Abstract
Free full text

Phospholipid scrambling by rhodopsin
Abstract
Rhodopsin has been intensively characterized in its role as a visual pigment and G protein-coupled receptor responsible for dim-light vision. We recently discovered that it also functions as an ATP-independent phospholipid scramblase: when reconstituted into large unilamellar vesicles, rhodopsin accelerates the normally sluggish transbilayer translocation of common phospholipids by more than 1000-fold, to rates in excess of 10,000 phospholipids transported per rhodopsin per second. Here we summarize the work leading to this discovery and speculate on the mechanism by which rhodopsin scrambles phospholipids. We also present a hypothesis that rhodopsin’s scramblase activity is necessary for the function of the ABC transporter ABCA4 that is responsible for mitigating the toxic accumulation of 11-cis-retinal and bis-retinoids in the retina.
Graphical Abstract
Rhodopsin as phospholipid scramblase – A perspective
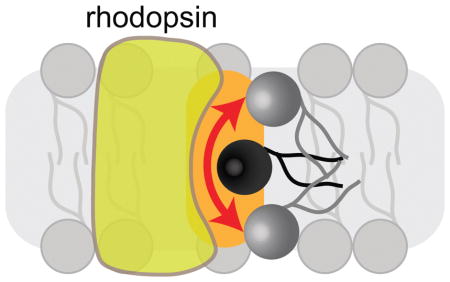
Introduction
Photoreceptor discs possess a remarkable assortment of lipid transporters, representing one each of the major classes of lipid transporters that are known to translocate phospholipids across cellular membranes (Figure 1). Thus disc membranes contain ABCA4 (previously known as Rim protein), an ATP-binding cassette (ABC) transporter that is posited to translocate phosphatidylethanolamine (PE) and N-retinylidene-PE (NRPE),1 as well as Atp8a2, a P4-ATPase that transports phosphatidylserine (PS) and PE.2 Both ABCA4 and Atp8a2 couple ATP hydrolysis to the movement of lipids from the luminal leaflet of the disc membrane to the cytoplasmic side. In addition to the ATP-driven transporters, discs possess an ATP-independent phospholipid scramblase that translocates all common phospholipids bidirectionally across the disc membrane. Disc scramblase activity was first observed by Wu and Hubbell3 in 1993, and confirmed by Hessel et al.4,5. Recent studies revealed, unexpectedly, that the scramblase activity is due to the photoreceptor protein rhodopsin,6,7 a prototypic G protein-coupled receptor (GPCR).8
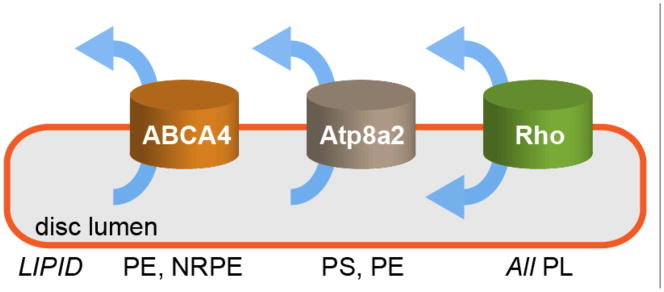
ABCA4 is an ABC transporter specific for PE and N-retinylidene-PE (NRPE; see text section ‘Biological function of lipid scrambling in discs’ for details); Atp8a2 is a P4-ATPase specific for PS and PE – however, it is not clear whether it is active in discs (see text); rhodopsin (Rho) is a scramblase that translocates common phospholipids (PL) in an ATP-independent manner. Arrows show the direction of lipid transport. ABCA4 is unusual amongst eukaryotic ABC transporters because it is the only one reported thus far that functions as an importer or flippase. In reconstituted systems, the ATP-driven transporters display turnover rates ranging from 1–100 s−1, whereas rhodopsin transports lipids at >10,000 s−1.
