Abstract
Free full text

In Vitro Synthesis of Modified mRNA for Induction of Protein Expression in Human Cells
Abstract
The exogenous delivery of coding synthetic messenger RNA (mRNA) for induction of protein synthesis in desired cells has enormous potential in the fields of regenerative medicine, basic cell biology, treatment of diseases, and reprogramming of cells. Here, we describe a step by step protocol for generation of modified mRNA with reduced immune activation potential and increased stability, quality control of produced mRNA, transfection of cells with mRNA and verification of the induced protein expression by flow cytometry. Up to 3 days after a single transfection with eGFP mRNA, the transfected HEK293 cells produce eGFP. In this video article, the synthesis of eGFP mRNA is described as an example. However, the procedure can be applied for production of other desired mRNA. Using the synthetic modified mRNA, cells can be induced to transiently express the desired proteins, which they normally would not express.

Introduction
In cells, the transcription of messenger RNA (mRNA) and the following translation of mRNA into desired proteins ensure the proper functioning of cells. Hereditary or acquired genetic disorders can lead to insufficient and dysfunctional synthesis of proteins and cause severe diseases. Thus, a new therapeutic approach to induce the production of missing or defective proteins is the exogenous delivery of synthetic modified mRNA into cells, which codes for the desired protein. Thereby, cells are triggered to synthesize functional proteins, which they normally cannot produce or would naturally not need. Using this approach, genetic disorders can be corrected by introduction of mRNA that codes for the defective or missing protein 1. The mRNA therapy can also be used for vaccination to synthetize protein antigens, which are expressed by tumor cells or pathogens. Thereby, host immune system can be activated to effectively eliminate tumor cells or prevent infections 2,3. Furthermore, in recent years, mRNA was successfully used to generate induced pluripotent stem cells (iPSCs). For this purpose, fibroblasts were transfected with mRNAs to induce the expression of reprogramming factors 4-6 and to convert them in iPSCs with an enormous potential in regenerative medicine.
Previously, the use of conventional mRNA was associated with low stability and strong immunogenicity. Thus, clinical applications of conventional mRNAs were limited. However, the replacement of cytidine and uridine by 5-methylcytidine and pseudouridine within the mRNA molecule by Kariko and colleagues rendered mRNA molecules stable in biological fluids and dramatically reduced immune activation 7-10, which now allows the clinical applicability of modified mRNAs.
Using synthetically produced modified mRNAs, desired gene transcripts can be temporarily delivered in vivo 11 or in vitro to induce protein expression. The introduced mRNA is translated under physiological conditions by the cellular translation machinery. Due to lack of integration into the host cell genome compared to viral gene therapy vectors, the risk of oncogenesis is prevented 12,13. Thus, therapy using modified synthetic mRNA will get better clinical acceptance in the future.
Here, we describe a detailed protocol for production of modified mRNA (Figure 1), transfection of cells with mRNA and the evaluation of protein expression in transfected cells.
Protocol
1. Augmentation of Plasmids Containing Required Coding DNA Sequences (CDS)
Pre-warm SOC medium, which is included in the transformation kit, to room temperature and the LB agar plates containing 100 µg/ml ampicillin to 37 °C. Equilibrate the water bath to 42 °C.
Thaw one vial of chemically component E. coli on ice.
Add 1-5 µl of the plasmid (10 pg to 100 ng) containing the CDS into the vial of chemically component E. coli and mix gently. Do not mix the cells by pipetting. After adding plasmids, mix by tapping the tube gently.
Incubate the mixture on ice for 30 min.
Heat-shock the E. coli for 30 sec at 42 °C in a water bath without shaking.
Place the vial on ice for 2 min.
Add 250 µl of pre-warmed SOC medium to the E. coli and shake the bacteria horizontally at 300 rpm for 1 hr at 37 °C in a bacterial incubator.
Spread 100 µl and 150 µl from transformation mix on pre-warmed LB agar plates, invert the plates and incubate overnight at 37 °C.
After colonies become visible, inoculate a single colony from each plate into a 15 ml culture tube containing 5 ml LB medium with 100 µg/ml ampicillin. Incubate them overnight at 37 °C with shaking at 300 rpm until culture is in late log or stationary phase.
For long-term storage of transformed E. coli, pipet 75 µl of 100% glycerol into the cryovial. Add 225 µl of the bacterial culture (frozen stock will contain 25% glycerol), mix well and store the cryovial at -80 °C.
Isolate the plasmids containing the required CDS using a plasmid purification kit according to manufacturer's instructions.
Determine the plasmid concentration using a spectrophotometer.
Prepare aliquots of 20 µl and store them at -20 °C for prolonged time.
