Abstract
Free full text

A Role for the Adaptor Proteins TRAM and TRIF in Toll-like Receptor 2 Signaling*
Abstract
Toll-like receptors (TLRs) are involved in sensing invading microbes by host innate immunity. TLR2 recognizes bacterial lipoproteins/lipopeptides, and lipopolysaccharide activates TLR4. TLR2 and TLR4 signal via the Toll/interleukin-1 receptor adaptors MyD88 and MAL, leading to NF-κB activation. TLR4 also utilizes the adaptors TRAM and TRIF, resulting in activation of interferon regulatory factor (IRF) 3. Here, we report a new role for TRAM and TRIF in TLR2 regulation and signaling. Interestingly, we observed that TLR2-mediated induction of the chemokine Ccl5 was impaired in TRAM or TRIF deficient macrophages. Inhibition of endocytosis reduced Ccl5 release, and the data also suggested that TRAM and TLR2 co-localize in early endosomes, supporting the hypothesis that signaling may occur from an intracellular compartment. Ccl5 release following lipoprotein challenge additionally involved the kinase Tbk-1 and Irf3, as well as MyD88 and Irf1. Induction of Interferon-β and Ccl4 by lipoproteins was also partially impaired in cells lacking TRIF cells. Our results show a novel function of TRAM and TRIF in TLR2-mediated signal transduction, and the findings broaden our understanding of how Toll/interleukin-1 receptor adaptor proteins may participate in signaling downstream from TLR2.
Introduction
Toll-like receptors (TLR)4 are a family of 13 (TLR1–13) transmembrane pattern-recognition receptors (1). TLRs recognize conserved molecular motifs found in microorganisms and induce the production of pro-inflammatory cytokines, type I interferons, and up-regulation of co-stimulatory molecules. TLR4 recognizes bacterial lipopolysaccharide (LPS), whereas TLR2 recognizes Gram-positive bacteria and bacterial lipoproteins, but other ligands are also suggested (2,–5). TLR2 discriminates between diacylated and triacylated lipoproteins by heterodimerization with TLR6 and TLR1, respectively. Viral single-stranded RNA and the base analog resiquimod (R848) are ligands for TLR7 and TLR8, whereas the dsRNA poly(I:C) is recognized by TLR3. Viral RNA is also recognized by TLR-independent pathways and can activate the cytoplasmic RNA helicases RIG-I and Mda-5 (6,–8).
Four TIR adaptor molecules mediate TLR4 signaling (1). Myeloid differentiation factor 88 (MyD88) and MyD88 adaptor-like protein (MAL/TIRAP) form one signaling pathway, leading to early NF-κB activation and induction of inflammatory cytokines. TLR2 signaling utilizes both MAL and MyD88 in a manner similar to TLR4. TIR domain-containing adaptor protein-inducing interferon β (TRIF/TICAM-1) and TRIF-related adaptor molecule (TRAM/TICAM-2), form the second signaling pathway mediated by TLR4, which activates interferon regulatory factor-3 (IRF-3), late NF-κB activation, and induction of type I interferons (IFN). Activation of the TLR4-dependent pathway via TRIF also induces up-regulation of co-stimulatory molecules, such as CD86, and production of the chemokine CCL5 (9, 10). TLR7/8/9 signaling is mediated by MyD88 alone, whereas TLR3 signaling requires only TRIF (2).
Here, we report a novel role for TRAM and TRIF in TLR2 signaling, as lipoprotein-triggered TLR2-mediated Ccl5 induction was abrogated in TRAM- and TRIF-deficient macrophages. Ccl5 release following lipoprotein challenge additionally involves the kinase Tbk-1 and Irf3, in addition to MyD88 and Irf1. Induction of the cytokines Ccl4 and Interferon-β (IFN-β) in response to TLR2 ligands was also partially dependent on TRIF. Combined, our results suggest involvement of the TRAM and TRIF in TLR2 signaling.
EXPERIMENTAL PROCEDURES
Ethics Statement
Experiments involving animals were conducted in accordance with the recommendations made the American Association for Laboratory Animal Science. The animal studies, covered under the protocol A2332, have been approved by the University of Massachusetts Medical School Institutional Animal Care and Use Committee (Animal Welfare Assurance number A3306-01).
Ligands
LPS from Escherichia coli O111:B4 (Sigma) was re-purified using two water/phenol extractions (11) or purchased from Invivogen (San Diego). LPS from Yersinia pestis pLpxL grown at 37 °C was purified as described (12). Poly(I:C) dsRNA was purchased purified from Invivogen. Synthetic diacylated (Pam2Cys-based) macrophage-activating lipopeptide 2 kDa (MALP-2), Pam3Cys-Ser-Lys4 (Pam3Cys), and fibroblast stimulating lipopeptide (FSL-1) were from EMC (Tübingen, Germany). R848 was from GLSynthesis (Worcester, MA) or Invivogen. Sendai virus (SV) Cantrell strain was from Charles River Laboratories (Wilmington, MA). Lipoteichoic acid (LTA) was a gift from Dr. Sonja von Aulock and Dr. Thomas Hartung (13).
Antibodies
Mouse anti-TLR2 mAb 6C2 was produced and purified as described (14). Mouse anti-CD86PE-Cy5 and control rat IgGPE-Cy5 antibody were from eBioscience (San Diego). TLR2 mAb 6C2 and rat IgG from Sigma or from Jackson ImmunoResearch (West Grove, PA) were labeled with an Alexa 488 kit from Molecular Probes (Eugene, OR).
Cells
Peritoneal macrophages were isolated and cultured as described previously (14). Human embryonic kidney 293 (HEK293) cells were cultured in 10% FBS/DMEM. HEK293 cell lines expressing TLR2YFP were cultured with the selection antibiotic G418 (0.5 mg/ml).
Plasmids
The following expression vectors were used: pcDNA3 (Invitrogen); pFLAG-CMV2 (Sigma). pcDNA3-Cherry, provided by Dr. Ø. Halaas (NTNU, Norway), was cloned into pcDNA3 from pRSET-B-mCherry, provided by Prof. R. Y. Tsien (University of California). TLR2 tagged with yellow fluorescent protein (YFP) (TLR2YFP) in pcDNA3 has been described (15). Murine TLR2YFP was made using murine TLR2 originally cloned from RAW264.7 (16). Cherry-tagged TLR2 (TLR2Cherry) was made by excising TLR2 from TLR2YFP and ligating it into pcDNA3-Cherry. HA-tagged TLR2 in pcDNA3 (TLR2HA) was from Dr. T. Fredsvik Gregers (University of Oslo, Norway). The following plasmids have been described: TRAM tagged with YFP (TRAMYFP); TRIF tagged with CFP (TRIFCFP); TRAMC117H (9); pEF-BOS, pEF-BOS-TRAMFLAG, pEF-BOS-MALFLAG, and pEF-BOS-TRIFFLAG (17); pEF-BOS-MyD88, MyD88-TIR, and TRIFΔNΔC (18); and early endosomal antigen-1 (Eea-1) tagged with CFP (Eea-1CFP) (19). CCL4 (MIP-1β) luciferase (pGL3–1062WT) (20) was kindly provided by Prof. M. S. Reitz, Jr. (University of Maryland, Baltimore, MD). p65-pcDNA3, p65-pcDNA3S536A, murine and human CCL5 (regulated on activation normal T cell expressed and secreted (RANTES)), and human ΔκB CCL5 luciferase reporter plasmids (CCL5-luc and ΔκB CCL5-luc) were kindly provided by Dr. Tom Maniatis (Harvard University, Cambridge, MA). pCMV-IRF3Flag and -IRF5Flag were from Prof. Paula M. Pitha (The Johns Hopkins University, Baltimore, MD). Renilla luciferase plasmid was from (Promega, Madison, WI). Transient transfections were performed using GeneJuiceTM transfection reagent (Novagen, Darmstadt, Germany) according to the manufacturer's instructions.
Mice
Wild-type (WT) C57Bl/6 and 129.B6 F2 mice were from the Jackson Laboratory (Bar Harbor, ME). myd88−/− mice (MyD88 knock-out), tirap−/− mice (denoted MAL−/−), ticam-1−/− (denoted TRIF−/−), ticam-2−/− mice (denoted TRAM−/−), and tlr2−/− and tlr4−/− mice (denoted TLR2−/− and TLR4−/−, respectively) were provided by Dr. Shizuo Akira (Osaka University, Japan) and have been described (21,–24). Mice were back-crossed for six to eight (TLR4, TLR2, and MyD88), five to six (MAL), or four to seven (TRIF and TRAM) generations onto the C57BL/6 strain. MAL−/−MyD88−/−, TRIF−/−MyD88−/−, and TRAM−/−TRIF−/− double knock-out animals were generated from the single knockouts (KO) mentioned above. TRIFLps2/Lps2 mice generated on a C57/Bl6 background have been described (25). TRIFLps2/Lps2 animals were originally purchased from The Jackson Laboratory and were provided by Dr. Ian Rifkin, Dr. Robin Ingalls, and Anjali Nair. Alternatively, mouse legs from these animals were received from Dr. K. Hoebe and Dr. B. Beutler. ikke−/− mice (denoted IKKϵ−/−) were from Millennium Pharmaceuticals (Cambridge, MA). ifnar−/− mice were obtained from Dr. J. Sprent (Scripps Institute, San Diego) and were backcrossed onto the C57BL/6 background. irf1−/−, irf3−/−, irf5−/−, and irf7−/− mice were provided by Dr. T. Taniguchi (University of Tokyo, Tokyo, Japan). tnfrsf1a−/− mice (denoted tnfrI−/− mice), tbk1−/−/tnfrI−/− mice and mouse embryonic fibroblasts (MEFs) from tbk1−/− and tbk1+/+ littermates were from Dr. T. Mak and W.-C. Yeh (University of Toronto, Canada) (26). Immortalized bone marrow-derived macrophages were generated with J2 recombinant retrovirus as described (27, 28).
Flow Cytometry Analysis and Cytokine Measurements
Cells were stimulated 16–20 h at 37 °C, 8% CO2 before supernatant was harvested and assessed for Tnf or Ccl5 content using ELISA kits from R&D Systems (Oxon, UK) or Pharmingen. Stimulated cells were labeled as described (14) and analyzed by flow cytometry analysis on a BD-LSR II instrument.
