Abstract
Free full text

Modeling infectious disease dynamics in the complex landscape of global health
Abstract
Despite some notable successes in the control of infectious diseases, transmissible pathogens still pose an enormous threat to human and animal health. The ecological and evolutionary dynamics of infections play out on a wide range of interconnected temporal, organizational and spatial scales, which even within a single pathogen often span hours to months, cellular to ecosystem levels, and local to pandemic spread. Some pathogens are directly transmitted between individuals of a single species, while others circulate among multiple hosts, need arthropod vectors, or can survive in environmental reservoirs. Many factors, including increasing antimicrobial resistance, increased human connectivity, and dynamic human behavior, raise prevention and control from formerly national to international issues. In the face of this complexity, mathematical models offer essential tools for synthesizing information to understand epidemiological patterns, and for developing the quantitative evidence base for decision-making in global health.
Introduction
Thirty-five years ago, there was a belief that the health burden of infectious diseases was close to becoming insignificant, as our means of defense and control, including hygiene, nutrition, drugs and vaccines, had brought about a steady decline in overall mortality (1). In recent decades, however, it has become clear that the threat persists in our rapidly changing world, and human mortality attributed to infection is now projected to remain at current levels of 13–15 million deaths annually until at least 2030 (2). Successes in eradicating smallpox and rinderpest have been isolated events in a landscape of endemic and epidemic infections (3). Newly emerging infectious agents represent a continuing challenge, for example HIV in the 20th century, more recently SARS- and MERS-coronavirus, West Nile Virus, Nipah virus, drug resistant pathogens, novel influenza-A strains, and a major Ebola outbreak in 2014. Most new infections enter the human population from wildlife or livestock, and the possibilities for emergence and spread in the coming decades are likely to increase due to population growth, increased urbanization and land changes, greater travel, and increased livestock production to meet demands from the world’s expanding population (4–8). In our modern world of instant communication the changing behavior of individuals, often in response to publicity about epidemics (as in the case of the SARS epidemic, H5N1 and H7N9 avian influenza, or recent Ebola and MERS outbreaks) or social media attention, is a key issue (9,10). Phylogenetic data shed light on an additional layer of complexity (11), as will an increased understanding of the human genome and how it influences factors such as susceptibility, infectiousness and its duration. At the same time, the development of effective new vaccines remains a difficult challenge, especially for antigenically very variable pathogens (e.g., HIV, falciparum malaria) and for pathogens that stimulate immunity that is only partly protective (e.g., Mycobacterium tuberculosis) or temporary (e.g., Vibrio cholerae).
In the face of this complexity, computational tools (Box 1) are essential for synthesizing information to understand epidemiological patterns, and for developing and weighing the evidence base for decision-making. Here, we review the contribution of these tools to our understanding of infectious disease dynamics for public health, concentrating both on selected illustrative examples and on current developments. We argue that experts on infectious disease dynamics and experts on prevention and control need to collaborate closely on a global scale, to improve decision making concerning veterinary and human public health as well as agricultural health, both for current infectious disease challenges and for those that lie ahead. Quantitative analysis should be part of all public health policy formulation when addressing the control of infectious diseases.
Models and public health policy formulation
The value of mathematical models to investigate public health policy questions was recognized at least 250 years ago when, in 1766, Daniel Bernoulli addressed an important public health controversy at the time and published a mathematical analysis of the benefits of smallpox inoculation (then called variolation) (12). Since then, and particularly over the past 50 years, the study of infectious disease dynamics has grown into a rich interdisciplinary field. It is driven both by the desire for fundamental understanding and the need to use that understanding to aid public health decision making. Decision making on vaccination strategies, for example, increasingly depends on model analyses, where infection dynamics must be combined with cost data (Box 2, Influenza: prevention & control). In recent decades, response to major infectious disease outbreaks including HIV, BSE, Foot and Mouth Disease (FMD), SARS, and pandemic and avian influenza, have shown both the need for and capabilities of models (Box 3, HIV: Test & treat strategy). Model-based analysis of such outbreaks also continually pinpoints areas for improvement in methodology and data, emerging from comparison of predictions of models with observed patterns.
For infectious agents important to public health, ‘case law’ has built up, for example regarding important factors for infection dynamics, concepts to aid understanding and communication of quantitative and qualitative insight, and regarding computational tools and modeling frameworks (Table 1 and Box 4). The basic reproduction number R0 is a central concept characterizing the average number of secondary cases generated by one primary case in a susceptible population. Such concepts serve to highlight what must be measured to interpret observed patterns and to quantify the impact of control strategies in a cycle of interaction between policy and modeling (Fig. 1).

