Abstract
Free full text

Priming a broadly neutralizing antibody response to HIV-1 using a germline-targeting immunogen
Associated Data
Abstract
A major goal of HIV-1 vaccine research is the design of immunogens capable of inducing broadly neutralizing antibodies (bnAbs) that bind to the viral envelope glycoprotein (Env). Poor binding of Env to unmutated precursors of bnAbs, including those of the VRC01-class, appears to be a major problem for bnAb induction. We engineered an immunogen that binds to VRC01-class bnAb precursors and immunized knock-in mice expressing germline-reverted VRC01 heavy chains. Induced antibodies showed characteristics of VRC01-class bnAbs, including a short light chain complementarity determining region 3 (CDRL3) and mutations that favored binding to near-native HIV-1 gp120 constructs. In contrast, native-like immunogens failed to activate VRC01-class precursors. The results suggest that rational epitope design can prime rare B cell precursors for affinity maturation to desired targets.
Introduction
We lack an effective vaccine against HIV, despite its identification more than 30 years ago. An HIV vaccine most likely will need to elicit antibodies capable of neutralizing the majority of the diverse strains circulating in the population. A minority of HIV infected individuals eventually do develop such bnAbs, but generally only after years of protracted viral/antibody co-evolution (1, 2). Although they fail to control virus in the individuals themselves, passive transfer of recombinant forms of such bnAbs can prevent infection in animal models (3–8). Hence there is an expectation that successful elicitation of bnAbs by vaccination prior to infection will be protective in humans, and developing such a bnAb-based vaccine is a major research goal.
The CD4 binding site (CD4bs) antibody VRC01 (9) and other VRC01-class bnAbs identified in at least seven different donors represent a response with distinguishing features that might be amenable to reproducible vaccine elicitation (10–15). In particular, VRC01-class bnAbs share a mode of binding that uses the immunoglobulin heavy (H) chain variable (V) gene segment VH1-2*02 to mimic CD4, in contrast to many antibodies that rely on the CDRH3 loop (10, 14, 16). The VH1-2*02 gene or suitable alternative alleles are present in ~96% of humans (17), and these genes are employed frequently, in ~3% of all human antibodies (18, 19), suggesting that the B cell precursors for a VRC01-class response are generally available for vaccine targeting.
However, several key challenges must be met to induce VRC01-class bnAbs. First, as is true for some but not all classes of HIV bnAbs, the predicted germline precursors of VRC01-class bnAbs lack detectable affinity for native HIV Envelope glycoproteins (Env) (10, 12, 17, 20–22). To address this problem, we and others have designed “germline-targeting” immunogens capable of binding and activating VRC01-class precursor B cells in vitro (17, 21). Whether these immunogens can activate precursors in vivo is an open question. Second, VRC01-class bnAbs carry light (L) chains with unusually short CDRL3s composed of 5 amino acid (aa) residues, typically within a CQQYEFF motif (14, 16). The short CDRL3 length is required to avoid clashing with gp120 Env loop D and V5, and amino acids within this motif make specific interactions to stabilize the antibody and to contact gp120 (10, 14, 16). CDRL3s with this length occur in only 0.6–1% of human kappa antibodies (fig. S1–2) (14, 16) and in 0.1% of mouse kappa antibodies (fig. S2), and the specific amino acid requirements described above will reduce the frequency of useful light chains further. Therefore, a germline-targeting immunogen must be capable of activating relatively rare VRC01-class precursors in the repertoire. Third, VRC01-class bnAbs, like most other HIV bnAbs, are heavily somatically mutated, as a result of chronic stimulation of B cells by successive HIV variants (9, 11, 12, 23). While engineering approaches can be used to develop less mutated bnAbs (24, 25), it remains clear that vaccine induction of bnAbs will require strategies to induce relatively high mutation levels. Most likely this will be achieved by a sequence of different immunogens that successively returns B cells to germinal centers to undergo repeated rounds of affinity maturation (1, 10, 11, 17, 21, 26–29). In this view, each immunogen in the sequence, while naturally inducing antibodies of increasing affinity to itself, must induce maturation in memory B cells that enables weak binding to the next immunogen in the sequence. This challenge is particularly acute for the priming step—the germline-targeting prime must not only activate VRC01-class precursors, it must induce mutations that enable binding to more native-like boost immunogens which themselves have no detectable affinity for the precursors.
To assess the feasibility of meeting the above challenges with a germline-targeting prime, we constructed a knock-in mouse in which the germline-reverted heavy chain of VRC01 pairs with native mouse light chains, and we conducted immunization experiments in this mouse with an improved version (eOD-GT8 60mer) of a previously described germline-targeting immunogen (17). Responses were interrogated by ELISA, hybridoma generation, and most importantly by antigen-specific B cell sorting to define the pool of memory B cells induced by the immunogens.