Peritoneal macrophages from wild-type mice were also pretreated with medium, a DMSO control (diluted 1:375 in medium), or Dynasore (80 μm; 1:375 dilution in medium) for 30 min prior to stimulation with medium, LPS (10 ng/ml), Pam3Cys (200 ng/ml), or FSL-1 (200 ng/ml) in 10% FBS/RPMI 1640 medium for 21 h. Supernatant was harvested and analyzed for Ccl5 and Tnf by ELISA.
In Vivo Experiments
WT or TRIFLps2/Lps2 mice were injected intraperitoneally with PBS or Pam3Cys (25 μg). Whole blood was collected after 4 h by terminal cardiac puncture, after isoflurane anesthesia. Serum was separated from whole blood using pedi-serum separator tubes (BD Biosciences), which were placed on ice for 30 min, prior to centrifugation at 900 × g for 60 s. Ccl5 content in the serum was analyzed by ELISA.
Confocal Microscopy
HEK293 cells, seeded on 35-mm glass bottom γ-irradiated tissue cell dishes (MatTek Corp., Ashland, MA), were transiently transfected with TLR2Cherry (1.25 μg/dish), TRAMYFP (0.25 μg/dish), and early endosome marker EEA-1CFP or TRIFCFP (0.25 μg/dish) for 48 h. Cells were treated with medium or FSL-1 (200 ng/ml) for 1 h before cells were fixed and observed by confocal microscopy using an Axiovert 100-M inverted microscope, equipped with an LSM 510 laser scanning unit and a ×63 1.4-NA plan Apochromat oil-immersion objective (Zeiss, Jena, Germany). Co-localization maps showing co-localization events of TLR2 and/or TRAM and Eea-1 were created in Imaris 5.0.2, 64-bit version (Bitplane AG, Zurich, Switzerland). In these images, white denotes co-localization events between two channels, and pixels above the threshold that failed to co-localize were set to zero (black). User-defined thresholds were set conservatively in a rectangular selection mode chosen above the apparent noise level for each channel.
Luciferase Assays
HEK293-TLR2YFP or HEK293 cells were plated in 96-well plates and transiently transfected with luciferase reporter genes (50 ng/well) and Renilla luciferase (10 ng/well) for 18–26 h prior to stimulation. In some experiments, HEK293 cells were transiently transfected with murine or human TLR2 (20 ng/well) and human CCL5 luciferase or human ΔkB CCL5 luciferase. In stimulation experiments, cells were stimulated in 10% FBS/DMEM for 18–20 h. In experiments where signaling molecules were overexpressed, cells were transfected with empty vector, MAL, MyD88, TRAM, and TRIF (2–20 ng/well) or the adaptor mutants MyD88-TIR, TRAMC117H, and TRIFΔNΔC (20 ng/well), or increasing concentrations of p65, p65S536A (0.1–1-10−20 ng/well), IRF3 (20–40-60–80 ng/well) or IRF5 (80 ng/well) for 24–48 h. Total amount of plasmid per well was kept constant. Following stimulation or overexpression, cells were lysed and assayed for luciferase reporter gene activity and Renilla activity. Luciferase results were normalized for Renilla activity and plotted as mean fold induction relative to medium/empty vector control.
Western
RAW264.7 mouse macrophages were stimulated with medium, FSL-1 (200 ng/ml) for 180, 90, 60, and 30 min, with LPS (100 ng/ml) for 90 or 60 min, or with poly(I:C) (50 μg/ml) for 60 min. Cells were lysed and run on a NuPAGE Novex 10% BisTris gel (Invitrogen) and transferred to a nitrocellulose membrane using the iBlot blotting system (Invitrogen). Membranes were blocked with 5% w/v BSA/TBS, 0.01% Tween 20 for 1 h before blots were incubated with antibodies against IRF3Ser-396 (4D4G) or total IRF3 (Cell Signaling Technology, Danvers, MA) or alternatively antibody against total IRF3 (Santa Cruz, CA) for 48–72 h in 5% w/v BSA/TBS, 0.01% Tween 20 at 4 °C. Blots were subsequently stained with HRP-conjugated anti-rabbit (DAKO, DK) and subjected to SuperSignal West Femto ECL substrate.
Gene Expression Analysis
Nanostring nCounter gene expression analysis (NanoString Technologies) of selected inflammatory genes was performed on peritoneal macrophages from wild-type (WT) or TRIF−/− mice treated with medium or FSL-1 lipopeptide for 4 h. Total RNA was isolated using RNeasy RNA isolation kit (Qiagen, Germany) and subjected to nCounter gene expression analysis of selected genes. Values are given as arbitrary units, normalized to internal GAPDH and hypoxanthine-guanine phosphoribosyltransferase standards. In other experiments, immortalized bone marrow macrophages (BMDM) from wild-type or from TRAM−/−-, TRIF−/−-, and TLR2−/−- deficient C57Bl/6 mice were seeded in 24-well plates in triplicate and treated with medium or FSL-1 (200 ng/ml) for 3 h before cells were lysed. Total RNA was isolated with the NucleoSpin 96 RNA kit (Macherey-Nagel, Germany) and processed by vacuum. RNA quantity and purity were checked using NanoDrop (Thermo Scientific). cDNA was made using Maxima first-strand cDNA synthesis kit (Thermo Scientific). qPCR for murine ifnb1 (IFN-β) and tnf was done with GAPDH as endogenous control using TaqMan Probes (Mm0043552_s1, Mm00443258_m1, and Mm99999915_g1; Applied Biosystems). Real time thermal cycling was done with StepOnePlus (Applied Biosystems) using Perfecta qPCR FastMix (Quanta Biosciences) with uracil N-glycosylase and ROX reference dye. Analysis was done with the StepOne software version 2.1.
Statistics
Analysis was performed with GraphPad Prism 6.0. The difference between the two groups was determined by the two-tailed t test or multiple t tests. Two or more groups were compared with one-way ANOVA with Tukey post-test, or two-way ANOVA with Bonferroni or Fischer's LSD post-test or multiple t-tests, as indicated.
RESULTS
TRIF Pathway Regulates LPS-induced TLR2 Expression and TLR2-mediated Ccl5 Induction
The level of surface expression of TLR2 may determine how efficiently cells respond to bacterial lipoproteins, or other pathogen-derived components, during an infection. We have previously shown that surface TLR2 is markedly up-regulated in response to a range of TLR ligands by an MyD88-dependent mechanism, except in response to LPS and poly(I:C), which induce TLR2 up-regulation independent of MyD88 (14). Because TLR4 utilizes the TIR adaptors TRAM and TRIF, we hypothesized that this pathway may play an important role in LPS-induced up-regulation of TLR2. We therefore assessed TLR2 expression on TRIF−/− peritoneal macrophages in response to LPS, as well as in response to Pam3Cys, R848, and Sendai virus. We found that LPS-induced up-regulation of surface TLR2 was only partially affected by the absence of TRIF, whereas TLR2 up-regulation was normal in response to Pam3Cys and R848 (Fig. 1A).
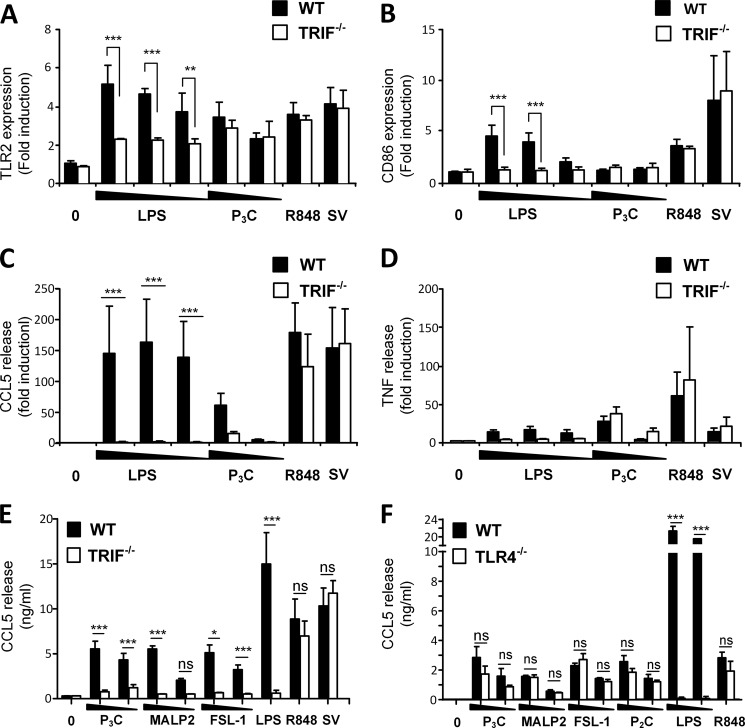
TRIF regulates LPS-induced TLR2 expression and TLR2-mediated Ccl5 induction. Peritoneal macrophages from wild-type (WT) and TRIF−/− mice were left untreated (0) or stimulated with LPS (100–10−1 ng/ml), Pam3Cys (P3C) (100–10 ng/ml), R848 (10 ng/ml), or SV (300 HAU/ml) for 16 h before surface expression of TLR2 (A) and CD86 (B) was assayed by flow cytometry, whereas cell supernatant was analyzed for Ccl5 (C) and Tnf (D) release by ELISA. The results were normalized to medium-treated WT cells and show mean ± S.D. of three independent experiments. Mean TNF release was 114 ± 72 and mean Ccl5 release was 96 ± 18 in medium-treated WT cells. ***, p < 0.001; **, p < 0.01 (two-way ANOVA with Bonferroni post-test). E, peritoneal macrophages from wild-type and TRIF−/− mice were left untreated (0) or stimulated with Pam3Cys (P3C) (1–0.1 μg/ml), MALP2 (100–10 ng/ml), FSL-1 (1–0.1 μg/ml), LPS (1 μg/ml), R848 (10 ng/ml), or SV (10 HAU/ml) for 18 h before supernatant was harvested and analyzed for Ccl5 by ELISA. Results show mean of duplicates from three independent experiments ± S.E. ***, p < 0.001; *, p < 0.01, and ns = not significant (p > 0.05) (multiple t tests). F, Ccl5 release in peritoneal macrophages from WT and TLR4−/− mice stimulated with medium (0), Pam3CysSK4 (P3C) (1–0.1 μg/ml), MALP2 (1–0.1 μg/ml), FSL-1 (1–0.1 μg/ml), Pam2CysSK4 (P2C) (1–0.1 μg/ml), LPS (100–10 ng/ml), or R848 (100 ng/ml) for 18 h before supernatant was harvested and analyzed for Ccl5 by ELISA. Results show mean ± S.D. of triplicates. ***, p < 0.001, and ns = not significant (p > 0.05) (multiple t tests).
