PMC
Go to all (103) article citations
Data
Data behind the article
This data has been text mined from the article, or deposited into data resources.
BioStudies: supplemental material and supporting data
GEO - Gene Expression Omnibus
- (2 citations) GEO - GSE57896
Similar Articles
To arrive at the top five similar articles we use a word-weighted algorithm to compare words from the Title and Abstract of each citation.
WY14643 combined with all-trans retinoic acid acts via p38 MAPK to induce "browning" of white adipocytes in mice.
Genet Mol Res, 14(2):6978-6984, 26 Jun 2015
Cited by: 3 articles | PMID: 26125906
Thermogenic activity of UCP1 in human white fat-derived beige adipocytes.
Mol Endocrinol, 29(1):130-139, 01 Jan 2015
Cited by: 58 articles | PMID: 25389910 | PMCID: PMC5414770
The emergence of cold-induced brown adipocytes in mouse white fat depots is determined predominantly by white to brown adipocyte transdifferentiation.
Am J Physiol Endocrinol Metab, 298(6):E1244-53, 30 Mar 2010
Cited by: 442 articles | PMID: 20354155
UCP1 protein: The molecular hub of adipose organ plasticity.
Biochimie, 134:71-76, 10 Sep 2016
Cited by: 22 articles | PMID: 27622583
Review
Funding
Funders who supported this work.
NIDDK NIH HHS (4)
Grant ID: R01 DK095384
Grant ID: R01DK097768
Grant ID: R01 DK097768
Grant ID: R01DK095384