ATP-independent phospholipid scramblases are required in the endoplasmic reticulum and bacterial cytoplasmic membrane for membrane growth, as well as for a variety of glycosylation pathways including protein N-glycosylation and synthesis of bacterial peptidoglycan.9 They also play a critical role in enabling activated blood platelets to promote blood coagulation and apoptotic cells to be engulfed by macrophages.10 In these cases, they are responsible for cell surface exposure of PS, a lipid that is normally sequestered in the cytoplasmic leaflet of the plasma membrane but whose presence at the cell surface represents an important physiological signal.11 Despite their key roles in cell physiology, the identity of the various scramblases remained elusive for decades until reports in 2011 revealed that rhodopsin is a phospholipid scramblase6 and the bacterial protein FtsW is the Lipid II scramblase required for peptidoglycan synthesis.12 These discoveries were based on the demonstration of scramblase activity by a purified protein reconstituted into lipid vesicles. Until this point, previous reports of lipid scramblases had fallen short of the critical biochemical test described above.13–17 Of relevance to our narrative, Nagata and coworkers18 showed that a member of the TMEM16 family of Ca2+-activated ion channels is intimately involved in lipid scrambling leading to PS exposure on blood platelets. However, as this protein was never purified and tested in a reconstituted system it remains to be determined whether it is indeed a scramblase.19 Nevertheless, two fungal homologues of the TMEM16 family could be purified and these proteins were shown recently to have Ca2+-dependent scramblase activity when reconstituted into phospholipid vesicles.20,21 Thus, the last several years have seen the identification of three biochemically verified, structurally dissimilar scramblases (rhodopsin, FtsW, TMEM16 family proteins), setting the stage for mechanistic analysis of the lipid translocation process. Here we focus on rhodopsin’s scramblase activity.
Phospholipid scrambling in isolated disc membranes and transbilayer lipid asymmetry of discs
Wu and Hubbell3 and Hessel et al.4,5 reported that spin-labeled PS, PE and phosphatidylcholine (PC) analogs flip rapidly and bidirectionally across purified bovine rod discs in an ATP-independent manner. Figure 2, adapted from the paper of Hessel et al.,4 illustrates how these measurements were made.
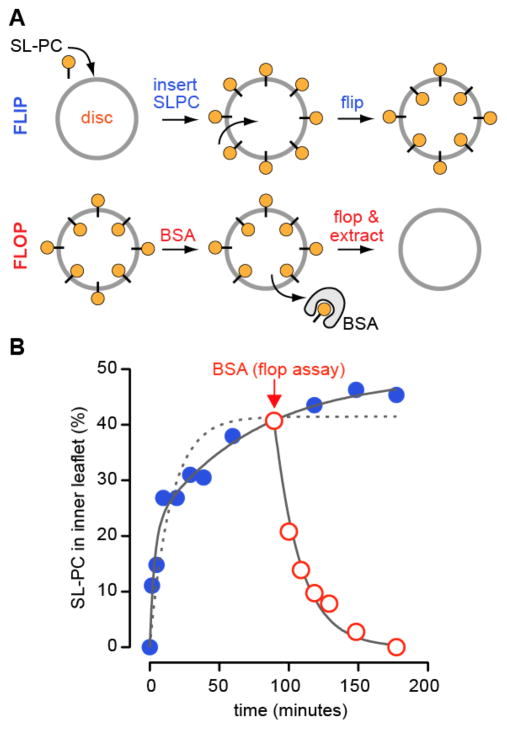
The figure is based on Figure 5A of Hessel et al..4 Measuring lipid flip (out-to-in translocation): The assay is illustrated in panel A (top). Discs are incubated at 25 °C with a trace quantity of spin-labeled phosphatidylcholine analog (SL-PC). SL-PC inserts into the outer leaflet of the discs within 30 s. At different time points, an aliquot of the labeled discs is withdrawn, chilled on ice to slow lipid flip-flop and treated with fatty acid-free bovine serum albumin (BSA) for 1 min. The treated aliquot is then centrifuged to pellet and wash the discs. The amount of SL-PC remaining in the discs (filled blue circles) is determined by electron paramagnetic resonance (EPR) spectroscopy. As BSA quantitatively extracts SL-PC from the outer leaflet, and lipid flipping is slow on the time-scale of extraction on ice, the amount of SL-PC remaining in the discs after BSA treatment corresponds to molecules that have flipped into the inner leaflet. The experimental results are shown in panel B (blue, filled circles). Although the data are best fit by a double exponential function (solid line), the authors use a single exponential with a t1/2~10 min to approximate the flip reaction (dotted line). Measuring lipid flop (in-to-out translocation): The assay is illustrated in panel A (bottom). Discs pre-equilibrated with SL-PC are incubated with fatty acid-free BSA at 25 °C (time of BSA addition indicated by arrow in panel B). Aliquots are withdrawn at different times and the discs are chilled, pelleted, washed and analyzed by EPR spectroscopy to determine the amount of remaining SL-PC (red open circles). The loss of SL-PC from the discs under conditions of continuous BSA back-extraction provides a measure of lipid flop. The data are well described by a mono-exponential decay process with t1/2~10 min (panel B, solid line through open red circles).