2. Amplification of Plasmid Inserts and Adding of Poly T-tail by Polymerase Chain Reaction (PCR)
NOTE: To obtain the DNA template for the in vitro transcription (IVT), the CDS of interest, here eGFP, is amplified using PCR. Simultaneously, a poly T-tail of 120 thymidines (T) is added to the insert by using a reverse primer with a T120 extension. Thereby, the generated mRNAs obtain a poly A-tail with a defined length after the IVT.
Prepare PCR mixture as detailed in Table 1.
Mix the reaction mix thoroughly. NOTE: PCR product can be used in 4-5 IVT reactions. To increase the DNA template amount for IVT, total volume of the PCR mixture can be increased. Several PCR tubes each containing 100 µl PCR mixture can be used to obtain a sufficient yield of PCR product.
Place the PCR tubes in a thermocycler and run the PCR using the PCR cycling protocol (Table 2).
Clean up the PCR reaction using a PCR purification kit according to manufacturer's instructions and elute the DNA using 20 µl nuclease-free water.
Measure the concentration of the DNA using a spectrophotometer. NOTE: The detected DNA concentration is approximately 300 µg/ml. Thus, the expected total amount should be ~6 µg. The amount of DNA for other proteins can differ depending on the length of CDS, amount of plasmid used for PCR, annealing temperature of primers, and cycle number.
Freeze the DNA at -20 °C for a long time or use it directly for IVT.
3. Control Step: Quality of PCR Product
Check the quality of the PCR product by DNA gel electrophoresis.
Mix the desired amount of agarose with 1x TBE in a flask. For a 1% agarose gel, add 0.5 g of agarose to 50 ml of 1x TBE.
Microwave until the agarose is dissolved. Do not let it boil. Ensure that the agarose is completely in solution.
Add 5 µl nucleic acid gel stain to 50 ml agarose gel.
Pour the gel into an agarose gel box, set the comb, and wait approximately 1 hr until the gel solidified.
Remove the comb and place the gel box in a gel tray.
Mix 2 µl (200 ng) of 80-10,000 bp DNA-Ladder and 200 ng of PCR product each with 2 µl 6x loading buffer in a total volume of 12 µl.
Carefully load DNA samples into the wells of the agarose gel.
Using 1x TBE as running buffer, run the agarose gel at 100 V for 1 hr.
Visualize the DNA bands on an imaging system (Figure 2).
4. In Vitro Transcription (IVT)
NOTE: After PCR, the plasmid inserts are amplified and a poly T-tail is added. During the IVT, genetic information is transcribed from DNA to RNA. The generated mRNA transcript is used to produce proteins in cells.
Clean the working area and pipettes with an RNase decontamination solution. Use PCR clean and sterile dual-filter pipette tips and frequently change gloves to minimize contamination of reaction mixes with RNases.
Prepare the NTP/cap analog mixture as described in Table 3.
Mix the NTP/cap analog mixture thoroughly by vortexing and spin down briefly.
Assemble the IVT reaction mixture as described in Table 4.
Mix the IVT reaction mixture thoroughly by gently pipetting up and down. Then centrifuge the PCR tube briefly to collect the mixture at the bottom of the tube.
Incubate at 37 °C for 3 hr in a thermomixer. NOTE: The optimum incubation time depends on the length of the inserts and transcriptional efficiency of given template. For short inserts (<500 nt), a longer incubation time may be advantageous. A time-course experiment can be done to determine the optimum incubation time for maximum yield. Therefore, set up IVT reactions, for example, for 1, 2, 3, 4, 6 hr, and overnight incubation and determine the mRNA amount. The kinetics of the in vitro transcription for generation of eGFP mRNA is shown in Figure 3.
To remove the template DNA, add 1 µl of DNase (2 U/µl) to the IVT reaction mixture, mix well and incubate 15 min at 37 °C.
Purify the reaction mixture using a RNA purification kit according to manufacturer´s instructions. Elute the modified mRNA from the spin column membrane twice with 40 µl nuclease-free water. NOTE: The detected RNA concentration is approximately 1,500 µg/ml. Thus, the expected total amount should be ~120 µg. The amount of synthesized mRNA for other proteins can differ depending on the length of CDS and the amount of PCR product used for IVT.
5. Treatment of Purified mRNA with Antarctic Phosphatase
NOTE: The generated mRNA is treated with phosphatase to remove 5' triphosphates, which can be recognized by RIG-I and lead to immune activation. Furthermore, the phosphatase treatment prevents re-circularization in a self-ligation reaction.
Add 9 µl of 10x Antarctic phosphatase reaction buffer to 79 µl of purified mRNA solution. Subsequently, add 2 µl of Antarctic phosphatase (5 U/µl) and gently mix the sample. Incubate the reaction mixture at 37 °C for 30 min.