Example of the process of modeling for public health, based on rubella. Policy questions are formulated; available data are brought to bear on the question, here illustrated by incidence of rubella following the introduction of vaccination in individuals aged less than 15 years, or 15 or more years in Costa Rica (127). Scientific understanding and subsequent policy advice is derived from model-based analyses of data, in this case using a non-linear age-structured SIR model (see Box 4), and can lead to collection of key missing data for model improvement. A plot illustrates insight where each square depicts a combination of birth rate and infant vaccine coverage (reflecting the situation in different countries, e.g. Somalia, diamond, Nepal, circle) for routine vaccination only. The color of the square indicates the confidence in the true value of R0 being higher (red) or lower (green) than a critical value that depends on birth rate and coverage. This translates into confidence that the public health burden caused by the rubella-complication of Congenital Rubella Syndrome (CRS) in newborns is likely to be reduced (green) compared to the situation without vaccination (128), suggesting that introduction of routine measles-rubella (MR) vaccine in Nepal is likely to succeed in bringing down CRS, while in Somalia routine MR vaccine would increase CRS burden without substantive improvements in the fraction that is vaccinated.
Table 1
As different infections have become the focus of public health attention, the modeling community has responded by developing improved concepts and methods. The table concentrates on the period since 1950. The first column lists the (classes) of infection, the second column lists factors whose importance to infection dynamics became particularly clear in relation to those infections; the third and fourth columns highlight concepts and methods that were developed in response. For each row, only a few typical references are given. Many factors, concepts and methods are relevant, in current use, and in continual development for much larger classes of infectious agents.
| Motivating studies | Important factors | Concepts | Methods |
|---|---|---|---|
| Malaria (1910s, 1950s onwards, 53,106,107) | Transmission via insect vectors; nonlinear dependence of transmission on mosquito biting rate; influence of environmental and climatic variables. | Threshold for control, basic reproduction number. | Models with two host species (host-vector models); using models to support and guide field campaigns; relating models to field data. |
| Childhood infectious diseases, e.g. measles. (1950s onwards, 54,108) | Immunizing infections; spatial and temporal heterogeneity; demography; age structure; household structure. | Critical community size and herd immunity; periodic outbreaks; fade out; vaccine efficacy. | SIR models; age-structured models; models with periodic forcing; spatial and stochastic models; metapopulation models; time-series models. |
| Macroparasites. (1970s onwards, 108) | Clumped infections, multi-strain and multi-species infections, cross immunity, concurrent infections. | Consequences of overdispersed distribution of parasite load (Figure 1) | Stochastic models, approximations including hybrid models and moment closure. |
| Sexually transmitted infections, e.g. HIV. (1980s onwards, 108,109) | High/low risk groups; non-random contact structure; partnerships; within-host strain diversity and evolution; time scale. | Incubation and infectious period distribution; core group; next-generation matrix and operator; partnership dynamics. | Statistical methods (e.g. back calculation); Models with (dis)assortative mixing; pair formation models; within-host dynamic models. |
| Veterinary outbreaks, e.g. BSE and FMD. (1990s onwards, 110,111) | Fixed spatial locations with changing contact networks. | Local versus long-range transmission; spatial intervention (ring vaccination/culling); conflict of priorities at different scales. | Individual based models and spatial simulations (FMD); data-driven real-time modeling; inference of transmission trees. |