Spin-labeled PC (SL-PC) is introduced into the outer leaflet of intact purified discs at time zero (Figure 2A, upper panel) and the sample is incubated at 25°C for a period of time. At different time points, an aliquot of the sample is withdrawn and incubated with fatty acid-free bovine serum albumin (BSA). Incubation with BSA serves to ‘back-extract’ SL-PC that is located in the outer leaflet and thereby to quantify the size of this pool. Back extraction is performed at 4°C, a precaution intended to minimize transbilayer movement during this analytical step as ongoing scrambling during back-extraction would result in an overestimate of the amount of SL-PC in the outer leaflet. At early times, SL-PC can be almost quantitatively extracted because the majority is still in the outer leaflet of the discs. Over time at 25°C, SL-PC flips to the inner leaflet such that eventually only ~50% of the lipid is extractable (Figure 2B, blue filled circles). Thus SL-PC equilibrates across the two leaflets of the disc membrane within the time scale of the experiment. Similar results were obtained for SL-PE, but for SL-PS equilibrium was reached with ~65% PS in the outer leaflet.4 To measure in-out movement (flop) SL-PC-equilibrated discs were incubated continuously with fatty acid-free BSA at 25°C (schematically shown in Figure 2A, lower panel). In this situation, all of the lipid can be eventually extracted, consistent with transport from the inner leaflet to the BSA-accessible outer leaflet of the discs (Figure 2B, red open circles; BSA was added 90 min after the initial addition of SL-PC to the discs as indicated by the arrow in order to first allow SL-PC to equilibrate across the disc membrane).
Both flip and flop occur with a t1/2 of ~10 min (Figure 2B), independent of ATP levels. As spontaneous flipping of phospholipids across pure lipid membranes is very slow (t1/2~tens of hours)22–24, the authors concluded that discs have a specific mechanism to facilitate rapid, bidirectional flip-flop. Hessel et al.4 speculated that a ‘solubilization zone’ around rhodopsin, essentially a zone of instability, could provide a locus for flip-flop. Wu and Hubbell3 tested whether the most abundant disc protein rhodopsin might be responsible for promoting phospholipid scrambling. We discuss their experiments in the next section.
The reports of Wu and Hubbell3 and Hessel et al.4,5 also indicated that disc membranes are asymmetric with respect to PS (65–80% in the cytoplasmic leaflet), and roughly symmetric in their transbilayer distribution of PC and PE. This asymmetry was predicted by the transbilayer coupling model of Hubbell,25 i.e., the combination of phospholipid scrambling and the asymmetric charge distribution (positive on the cytoplasmic face of discs) created by the large number of oriented rhodopsin molecules in the disc membrane, explains why negatively charged PS is mainly located on the cytoplasmic side at steady state whereas zwitterionic PC and PE are symmetrically distributed across the disc membranes. Interestingly, the transbilayer asymmetry of PS changes reversibly in response to light, presumably because of associated changes in transbilayer charge asymmetry.5 Given the transbilayer coupling model,25 it is not necessary to invoke the activity of the aminophospholipid-specific transporter Atp8a2 (Figure 1) to explain PS asymmetry. Indeed the function of Atp8a2 in discs is enigmatic, as its activity would needlessly deplete ATP. We speculate that Atp8a2 may be silent in disc membranes because of unrelieved auto-inhibition, or because of regulation by phosphorylation as shown for other lipid-translocating P-type ATPases.26,27 Atp8a2 may instead play a functional role in the secretory pathway28,29 by regulating protein traffic to the disc membrane.