Purify the reaction mixture using a RNA purification kit according to manufacturer´s instructions. Elute the modified mRNA from the spin column membrane twice with 50 µl nuclease-free water.
Measure the concentration of the modified mRNA using a spectrophotometer. Check that the ratio of absorbance at 260 nm/280 nm (A260/A280) is ≥ 1.8 and the ratio of A260/A230 is 2.0, which indicates purity. NOTE: The expected total yield should be approximately 90 µg, which is sufficient for approximately 36 mRNA transfections.
Adjust the concentration of mRNA to 100 ng/µl by adding nuclease-free water.
Aliquot the modified mRNA into single-use aliquots required for transfections and store at -80 °C. Avoid multiple freeze and thaw cycles. Properly prepared and stored mRNA is stable for many years. During working, always keep the modified mRNA on ice.
6. Control Step: Quality of Synthesized Modified mRNA
Check the quality of modified mRNA by RNA gel electrophoresis.
Mix the desired amount of agarose with 1x TBE in a flask. For a 1% agarose gel, add 0.5 g of agarose to 50 ml of 1x TBE.
Microwave until the agarose is dissolved. Do not let it boil. Ensure that the agarose is completely in solution.
Add 5 µl nucleic acid gel stain to 50 ml agarose gel.
Pour the gel into a agarose gel box, set the comb, and wait approximately 1 hr until the gel is solidified.
Remove the comb and place the gel box in a gel tray.
Mix 3 µl (3 µg) of 0.5-10 kb RNA Ladder and 200 ng of synthesized modified mRNA each with 7 µl loading buffer in a total volume of 10 µl. The composition of the loading buffer is described in Table 5.
Denature the ladder and the mRNA samples at 70 °C for 10 min.
Carefully load the ladder and the mRNA samples on the 1% agarose gel.
Perform electrophoresis at 100 V for 1 hr using 1x TBE as running buffer.
Visualize the ladder and mRNA bands on a UV transilluminator and photograph using a gel documentation system (Figure 4).
7. Preparing of Cells for Transfection
Plate 2 x105 cells (HEK293 cells) per well of 12-well plate.
Incubate the cells overnight at 37 °C in a cell incubator. NOTE: At the day of transfection, the cells should have reached 80-90% confluence.
8. Performing mRNA Transfection of Cells
Thaw the modified mRNA.
Generate the lipoplexes for transfection.
Add 25 µl (2.5 µg) of modified mRNA and 2 µl of cationic lipid transfection reagent to 473 µl Opti-MEM (Minimal Essential Medium) I reduced serum medium to generate the transfection mixture for transfection of one well of a 12-well plate. Scale up the volumes according to the number of wells to be transfected. As negative control, prepare a transfection mixture without mRNA.
Mix the components gently by pipetting. Incubate the transfection mixture at room temperature for 20 min to generate lipoplexes for transfection. NOTE: The transfection mixture contains no antibiotics. Thus, take care to ensure the sterility while handling cells.
Wash cells with 500 µl DPBS/well.
Add 500 µl transfection mixture to one well of a 12-well plate.
Incubate the cells for 4 hr at 37 °C and 5% CO2.
Aspirate the transfection mixture and add 1 ml complete cell culture medium to the cells.
Incubate the cells for 24 hr in the cell incubator.
Perform fluorescence microscopic analyses using a fluorescence microscope (Figure 5). NOTE: To examine the expression of other proteins in cells, which are produced by transfection with other mRNA than eGFP coding mRNA, the cells can be cultivated on coverslips. Then the transfection can be performed and the cells can be stained with appropriate antibodies and analyzed by fluorescence microscopy.
9. Flow Cytometric Analyses of eGFP Expression in Cells
Remove cell culture medium from the wells and wash each well with 1 ml DPBS (without calcium and magnesium). Add 500 µl trypsin/EDTA per well to detach the cells from the surface of cell culture plates. Subsequently, add 500 µl TNS per well to inactivate trypsin.
Centrifuge the detached cells for 5 min at 300 x g at room temperature. Remove the supernatant leaving only the pellet. Resuspend the cell pellet in 150 µl cell fixation solution and transfer the cell suspension into a flow cytometry tube.
Analyze the percentage of eGFP expressing cells and the fluorescence intensity using Geo Mean (geometric mean of fluorescence intensity) (Figure 6).
10. Measurement of Protein Expression Over Time
Perform mRNA transfection of cells in 3 wells of a 12-well plate as described in step 8. Additionally, incubate 3 wells of HEK293 cells with the transfection mixture without the addition of mRNA.
Measure the expression of eGFP 1, 2, and 3 days after transfection to evaluate the duration of protein expression (Figure 7).