We proceeded to investigate whether LPS-induced TLR2 surface expression may be regulated by a mechanism similar to the regulation of the co-stimulatory molecule CD86 or the release of the cytokine CCL5, because TRIF is shown to play a role in these responses. We observed that surface CD86 was up-regulated in response to LPS and Sendai virus (Fig. 1B) but not in response to Pam3Cys or R848 (Fig. 1B). The up-regulation of TLR2 was induced by all these ligands (Fig. 1A), suggesting differences in the regulation of surface TLR2 and CD86 molecules in response to different TLR ligands.
Upon assaying Ccl5, we found that all tested ligands induced Ccl5 release in wild-type peritoneal macrophages (Fig. 1C) (14), including TLR2 ligand Pam3CysSK4 (Pam3Cys). Interestingly, TRIF−/− macrophages were impaired in their ability to induce Ccl5 in response to Pam3Cys (Fig. 1C), suggesting a role for TRIF in TLR2-mediated Ccl5 induction. In contrast, TNF release was normal in TRIF−/− macrophages in response to Pam3Cys (Fig. 1D). Ccl5 and TNF induction in response to R848 and SV was normal in TRIF−/− macrophages (Fig. 1, C and D), implying no role for the TRIF pathway in these responses, whereas LPS-induced Ccl5 was effectively abolished in the absence of TRIF (Fig. 1C), in agreement with previous reports (6,–9).
To verify a potential role for TRIF in Ccl5 release in response to TLR2 ligands, WT or TRIF−/− macrophages were stimulated with the TLR2/6 ligands macrophage-activating lipopeptide 2 (MALP-2), fibroblast-stimulating lipopeptide 1 (FSL-1), or the TLR2/1 ligand Pam3CysSK4 for 18 h. Ccl5 release in the supernatant was subsequently assessed by ELISA. We found that all tested TLR2 ligands induced Ccl5 (Fig. 1E). Furthermore, Ccl5 induction was impaired in TRIF−/− cells in response to all tested TLR2 ligands (Fig. 1E), inferring a role for TRIF in mediating Ccl5 induction in response to TLR2 ligands. TNF induction in the same supernatant was normal (data not shown), implying that TNF induction in TLR2-mediated TNF release does not require TRIF.
TLR4 and TLR3 are known to induce CCL5 in a TRIF-dependent manner. Because it has recently been shown that TLR2 and TLR4 ligands can heterodimerize (29), we assessed Ccl5 induction in WT and TLR4−/− macrophages in response to TLR2 ligands to determine the role of TLR4 in this response. We found that Ccl5 induction was induced in TLR4−/− cells in response to the tested TLR2 ligands (Fig. 1F), suggesting that TLR4 is not a major contributor to Ccl5 release in response to TLR2 ligands.
TIR Adaptors TRAM and TRIF Mediate Ccl5 Release Induced by TLR2 Ligands
We proceeded to test the role for TRAM in response to different TLR2 ligands. Ccl5 and Tnf induction was assessed in MyD88−/−-, TRAM−/−-, and Tlr2−/−-deficient macrophages, in response to MALP-2, FSL-1, and LTA, as well as TLR2/1 ligand Pam3Cys. We found that Ccl5 release was markedly reduced in TRAM−/− macrophages in response to all TLR2 ligands tested (Fig. 2A), whereas Tnf induction in TRAM−/− macrophages was normal (Fig. 2B). In contrast, the lack of MyD88 led to a sharp decline of both Tnf and Ccl5 release in response to all tested TLR2 ligands (Fig. 2, A and B). Tlr2-mediated Ccl5 induction was also impaired in MAL−/− macrophages (data not shown). Ccl5 and Tnf release in response to Pam3Cys, FSL-1, and LTA was completely abrogated in Tlr2−/− macrophages, confirming the dependence on TLR2 in mediating these responses and indicating that the TLR2 ligands have high purity (Fig. 2, A and B). As TLR4 does not mediate the lipoprotein-induced Ccl5 (Fig. 1F), this promotes the view that TLR2 alone, and not TLR2/TLR4 heterodimers, triggers responses via TRIF and TRAM. LPS and SV induced normal levels of TNF and Ccl5 in TLR2−/− macrophages (Fig. 2C).
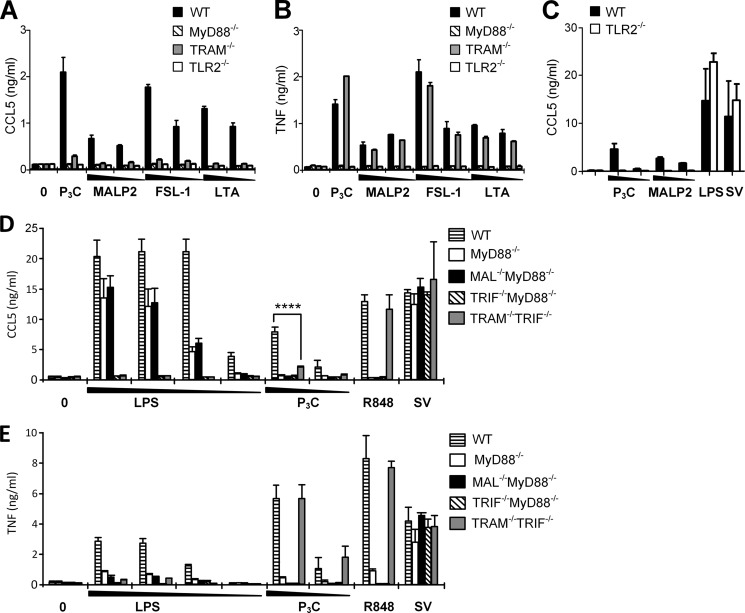
TRAM and TRIF mediate Ccl5 release in response to TLR2 ligands. Peritoneal macrophages from wild-type (WT), MyD88−/−, TRAM−/−, and TLR2−/− mice were exposed to medium (0), Pam3Cys (P3C) (100 ng/ml), or decreasing concentrations of MALP-2 (10–1 ng/ml), FSL-1 (20–2 ng/ml), or LTA (10–1 μg/ml) 18 h before supernatant was analyzed for Ccl5 content (A) and Tnf content (B) by ELISA. The results show mean with range of duplicates and are representative of three independent experiments. C, responses induced by TLR2 ligands are abolished in TLR2−/− mice. Peritoneal macrophages from wild-type (WT) and TLR2−/− mice were stimulated with medium or decreasing concentrations of Pam3Cys (P3C) (100–10 ng/ml), MALP-2 (100–1 ng/ml), LPS (100 ng/ml), or Sendai virus (SV) (100 HAU/ml) for 20 h before supernatant was harvested and analyzed for Ccl5 content by ELISA. The results show mean of quadruplicates ± S.E. Ccl5 (D) and Tnf (E) content in supernatant from macrophages from wild-type (WT), MyD88−/−, and double knock-out MAL−/−MyD88−/−, TRIF−/−MyD88−/−, and TRAM−/−TRIF−/− mice stimulated with medium (0) or decreasing concentrations of LPS (100–10−1 to 0.1 ng/ml), Pam3Cys (P3C) (100–10 ng/ml), R848 (100 ng/ml), or SV (100 HAU/ml) for 18 h before supernatant was analyzed by ELISA. Results show mean of triplicates ± S.D. ****, p < 0.0001 (two-way ANOVA with Bonferroni post-test).
Next, we assessed Ccl5 induction in response to TLR2 ligands in macrophages from MyD88−/−, MAL−/−MyD88−/−, TRIF−/−MyD88−/−, and TRAM−/−TRIF−/− double-deficient mice. We observed that Ccl5 induction was impaired in TRAM−/−TRIF−/− cells in response to Pam3Cys (Fig. 2D), confirming our observations in TRIF−/− and TRAM−/− macrophages (Figs. 1C and and22A). Ccl5 induction was also impaired in all the tested adaptor-deficient cells in response to the TLR2/TLR6 ligand MALP-2 (data not shown). Importantly, TRAM−/−TRIF−/− macrophages displayed normal Tnf expression in response to both Pam3Cys and R848 (Fig. 2E). Combined, our results imply that the TRAM/TRIF-dependent pathway is involved in TLR2-induced Ccl5 release (Figs. 1C and and2,2, A, C, and D), but not in TNF release (Figs. 1D and and2,2, B and E). LPS-induced Ccl5 was completely abrogated in both TRIF−/− MyD88−/− and TRAM−/−TRIF−/− macrophages (Fig. 2D), emphasizing the recognized TRAM/TRIF dependence of this response. TNF release following LPS challenge was greatly reduced in both MAL−/−MyD88−/− and TRAM−/−TRIF−/− macrophages (Fig. 2E), implying that both of the signaling branches contribute considerably to LPS-induced TNF release. Responses toward SV were normal in TRIF−/−MyD88−/− macrophages (Fig. 2, C and D), showing that SV recognition is TLR-independent in these cells, as reported previously (6,–8).
TLR2 and TRAM Co-localize in Endosomes, and TLR2-mediated CCL5 Induction Is Dependent on Endocytosis
We proceeded to investigate the subcellular expression of TLR2 and TRAM to verify the involvement of TRAM in TLR2 signaling. TLR2 is expressed in early endosomes, as well as in recycling Rab11a-positive endosomes in human monocytes (30). TRAM has been reported to be expressed at the plasma membrane and in Rab5-positive endosomes (46). We therefore proceeded to investigate whether TLR2 co-localizes with TRAM in early endosomes. Overexpression of fluorescently tagged TLR2Cherry, TRAMYFP, and the early endosome marker Eea-1CFP in HEK293 cells revealed that TLR2 and TRAM indeed do co-localize, both at the plasma membrane and in a portion of Eea-1-positive endocytic vesicles (Fig. 3A). Upon stimulation with FSL-1, TLR2 and TRAM accumulated and co-localized in the perinuclear area (Fig. 3B). Overexpression of TRAMYFP and TRIFCFP in HEK293 showed particularly good co-localization in endocytic vesicles (Fig. 3C). Minimal concentrations of plasmid were used to avoid cell activation because expression of high levels of these adaptor molecules alone can drive NFκB activation. However, we cannot exclude the possibility that the strong co-localization between TRAM and TRIF may partly be a consequence of transfection-driven cell activation. Nevertheless, we also observed TLR2 co-localizing with TRIF in TRAM-positive vesicles in HEK293 overexpressing TLR2Cherry, TRIFCFP, and TRAMYFP (Fig. 3C), implying that TLR2 is recruited to a TRAM- and TRIF-containing complex.