VRC01 gH knock-in mice
The true germline precursor is not known for VRC01 or other VRC01-class bnAbs (30). In the knock-in mouse, we approximated the true heavy chain precursor with a VRC01 germline-reverted heavy chain (VRC01 gH) composed of the VH1-2*02 and IGHJ1*01 genes assigned by JoinSolver (31) and supported by recent longitudinal analysis of the VRC01 lineage (30), along with the CDRH3 from VRC01 with a single mutation to remove an unpaired cysteine (fig. S3). While our use of the VRC01 CDRH3 in VRC01 gH (necessary because the germline D gene and V-D and D-J junctions cannot be inferred with confidence) is likely a departure from the (unknown) true germline precursor, the VRC01 CDRH3 plays a relatively minor role in epitope recognition, accounting for only 13.7% of the area buried on the heavy chain in the VRC01 interaction with gp120 (10) or 10.2% of the area buried on germline-reverted VRC01 in its interaction with eOD-GT6 (17). Furthermore, the CDRH3 in VRC01 is disulfide-bonded to an affinity-matured cysteine in CDRH1, which may serve to stabilize the antibody conformation and increase affinity for gp120, but this disulfide is not included in VRC01 gH. Thus, use of this CDRH3 is unlikely to strongly bias the VRC01 gH mouse toward favorable interactions with gp120- or eOD-based immunogens, and we believe the VRC01 gH sequence is a reasonable approximation for the true germline, for the purpose of evaluating germline-targeting immunogens.
Testing for the ability to stimulate VRC01-class precursor B cells could not be carried out directly in wild-type mice or other small animals, as none are known to have a VH gene with sufficient similarity to the human VH1-02 germline gene (16, 17). To overcome this limitation, we engineered mice to express a VRC01 gH-chain exon under the control of a mouse VH promoter, introduced by gene targeting into the Igh locus (fig. S4). This targeting to the physiological locus allows normal regulation of H-chain expression, antibody class switching and somatic mutation. These VRC01 gH mice have similar frequencies of CD19+/B220+ B cells as wild-type littermates (siblings of knock-in mice that lack the knock-in gene by random chance in breeding male heterozygous knock-in mice with wild-type females) (Fig. 1A). By next-generation sequencing, the VRC01 gH-chain gene was expressed by ~80% of B cells (Fig. 1B, figs S5–6), and was paired with random mouse L-chains generated in the course of normal B cell development (fig S7). The L-chains have similar V gene usage and CDRL3 length distributions as those of wild type littermates (fig S8). Thus, VRC01 gH mice carry germline-reverted bnAb precursor B cells at a frequency appropriate for testing of germline-targeting VRC01-class immunogens, including eOD-GT8 60mer.
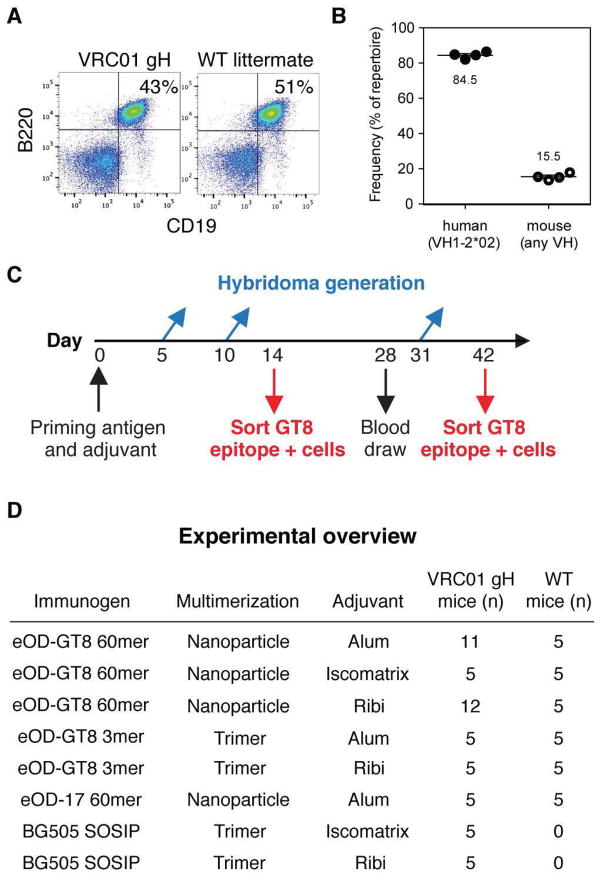
(A) Flow cytometry analysis of spleen cells showing B cell frequencies in VRC01 gH mice and WT littermates. (B) Next-generation sequencing of splenic cDNA from VRC01 gH mice revealed VH1-2*02 usage compared to mouse VH gene usage. (C) Summary of the time course for experiments and analysis. Mice were given a single prime of immunogen in adjuvant and then serum immune responses were evaluated at days 14, 28, and 42 post-immunization. Two of five mice per group were sacrificed for B cell sorting analysis at day 14, and the remaining three of five mice were sacrificed at day 42. In other animals, splenic B cells were collected at day 5, day 10, or day 31 for hybridoma generation. (D) Overview of the immunization groups listed by immunogen (eOD-GT8 60mer, eOD-GT8 3mer, eOD17 60mer, and BG505 SOSIP), multimeric state (nanoparticle or trimer), and adjuvant (Alum, Iscomatrix, and Ribi), along with the number of mice used to test each group. All groups were tested in both VRC01 gH and wild-type (WT) mice, except for BG505 SOSIP which was tested only in VRC01 gH mice. 5 mice per group were used for B cell sorting and/or ELISA, and an additional 13 VRC01 gH mice were employed for hybridoma generation after immunization with eOD-GT8 60mer (6 for Alum, 7 for Ribi).