Representative Results
Using a pcDNA 3.3 plasmid containing the CDS of eGFP, the synthesis of modified eGFP mRNA was established (Figure 1). After insert amplification by PCR and poly T-tailing, a clear band with a length of approximately 1,100 bp is detected (Figure 2). Increasing the IVT time augmented the yield of mRNA (Figure 3). After the IVT, a clear mRNA band with a length of approximately 1,100 bp was detected, which corresponds to the length of eGFP mRNA to be produced (Figure 4).
The functionality of the generated eGFP mRNA was tested by transfection of HEK293 cells. For this purpose, transfection complexes (lipoplexes) were generated using a cationic lipid transfection reagent. The transfection was performed with 2 x 105 cells per well of 12-well plate. The production of eGFP in the cells was detected 24 hr after transfection using fluorescence microscopy (Figure 5) and flow cytometry (Figure 6).
HEK293 cells were transfected with eGFP mRNA. eGFP expression was determined 1, 2, and 3 days after transfection to evaluate the duration of protein expression in the cells (Figure 7). After 24 hr, the protein expression was highest in the cells. The amount was reduced 1.6-fold every next day. Even after 3 days, the cells contained eGFP.
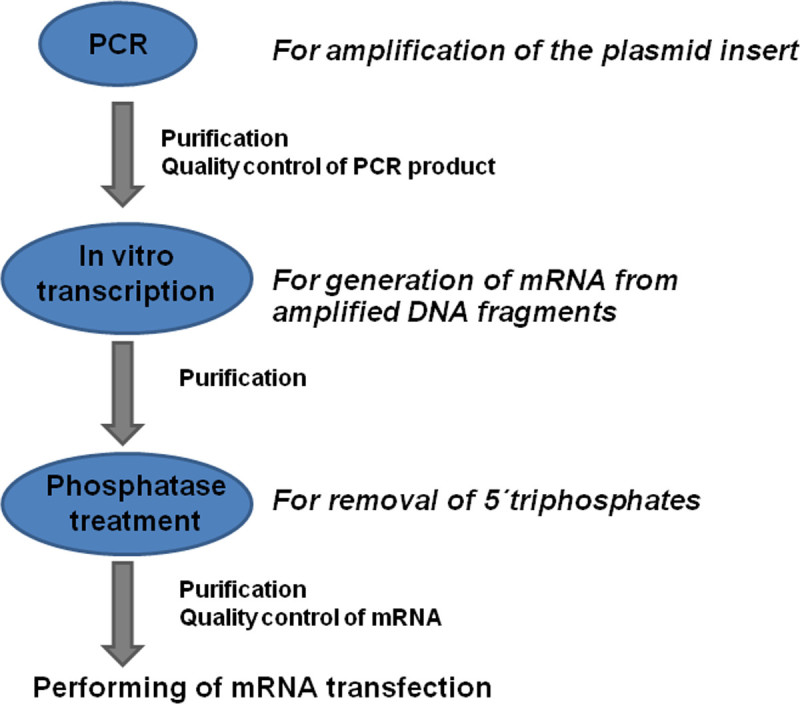
Figure 1: Overview of the modified mRNA production process. Coding DNA sequences (CDS) with known flanking sequences are amplified by PCR using specific primers. The PCR product is purified and the quality of the generated DNA is determined. The mRNA is generated from DNA product using the in vitro transcription process. The product is purified and treated with phosphatase to remove 5'-triphosphates. After the additional purification and quality control of generated mRNA, the mRNA transfections can be performed.
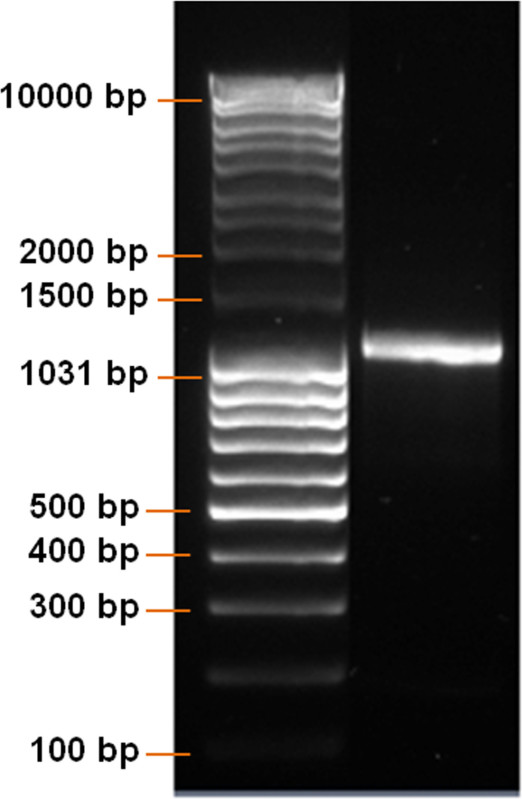
Figure 2: Analysis of DNA product after PCR. DNA ladder and PCR product were run on a 1% agarose gel. A clear DNA band with a length of approximately 1,100 bp should be detected.