Rhodopsin is a phospholipid scramblase
What is the molecular basis of phospholipid scrambling in discs? We had previously developed methods to assay phospholipid scrambling in proteoliposomes generated from detergent-solubilized ER membrane proteins30,31 and we applied these techniques to identify the disc scramblase.6 Our approach is illustrated in Figure 3A. We reconstitute large unilamellar vesicles (LUV; ~175-nm diameter) from egg PC and a trace amount of a phospholipid that is acyl-labeled with a 7-nitro-2,1,3-benzoxadiazol (NBD) fluorophore. On treating these vesicles with dithionite, a membrane-impermeant dianion, NBD-phospholipids (NBD-PLs) in the outer leaflet of the vesicles are chemically reduced to non-fluorescent 7-amino-2,1,3-benzoxadiazol (ABD) phospholipids. This results in loss of half the fluorescence signal (Figure 3B, ‘liposome’). All fluorescence is lost if the vesicles are disrupted with detergent which permits dithionite access to NBD-PLs in the inner leaflet. When we reconstituted detergent-solubilized disc membrane proteins into such vesicles we observed an increase in the extent of fluorescence reduction beyond the 50% level seen for protein-free liposomes (Figure 3B, ‘proteoliposome’). As the transbilayer distribution of NBD-PLs in the reconstituted proteoliposomes was symmetric based on iodide collisional quenching experiments,6,7 this result suggested that either NBD-phospholipids were being translocated from the inner to the outer leaflet or that dithionite was entering the proteoliposomes. The latter possibility was ruled out by showing that NBD-glucose trapped within the vesicles was protected from reduction.7 Thus, the disc extract contains phospholipid scramblase activity. The time scale of fluorescence reduction in both liposomes and proteoliposomes was similar (Figure 3B), indicating that transbilayer phospholipid movement in proteoliposomes occurred at the same rate as the dithionite-mediated reduction of NBD fluorophores, or faster. Curiously, we never observed the expected 100% reduction for proteoliposomes (Figure 3B, ‘ideal’), even when reconstitutions were done using high enough amounts of protein to ensure that every vesicle should have multiple copies of scramblase on average. Although the reasons for this are not clear, the phenomenon of ‘incomplete reconstitution’ is quite general: many reconstitution studies of transport proteins find that a small population of vesicles (typically ~15–20% of the total vesicle pool) generated during reconstitution by different procedures appears incapable of incorporating functional transporters7,20,32–35 and is consequently silent in transport assays.