Analysis of antibody responses to different priming immunogens
VRC01 gH mice were immunized with a single injection of eOD-GT8 60mer, a self-assembling nanoparticle composed of an engineered outer domain from HIV gp120 fused to a lumazine synthase protein (fig S9). To assess if VRC01-like germline precursors were indeed primed, we followed antibody responses and sequenced antibody genes of eOD-GT8-reactive B cells that were captured as hybridomas or by cell sorting of eOD-GT8-binding IgG B cells (Fig. 1C, fig S10). To investigate the effect of multimeric state, we compared responses to 60 subunit nanoparticles (eOD-GT8-60mers) and trimers (eOD-GT8-3mers). To probe for adjuvant effects, antigens were delivered in three different adjuvants: alum, Iscomatrix (“Isco”, 40 nm diameter cage-like structures composed of phosopholipids, cholesterol, and saponin that traffic to lymph nodes and can heighten both antibody and T cell responses but contain no known Toll-like receptor (TLR) agonist activity (32)), or Sigma Adjuvant system (“Ribi”, an oil-in-water emulsion containing synthetic trehalose dicorynomycolate and the TLR4 agonist Monophosphoryl Lipid A). The alum and Ribi immunizations given by intraperitoneal injection, and the Isco immunizations were delivered subcutaneously per the manufacturer’s recommendations (Fig. 1D). To evaluate if immunogens bearing an unmodified CD4bs could activate VRC01-like precursors, we tested responses to both the native-like trimer BG505 SOSIP.664 (33–36) and also to eOD17-60mers, nanoparticles presenting a native-like and non-germline-targeting CD4bs on an eOD protein similar to eOD-Base (17) with all glycosylation sites intact.
eOD-GT8-60mer challenge elicited a CD4bs response in VRC01 gH mice, as their immune serum IgG bound more strongly to eOD-GT8 than to eOD-GT8-KO, a mutant designed to block germline VRC01 binding (D368R, N279A and mutations to restore the N276 glycosylation site) (Fig. 2A, fig. S11). The IgG response of WT mice, in contrast, was mainly to non-CD4bs epitopes. eOD-GT8 immunogens given in all three adjuvants supported a serum IgG response to CD4bs, though eOD-GT8-60mers were stronger than eOD-GT8-3mers as assessed by an ELISA area-under-the-curve analysis (i.e., area under the eOD-GT8 reactivity curve minus area under the eOD-GT8-KO curve; Fig. 2B) and by frequencies of IgG+ memory phenotype B cells that bound eOD-GT8 but not eOD-GT8-KO (eOD-GT8(+)/eOD-GT8-KO(−)) identified by cell sorting (Fig. 2C). eOD-GT8 60mers induced lower frequencies of (non-CD4bs) IgG+ memory phenotype B cells that bound both eOD-GT8 and eOD-GT8-KO (eOD-GT8(+)/eOD-GT8-KO(+)), suggestive of an epitope-specific response (fig. S12). Both BG505 SOSIP.664 trimer and eOD17-60mers elicited weak responses by ELISA and antigen-specific B cell frequencies (Fig. 2B, C).