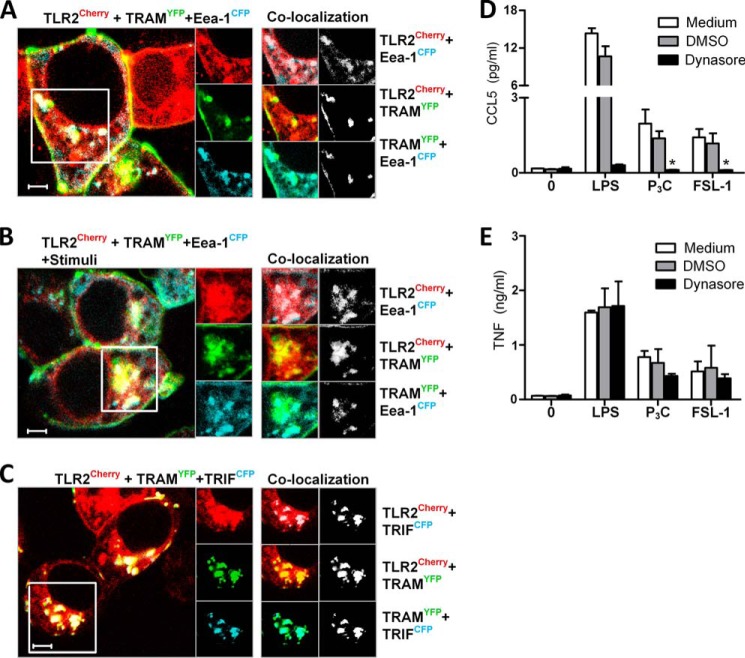
TLR2 and TRAM co-localize in endosomes, and TLR2-mediated Ccl5 is impaired upon inhibition of endocytosis. HEK293 cells were transiently transfected with TLR2Cherry (red), TRAMYFP (green), and early endosome marker Eea-1CFP (blue) (A and B) for 48 h prior to treatment with medium (A) or FSL-1 (B) (200 ng/ml) for 1 h before cells were fixed and observed by confocal microscopy. Overlays of TLR2Cherry (red), TRAMYFP (green), and early endosome marker Eea-1CFP (blue) are shown in images to the left. C, TLR2 co-localizes with TRAM and TRIF. HEK293 cells were transiently transfected with TLR2Cherry (red), TRAMYFP (green), and TRIFCFP (blue) for 48 h. Images to the right show single tracks of enlargements of the areas denoted by the square in the main picture. Co-localization between channels in this area are shown to the far right, both in color and by using co-localization maps. Maps show co-localization events between the two respective channels (white). Measure bar is 5 μm. D, Ccl5 and E, Tnf release in peritoneal macrophages from wild-type mice pretreated with medium, a DMSO control, or Dynasore (80 μm) for 30 min prior to stimulation with medium (0), LPS (10 ng/ml), Pam3Cys (P3C) (200 ng/ml), or FSL-1 (200 ng/ml) for 21 h. Supernatant was analyzed for cytokines by ELISA. Results show mean of triplicates ± S.D.
Because we observed TLR2 and TRAM co-localizing in endocytic compartments, we investigated the importance of endocytosis for Ccl5 induction by pretreating peritoneal macrophages with the dynamin GTPase inhibitor Dynasore prior to stimuli with LPS and TLR2 ligands. Dynasore efficiently inhibited CCL5 release induced by TLR2 ligands (Fig. 3D) but not TNF release (Fig. 3E), suggesting that endocytosis of ligand is required for Ccl5 induction in response to these ligands. These results suggest that signaling leading to Ccl5 induction likely occurs from early endosomes, where TLR2 and TRAM co-localize.
TLR2-mediated CCL5 Induction Is Impaired in TRIFLps2/Lps2 Mice in Vitro and in Vivo
TRIFLps2/Lps2 mice are derived from a C57Bl/6 background and contain a chemically induced base pair-deletion/frameshift in the gene encoding TRIF (25). These animals mimic mice with a gene targeted TRIF deletion. We observed that macrophages from TRIFLps2/Lps2 mice displayed reduced Ccl5 release in response to TLR2 ligands MALP-2 and FSL-1 (Fig. 4A). In contrast, Tnf release was not impaired in response to these TLR2 ligands (Fig. 4B), which is consistent with previous reports (25).Ccl5 and Tnf release were normal in response to R848 (Fig. 4, A and B).
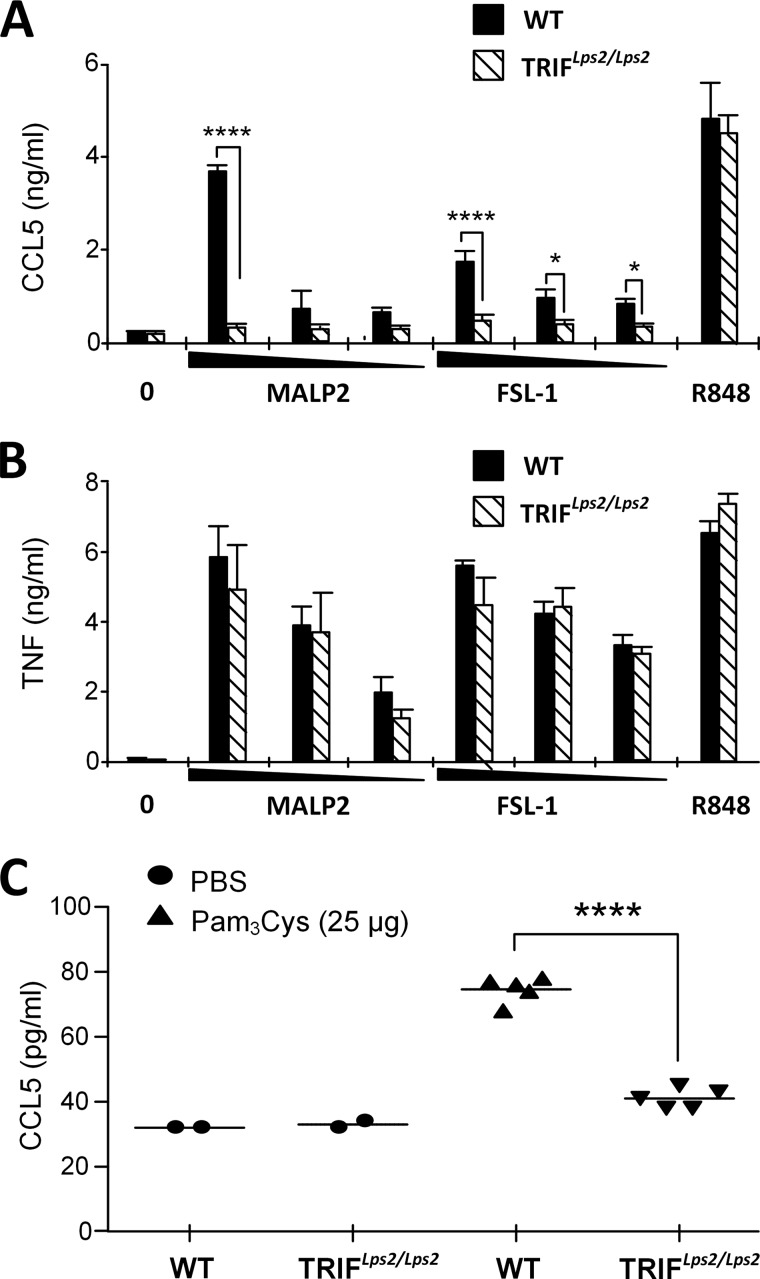
TLR2-mediated Ccl5 induction is impaired in TRIFLps2/Lps2 mice in vitro and in vivo. Peritoneal macrophages from wild-type (WT) or TRIFLps2/Lps2 mice were treated with medium (0), and MALP-2 (1–0.1–0.01 μg/ml), FSL-1 (1–0.1–0.01 μg/ml), or R848 (10 ng/ml) for 20 h before supernatant was harvested and analyzed for Ccl5 (A) and TNF (B) release by ELISA. Results show mean of triplicates with standard deviations. ****, p < 0.0001; *, p < 0.05 (two-way ANOVA with Bonferroni post-test). C, in vivo induction of Ccl5 in the serum of wild-type mice (WT) or TRIFLps2/Lps2 mice injected intraperitoneally with PBS or Pam3Cys (25 μg). Blood was collected by cardiac puncture 4 h after injection, and serum was analyzed for Ccl5 content by ELISA. Results show mean values of two PBS-injected mice and five Pam3Cys-injected mice. ****, p < 0.0001 (unpaired Student's t test).
To determine whether TLR2 ligands can induce Ccl5 in a TRIF-dependent manner in vivo, WT or TRIFLps2/Lps2 mice received an intraperitoneal injection with Pam3Cys. Serum was collected after 4 h and assayed for Ccl5. We found that Ccl5 levels were elevated in wild-type mice following Pam3Cys injection (Fig. 4C). This Ccl5 induction in response to Pam3Cys injection was impaired in TRIFLps2/Lps2 mice (Fig. 4C), confirming our in vitro findings and supporting a role for TRIF in TLR2-mediated Ccl5 induction.
TLR2-mediated CCL5 Activation Is Impaired upon Overexpression of Signaling Defective Mutant Versions of TRAM and TRIF
Our findings in peritoneal macrophages were verified by transiently transfecting HEK293 cells stably expressing TLR2 with a murine ccl5 luciferase reporter gene. Stimulation of TLR2-expressing HEK293 cells with TLR2 ligands activated the ccl5 promoter (Fig. 5A). Similar results were obtained using a human CCL5 luciferase reporter gene (data not shown). The different TIR-adaptor proteins were also assessed for their ability to activate the CCL5 promoter upon overexpression. This was done by transfecting HEK293 cells with TLR2 and the CCL5 luciferase and co-transfecting with MAL, MyD88, TRAM, or TRIF or the functionally defective adaptor mutants MyD88TIR, TRAMC117H, and TRIFΔNΔC. Transfection of MyD88, TRAM, or TRIF all resulted in activation of the CCL5 reporter in this system (Fig. 5B), whereas overexpression of TRAM C117H, TRIFΔNΔC, or MyD88TIR impaired CCL5 induction in response to TLR2 ligands Pam3Cys and FSL-1 (Fig. 5C). Combined, these results confirm that TLR2 ligands induce CCL5 and that TRAM and TRIF mediate CCL5 induction in response to TLR2 ligands.