| Novel emerging infections, e.g. SARS, Nipah virus, MERS. (2000 onwards, 6,112–115) | Behavior change; global inter-connectedness and international cooperation in control; responses in absence of biomedical measures; animal reservoirs. | Zoonotic spillover; stuttering chains; importance of index case; super-spreaders; unobserved dynamics in an animal reservoir; super-shedding | Contact tracing; modeling international spread and control; quarantine and case isolation; individual heterogeneity in infectiousness, incubation and latency period. |
| Influenza including avian influenza. (Present, 27,28,116–118) | Distribution of prior immunity; within-population and species strain differences, virus evolution and interaction; role of wildlife and farm animals. | Pandemics; spillover between wild birds, and farmed birds; phylodynamics. | Interaction between immunological and epidemiological dynamics; integrating phylogenetic and epidemic methods and models. |
| Vector-borne diseases, e.g. dengue, malaria. (Present, 119–122) | The influence of climate and environment on vector and pathogen development; animal reservoirs; interaction between strains within-host and between-host. | Dilution effect and role of biodiversity in infectious disease dynamics; re-emerging infections. | Evolutionary impact of vaccines/other interventions; synthesis of data from ecology and epidemiology; elimination modeling; statistical modeling of environmental vector suitability. |
| Bacterial infections, e.g. pneumococcal disease, MRSA and tuberculosis. (Present, 39,123–126) | Antibiotic/drug resistance; adaptive dynamics. | Vaccine effectiveness; interacting natural immunity and vaccine boosting. | Modeling interacting and emerging strains; stochastic models in small populations. |
Two fundamental properties of the world that shape infectious disease dynamics make computational tools key for understanding reality. The world is essentially a stochastic and highly non-linear system. The non-linearity not only derives from the complex interaction between factors involved in transmission are complicated, but also from the influence that the infection process has on the distribution of important characteristics at various temporal and spatial scales. For example, the age-related nature of infection and mortality in HIV changes the age distribution of the population, and previous exposure to strains of influenza alters the distribution of influenza susceptibility. Such feedback mechanisms contribute to the high nonlinearity of infection processes. Non-linearity leads to counterintuitive phenomena (Fig. 2), and the inability simply to extrapolate experience from one situation to another (for example when deciding whether to implement a vaccination policy in different countries, Fig. 1). Mathematical tools, relating to data and processes on a large range of interacting scales, have become essential to explore, anticipate, understand and predict the effects of feedbacks within such complex systems, including changes caused by intervention.
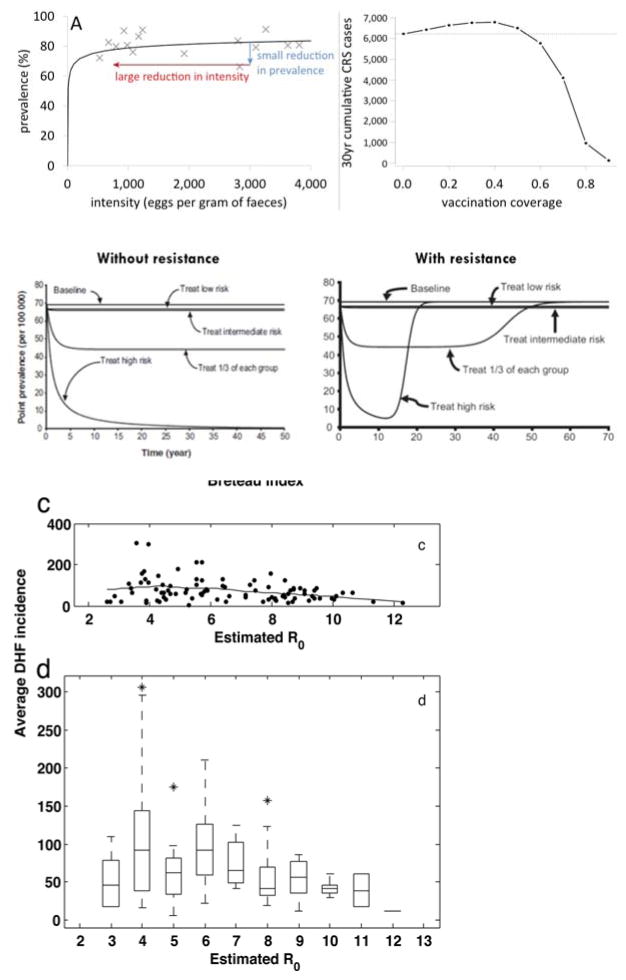
Examples of counter-intuitive effects of non-linear infection dynamics. Upper left graph: Non-linear interaction between prevalence of a helminth infection and infection pressure (as measured by the mean intensity of existing infections) means that control measures must have a disproportionately large impact on intensity before prevalence is reduced. This effect is predicted by a mathematical model (solid line) and corroborated by field data (crosses) (129). Upper right graph (adapted from 130): Non-linear relation between total number of cases of congenital rubella syndrome (CRS) and rubella vaccine coverage, showing that sub-optimal levels of vaccine coverage cause worse health outcomes than no vaccination. The line shows model predictions; similar effects have been documented for real rubella control situations (131). Middle graphs (from 132): Modeling results of rebound of gonorrhea transmission with different treatment strategies without (left panel) and with (right panel) antimicrobial resistance developing. In the presence of resistance, focusing treatment on the high-risk core group leads to an increase to un-treated baseline prevalence, after initially strong decline for more than a decade. Bottom graphs (from 133): Field data and box plot of a non-linear relation between R0 for dengue transmission and average dengue hemorrhagic fever incidence across Thailand, showing that starting control that brings down transmission from a situation with high R0 may paradoxically increase cases of DHF.