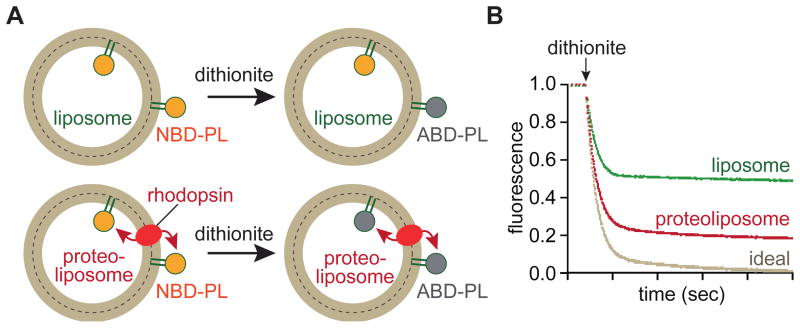
A. Principle of the assay. Large unilamellar vesicles (LUVs) are reconstituted with a trace amount of a fluorescent 7-nitro-2,1,3-benzoxadiazol (NBD) acyl-labelled phospholipid (NBD-PL) and rhodopsin to generate proteoliposomes. Control LUVs (liposomes) are reconstituted similarly but without protein. On adding dithionite (S2O42−), a membrane impermeant dianion, NBD-PL molecules in the outer leaflet of the vesicles become non-fluorescent because dithionite reduces the NBD-fluorophore to a non-fluorescent amino derivative (ABD). For liposomes, this results in ~50% loss in fluorescence as NBD-PL molecules in the inner leaflet are protected. For rhodopsin-containing proteoliposomes, all NBD-PL molecules should be reduced since those in the inner leaflet are translocated to the outer leaflet. Dithionite does not enter the vesicles as evinced by the observation that NBD-glucose that is trapped within the vesicles cannot be reduced unless the vesicles are disrupted with detergent.7 B. Example of data. The graph shows time-dependent recordings of fluorescence of liposome and proteoliposome samples, in addition to an idealized trace for proteoliposomes. Dithionite is added as indicated, resulting in a rapid decrease in fluorescence. Fluorescence of liposomes drops to ~50% of the starting value as expected, whereas that of proteoliposomes is reduced by ~80%. The expected 100% reduction (trace labeled ‘ideal’) is never seen, even when reconstitutions are done such that every vesicle can be expected to have multiple copies of rhodopsin on average. The reasons for this are not clear but it has been noted in numerous systems that a population of vesicles is refractory to reconstitution.7 The rate of fluorescence reduction is the same for both liposomes and proteoliposomes, indicating that the dithionite reaction is rate limiting. Thus only a lower limit can be set for the rate of lipid flipping in this assay.
By generating proteoliposomes using different amounts of disc extract, we could vary the fraction of vesicles that contained an active scramblase. Systematic measurement of the extent of fluorescence reduction versus the protein/phospholipid ratio of the proteoliposomes allowed us to estimate the abundance of the scramblase in the crude extract of disc membrane proteins. The abundance we deduced was surprisingly high, >60%.6 As the only protein with such a high abundance in disc membranes is rhodopsin, this result strongly indicated that the observed scramblase activity was due to rhodopsin. To determine that this was the case, we tested scramblase activity of disc extracts that had been treated with antibodies to remove rhodopsin specifically and quantitatively. The treated extracts had no activity.6 Conversely, we found robust activity when we reconstituted rhodopsin that had been purified from disc membranes or after heterologous expression in COS cells or HEK293 cells.6 Proteoliposomes containing purified rhodopsin were able to scramble a variety of NBD-labeled lipids, including PC, PE, PS, and sphingomyelin, as well as a natural, i.e. non-fluorescent, glycosylated phosphatidylinositol.6 Of the lipids tested, the only lipid that was not scrambled was Man5GlcNAc2-PP-dolichol, an intermediate in the dolichol pathway of protein N-glycosylation.6,9 The inability to scramble this dolichol-based diphosphate lipid will inevitably provide clues about the scramblase mechanism (see below).