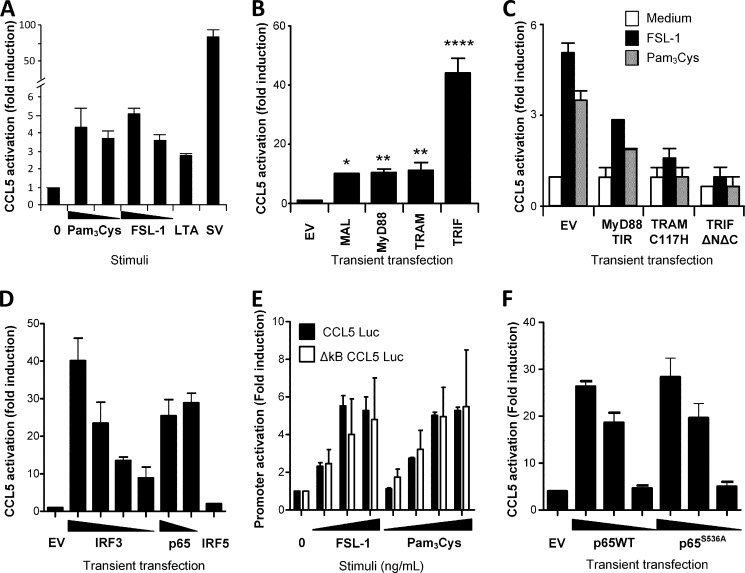
TLR2-mediated CCL5 activation is impaired upon overexpression of TRAM and TRIF. A, HEK293-TLR2YFP cells were transiently transfected with a murine Ccl5 luciferase reporter gene for 24 h prior to stimulation with medium, Pam3Cys (100–10 ng/ml), FSL-1 (100–10 ng/ml), LTA (10 μg/ml), or Sendai virus (SV) (50 HAU/ml) for 18 h and assayed for luciferase reporter gene activity. Results show fold induction relative to the medium control. B, HEK293 cells were transiently transfected with CCL5 luciferase reporter gene and co-transfected with empty vector (EV), MAL, MyD88, TRAM, or TRIF (20 ng/well) for 48 h and assayed for luciferase activity. ****, p < 0.0001; **, denotes p < 0.01; *, p < 0.05 (one-way ANOVA with Tukey post-test). C, HEK293 cells were transiently transfected with CCL5 luciferase, TLR2, and the adaptor mutants MyD88TIR, TRAMC117H, and TRIFΔNΔC for 48 h. Cells were then stimulated with medium, Pam3Cys (100 ng/ml), or FSL-1 (200 ng/ml) for 16 h before luciferase reporter gene activity was assayed. D, HEK293 cells were transfected with CCL5 luciferase and empty vector (EV), IRF3-FLAG (80–60–40–20 ng), NF-κB subunit p65 (20–10 ng), or IRF5-FLAG (80 ng) for 26 h, before cells were lysed and assayed for luciferase activity. Activation of the CCL5 promoter occurs independent of NF-κB p65S536. E, HEK293 cells were transfected with TLR2 and a CCL5 luciferase reporter gene (CCL5 Luc) or a CCL5 luciferase reporter gene with mutated NF-κB sites (ΔκB CCL5 Luc) overnight. Cells were stimulated with medium (0), FSL-1 (1–10–100 ng/ml), or Pam3Cys (1–10–100–200 ng/ml) for 18 h before cells were lysed and assayed for luciferase activity. F, CCL5 luciferase activation upon overexpression of NF-κB subunit p65 (10–1–0.1 ng) or mutant p65S536A (10–1–0.1 ng) for 27 h before cells were lysed and assayed for luciferase activity. All results show firefly luciferase values normalized to Renilla activity and display mean of triplicates ± S.D. Results are representative of three independent experiments.
The promoters of CCL5, CXCL10, as well as IFN-β contain transcription factor-binding elements for both NF-κB and IRFs (31, 32). We observed that overexpression of NF-κB p65 and IRF3, but not of IRF5, efficiently induced activation of the CCL5 promoter in HEK293 cells co-transfected with a CCL5 luciferase reporter (Fig. 5D). TLR2 ligands are considered poor activators of IRF3 (9, 33), so we initially investigated whether the observed TLR2-mediated CCL5 response could be mediated by NF-κB alone. Using HEK293 cells transiently expressing TLR2 and a CCL5 luciferase reporter containing mutated NF-κB sites (ΔκB CCL5 luciferase), we found that the fold induction activation of this reporter was similar to the activation of WT CCL5 luciferase reporter in response to TLR2 ligands (Fig. 5E). Overexpression of mutant NF-κB p65S536A in HEK293 cells also activated the CCL5 luciferase reporter to the same extent as wild-type p65 (Fig. 5F). These results indicate that phosphorylation of p65Ser-536 is not essential for TLR2-mediated CCL5 induction and prompted us to investigate a role for IRF3 in TLR2-mediated CCL5.
TLR2 Ligands Activate IRF3, and CCL5 Induction Is impaired in IRF3−/− and IRF1−/−Macrophages in Response to TLR2 Ligands
To investigate whether TLR2 ligands can activate IRF3, we initially applied a GAL4 luciferase reporter assay and a vector expressing yeast Gal4 DNA binding domain fused to IRF-3 lacking its own DNA binding domain (IRF3-Gal4). Activation of IRF3 in this assay leads to the expression of the GAL4 luciferase reporter (34, 35). We observed that TLR2 ligands activated IRF3 measured by this reporter system, and that the activation was impaired upon overexpression of the dominant negative version of TRAM, TRAMC117H (Fig. 6A). The FSL-1 lipopeptide also induced phosphorylation of IRF3Ser-396 in RAW264.7 cells (Fig. 6B), confirming that TLR2 ligands can activate IRF3, although to a lesser extent than LPS and poly(I:C) (Fig. 6B). IRF1 also regulates TLR-dependent induction of type I interferon responses in myeloid dendritic cells and macrophages (36). We further investigated the role for IRF3 and IRF1, as well as IRF5 and IRF7, in TLR2-mediated CCL5 induction. Peritoneal macrophages from wild-type (WT), irf1−/−-, irf3−/−-, irf5−/−-, or irf7−/−-deficient mice were stimulated with TLR2 ligands, as well as LPS, poly(I:C), R848, or SV prior to assaying Ccl5 and Tnf release in these cells by ELISA. We found that Ccl5 release was impaired in peritoneal macrophages from both Irf1- and Irf3-deficient mice in response to the TLR2 ligands Pam3Cys, Pam2Cys, FSL-1, and MALP-2 (Fig. 6C). Tnf was, however, also partially impaired in irf1−/− macrophages in response to TLR2 ligands (Fig. 6D), suggesting that irf1 plays a role in both Tnf and Ccl5 release in response to lipopeptides. LPS- and poly(I:C)-induced Ccl5 was normal in irf1−/− macrophages, in line with previous reports (37). Importantly, IRF3 was found to be a key player in mediating the release of Ccl5 (Fig. 6C), but not Tnf (Fig. 6D), in response to TLR2 ligands. This correlates with our observations in TRAM−/− and TRIF−/− macrophages (Figs. 1 and and2).2). Ccl5 induction in response to TLR2 ligands was found to be normal in irf5−/− peritoneal macrophages, whereas both Ccl5 and TNF were partially impaired in irf7−/− cells in response to TLR2 ligands (Fig. 6, C and D). These results show that TLR2 ligands activate Irf3, and TLR2-mediated Ccl5 is mediated by Irf3, as well as by Irf1, and they suggest that TLR2-induced Irf3 activation is mediated by the TRAM/TRIF pathway.
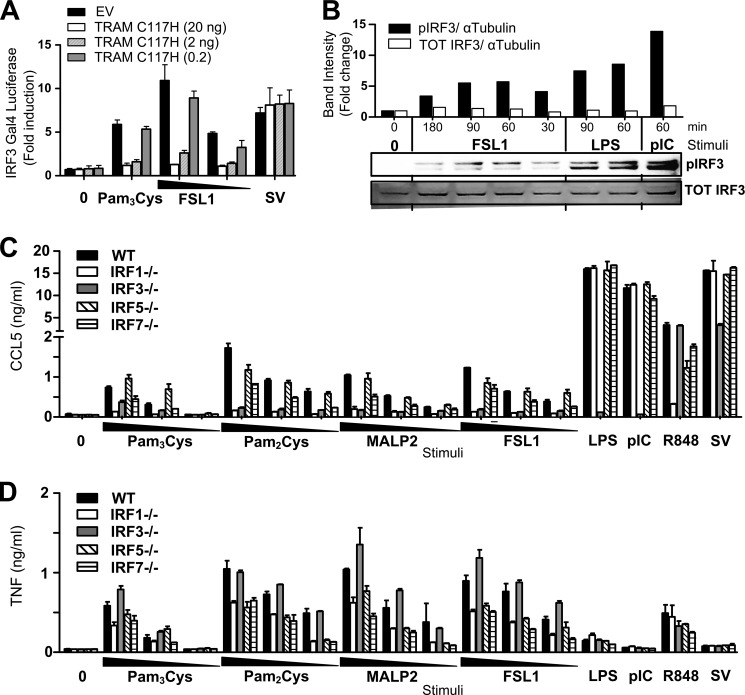
TLR2 ligands activate IRF3, and TLR2-mediated Ccl5 release is impaired in IRF3−/− cells. A, HEK293 cells were transiently transfected overnight with empty vector (EV) or TRAMC117H (20–2-0.2 ng), and co-transfected with TLR2 and IRF3-Gal4 and a luciferase reporter containing the Gal4 activation sequence (Gal4 luciferase). Cells were subsequently stimulated with Pam3Cys (200 ng/ml), FSL-1 (200–20 ng/ml), or Sendai virus (SV) (100 HAU/ml) for 16 h before cells were assayed for luciferase activity. Results are normalized for Renilla activity and show mean fold induction ± S.D. of triplicates. B, Western blots of lysate of RAW264.7 stimulated with medium (0), with FSL-1 (200 ng/ml) for 180–90-60 and 30 min, with LPS (100 ng/ml) for 90 and 60 min, or poly(I:C) (50 μg/ml) for 60 min and stained with anti-IRF3Ser-396 (pIRF3) or anti-Total-IRF3. Graph shows quantification of band intensities of blot shown below stained with anti-IRF3Ser-396 (pIRF3) (top blot) and total IRF3 (TOT IRF3) (bottom blot) relative to α-tubulin staining. TLR2-mediated Ccl5 induction is impaired in IRF1−/− and IRF3−/− mice. Peritoneal macrophages from wild-type (WT), irf1−/−, irf3−/−, irf5−/−, and irf7−/− mice were exposed to medium (0), Pam3Cys (1000–100–10 ng/ml), Pam2Cys (1000–100–10 ng/ml), MALP-2 (1000–100–10 ng/ml), FSL-1 (1000–100-10 ng/ml), LPS (10 ng/ml), poly(I:C) (pIC) (50 μg/ml), R848 (100 ng/ml), or Sendai virus (SV) (100 HAU/ml) for 18 h before supernatant was harvested and analyzed for Ccl5 content (C) and Tnf content (D) by ELISA. Results show mean with range of duplicate samples and are representative of two experiments.