Current and future opportunities for models in public health
Over the last decade, key public health questions, ranging from emergence to elimination, have posed a range of challenges for modeling infectious disease dynamics, many of which rest on leveraging novel data sources, and integrating data from a range of scales - from sequence data to global circulation. Given commonalities in processes across pathogens, progress made in one area can lead to advances in another, and progress in the areas described above all build on and inform each other, making this a dynamic time for research in the discipline (13). A few themes are chosen to illustrate current trends in model development and public health application.
The difficulties of real-time outbreak modeling: the Ebola 2014 outbreak
The ongoing (at the time of writing) outbreak of Ebola in West Africa serves to highlight both opportunities and challenges in modeling for public health. In the initial phase of an outbreak, real-time estimates of the reproduction number or simple exponential extrapolation (14) allow short-term predictions of epidemic growth that can be used, for example, to plan for necessary bed capacity. Also in an early phase, phylogenetic quantitative tools can use samples from initial victims to provide important estimates of outbreak origin (15). Later in an outbreak, these methods are replaced by more detailed mechanistic models that explicitly take into account depletion of susceptibles, as well as specifics such as different transmission routes or settings, to provide more accurate and fine-grained predictions of the impact of public health measures. For Ebola, such methods make it possible to distinguish between transmission at funerals, in health-care settings and in the community (16,17) to predict the effect of increasing bed capacity or reducing funeral transmission. Determining which combinations of measures are most effective in bringing down the reproduction number call for close and fast interaction between modelers and policy makers (18). It is likely that disease-dynamic models will also be at the forefront of a debate on the optimal deployment of initially scarce Ebola vaccines, once such vaccines become available.
With the opportunities of real-time modeling for public health come specific challenges. The imperative to produce reliable and meaningful analysis for those at the forefront of battling the outbreak has to be balanced against the pressures and delays of scientific publication. In an ongoing outbreak, data can be patchy and reporting delayed, and different data sources are not always synthesized. Particularly, data are lacking on the effect on transmission dynamics of the various control measures operating simultaneously in the hectic circumstances of the most severely hit areas. Also, under-reporting is a critical challenge for ongoing assessment of the epidemic and has enormous impact on predictions of outbreak size, but also of outbreak impact, for example in terms of the case fatality ratio. This estimate of severity suffers early in an outbreak from imprecise information on both the numerator (deaths confirmed as being caused by the infection) and the denominator (under-reporting and not all suspected cases are laboratory-confirmed infections). This caused problems early in the H1N1 influenza outbreak starting in Mexico in 2009, as well as in the current Ebola outbreak. While levels of under-reporting can be estimated after the outbreak from retrospective serological studies, it is usually not identifiable in real-time data.
These limitations make it almost impossible to make reliable long-term predictions. Modeling results are therefore often based on scenarios in which a pathogen spreads unaltered by behavioral changes or the public health response. In reality this is rarely the case, especially in a devastating outbreak such as the one of Ebola in West Africa, where the situation constantly changes due to growing awareness in the community, as well as national and international intervention. Careful communication of findings is key, and data and methods of analysis (including code) must be made freely available to the wider research community. Only in this way can reproducibility of analyses and an open exchange of methods and results be ensured for maximal transparency and, consequently, benefit to public health.
Emergence of novel human pathogens
There is an ever-present hazard that novel human pathogens emerge from livestock and wild mammal and bird reservoirs. Research on potential emerging zoonoses draws on concepts from across the spectrum of infectious disease dynamics, disease ecology, microbiology and phylogenetic analysis. Particular challenges include estimating human-to-human transmissibility against a backdrop of on-going zoonotic spillover, detecting anomalous outbreaks, and assessing the risk that more dangerous strains may arise through pathogen evolution.
The recently identified gap in methodology for zoonoses with weak human-to-human transmission (6) is being filled, with new approaches to estimating R0 and other transmission-related quantities from subcritical outbreak data (19–21). These studies address key public health concerns, but rely on strong assumptions regarding the quality and completeness of case observations. Better information on surveillance program efficacy could be gained through serological surveys (where blood and saliva samples reveal evidence of past and present infections) or sociological study, and modeling studies can help to design and characterize efficient surveillance programs (22). Given the predominance of zoonotic pathogens among emerging infections, models for transmission dynamics and evolution in multispecies ecosystems and food webs, consisting of host species and non-host species interacting ecologically and epidemiologically, are a crucial area for future development (6,23). The greatest challenge – and the greatest prize – in modeling emerging zoonoses is to assess which diseases pose the most risk to humans and how these might change over time and in different localities (24). Such tasks, which will join molecular studies to experimental infections to epidemiological and ecological surveys, will drive empirical and theoretical efforts for decades to come.