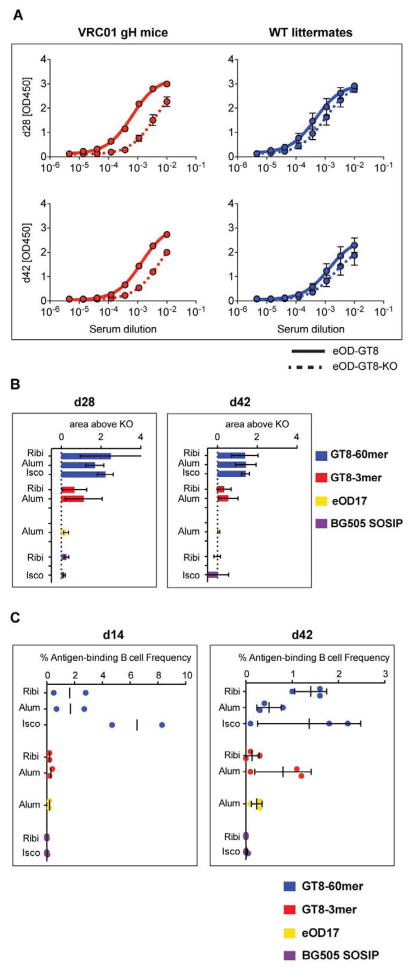
(A) Serum binding titers of VRC01 gH or WT littermate mice immunized with eOD-GT8-60mer nanoparticles in Ribi were measured by enzyme-linked immunosorbent assay (ELISA). Sera were titrated for binding to monomers of eOD-GT8 or eOD-GT8-KO. Plotted values represent the mean of OD450 measurements from 3 different mice for the indicated serum time point (day 28, top; day 42, bottom) at the listed dilutions. Error bars are standard error of the mean (SEM). (B) To determine differences in the level of specificity of antibody responses, the differences between the areas under the eOD-GT8 and eOD-GT8-KO ELISA binding curves were calculated for day 28 and 42 sera. Mean and standard deviation for 3 animals are shown. (C) Frequencies of epitope- or antigen- specific memory phenotype B cells sorted by flow cytometry for each immunization group. The frequency of eOD-GT8(+)/eOD-GT8-KO(−) cells among all memory phenotype B cells is shown for all groups except for BG505 SOSIP, for which the frequency of BG505 SOSIP+ cells among all memory phenotype B cells is shown. Each point represents a mouse sacrificed at day 14 (left) or day 42 (right). Mean (day 14), or mean and standard deviation (day 42, N=3), are indicated by bars.
Selection of light chain partners by the priming immunogen
Priming of the VRC01-class response was revealed in the sequencing data from sorted B cells and hybridomas. B cell sorting recovered 177 IgG heavy/light paired sequences from day 14 and 42, 167 of which utilized the VRC01 knock-in H-chain (some were unmutated and others had mutations in either or both of the H or L-chains, as discussed below), and 95 (IgG or IgM) hybridomas were recovered, all of which used the knock-in H-chain. Among IgG B cells, this H-chain was paired with κ L-chain partners of highly restricted CDRL3 length and Vκ-gene usage (Fig. 3). 92% (154/167) of eOD-GT8(+)/eOD-GT8-KO(−) sorted IgG B cells using the VRC01 gH had L-chains with CDRL3 length of 5 aa (Fig 3A, table S1–2), whereas only ~0.1% of naive (non-immunized) VRC01 gH B cells or wild-type mouse B cells had a κ L-chain CDRL3 length of 5 aa (fig. S2). None of the 10 sorted B cells that utilized an endogenous mouse VH gene contained a 5 aa CDRL3. Among IgG hybridomas, which were captured as early as day 5 of the response, 6 of 7 hybridomas carried a κ L-chain with a 5 aa CDRL3, each isolated from a different mouse (table S3–6). In contrast, among 88 IgM hybridomas recovered following eOD-GT8 60mer immunization (for which eOD-GT8 affinity was weaker than 100 μM for all but two according to SPR), only one had this CDRL3 signature; this suggests that the initial selection for the unusual CDRL3 length occurred upon class switching. Priming was reproducible, as IgG B cells with 5 aa CDRL3s were isolated from 20 of 22 mice immunized with eOD-GT8 60mer (14 of 15 mice analyzed by sorting and 6 of 7 mice that produced IgG hybridomas) (table S1). eOD-GT8-binding IgGs preferentially used Vκ genes with a QQY motif at the start of CDRL3 common to mature VRC01-class bnAbs (CQQYEFF) (Fig. 3B, C; fig. S13). In contrast, IgM hybridomas used a broad distribution of Vκ and Vλs (table S6), again indicating selection at the class-switch stage. In summary, eOD-GT8-60mer immunization successfully recruited VRC01-like precursors into the T cell-dependent response and promoted the selective IgG class switching of cells carrying desirable L-chain features.