TLR2-mediated Ccl5 Induction Is Impaired in TBK1-deficient Cells
The noncanonical IκB kinases IKKϵ and TBK1 are important mediators of TRIF-dependent IRF3 activation leading to type I IFN and CCL5 induction in response to viral components and LPS (38). To determine the role of IKKϵ, as well as the type I interferon receptor I (IFNAR) in TLR2-mediated Ccl5, we assayed Ccl5 induction in response to TLR2 ligands in peritoneal macrophages from WT, ikke−/− (IKKϵ) and ifnar−/− mice, as well as irf3−/− mice. Ccl5 release in response to TLR2 ligands was normal in IKKϵ-deficient cells after 18 h of stimulation (Fig. 7A). The receptor for type I IFNs, Ifnar, did not appear to be important for TLR2-mediated Ccl5 release either (Fig. 7A), in contrast to Irf3 (Fig. 7A), indicating that TLR2-mediated Ccl5 induction is a primary response that occurs independent of type I IFN signaling. TLR2-induced ccl5 mRNA induction was also unaffected by inhibition of protein synthesis with cycloheximide (data not shown), further suggesting that this Ccl5 induction is a primary response.
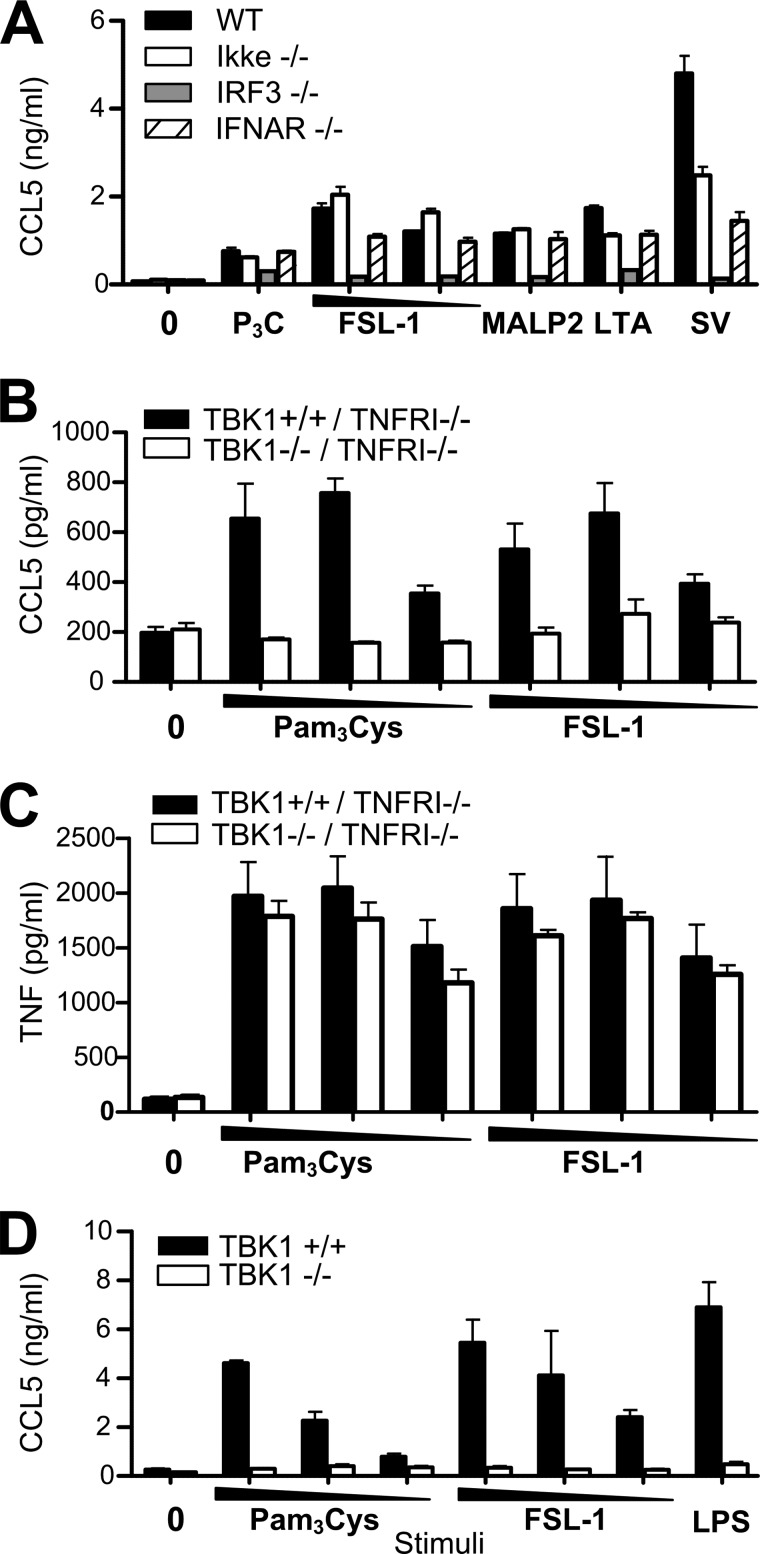
TLR2-mediated Ccl5 induction is impaired in tbk1-deficient cells. A, peritoneal macrophages from wild-type (WT), ikkϵ−/−, irf3−/−, and ifnar1−/− mice were exposed to medium (0), Pam3Cys (P3C) (100 ng/ml), FSL-1 (100–10 ng/ml), MALP-2 (100 ng/ml), LTA (10 μg/ml), or Sendai virus (SV) (10 HAU/ml) for 18 h before supernatant was harvested and analyzed for Ccl5 content by ELISA. Results show mean with range of duplicate samples. Ccl5 (B) and Tnf (C) induction in immortalized BMDM from tnfrI−/−-deficient or tbk1−/−/tnfrI−/− double-deficient mice stimulated with medium (0), Pam3Cys (100–10−1 ng/ml), or FSL-1 (100–10−1 ng/ml) for 6 h. Supernatant was harvested and analyzed for Ccl5 and Tnf by ELISA. Results show mean ± S.D. of quadruplicates. D, Ccl5 induction in tbk1+/+ or tbk1−/− MEFs stimulated for 18 h with medium (0), Pam3Cys (200–100–50 ng/ml), FSL-1 (200–100-50 ng/ml), or LPS (100 ng/ml) before supernatant was assayed for Ccl5 by ELISA. Results show mean ± S.D. of triplicate samples.
IKKϵ and TBK1 have both been shown to participate in mediating TRIF-IRF3 signaling (17), so we proceeded to investigate the role of TBK1 in TLR2-mediated CCL5. Homozygous deletion of tbk1 is lethal in C57BL/6, but because this embryonic lethality is TNF-dependent, mice survive upon deletion of tnfrsf1a (tnfrI) (17). We consequently assayed Ccl5 induction in response to TLR2 ligands in immortalized BMDM from tbk1−/−/tnfrI−/− double-deficient and tnfrI−/−-deficient mice. tbk1−/−/tnfrI−/− cells displayed impaired Ccl5 induction in response to TLR2 ligands at early time points, following 6 h of stimulation (Fig. 7B), whereas the same cells displayed similar TNF release (Fig. 7C). After longer stimulation (>12 h) TLR2-mediated Ccl5 induction appeared normal in tbk1−/−/tnfrI−/− BMDM (data not shown), possibly due to redundancy between IKKϵ and TBK1 in their ability to activate IRF3 (39, 40). The role for TBK1 in mediating TLR2-induced Ccl5 was confirmed in tbk1−/− MEFs (Fig. 7D). LPS-induced Ccl5 was also impaired in tbk1−/− MEFs (Fig. 7D), in accordance with previous reports (38). These data are consistent with a role for TBK1 in TLR2-mediated CCL5 induction.
TLR2 Ligand-induced Ccl4 and IFN-β Are Partially Impaired in the Absence of TRIF−/−
To determine whether the TRAM/TRIF pathway is involved in the regulation of other genes in response to TLR2 ligands, we performed gene expression analysis of selected inflammatory genes on total RNA from peritoneal macrophages from WT or TRIF−/− mice treated with medium or FSL-1 lipopeptide for 4 h. The results confirmed that Ccl5 is induced in response to TLR2 ligand FSL-1 and that this response is impaired in TRIF−/− macrophages (Fig. 8A). The interferon-inducible genes cxcl10, ifit1, and ifit2 were very weakly induced in response to FSL-1, although FSL-1 failed to induce ifnβ1 induction above the detection limit in these cells using this nonamplified system (Fig. 8A). The induction of the chemokine ccl4 was partially impaired in TRIF−/− macrophages in response to FSL-1 (Fig. 8B), suggesting that TRIF participates in mediating this response. We proceeded to evaluate Ccl4 protein induction in response to TLR2 ligands by stimulating peritoneal macrophages from TRIF−/− mice with lipopeptides and LPS, poly(I:C), or Sendai virus. TLR2-induced Ccl4 protein was found to be partially impaired in TRIF−/− cells after 20 h of stimulation (Fig. 8C). Ccl4 release was also impaired in TRIF−/− cells in response to LPS and poly(I:C), but not in response to Sendai virus (Fig. 8C). Overexpression of TRAM and TRIF, as well as MAL and MyD88, in HEK293 cells activated the CCL4 promoter-driven reporter luciferase (data not shown), supporting the view that these adaptors are involved in the regulation of CCL4. Overexpression of MAL and MyD88 induced a more potent activation of the CCL4 reporter luciferase (data not shown), suggesting that the MAL/MyD88 pathway is more important in regulating CCL4, and possibly explaining why TLR2-mediated Ccl4 induction in TRIF−/− macrophages was only partial impaired (Fig. 8, B and C). To further confirm a role for TRAM in the regulation of CCL4, HEK293 cells were transiently transfected with TLR2, CCL4 luciferase reporter, and decreasing concentrations of dominant-negative TRAMC117H, prior to stimulation with lipopeptides or Sendai virus. TLR2 ligands potently induced the CCL4 reporter luciferase, and this induction was impaired upon overexpression of mutant adaptor TRAMC117H (Fig. 8D), indicating a role for TRAM in mediating CCL4. In contrast, CCL4 induction in response to Sendai virus was normal upon overexpression of TRAMC117H (Fig. 8D).