The rising availability of pathogen genome sequence data, coupled with new computational methods, presents opportunities to identify with precision ‘who infects whom’ and the networks of infection between humans and reservoirs (25). Full realization of this potential, though, will require denser and more systematic whole genome sampling of pathogens coupled with associated epidemiological data, as well as baseline information on genetic diversity and evolutionary rates, especially in animal hosts (26).
Pathogen evolution and phylodynamics
As pathogen genetic data become increasingly available, modelers are finding ways to synthesize these new data streams with more traditional epidemiological information in phylodynamic tools (27,28). However, current frameworks employ compartmental epidemiological models, which do not make efficient use of individual-level epidemiological data. Although sampling theory is well developed for standard surveillance data, the relationship between a set of pathogen sequences and the phylogeny inferred from a population sample is more complex (11). Many-toone mapping possibilities between, on the one hand, combinations of epidemiological, immunological and evolutionary processes shaping sequences and, on the other hand the inferred phylogeny, demand the integration of diverse data sources and an increased focus on systematic sampling.
Phylodynamic studies to date have largely focused on fast-evolving RNA viruses, driven by the large amount of data generated for clinical (e.g. Hepatitis C Virus (HCV), HIV) or surveillance (e.g. influenza) purposes (11). Replicating these efforts on an expanded array of pathogens, including DNA viruses, bacteria, fungi, protozoa (e.g. malaria) and helminths is a promising avenue for future research (29). It is of particular importance in the context of the evolution and spread of drug resistant variants and escape mutants from vaccine protection. However, genome-wide pathogen data also present challenges, in particular in relation to accommodating recombination, re-assortment, and mobile genetic elements. Analysis of bacterial genomes usually considers only those genes that are shared across taxa, but there are good reasons to believe that non-core genes play an important role in bacterial evolution, including the evolution of antibiotic resistance (30).
While sequence data are extremely valuable, in order to link these data fully to disease dynamics a key advance will be determining how sequence changes may affect functions related to pathogen fitness, such as replication rate, transmissibility, and immune recognition (genotype to phenotype). Molecular epidemiological studies often treat pathogen genetic variation as simply reflecting the underlying transmission process, whereas in reality such variation may play an important role in determining transmission dynamics, as exemplified by the escape from herd immunity by influenza A virus (31).
‘Deep’ sequencing of pathogens within individual hosts generates information on within-host diversity, resulting from evolution within the host (often in response to drug treatment), or multiple infections. To tackle within-host diversity, models that embed pathogen evolution within a transmission tree are needed. Such models, which cross the within- and between-host scales, are only just becoming analytically and computationally feasible despite being proposed several years ago (32). Similarly, while progress has been made in scaling inference from genes to genomes (33), scaling inference to large numbers of sequences is lagging far behind.
Multiple infections
Infectious disease epidemiology evolved by focusing on interactions between a single host species and a single infectious agent. It is becoming increasingly clear that multiple agents simultaneously infecting the same host populations and individuals significantly add to public health burden and complicate prevention and control. Co-infections in relation to HIV, for example tuberculosis and HCV, or co-infection of different strains of influenza A virus raise important public health and evolutionary issues. Multiple agents infecting the same host individual have been shown to influence each other by increasing or decreasing susceptibility and/or infectivity of that individual, thereby influencing the population dynamics of these agents in ways that we have yet to explore and understand (34,35).
Multiple infections of the same individual with closely related pathogens occur when infection elicits no, or only a partial, immune response. Macroparasites including many of the important human helminth infections are good examples of pathogens that evade human immune responses permitting repeated infection of the same host (36). Biological mechanisms giving rise to such multiple infections include sequential reinfections caused by antigenic drift in influenza, antigenic variation in respiratory syncytial virus (RSV) and waning (slow loss of) immunity in pertussis while lack of cross-protection in many colonizing microparasites, for example pneumococcus and human papilloma virus (HPV), allows for multiple concurrent infections. Although the existence of reinfections is a clinical fact, population-level data are scarce as reinfections are often subclinical and individual-based longitudinal infection-histories only anecdotal. Results from new analytic approaches relating to deep sequencing and neutralization tests covering multiple antigens are being utilized (37).