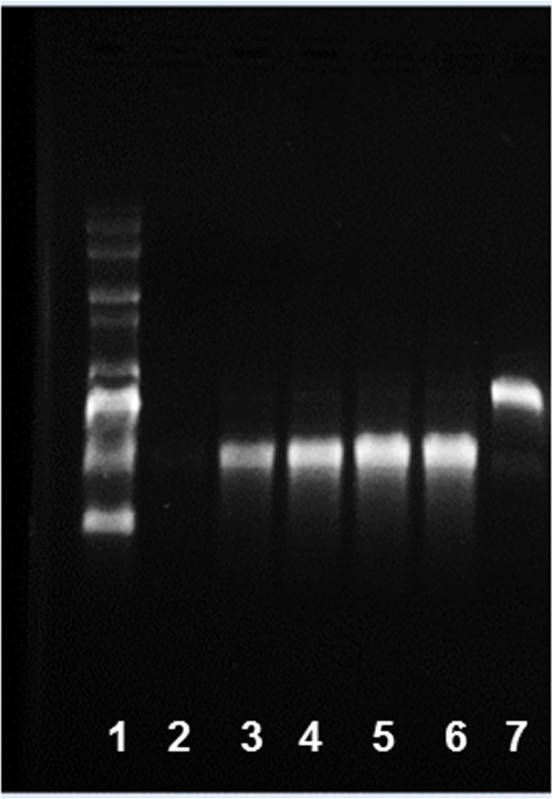
Figure 3: Kinetics of in vitro transcription for generation of eGFP mRNA. 1) RNA ladder and IVT products after 2) 0 min, 3) 10 min, 4) 30 min, 5) 180 min, 6) 360 min, and 7) DNA template alone were run on a 1% agarose gel.
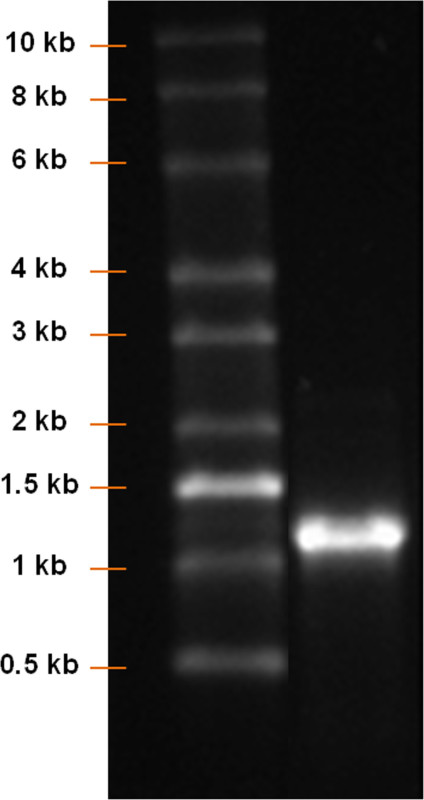
Figure 4: Analysis of mRNA product after IVT. RNA ladder and IVT product were run on a 1% agarose gel. A clear mRNA band with a length of approximately 1,100 bp should be detected.
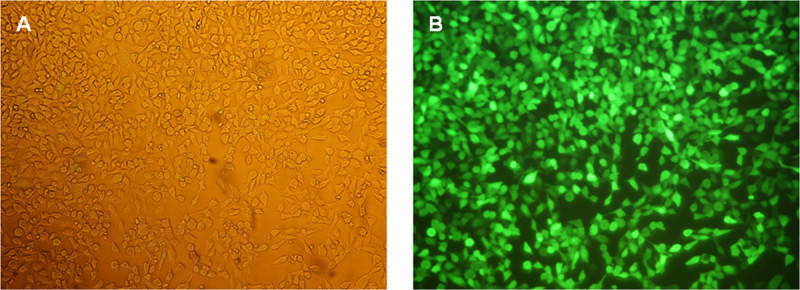
Figure 5: Fluorescence microscopic analyses of HEK293 cells 24 hr after eGFP mRNA transfection. (A) Phase contrast image of the cells at a magnification of 100X. (B) Fluorescence images of the cells at a magnification of 100X.