A question that is often asked is whether rapid scrambling occurs in vesicles simply because of the reconstitution of any membrane protein. The answer is clearly ‘no’. Thus, a variety of purified polytopic membrane proteins such as TMEM16A (a Ca2+-activated Cl− channel), CLC-ec1 (a CLC-type H+/Cl− exchanger), and GltPh (a glutamate transporter) do not facilitate scrambling when reconstituted into LUVs6,20,21 and the fungal TMEM16 homologs, afTMEM16 and nhTMEM16 scramble lipids rapidly in LUVs only in the presence of Ca2+.20,21 Also of note is the fact that ABC transporters such as MsbA, Pgp, Cdr1 and ABCA4 do not scramble phospholipids when reconstituted into LUVs, but are able to flip them undirectionally on ATP addition.36–38 Despite these examples showing that scrambling does not occur generically in LUVs that are reconstituted with any membrane protein, there is the possibility that scrambling in specific instances could be due to protein-associated detergent molecules that are not completely removed during reconstitution. This possibility was ruled out in the case of rhodopsin as follows: when the protein was solubilized in β-octylglucoside and reconstituted by extensive dialysis, scramblase activity was readily observed even though the amount of residual detergent in the final vesicle preparation was sub-stoichiometric relative to rhodopsin (molar ratio of β-octylglucoside: rhodopsin: phospholipid ~1: 5: 2x106).6 The reconstitution-based studies described above clearly establish that rhodopsin is a phospholipid scramblase. We estimate that lipid scrambling in vesicles with one or only a few rhodopsin molecules occurs at rates in excess of 10,000 phospholipids per second, corresponding to at least a 1000-fold increase over the rate of spontaneous flipping in pure lipid vesicles.6,7
Using a reconstitution approach similar to the one described above, Wu and Hubbell3 came to a different conclusion, i.e. that rhodopsin would not facilitate rapid flip-flop. They reconstituted purified rhodopsin into small unilamellar vesicles (SUV) at a high concentration (100:1 molar ratio of egg PC to rhodopsin, corresponding to ~80 rhodopsin molecules per 35-nm diameter vesicle) and tested the ability of the vesicles to promote flipping of spin-labeled (SL)-PC by using ascorbate to reduce the pool of spin label exposed in the outer leaflet of the vesicles. As they had done in their measurements of SL-PC scrambling in intact discs, the reduction reaction was carried out at 1°C in an attempt to minimize both ascorbate leakage into the vesicles and SL-PC scrambling. When SL-PC was added exogenously to proteoliposomes and allowed to equilibrate for 2 hours at 39°C, all the spin label was reduced by ascorbate in a single rate process. The authors considered that this could be due to the absence of scrambling at 39°C, i.e. all the SL-PC remained in the outer leaflet, or the rapid leakage of ascorbate into the vesicles at 1°C. To resolve these possibilities, the authors included SL-PC prior to vesicle formation so that it was incorporated into both leaflets of the bilayer. On treatment of these proteoliposomes with ascorbate at 1°C, ~80% of the SL-PC was reduced over a period of ~30 min leaving a significant protected pool of ~20% SL-PC (it was noted that the vesicles were leaky to ascorbate, but the rate of leakage was sufficiently slow that it could be distinguished from the much faster rate of reduction of the bulk of the spin label and that an appropriate correction could be made). In a situation where scrambling does not occur, an unbiased distribution of SL-PC in a 35-nm diameter SUV would be expected to be 66:34, out:in, reflecting the relative surface area of the outer and inner leaflet.3 The authors claimed that an unbiased distribution would be unlikely because of the high concentration of rhodopsin in the vesicles and its likely asymmetric reconstitution. Because they had obtained a stable protected pool of SL-PC in rhodopsin-SUVs they concluded that rhodopsin does not scramble phospholipids.