The immunodynamics of influenza have clear policy implications for the identification of high-risk groups in connection with pandemic planning (38) while the dynamics of waning immunity are key to the current concerns about immunization level for pertussis (39). Multivalent vaccines covering only a targeted subset from the circulating strains of pneumococcus and HPV pose important new applied problems (40). The spread of recombinant viruses implies the existence of multiple infections. An example is the Sydney 2012 strain of norovirus, but how this can occur in an acute infection remains to be understood, as the time window for multiple exposure is limited. Mathematical models may help to explore how, for example, a subpopulation with chronic infection or a hypothesized environmental reservoir may contribute to the dynamics of multiple infections.
Behavior of hosts
Human behavior is a fundamental determinant of infectious disease dynamics, whether by affecting how people come in contact with each other, vaccination coverage, reporting biases, or adherence to treatment. Traditional epidemic models have tended to ignore heterogeneity in contact behavior (although early HIV models addressed heterogeneity in sexual behavior by necessity (41)), however increasing sophistication of contact network models (42), together with data on epidemiological contacts, creates opportunities for understanding and controlling transmission at a fundamental level (43), and opens up the possibility of independent study of relevant social factors (10). Recent years have seen exciting developments in the measurement of contact patterns and ‘who might infect whom’ through advances in individual electronic identification technology. This is a promising avenue for linking pathogen genetic data and human behavior.
Contact patterns are not static, and can shift during outbreaks as individuals change their behavior in response to perceived risk and public-health interventions (44). Modeling has illuminated this process, for example incorporating peer influence on vaccination behavior into models of infectious disease dynamics (45,46). Analysis of data from online social networks has also created promising opportunities to validate such approaches with empirical observations (47,48).
Movement and travel are tightly linked to the spread of infection, and have been explored through models to highlight commuting and agricultural migration driving local disease transmission (49), and global disease patterns through air travel (50). These processes are now being investigated to gain insights into the more complex case of vector-borne diseases such as malaria and dengue, where both host and vector movement can interact to drive local (51) and large-scale dynamics (52).
Elimination/eradication
Modeling has long provided support for elimination efforts – vector control (53), critical community size (54), herd immunity and critical vaccination threshold (55,56) were all powerful insights from models framed in relatively simple and homogenous terms. Subtleties and complexities in many current eradication programs, as well as the availability of novel data sources, have called for a range of extensions in the theory. As we approach elimination targets, disease dynamics have changed in ways that were largely predicted by models, but also in unanticipated ways due to ignorance about key epidemiological processes.
Incentives for control efforts also change, both at the individual level (passive or active refusal can develop (57)) and at the country level (58). This reinforces the call for development of models of human behavior and its interaction with infectious disease dynamics (9), potentially drawing on new data sources (e.g. from social media (59,60)), as well as for models that can capture national and non-governmental motivations, interactions and competition, economical or otherwise. Long-term control puts pathogens under strong selection for resistance, calling for evolution-proof control methods (61) and novel vaccine technologies and their optimized delivery (62).
Finally, since the era of smallpox eradication, patterns of global disease circulation have changed radically. Human mobility and migration are increasing global connectivity, strengthening the need for cooperation and international synchronization of efforts (as illustrated by polio). Techniques for analysis of novel data-sources are again key here, e.g., Call Data Records provide unique opportunities to understand disease source-sink dynamics (52).
Computational statistics, model fitting and big data
By definition and design, models are not reality. The properties of stochasticity and non-linearity strongly influence the accuracy of absolute predictions over long time horizons. Even if the mechanisms involved are broadly understood and relevant data are available, predicting the exact future course of an outbreak is impossible due to changes in conditions in response to the outbreak itself, and due to the many chance effects in play. These stochastic effects dominate developments in situations with relatively few infected individuals (such as at emergence, approaching the threshold for sustained host-host spread, or approaching elimination/eradication). This makes it virtually impossible to predict which infectious disease agent is going to emerge/evolve next and where, or to predict when and where the next or last case in an outbreak will occur. There is, typically in complex systems, a fundamental horizon beyond which accurate prediction is impossible. The field has yet to explore where that horizon is and whether computational tools and additional data (and if so which data) can stretch predictions to this limit. In contrast, ‘what-if’ scenarios for public health intervention can provide qualitative (and increasingly semi-quantitative) insight into their population consequences.