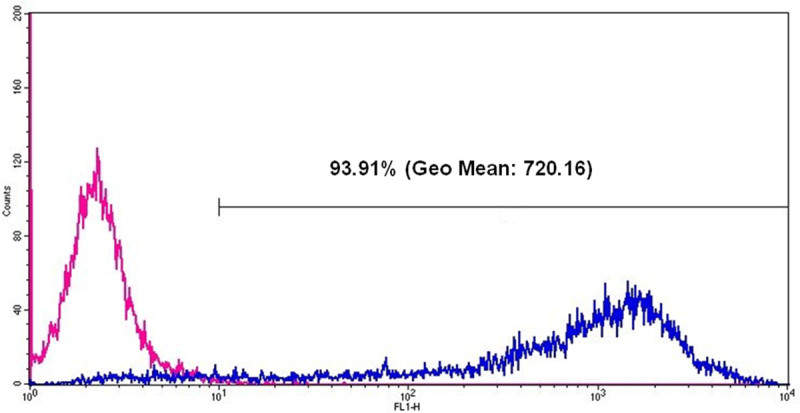
Figure 6: Flow cytometric analyses of eGFP expression in HEK293 cells 24 hr after eGFP mRNA transfection. The pink line represents cells without mRNA transfection and the blue line represents eGFP positive cells after eGFP mRNA transfection. After eGFP mRNA transfection, 93.91% of all measured cells are positive and the Geo Mean (geometric mean of fluorescence intensity) is 720.16.
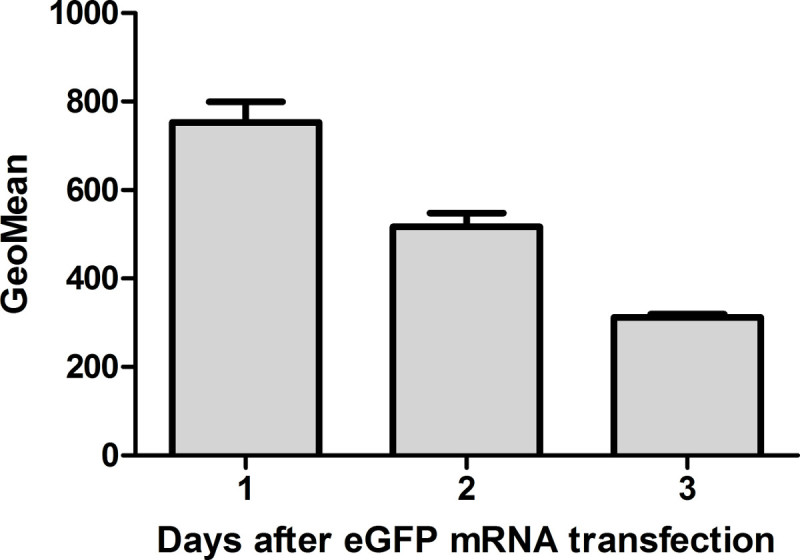
Figure 7: Flow cytometric analyses of eGFP expression in HEK293 cells 1, 2, and 3 days after eGFP mRNA transfection. The protein expression is highest 24 hr after mRNA transfection. Thereafter, the amount is reduced 1.6-fold every day (n = 3).
Table 1: Composition of PCR mixture.
| Component | Final concentration | Amount (µl) |
| Forward Primer | 0.7 µM | 7 |
| Reverse Primer | 0.7 µM | 7 |
| 5x Q-Solution | 1x | 20 |
| 5x HotStar HiFidelity PCR Buffer | 1x | 20 |
| Plasmid DNA | 50 ng / 100µl | Variable |
| HotStar HiFidelity DNA Polymerase (2.5 U/µl) | 2.5 U | 1 |
| Nuclease-free water | Variable | |
| Total volume | 100 |
Table 1: Composition of PCR mixture.
| Cycle Number | Time | Temperature (°C) | |
| Initial denaturation step | 1 | 5 min | 95 |
| 3-step cycling | 2-25 | ||
| · Denaturation | 45 sec | 95 | |
| · Annealing | 1 min | 55 | |
| · Extension | 1 min | 72 | |
| Final extension step | 26 | 10 min | 72 |
| End of PCR cycling | Indefinite | 4 |
Table 2: PCR cycling protocol.
| Component | Stock concentration (mM) | Final concentration (mM) | Volume (µl) |
| ATP (from MEGAscript T7 Kit) | 75 | 7.5 | 4 |
| GTP (from MEGAscript T7 Kit) | 75 | 1.875 | 1 |
| Me-CTP (from Trilink) | 100 | 7.5 | 3 |
| Pseudo-UTP (from Trilink) | 100 | 7.5 | 3 |
| 3´-O-Me-m7G(5´)ppp(5´)G RNA cap structure analog | 10 | 2.5 | 10 |
| Total volume | 23 |
Table 3: Composition of NTP/cap analog mixture.
| Component | Final concentration | Amount (µl) |
| Nuclease-free water | Variable | |
| RNase Inhibitor | 40 U | 1 |
| NTP/cap analog mixture (from step 4.3) | 23 | |
| PCR product | 1 µg | Variable |
| 10x reaction buffer | 1x | 4 |
| 10x T7 RNA polymerase enzyme mix | 1x | 4 |
| Total volume | 40 |
Table 4: Composition of in vitro transcription (IVT) reaction mixture.