(A) Mouse light chains from sorted antigen-specific IgG+ memory phenotype B cells (red) and from hybridomas (blue) as well as mouse light chains from IgM+ antigen-specific hybridomas (orange) were sequenced to identify CDRL3 lengths and mutations from germline mouse kappa chains. The distribution of CDRL3 lengths is shown in a histogram compared to known VRC01-class antibodies (black) and to the naïve (un-immunized) VRC01 gH mouse antibody repertoire (white). This analysis is based on all sequences using the VRC01 gH-chain from all mice immunized with eOD-GT8 60mers (from all hybridoma or sorting timepoints and all adjuvant groups listed in Fig. 1). (B) Gene usage is shown for all Vκ genes in antibodies using the VRC01 gH-chain and a 5aa CDRL3 recovered by sorting IgG+ eOD-GT8+/eOD-GT8-KO(−) memory phenotype B cells at day 14 or 42 from all mice immunized with eOD-GT8 60mers in all adjuvants (table S7). (C) Comparison of the VRC01 CDRL3 sequence with sequences of 5 aa CDRL3s recovered from VRC01 gH mice. Sequences are depicted as sequence logos at the indicated positions, with the size of each letter corresponding to the prevalence of that residue at that position. The “VRC01” sequence logo shows the sequence of the VRC01 CDRL3; the “Naïve Repertoire” sequence logo represents all 1,653 sequences with 5 aa CDRL3 found by deep sequencing of 4 unimmunized VRC01 gH mice (these sequences amount to 0.14% of all 1,169,886 sequences from those mice); the “Unmutated Abs” and “Mutated Abs” sequence logos represent the sets of unmutated (N=84) or mutated (N=70) antibodies, respectively, using the human VH1-2*02 gene and a 5aa CDRL3, isolated from VRC01 gH mice at days 14 or 42 after immunization with eOD-GT8 60mer and Alum, Isco, or Ribi (the red bar at CDRL3 length = 5 in Fig 3A corresponds to these 154 sequences, and see table S7).
Somatic mutation patterns
A bnAb priming immunogen must not only expand precursor numbers, but also promote somatic mutations that allow binding to boosting antigens with closer similarity to HIV Env. We found many somatic mutations among IgG memory phenotype B cells responding to eOD-GT8 60mers and containing a 5aa CDRL3, including some L-chain mutations shared with mature VRC01-class antibodies (Fig. 3C). On the L-chain, many sequences isolated at day 42 of the response to eOD-GT8-60mer/Ribi achieved a D or E in the VRC01 CQQYEF sequence motif. 16 of 47 analyzed IgG memory phenotype B cells had a T-to-G nucleotide mutation in their 5 aa CDRL3s to introduce a D at position 4 (fig. S14), and several cells had an E at that position.
As the VRC01 gH-chain sequence was known, H-chain mutations were readily identified and could be compared directly to mature VRC01 to identify favorable mutations. To focus exclusively on VRC01-class antibodies, our heavy chain analysis only included the VH region of Abs that derived from the VRC01 gH-chain and contained a 5aa CDRL3. Nearly all VRC01-class Abs from day 14 were unmutated. By day 42, however, 53 of 98 VRC01-class Abs contained at least one coding mutation from the starting heavy chain sequence (table S7). Among all VRC01-class Abs from day 14 and day 42 with at least one coding mutation on the heavy chain, 55 of 61 contained at least one mutation that is identical to VRC01 (fig. S15), and ≥50% of the mutations in 49 of 61 such Abs were identical to those in one of 6 VRC01-class bnAbs (12a21, 3BNC60, PGV04, PGV20, VRC-CH31, or VRC01) (Fig. 4A, table S7). In one case, all 6 coding mutations were identical to mutations found in VRC01-class bnAbs. One particular mutation (H35N) was found in >80% of B cells that had at least one mutation, including cells from 12 different mice and all adjuvant groups (table S2), and including both sorted IgG cells and hybridomas. Examination of the eOD-GT6/GL-VRC01 complex structure (PDBID:4jpk) and the gp120/VRC01 complex structure (PDBID:3ngb) revealed that the H35N mutation enables a favorable hydrogen bonding interaction with an asparagine on CDRH3 (fig S16). We also noted differences in mutation levels in different adjuvant groups—among the day 42 sequences, the percentages of Abs with at least one heavy chain coding mutation were 44% (8/18) for alum-immunized mice, 19% (3/16) for Isco-immunized mice, and 67% (42/63) for Ribi-immunized mice (Fig. 4B). Overall, the strong selection of mutations is suggestive of a VRC01-class response, with many mutations identical to those in VRC01-class bnAbs that may help primed cells become cross-reactive to more native-like gp120 molecules. Thus, priming with the eOD-GT8 60mer selected antibody features predicted to improve binding to the CD4bs of Env.