With growing applications in public health there is an increasing demand to validate models by making model predictions consistent with observed data. The development of evermore-powerful computers is accompanied by new techniques utilizing this power, notably for statistically rigorous parameter estimation and model comparison. Techniques such as Markov Chain Monte Carlo (MCMC) have become firmly established tools for parameter estimation from data in infectious disease models (e.g. 63), and Monte Carlo based methods will play a pivotal role in addressing the challenges that lie in reconciling predictions and observations (64,65). Other techniques, such as so-called particle filters, approximate Bayesian computation, emulation, and their combinations with MCMC (e.g. 66), are rapidly developing and allow matching stochastic models that explicitly account for incomplete observations to time series of cases, giving insights into scenarios as diverse as cholera in Bangladesh (67) and influenza (68,69). The need to integrate multiple data sources (70,71) as well as to include uncertainty in model parameters and/or structure has led to an increase in the popularity of Bayesian approaches.
The rapid expansion of infectious disease models and their application over the past decade has coincided with an increase in open access datasets available from a variety of sources, but progress in data capture needs to be accelerated. While some of these technologically advanced data streams have been incorporated into models, for example to track the incidence of influenza in the USA (72), to elucidate the spatial dynamics of measles and malaria in Africa (53,73) and to chart the spread of dengue globally (74), much more remains to be done to leverage data collected from different sources (e.g. demographic, genetic, epidemiological, treatment and travel patterns) and at different temporal and spatial scales.
Concluding remarks
Infectious diseases are an important frontier in public health, and their prevention and control call for global, rather than national or regional, coordinated efforts (75–78). The success of smallpox and rinderpest eradication campaigns shows the possibilities; the global spread of newly emerged pathogens (recently avian influenza strains and MERS coronavirus), the difficulties in curbing the spread of antibiotic resistance, the upsurge of polio towards the ‘end-phase’ of its eradication, and the recent unprecedented spread of Ebola virus, are examples that show the need for international coordination and collaboration. Non-linearity in infectious disease dynamics and global connectivity cause sub-optimal national decisions on control and prevention to have regional and even global repercussions.
Given the mismatch with regions where most expertise on infectious disease dynamics is concentrated, it is important to empower local scientists and policy makers in regions where the burden of disease is heaviest about problems facing their own countries and the consequences of local actions. It is essential to make expertise, data, models, statistical methods and software widely available by open access. There are several initiatives (e.g. Thiswormyworld.org, Garkiproject.nd.edu, EDENextdata.com, and the Malaria Atlas Project), but much more needs to be done. Modeling tools and software for data analysis are beginning to become open source, such that findings can be replicated, additional scenarios can be evaluated, and others can incorporate methods for data analysis or simulation. Ultimately, sharing models guarantees more reproducible results, while maximizing model transparency.
Making data sets widely available is also crucial, for example to support replication of findings and broader comparative analyses (79). As models become open access, so should much of the data collected by governments, international agencies and epidemiology research groups. Two outbreaks never occur in exactly matching circumstances, even for the same infectious agent, so there is potential to study many outbreaks in parallel to gain insight into the determinants of outbreak pattern and severity. Looking forward, there is a major opportunity to design experiments, clinical trials (for example for vaccines (80)), and surveillance protocols to test model predictions or assumptions, and to help reduce or better target the enormous costs involved. By integrating modeling approaches throughout the full life cycle of infectious disease policies, including economic considerations (58, 70, 81), health outcomes can be improved and scientific understanding can be advanced.
At present, the evidence provided by infectious disease models is not considered on the GRADE scale (www.gradeworkinggroup.org) alongside that of conventional studies such as clinical trials. Regardless, models are essential when diverse sources of data (including GRADE-scale evidence) need to be combined and weighed. In many cases, the definitive trial cannot be performed and models are needed to catalyze insight and extract maximum value from data that are available. In recent years, uniformity of practice and quality control for models has received more attention, resulting in initial attempts to characterize Good Modelling Practice for infectious diseases (82,83).
The optimal use of models to inform policy decisions requires a continuous dialogue between the multidisciplinary infectious disease dynamics community and decision makers. This is increasingly understood by governments in developed countries, in non-governmental agencies and by large funding bodies such as the Bill and Melinda Gates Foundation. This dialogue will help to reduce the burden from infectious diseases by providing better-informed control strategies. Mathematical models will allow us to capitalize on new data streams, and lead to an ever-greater ability to generate robust insight and collectively shape successful local and global public health policy.