We re-examined the reconstitution data of Wu and Hubbell3 in light of our observations that (1) NBD-PC is scrambled rapidly in rhodopsin-containing LUVs at 0°C (B. Ploier and A.K. Menon, unpublished observation), consistent with previous observations of lipid scrambling at 0°C in LUVs reconstituted with endoplasmic reticulum membrane proteins,39,40 and (2) a fraction of the rhodopsin-containing vesicles (~10–20% of the sample) is silent in the scrambling assay (see above and Figure 3B). We suggest that when SL-PC is added to rhodopsin-LUVs and allowed to equilibrate at 39°C, the entire vesicle population is labeled with the lipid probe. In the majority of these vesicles, the SL-PC equilibrates across both leaflets whereas in the ‘silent’ vesicles (minority fraction) it remains in the outer leaflet. Thus, when ascorbate is added to these vesicles at 1°C, all SL-PC is reduced because it can scramble to the outer leaflet in the majority population even at low temperature (see above) and is located anyway in the outer leaflet of the minority population. For rhodopsin-LUVs generated in the presence of SL-PC, the majority of the vesicles are fully detected because SL-PC scrambling occurs at 1°C, whereas the minority fraction (up to 20% of the total) is fully or partly refractory to detection. We recreated this scenario using NBD-PC labeled rhodopsin-LUVs that were probed with dithionite at 23°C. The data, shown in Figure 4, resemble those shown by Wu and Hubbell3 in their Figures 8 and 9, and can be readily explained as described above. A final point is that Wu and Hubbell detected ~80% of the SL-PC in rhodopsin-SUVs that had been reconstituted in the presence of the lipid probe, whereas only 66% would have been detected had no scrambling occurred during ascorbate treatment at 1°C. They argued that because the ensemble of rhodopsin molecules within an SUV was likely to be asymmetric, an unbiased distribution of SL-PC could not be expected. In case of an asymmetric distribution of rhodopsin, the wider cytoplasmic side of rhodopsin is likely to face towards the outside of the vesicle, thus reducing the area for lipids disproportionally in the outer vs. inner leaflet, and arguing for a more even distribution of SL-PC. As a zwitterion, SL-PC would also not respond to any transbilayer charge asymmetry generated by rhodopsin. Thus, consistent with the arguments presented above, we suggest that Wu and Hubbell were measuring scrambling at 1°C when they detected more SL-PC (80% versus 66% or less) than would be found simply in the outer leaflet of the 35-nm diameter rhodopsin-SUVs.
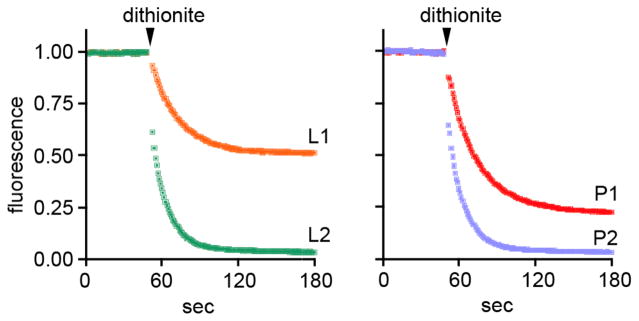
Dithionite reduction assays (see Figure 3) were carried out at 23°C with protein-free liposomes (L1, L2) and rhodopsin-proteoliposomes (P1, P2). The average diameter of the vesicles was ~150 nm. The lipid to protein ratio of the proteoliposomes was ~25,000:1 (~8 rhodopsins per vesicle). NBD-PC was included during the reconstitution of L1 and P1, whereas for L2 and P2 NBD-PC was inserted into the outer leaflet after vesicle formation and allowed to equilibrate for 15 min (insertion of NBD-PC into vesicles is rapid and occurs within 2 minutes37; scrambling at 23°C is also complete in <2 min6,7). The dithionite reduction assays for L1 and P1 yield ~50% and ~80% reduction, similar to the results shown in Figure 3B. For L2 and P2 reduction is quantitative. See text for further explanation. I. Menon and A.K. Menon, unpublished observations.