| Component | Amount (µl) |
| Formamide | 3.3 |
| 37% formaldehyde | 1 |
| MEN buffer (10x) | 1 |
| 6x loading buffer (supplied with the peqGOLD Range Mix DNA-Ladder) | 1.7 |
| Total volume | 7 |
Table 5: Preparation of loading buffer for RNA gel electrophoresis.
| Cell Culture Medium and Buffer | |
| HEK-293 cell culture medium | Add 25 ml of FCS, 2.5 ml of penicillin/streptomycin, 2.5 ml of L-glutamine in 220 ml DMEM high glucose. Store the medium at 4 °C and use it within 2 weeks. |
| TBE buffer (10x) | Dissolve 0.9 M Tris base, 0.9 M boric acid and 20 mM EDTA in 1 L water (Ampuwa). The pH of the buffer is 8. |
| MEN buffer (10x) | Dissolve 200 mM MOPS, 50 mM NaOAc, 10 mM EDTA in 1 L water (Ampuwa). Adjust the pH value with NaOH to 7. |
Table 6: Cell culture medium and buffers.
Discussion
The mRNA therapy has tremendous potential in the field of regenerative medicine, treatment of diseases and vaccination. In this video, we demonstrate the production of a stabilized, modified mRNA for induction of protein expression in cells. Using this protocol, other desired mRNA can be generated. The in vitro synthesis of modified mRNA allows the transfection of cells with desired mRNAs to induce expression of target proteins. Thereby, the desired protein is expressed transiently under physiological conditions until the exogenously delivered mRNA is completely degraded.
In this video, the expression of eGFP was demonstrated for 3 days after a single transfection of HEK293 cells with eGFP mRNA. mRNA molecules coding other proteins could lead to a shorter protein expression period. The expression of the protein decreases due to degradation of the exogenously delivered mRNA. Therefore, the limitation of this technique for some applications might be the transient induction of protein expression. Thus, to maintain protein expression in cells for a longer period, repeated delivery of mRNA is required. Although, mRNA transfection has the advantage of non-integration into the host genome, which prevents insertional mutagenesis and the development of cancer and leukemia compared to viral vectors, the in vivo transfection efficiency could be less than using viral vectors.
The required mRNA concentration and amount of transfection reagent should be optimized for each different cell type14, which are the target cells for exogenous delivery of mRNA to produce the missing protein.
Repeated freezing and thawing of mRNA should be avoided to maintain the stability of the produced mRNA. Therefore, working aliquots can be prepared. After PCR and IVT, only a single specific band should be detected. Otherwise, the number of PCR cycles, primer annealing temperature, and/or amount of plasmid DNA should be optimized to obtain the specific DNA product for IVT. Furthermore, the IVT time and the amount of DNA template for IVT can be optimized to obtain mRNA of a specific length in sufficient amounts.
Disclosures
The authors declare that they have no competing financial interests.
Acknowledgments
This project was funded by the European Social Funds in Baden-Wuerttemberg, Germany.
References
- Bangel-Ruland N, et al. CFTR-mRNA delivery: a novel alternative for cystic fibrosis "gene therapy" The journal of gene medicine. 2013. [Abstract]
- Benteyn D, et al. Design of an Optimized Wilms" Tumor 1 (WT1) mRNA Construct for Enhanced WT1 Expression and Improved Immunogenicity In Vitro and In Vivo. Molecular therapy Nucleic acids. 2013;2:134. [Europe PMC free article] [Abstract] [Google Scholar]
- Petsch B, et al. Protective efficacy of in vitro synthesized, specific mRNA vaccines against influenza A virus infection. Nature. 2012;30:1210–1216. [Abstract] [Google Scholar]
- Mandal PK, Rossi DJ. Reprogramming human fibroblasts to pluripotency using modified mRNA. Nature protocols. 2013;8:568–582. [Abstract] [Google Scholar]
- Warren L, et al. Highly efficient reprogramming to pluripotency and directed differentiation of human cells with synthetic modified mRNA. Cell stem cell. 2010;7:618–630. [Europe PMC free article] [Abstract] [Google Scholar]
- Yakubov E, Rechavi G, Rozenblatt S, Givol D. Reprogramming of human fibroblasts to pluripotent stem cells using mRNA of four transcription factors. Biochemical and biophysical research communications. 2010;394:189–193. [Abstract] [Google Scholar]
- Anderson BR, et al. Incorporation of pseudouridine into mRNA enhances translation by diminishing PKR activation. Nucleic acids research. 2010;38:5884–5892. [Europe PMC free article] [Abstract] [Google Scholar]