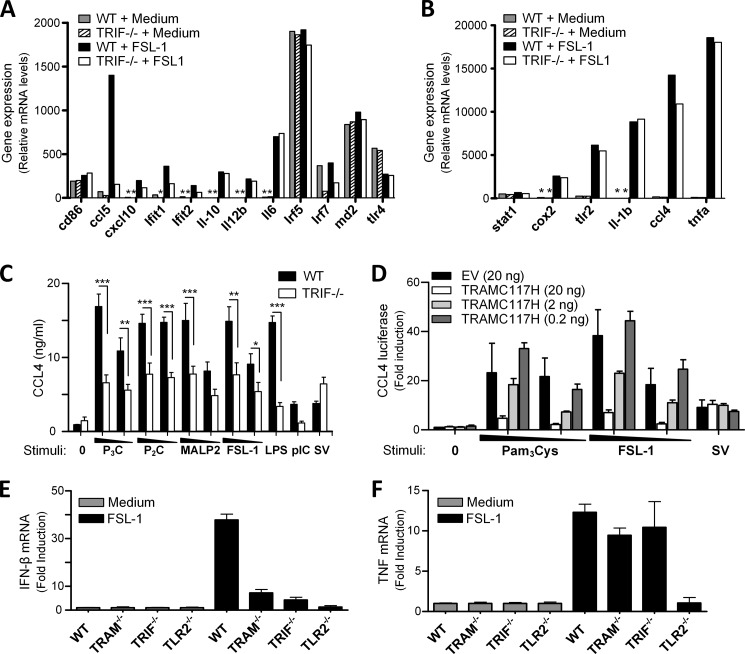
Ccl4 and IFN-β are partially impaired in TRIF−/− macrophages in response to TLR2 ligands. A and B, Nanostring nCounter gene expression analysis of wild-type (WT) or TRIF−/− peritoneal macrophages stimulated with medium (0) or FSL-1 (200 ng/ml) for 4 h before total RNA was isolated and assayed for the mRNA copy number of selected genes by Nanostring nCounter gene expression analysis. Results in A and B are from the same experiments. Results are representative of three experiments. * denotes below detection limit. TLR2-mediated Ccl4 activation is partially mediated by the TRAM/TRIF pathway. C, Ccl4 induction in wild-type (WT) or TRIF−/− peritoneal macrophages treated with medium (0), Pam3Cys (P3C) (1–0.1 μg/ml), Pam2Cys (P2C) (1–0.1 μg/ml), MALP-2 (1–0.1 μg/ml), FSL-1 (1–0.1 μg/ml), LPS (100 ng/ml), poly(I:C) (5 μg/ml), or Sendai virus (10 HAU/ml) for 20 h. Supernatant was analyzed for Ccl4 by ELISA. Results show mean ± S.E. of duplicates from two independent experiments. ***, p < 0.001; **, p < 0.01; *, p < 0.05 (one-way ANOVA with Bonferroni post-test). D, HEK293 cells were transfected with TLR2 and CCL4 reporter luciferase and co-transfected with empty vector (EV) and decreasing amounts of TRAMC117H (20–2–0.2 ng) overnight. Cells were subsequently stimulated with Pam3Cys (100–10 ng/ml), FSL-1 (100–10 ng/ml), or Sendai virus (SV) (300 HAU/ml) for 12 h before cells were lysed and assayed for luciferase activity. Results were normalized for Renilla activity and show mean ± S.D. of triplicates. F, IFN-β induction in BMDM in response to FSL-1 is mediated by TRAM and TRIF. Induction of Ifn-β (E) and Tnf (F) mRNA in BMDM from C57BL/6 wild-type (WT), TRAM−/−, TRIF−/−, and TLR2−/− mice that were left untreated (gray bars) or stimulated with FSL-1 (10 ng/ml) (black bars) for 3 h. Ifnb1 and tnf mRNA in the samples were determined by RT-qPCR and are presented as relative induction with each of the nontreated cell lines as reference sample. Gapdh served as internal control. Results show mean fold induction ± S.D. of triplicates and are representative of five experiments.
TLR2 has been suggested to induce IFN-β in an IRF-3-dependent manner in inflammatory monocytes (41). We consequently assayed immortalized bone marrow-derived macrophages from WT, TRAM−/−, TRIF−/−, and TLR2−/− mice stimulated with FSL-1 by quantitative RT-PCR to determine whether these cells can induce IFN-β in a TRAM/TRIF-dependent manner. Induction of ifn-β was observed in wild-type cells following 3 h of stimulation (Fig. 8E). This ifn-β induction was further found to be partially impaired in TRAM−/− and TRIF−/− cells (Fig. 8E), implying a role for TRAM and TRIF in TLR2-mediated IFN-β induction. TNF induction was normal in TRAM−/− and TRIF−/− cells in response to FSL-1 (Fig. 8F). These results suggest that the TRAM/TRIF signaling pathway plays an important role in certain TLR2-mediated responses.
DISCUSSION
TLR4 has been considered the only TLR that can activate both the MyD88-dependent pathway and the TRAM/TRIF-dependent signaling pathway in macrophages. In this study, we show a new role for TRAM and TRIF in TLR2 signaling. In resemblance to TLR4 signaling, we propose that TLR2 can utilize the TRAM/TRIF signaling pathway, in addition to the MyD88-dependent pathway, to mediate certain responses.
We initially observed that LPS-induced up-regulation of surface TLR2 was partially impaired in TRIF−/− macrophages, suggesting that TRIF contributes to this response. TLR2 has previously been shown to be differentially regulated by both TRIF and MyD88 in response to LPS at the gene level by microarray (42). Here, we show that the TRIF signaling branch appears to be more important for correct regulation of surface TLR2 in response to LPS, because this expression was impaired in the absence of TRIF but not in the absence of MyD88 (14). The mechanisms regulating LPS-induced TLR2 expression have yet to be elucidated, but they appear to differ from LPS-induced CD86 surface expression, which has been shown to be TRIF-, type I IFN-, and IFNARI-dependent (25).
Interestingly, we observed that TLR2-mediated induction of Ccl5, Ccl4, and IFN-β are impaired in TRIF-deficient macrophages. TLR2-dependent induction of IFN-β has previously been reported in murine inflammatory monocytes in response to vaccinia virus (41), although this response was not found to be regulated by a TRIF-dependent mechanism. Bacterial lipopeptides were later shown to induce IFN-β, as well as the IFN-inducible genes cxcl10, mx-2, il-6, and nos2 (inos) in a TLR2-dependent manner in BMDM at high concentrations of ligand (43). The IFN-β induction observed by Dietrich et al. (43) in response to TLR2 ligands was shown to be a primary response mediated by MyD88, by a mechanism resembling TLR7/9 signaling. In this report we confirm that TLR2 ligands induce IFN-β in macrophages. We further show that TLR2-mediated IFN-β, Ccl5, and Ccl4 are TRAM- and TRIF-dependent, in addition to MyD88-dependent. The TLR2-mediated Ccl5 induction we observed was also a primary response occurring independent of IfnarI and protein synthesis. We observed that high concentrations of 0.1–1 μg/ml lipopeptide were required to induce Ccl5, whereas Tnf release is induced at considerably lower concentrations of ligand. Involvement of TRAM and TRIF was observed regardless of the magnitude of Ccl5 that was induced by either TLR4 or TLR2 ligands. Importantly, Tnf release in response to TLR2 ligands was not inhibited in TRIF−/− or TRAM−/−TRIF−/− macrophages, consistent with previous reports (25, 44).
We have previously shown that TLR2 localizes to early and recycling endosomes (30, 45) and that inhibiting endocytosis did not affect NF-κB activation and TNF release. Dietrich et al. (43) found that IFN-β induction in response to TLR2 ligands was efficiently blocked by inhibiting endocytosis. We also observe that TRAM/TRIF-dependent TLR2-mediated Ccl5 induction is impaired upon inhibition of endocytosis, in line with Dietrich et al. (43), whereas TNF release occurs independent of ligand internalization as we previously reported (30). We further show that TLR2 co-localizes with TRAM in early endosomes, in a manner resembling TLR4 (46). TLR2 and TRIF co-localization was observed in TRAM-positive vesicles suggesting that endocytosis is required to deliver TLR2 to endosomes where TRIF resides. Subsequently, TRAF3 may be recruited to initiate downstream signaling leading to IRF3 activation in resemblance with TLR4 (46). Expression of TRAF3 at the plasma membrane has indeed been shown to potentiate TLR2-mediated IFN-β induction from the plasma membrane (46).