Acknowledgments
HH conceived and wrote the paper; RA, VA, SB, DDA, CD, KE, JE, SFr, SFu DH, TH, VI, PK, JL JLS, JM, DM, JP, LP, MR and CV provided text and edited the manuscript; JL, JM and DH produced the figures; the Isaac Newton Institute IDD Collaboration jointly produced and discussed ideas for the outline and content (all of the above plus NA, FB, TB, JG, BG, AL, AM, PON, CP, SR, GST, PT, JW). This paper was conceived and developed at a program on Infectious Disease Dynamics at the Isaac Newton Institute, Cambridge, UK, August 19 – September 13, 2013, and May 19 – June 6, 2014 (www.newton.ac.uk). We gratefully acknowledge financial and infrastructural support from the Isaac Newton Institute, fundamental to the success of this program. We are also grateful for the financial support the program received from the Research and Policy for Infectious Disease Dynamics (RAPIDD) program of the Science and Technology Directorate, U.S. Department of Homeland Security, and the Fogarty International Center, NIH.
References and Notes
Full text links
Read article at publisher's site: https://doi.org/10.1126/science.aaa4339
Read article for free, from open access legal sources, via Unpaywall:
https://dspace.library.uu.nl/bitstream/handle/1874/330568/aaa4339.full.pd.pdf?sequence=1&isAllowed=y
Citations & impact
Impact metrics
Article citations
A conceptual health state diagram for modelling the transmission of a (re)emerging infectious respiratory disease in a human population.
BMC Infect Dis, 24(1):1198, 24 Oct 2024
Cited by: 0 articles | PMID: 39448915 | PMCID: PMC11515510
Managing spatio-temporal heterogeneity of susceptibles by embedding it into an homogeneous model: A mechanistic and deep learning study.
PLoS Comput Biol, 20(9):e1012497, 30 Sep 2024
Cited by: 0 articles | PMID: 39348420 | PMCID: PMC11476686
Hospital crisis management in the epidemic: A qualitative study.
Health Sci Rep, 7(10):e70059, 03 Oct 2024
Cited by: 0 articles | PMID: 39372333 | PMCID: PMC11449806
Using real-time modelling to inform the 2017 Ebola outbreak response in DR Congo.
Nat Commun, 15(1):5667, 06 Jul 2024
Cited by: 1 article | PMID: 38971835 | PMCID: PMC11227569
Host behaviour driven by awareness of infection risk amplifies the chance of superspreading events.
J R Soc Interface, 21(216):20240325, 24 Jul 2024
Cited by: 1 article | PMID: 39046766 | PMCID: PMC11268441
Go to all (279) article citations
Similar Articles
To arrive at the top five similar articles we use a word-weighted algorithm to compare words from the Title and Abstract of each citation.
The global one health paradigm: challenges and opportunities for tackling infectious diseases at the human, animal, and environment interface in low-resource settings.
PLoS Negl Trop Dis, 8(11):e3257, 13 Nov 2014
Cited by: 112 articles | PMID: 25393303 | PMCID: PMC4230840
Review Free full text in Europe PMC
Public health implications of emerging zoonoses.
Rev Sci Tech, 19(1):310-317, 01 Apr 2000
Cited by: 27 articles | PMID: 11189723
Review
Emerging zoonoses--yesterday, today and tomorrow.
Acta Vet Scand Suppl, 100:55-58, 01 Jan 2003
Cited by: 0 articles | PMID: 16429810
Review
Modeling spillover dynamics: understanding emerging pathogens of public health concern.
Sci Rep, 14(1):9823, 29 Apr 2024
Cited by: 0 articles | PMID: 38684927 | PMCID: PMC11058258
Funding
Funders who supported this work.
Engineering and Physical Sciences Research Council (3)
Grant ID: EP/J002437/2
Grant ID: EP/L001950/1
Grant ID: EP/K026550/2
Isaac Newton Institute for Mathematical Sciences
Medical Research Council (3)
Evidence Synthesis to inform health related decision making
Professor Daniela De Angelis, MRC Biostatistics Unit
Grant ID: MC_U105260556
Grant ID: MR/K010174/1B
The dynamics and control of infectious diseases close to elimination
Professor Sebastian Funk, London Sch of Hygiene & Tropic. Medicine
Grant ID: MR/K021680/1
NICHD NIH HHS (1)
Grant ID: P2C HD047879
NIGMS NIH HHS (1)
Grant ID: U01 GM110721
Research and Policy for Infectious Disease Dynamics (RAPIDD)
World Health Organization (1)
WHO generic grant number for open-access policy
World Organization
Grant ID: 001