- Kariko K, Buckstein M, Ni H, Weissman D. Suppression of RNA recognition by Toll-like receptors: the impact of nucleoside modification and the evolutionary origin of RNA. Immunity. 2005;23:165–175. [Abstract] [Google Scholar]
- Kariko K, et al. Incorporation of pseudouridine into mRNA yields superior nonimmunogenic vector with increased translational capacity and biological stability. Molecular therapy : the journal of the American Society of Gene Therapy. 2008;16:1833–1840. [Europe PMC free article] [Abstract] [Google Scholar]
- Kariko K, Weissman D. Naturally occurring nucleoside modifications suppress the immunostimulatory activity of RNA: implication for therapeutic RNA development. Current opinion in drug discovery & development. 2007;10:523–532. [Abstract] [Google Scholar]
- Kormann MS, et al. Expression of therapeutic proteins after delivery of chemically modified mRNA in mice. Nature biotechnology. 2011;29:154–157. [Abstract] [Google Scholar]
- Hacein-Bey-Abina S, et al. Insertional oncogenesis in 4 patients after retrovirus-mediated gene therapy of SCID-X1. The Journal of clinical investigation. 2008;118:3132–3142. [Abstract] [Google Scholar]
- Hacein-Bey-Abina S, et al. Efficacy of gene therapy for X-linked severe combined immunodeficiency. The New England journal of medicine. 2010;363:355–364. [Europe PMC free article] [Abstract] [Google Scholar]
- Avci-Adali M, et al. Optimized conditions for successful transfection of human endothelial cells with in vitro synthesized and modified mRNA for induction of protein expression. Journal of biological engineering. 2014;8:8. [Europe PMC free article] [Abstract] [Google Scholar]
Articles from Journal of Visualized Experiments : JoVE are provided here courtesy of MyJoVE Corporation
Full text links
Read article at publisher's site: https://doi.org/10.3791/51943
Read article for free, from open access legal sources, via Unpaywall:
https://europepmc.org/articles/pmc4354090?pdf=render
Citations & impact
Impact metrics
Citations of article over time
Alternative metrics
Smart citations by scite.ai
Explore citation contexts and check if this article has been
supported or disputed.
https://scite.ai/reports/10.3791/51943
Article citations
Modified mesenchymal stromal cells by in vitro transcribed mRNA: a therapeutic strategy for hepatocellular carcinoma.
Stem Cell Res Ther, 15(1):208, 11 Jul 2024
Cited by: 1 article | PMID: 38992782 | PMCID: PMC11241816
mRNA-based therapeutic strategies for cancer treatment.
Mol Ther, 32(9):2819-2834, 03 May 2024
Cited by: 1 article | PMID: 38702886
Review
A Novel Strategy for the Treatment of Aneurysms: Inhibition of MMP-9 Activity through the Delivery of TIMP-1 Encoding Synthetic mRNA into Arteries.
Int J Mol Sci, 25(12):6599, 15 Jun 2024
Cited by: 0 articles | PMID: 38928311
Improved tropoelastin synthesis in the skin by codon optimization and nucleotide modification of tropoelastin-encoding synthetic mRNA.
Mol Ther Nucleic Acids, 33:642-654, 02 Aug 2023
Cited by: 2 articles | PMID: 37650117 | PMCID: PMC10462787
The Prodigious Potential of mRNA Electrotransfer as a Substitute to Conventional DNA-Based Transient Transfection.
Cells, 12(12):1591, 08 Jun 2023
Cited by: 1 article | PMID: 37371061 | PMCID: PMC10297312
Go to all (25) article citations
Similar Articles
To arrive at the top five similar articles we use a word-weighted algorithm to compare words from the Title and Abstract of each citation.
Easy In Vitro Synthesis of Optimised Functioning Reporter mRNA from Common eGFP Plasmid.
Mol Biotechnol, 60(10):762-771, 01 Oct 2018
Cited by: 0 articles | PMID: 30120676
Optimized conditions for successful transfection of human endothelial cells with in vitro synthesized and modified mRNA for induction of protein expression.
J Biol Eng, 8(1):8, 03 Mar 2014
Cited by: 22 articles | PMID: 24581116 | PMCID: PMC3975882
Method for efficient transfection of in vitro-transcribed mRNA into SK-N-AS and HEK293 cells: difference in the toxicity of nuclear EGFP compared to cytoplasmic EGFP.
Int J Mol Med, 17(6):1011-1016, 01 Jun 2006
Cited by: 5 articles | PMID: 16685409
Concise Review: Application of In Vitro Transcribed Messenger RNA for Cellular Engineering and Reprogramming: Progress and Challenges.
Stem Cells, 35(1):68-79, 20 Jun 2016
Cited by: 31 articles | PMID: 27250673
Review