(A) A total of 61 mutated heavy chain sequences from day 14 and 42 eOD-GT8 60mer-immunized VRC01 gH-chain mice (table S7) were evaluated for the number of amino acids that match the mutations found in VRC01-class bnAbs (12a12, 3BNC60, PGV04, PGV20, VRC-CH31 and VRC01) compared to total heavy chain amino acid mutations from germline. Each circle represents a single heavy chain sequence that was isolated by antigen-specific memory phenotype B cell sorting. (B) The total number of amino acid mutations observed in the heavy chains of antibodies isolated by antigen-specific memory phenotype B cell sorting are listed by adjuvant (Alum, Isco, or Ribi) for spleen and lymph node samples harvested at 14 or 42 days post-priming immunization. Bar graphs are divided by unmutated (white) vs mutated (colored). The mutated bars are divided into Abs with 1–2 coding mutations (red), 3–4 coding mutations (blue) or 5–6 coding mutations (orange). The number of mice and the number of antibodies used to compute the frequencies in each bar are listed at the top of the graph.
Antibody affinity for the germline-targeting prime and candidate boost immunogens
T cell-dependent immune responses promote somatic hypermutation and selection for B cells with improved affinity for immunogen, but an additional requirement for an effective bnAb HIV priming immunogen is to promote enhanced affinity for the presumed HIV boosting antigen(s). To assess this aspect of the efficacy of eOD-GT8 60mer priming, we expressed 115 H/L paired sequences that utilized the VRC01 knock-in H-chain and contained a 5aa CDRL3 from eOD-GT8(+)/eOD-GT8-KO(−) IgG memory phenotype sorted B cells (table S7), and we then evaluated their binding to eOD-GT8, eOD-GT8-KO and to candidate boosting antigens by SPR. Of the 115 Abs, 72 contained no H- or L-chain mutations from germline. These unmutated Abs bound eOD-GT8 with a median KD of 32 nM (Fig. 5A–B, table S8). Few mutations were required to promote high affinity—most Abs with >3 coding mutations had an affinity too high to measure accurately (KD <16 pM, Fig. 5A). Confirming epitope-specificity, antibodies for which we could measure a KD for eOD-GT8 showed reduced affinity for eOD-GT8-KO by factors of 36 to 200. We observed intriguing differences among adjuvant groups, with Ribi-immunized mice producing both more Abs (recoverable by sorting) and higher affinity Abs compared to Alum or Isco.

(A) eOD-GT8 dissociation constants measured by SPR for 115 eOD-GT8 60mer-elicited antibodies isolated by antigen-specific B cell sorting (table S7). Antibodies were captured on the sensor chip and eOD-GT8 monomer was analyte. Data are shown for 42 antibodies from day 14 and 73 antibodies from day 42 following immunization of VRC01 gH mice with eOD-GT8 60mer. Each point is colored to indicate the type of adjuvant used (Alum, Iscomatrix, and Ribi) in the immunizations. The scale on the y-axis spans from the smallest dissociation constant (16 pM) measureable by our SPR instrument (as stated by the manufacturer) to the highest dissociation constant (10 μM) measureable based on the analyte concentration used in the experiment. (B) Dissociation constants measured by SPR between selected eOD-GT8 60mer-elicited antibodies and candidate boost immunogens. Among the 115 Abs in (A), the 29 antibodies with highest affinity for eOD-GT8 (KD < 1 nM), along with 8 unmutated antibodies with lower affinity for eOD-GT8, were selected for binding to candidate boosting immunogens (HxB2 core-e 2CC N276D and core BG505 N276D) by SPR. High analyte concentration was used to determine KDs up to 100 μM. HxB2 core-e 2CC N276D 60mer nanoparticles were also assayed, with values presented as apparent affinity, due to the avidity between particles and IgG. Mutated antibodies are shown as green open squares while germline antibodies are shown as black open diamonds.
The eOD-GT8 60mer was designed both to prime germline VRC01-class precursors and to select for mutations that confer cross-reactivity to more native-like gp120 (17). To test if the latter was effective, we selected the 29 Abs that bound eOD-GT8 with subnanomolar affinity as well as 8 unmutated variants (with average KDs for eOD-GT8) and screened them for binding to more native-like gp120 constructs in both monomer and 60mer form. The Abs with more than 3 coding mutations not only had improved affinity for eOD-GT8 but in many cases showed affinity for core-e-2CC HxB2 N276D, a conformationally stabilized core gp120 monomer with a near-native CD4bs from strain HxB2 that combines the loop and termini trimming of the “coreE” design (10, 14) with the disulfides and space-fill mutations of the “2CC” design (37) but also lacks the N276 glycan. In total, 23 of 29 Abs that bound with high affinity to eOD-GT8 showed detectable binding to core-e-2CC HxB2 N276D (KD < 100 μM), while none of unmutated Abs did (Fig. 5B, table S8). 60mer nanoparticles of core-e-2CC HxB2 N276D bound to 24 of 29 mutated Abs more strongly than to the monomer by a factor of ~100 due to avidity, but the 60mers also showed no binding to the unmutated Abs. We conclude that priming with eOD-GT8 60mers promotes clonal expansion and facilitates recognition of molecules presenting a near native CD4bs.