Our results indicate that TLR2 appears to resemble TLR4 with regard to some aspects of localization and the requirement for ligand internalization to activate the TRAM/TRIF pathway. Important differences between TLR2 and TLR4 were, however, also observed in our experimental settings. In contrast to TLR4-mediated Ccl5, which is heavily TRIF-dependent, TLR2-mediated Ccl5 is dependent on both the MyD88- and TRIF-dependent pathway. TLR4-mediated Tnf release is controlled by both signaling branches, whereas TLR2-mediated Tnf release is strictly MyD88-dependent and is not influenced by TRAM and TRIF. Cross-talk between the MyD88- and the TRIF-dependent pathway appears to be regulated differently downstream of TLR2 and TLR4. TLR2 ligands also induce lower levels of Ccl5 and IFN-β than TLR4 ligands, and the induction of the interferon-inducible genes cxcl10, ifit1, and ifit2 were only weakly induced in response to FSL-1. More studies are needed to investigate the role of TRIF or TRAM in these responses. The levels of IFN-β induced by TLR2 ligands may be too low to induce these genes in peritoneal macrophages, although BMDM may induce higher levels of IFN-β (43). It was recently reported that the TLR2 ligand Pam3Cys induces cxcl10, ifit1, and type I IFNs in a TRIF-dependent manner (47). In that report, Pam3Cys was, however, also shown to induce Tnf, Il6, and Il10 by a TRIF-dependent mechanism, which is different from our results. Notably, the latter is in contradiction with early studies of TRIFLps2/Lps2 mice, which contain a loss-of-function mutation in the gene encoding TRIF (25). We consistently observed normal induction of Tnf, as well as Il6 and Il10 in response to TLR2 ligands in TRIF-deficient cells, which is in line with Hoebe et al. (25). Our results support however the finding by Petnicki-Ocwieja et al. (47), showing that TLR2 ligands induce type I IFNs in a TRIF-dependent manner, but we observed that only a subset of genes, including Ccl4, Ccl5, and IFN-β, were impaired in TRIF−/− cells in response to TLR2 ligands.
We speculate that some of the differences between TLR2 and TLR4 signaling could be due to the relative subcellular location of the receptors. IFN-β induction in response to LPS is proposed to originate in endosomes where TRAM is expressed, whereas MyD88 activation and NF-κB activation are proposed to originate at the plasma membrane. We have shown that TLR2 is expressed in early endosomes where the receptor co-localizes with TRAM, and that Ccl5 release is impaired upon inhibition of endocytosis, in likeness with TLR4. In contrast to TLR4, which is down-regulated at the cell surface and targeted to endosomes (48), we observe that TLR2 is up-regulated at the plasma membrane. Although TLR2 is expressed in endosomes, we speculate that up-regulation of TLR2 at the plasma membrane in response to stimuli could still limit the amount of TLR2 delivered to the endosomes where activation of TRAM and TRIF seems to occur. This could, in turn, lead to lower levels of TRIF-mediated responses induced by TLR2 ligands, relative to TLR4 ligands. The precise mechanisms differentiating TLR2 and TLR4 signaling and responses have yet to be elucidated.
The IFN-β induction observed by Dietrich et al. (43) in response to TLR2 ligands was shown to be mediated by MyD88, Irf1, and Irf7 in BMDM, in a manner resembling TLR7/9 signaling. Irf3 and Irf7 were, however, required for the TLR2-dependent induction of IFN-β in response to vaccinia virus, which was observed by Barbalat et al. (41). IFN induction has been proposed to be controlled by different IRFs in different cell types (36), possibly explaining these variations. We observed that IFN-β induction was TRAM- and TRIF-dependent, in addition to MyD88-dependent. TLR2-mediated Ccl5 induction was further found to require Irf3, in addition to Irf1, and to some extent Irf7, but not Irf5, in peritoneal macrophages. Thus, TLR2-induced Ccl5 induction mimics TLR4-induced Ccl5 induction with regard to Irf3 dependence (9); however, similarities between TLR2 and TLR7/9 signaling were also observed with regard to Irf1 dependence in this response. IRF1 has been reported to be involved in TLR7/9-MyD88-mediated IFN-β, CCL5, and IL-12 induction in dendritic cells, but not in the induction of TNF. We observed that TNF release was also partially impaired in response to TLR2 ligands in IRF1−/− cells. The role of IRF1 in TLR signaling is still incompletely understood, although it has been suggested that IRF1 can cooperate with STAT1. It has also been proposed that IRF1 can be recruited to promoter elements of TNF, IL6, IL-12, and other inflammatory genes following cellular exposure to LPS (45). Our results show that TLR2 can utilize both the MyD88-dependent pathway and the TRAM/TRIF pathway leading to induction of cytokines such as CCL5 and IFN-β. We cannot exclude the possibility that the relative importance of the adaptor molecules in signaling may differ somewhat between different cell types.
In addition to Ccl5 and IFN-β, we also observed that TLR2 ligands induce Ccl4 in a manner partially dependent on TRIF, suggesting that TRIF plays a role in several TLR2-mediated responses. LPS-induced Ccl4 was also partially impaired in TRIF−/− macrophages, suggesting that both the MyD88 and the TRIF pathways contribute to this response.
A role for TRIF in TLR5 signaling in intestinal epithelial cells has also been proposed (49). In that study TRIF was found to mediate NF-κB and mitogen-activated protein kinases (MAPK) activation and the induction of Il6, Cxcl1, and Ccl20. We did not observe impaired IL-6 induction in response to TLR2 ligands in TRIF−/− cells or impaired Cxcl1 release (data not shown), suggesting differences in the role of TRIF in TLR5 and TLR2 signaling. Choi et al. (49) also reported that flagellin failed to activate IRF3 and did not induce IFN-β, while we found that TLR2 ligands can activate IRF3 and can induce IFN-β in a TRAM/TRIF-dependent manner in BMDM.
Both the TRIF and the MyD88 pathway mediate TLR2-induced Ccl5 release in macrophages, in contrast to Tnf induction, which is tightly controlled by the MyD88-dependent pathway. The reason for this redundancy is still unclear, but it is likely important for induction of certain responses, even when the MyD88 pathway is compromised. Cytokine induction by whole microorganisms are typically mediated by several pattern-recognition receptors. It has recently been reported that a mutant strain of Listeria monocytogenes induces IFN-β by a TLR2-TRIF-dependent mechanism (50); however, this response was also shown to be mediated by TLR3, which is known to utilize the adaptor TRIF.
In conclusion, our results provide new insight into the contribution of the MyD88- and TRIF-dependent signaling pathways in response to different TLR ligands, and they show a novel role for TRAM and TRIF in TLR2 signaling. Induction of gene expression by both MyD88 and TRIF pathways may be necessary for optimal host responses toward certain infections. These results have implications for our understanding of TLR-mediated innate immune responses against infectious organisms.
Acknowledgments
We thank Dr. S. Akira (Osaka University, Japan) for MyD88−/−, MAL−/−, TRIF−/−, TRAM−/−, TLR2−/−, and TLR4−/− mice. We also thank Dr. I. Rifkin, Dr. R. Ingalls, A. Nair, Dr. K. Hoebe, and Dr. B. Beutler for help with TRIFLps2/Lps2 mice and cells.
*This work was supported, in whole or in part, by National Institutes of Health Grants R01 AI57538 (to E. L.) and AI095213 (to G. I. V.). This work was also supported by the Research Council of Norway, the Norwegian Cancer Society, Commission of the European Communities Grant QLK2-2000-00336 (RTD Program “Quality of Life and Management of Living Resources” and “Hospath”), and by the Cancer Fund from St. Olavs Hospital, Trondheim, Norway.
4The abbreviations used are:
- TLR
- Toll-like receptor
- IRF
- interferon regulatory factor
- TIR
- Toll/interleukin-1 receptor
- SV
- Sendai virus
- LTA
- lipoteichoic acid
- CFP
- cyan fluorescent protein
- MEF
- mouse embryonic fibroblast
- BisTris
- 2-[bis(2-hydroxyethyl)amino]-2-(hydroxymethyl)propane-1,3-diol
- TRIF
- TIR domain-containing adaptor protein-inducing interferon β
- TRAM
- TRIF-related adaptor molecule
- ANOVA
- analysis of variance
- BMDM
- bone marrow macrophage
- IFNAR
- interferon receptor I
- qPCR
- quantitative PCR
- HAU
- hemagglutination unit.
REFERENCES
Articles from The Journal of Biological Chemistry are provided here courtesy of American Society for Biochemistry and Molecular Biology
Citations & impact
Impact metrics
Article citations
Intestinal barrier permeability: the influence of gut microbiota, nutrition, and exercise.
Front Physiol, 15:1380713, 08 Jul 2024
Cited by: 3 articles | PMID: 39040079 | PMCID: PMC11260943
Review Free full text in Europe PMC
Neurosteroids mediate and modulate the effects of pro-inflammatory stimulation and toll-like receptors on hippocampal plasticity and learning.
PLoS One, 19(6):e0304481, 14 Jun 2024
Cited by: 0 articles | PMID: 38875235 | PMCID: PMC11178232
Molecular Mechanisms of Cachexia: A Review.
Cells, 13(3):252, 29 Jan 2024
Cited by: 3 articles | PMID: 38334644 | PMCID: PMC10854699
Review Free full text in Europe PMC
Recurrent urinary tract infection genetic risk: a systematic review and gene network analysis.
Int Urogynecol J, 35(2):259-271, 02 Nov 2023
Cited by: 0 articles | PMID: 37917182
Review
Re-identification and characterization of grass carp Ctenopharyngodon idella TLR20.
Fish Shellfish Immunol Rep, 5:100119, 07 Oct 2023
Cited by: 0 articles | PMID: 37841419 | PMCID: PMC10568090
Go to all (66) article citations
Similar Articles
To arrive at the top five similar articles we use a word-weighted algorithm to compare words from the Title and Abstract of each citation.
TRAM is required for TLR2 endosomal signaling to type I IFN induction.
J Immunol, 193(12):6090-6102, 10 Nov 2014
Cited by: 73 articles | PMID: 25385819 | PMCID: PMC4258402
LPS-TLR4 signaling to IRF-3/7 and NF-kappaB involves the toll adapters TRAM and TRIF.
J Exp Med, 198(7):1043-1055, 29 Sep 2003
Cited by: 719 articles | PMID: 14517278 | PMCID: PMC2194210
CD14 dependence of TLR4 endocytosis and TRIF signaling displays ligand specificity and is dissociable in endotoxin tolerance.
Proc Natl Acad Sci U S A, 112(27):8391-8396, 23 Jun 2015
Cited by: 80 articles | PMID: 26106158 | PMCID: PMC4500272
The family of five: TIR-domain-containing adaptors in Toll-like receptor signalling.
Nat Rev Immunol, 7(5):353-364, 01 May 2007
Cited by: 1593 articles | PMID: 17457343
Review
Funding
Funders who supported this work.
BLRD VA (1)
Grant ID: I01 BX002607
NIAID NIH HHS (4)
Grant ID: T32 AI095213
Grant ID: L40 AI057538
Grant ID: R01 AI57538
Grant ID: AI095213
National Institutes of Health (2)
Grant ID: AI095213
Grant ID: R01 AI57538