Discussion: The priming problem
A vital goal of rational vaccine design is to understand how to prime naturally subdominant antibody responses in a reproducible manner. Germline-targeting offers one potential strategy to achieve this goal. Here, we have demonstrated using a germline-reverted VRC01 H-chain knock-in mouse model that a germline-targeting immunogen (eOD-GT8 60mer) can activate relatively rare VRC01-class precursors, select productive mutations, and create a pool of memory phenotype B cells that are likely to be susceptible to boosting by more native-like immunogens. In contrast, we found that immunogens bearing a native-like CD4 binding site, including both the eOD17-60mer and the well-ordered BG505 SOSIP.D664 trimer, failed to achieve these goals. These results illustrate the value of an engineered priming immunogen to initiate the development of bnAb lineages by vaccination.
The data in the VRC01 gH mouse model described here have strong potential relevance to human vaccination. Given that: (i) VH1-2*02 is expressed in ~80% of B cells in this mouse compared to ~3% of human B cells (18, 19); (ii) the frequency of 5aa CDRL3 L chains is 0.1% in VRC01 gH B cells compared to 0.6–1% in humans (fig. S1–2) (16), and (iii) the CDRH3 requirements are modest for VRC01-class bnAbs (10, 14, 16, 38) and appear to be minimal for VRC01-class precursors, perhaps requiring a length of 11–18 (75% of human Abs (fig. S17)), it is possible that VRC01-class precursors are less frequent in humans compared to the VRC01 gH mouse by a factor of only ~5 (= 80/3 × 0.1/0.6 × 1/0.75). Even if this estimate is off by an order of magnitude or two due to unknown factors, it is also true that humans have orders of magnitude more B cells than mice, hence more potential targets. Therefore we believe that this study provides strong support for the idea of human clinical testing of the eOD-GT8 60mer, to assess whether this germline-targeting prime can perform similarly in diverse humans. Moreover, the differences observed with different adjuvants in this mouse model — in serum titers, B cell frequencies, selection of favorable mutations and generation of high affinity Abs — indicate that testing different adjuvants should be considered in the design of human clinical experiments probing activation of specific classes of precursor B cells.
Having demonstrated that eOD-GT8 60mer immunization initiates a VRC01-class response in this mouse model, several additional developments are likely needed to induce broad neutralizing activity. The eOD-GT8 60mer contains a modified CD4bs to confer germline-reactivity and as such is probably not capable of selecting all of the heavy and light chain mutations required for bnAb activity against the native CD4bs. Indeed, no neutralizing activity was detected for any of the 8 eOD-GT8 60mer-induced Abs (all with high affinity [KD < 1 nM] for eOD-GT8 and low affinity [1 μM < KD < 100 μM] for core-e-2CC HxB2 N276D) that we tested against a panel of four viruses from clades A and B that included both wild-type and N276A mutant viruses with increased sensitivity to VRC01-class bnAbs (fig. S18). One design feature of eOD-GT8 is that it lacks the N276 glycan—removal of this glycan is a requirement for germline-reactivity (17, 21). However, the N276 glycosylation site is conserved in 94.5% of HIV strains, according to analysis of 3,796 sequences from the Los Alamos HIV database (http://www.hiv.lanl.gov/). Induction of broad neutralization will likely require one or more boosting immunogens bearing a glycan at N276 so as to select mutations to accommodate that glycan (17). On the H chain of VRC01-class bnAbs, mutations in the CDR2, CDR1, FW1 and FW3 are likely required for maximum potency and breadth (24, 39), and native-like Env immunogens will probably be needed to select for these. In sum, boosting with a sequence of increasingly native-like antigens, and potentially including cocktails of different antigens within each boost to mimic the antigenic diversity of the CD4bs, will likely be needed to select the mutations required for VRC01-class bnAb activity. The mouse model presented here, as well as other newly developed VRC01-class knock-in mouse models (40), should aid us to test this notion and can be used to identify the antigens and boosting strategies that work best. Of note, we demonstrated here that a single immunization with the eOD-GT8 60mer induces VRC01-class antibodies with modest affinity for the core-e-2CC HxB2 N276D monomer and 60mer, so these molecules represent promising candidates for the first boost. We are thus mapping the first steps in a sequential strategy for the rational induction of bnAbs against HIV.
Acknowledgments
The authors thank T.R. Blane, S. Kupriyanov and G.S. Martin for outstanding technical assistance. The data presented in this manuscript are tabulated in the main paper and in the supplementary materials. This work was supported by the International AIDS Vaccine Initiative Neutralizing Antibody Consortium and Center (W.R.S., D.R.B.), CAVD funding for the IAVI NAC Center (W.R.S., D.R.B.), the Ragon Institute of MGH, MIT and Harvard (D.R.B. and W.R.S.), the Helen Hay Whitney Foundation (J.G.J.), and National Institute of Allergy and Infectious Diseases grants: R01-AI073148 (D.N.), P01AI081625 (W.R.S.), CHAVI-ID 1UM1AI100663 (W.R.S., D.R.B.). International AIDS Vaccine Initiative (IAVI) and the Scripps Research Institute are filing a patent relating to the eOD-GT8 immunogens in this manuscript, with inventors JGJ, DWK, SM, WRS. Materials and information will be provided under an MTA.
Footnotes
References and Notes
Full text links
Read article at publisher's site: https://doi.org/10.1126/science.aac5894
Read article for free, from open access legal sources, via Unpaywall:
https://science.sciencemag.org/content/sci/349/6244/156.full.pdf
Citations & impact
Impact metrics
Citations of article over time
Alternative metrics
Smart citations by scite.ai
Explore citation contexts and check if this article has been
supported or disputed.
https://scite.ai/reports/10.1126/science.aac5894
Article citations
Advanced technologies for the development of infectious disease vaccines.
Nat Rev Drug Discov, 21 Oct 2024
Cited by: 0 articles | PMID: 39433939
Review
Direct restoration of photocurable cross-linkers for repeated light-based 3D printing of covalent adaptable networks.
Mater Horiz, 08 Oct 2024
Cited by: 0 articles | PMID: 39376135 | PMCID: PMC11459227
Mosaic and mixed HIV-1 glycoprotein nanoparticles elicit antibody responses to broadly neutralizing epitopes.
PLoS Pathog, 20(10):e1012558, 03 Oct 2024
Cited by: 0 articles | PMID: 39361585 | PMCID: PMC11449375
Short CDRL1 in intermediate VRC01-like mAbs is not sufficient to overcome key glycan barriers on HIV-1 Env.
J Virol, 98(10):e0074424, 06 Sep 2024
Cited by: 0 articles | PMID: 39240111 | PMCID: PMC11495006
Isolation and characterization of IgG3 glycan-targeting antibodies with exceptional cross-reactivity for diverse viral families.
PLoS Pathog, 20(9):e1012499, 18 Sep 2024
Cited by: 0 articles | PMID: 39292703 | PMCID: PMC11410209
Go to all (286) article citations
Data
Data behind the article
This data has been text mined from the article, or deposited into data resources.
BioStudies: supplemental material and supporting data
Protein structures in PDBe (2)
-
(1 citation)
PDBe - 4jpkView structure
-
(1 citation)
PDBe - 3ngbView structure
Similar Articles
To arrive at the top five similar articles we use a word-weighted algorithm to compare words from the Title and Abstract of each citation.
Short CDRL1 in intermediate VRC01-like mAbs is not sufficient to overcome key glycan barriers on HIV-1 Env.
J Virol, 98(10):e0074424, 06 Sep 2024
Cited by: 0 articles | PMID: 39240111 | PMCID: PMC11495006
HIV-1 envelope glycan modifications that permit neutralization by germline-reverted VRC01-class broadly neutralizing antibodies.
PLoS Pathog, 14(11):e1007431, 05 Nov 2018
Cited by: 26 articles | PMID: 30395637 | PMCID: PMC6237427
Glycan Masking Focuses Immune Responses to the HIV-1 CD4-Binding Site and Enhances Elicitation of VRC01-Class Precursor Antibodies.
Immunity, 49(2):301-311.e5, 31 Jul 2018
Cited by: 87 articles | PMID: 30076101 | PMCID: PMC6896779
HIV Broadly Neutralizing Antibodies: VRC01 and Beyond.
Adv Exp Med Biol, 1075:53-72, 01 Jan 2018
Cited by: 7 articles | PMID: 30030789
Review
Funding
Funders who supported this work.
Helen Hay Whitney Foundation
IAVI NAC
IAVI Neutralizing Antibody Consortium and Center
MGH
MIT
NIAID NIH HHS (10)
Grant ID: R01 AI073148
Grant ID: R01 AI081625
Grant ID: P01AI081625
Grant ID: R01-AI073148
Grant ID: P01 AI094419
Grant ID: T32 AI007244
Grant ID: UM1 AI100663
Grant ID: 1UM1AI100663
Grant ID: R01 AI128836
Grant ID: R37 AI059714
National Institute of Allergy and Infectious Diseases (3)
Grant ID: P01AI081625
Grant ID: CHAVI-ID 1UM1AI100663
Grant ID: R01-AI073148