Mechanism of phospholipid scrambling
As noted above, the mere presence of a membrane protein in a bilayer is not sufficient to promote rapid phospholipid flip-flop6,9,20,36 and so there must be something unusual about rhodopsin’s structure and/or its dynamical behavior in the membrane that enables it to function as a scramblase. When reconstituted into vesicles, rhodopsin translocates lipids at a rate >104 per second,6,7 and this occurs independently of its conformational state and whether it contains retinal. Thus different conformations of opsin and rhodopsin, including a mimic of the active metarhodopsin II state, are all capable of scrambling phospholipids rapidly.7 This suggests that scrambling is unlikely to be accomplished by the ‘alternating-access’ mechanism used by many solute transporters where turnover numbers, dictated by large-scale conformational changes, typically reach only ~102 per second.41,42 Thus, a transverse diffusion mechanism is likely, where the lipid headgroup moves through a presumably solvated environment as the lipid re-orients across the bilayer. Although it was originally suggested that a possible lipid translocation pathway might be provided by the central, water-lined core of the protein,6 this was ruled out as access to the core is occluded by a loop (the E2-loop) on the exoplasmic side and the core itself is blocked by retinal in rhodopsin and metarhodopsin II, both of which are active as scramblases.7 A suitable environment for lipid translocation could be provided instead by specific residues on the surface of the rhodopsin transmembrane (TM) helical bundle or by membrane packing defects that are specifically generated in the vicinity of rhodopsin (Figure 5A). As other Class A (rhodopsin class) GPCRs, like the β1- and β2-adrenergic receptors and the A2A adenosine receptor also scramble lipids,6,7 a structural element common amongst these proteins (see for example ref.43) likely contributes to the scramblase activity. GPCRs contain an intracellular amphipathic helix 8 that immediately follows the last transmembrane helix, is often terminated by one or two palmitoylated cysteines and runs parallel to the membrane. This helix has the potential to disturb bilayer structure on one side of the membrane, but propagation of the disturbance across the bilayer may contribute to bidirectional scrambling.
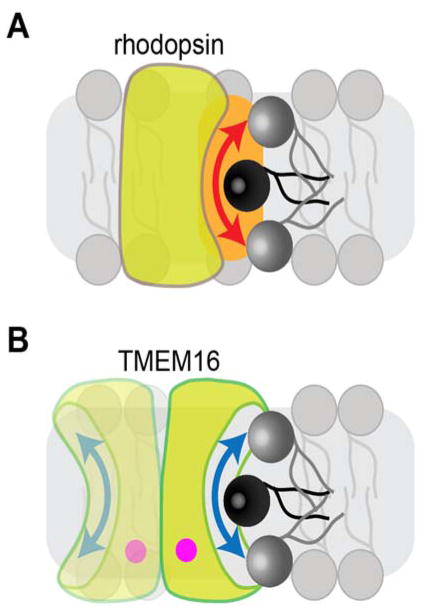
A possible mechanism for rhodopsin-mediated phospholipid scrambling (A) is shown alongside a distinct mechanism that likely explains the mechanism of action of Ca2+-dependent TMEM16 scramblases (B). The mechanism of rhodopsin-mediated scrambling is poorly understood and may involve a specific perturbation of the membrane (orange zone) in the vicinity of the protein. See text for further explanation. The TMEM16 scramblase is shown in panel B as a homodimer, with each monomer (containing bound Ca2+, indicated by the pink dot) capable of scrambling lipids via a hydrophilic, membrane-facing groove,21 akin to swiping a credit card through a card reader.82
An interesting proposal by Kol et al.44,45 suggests that ATP-independent scramblases operate by creating ‘flip-flop’ sites near particular TM helices as a result of dynamic interactions of the helices with the membrane and properties of the helices themselves. These sites may be packing defects in the bilayer induced by helix movement, localized thinning of the membrane, and even transient penetration of water into the bilayer from both sides. As a result of the discontinuity in lipid–lipid interactions at the lipid–helix interface, a lipid may ‘slip’ into the hydrophobic interior of the membrane with its head-group in a solvated state, and ‘pop’ out on the other side. It is likely that a ‘slip-pop’ mechanism would depend on the primary sequence of TM helices, and helix dynamics. These ideas are supported by recent analyses of lipid scrambling by low complexity synthetic TM peptides.46
Scrambling is significantly slower in intact discs than in lipid vesicles reconstituted with very low amounts of rhodopsin (Figure 2B versus versus3B).3B). The reasons for this are not clear, but the possibility that rhodopsins can multimerize47–50 may be a contributing factor. Thus fewer protein–membrane interfaces are present when rhodopsins contact each other, and multimerization might affect rhodopsin dynamics and thus slip-pop processes as discussed above.
It is likely that the bulk properties and composition of the membrane influence scrambling. Rhodopsin is found in the plasma membrane of photoreceptor outer segments, yet these membranes sequester PS in the inner leaflet as evinced by lack of staining by the PS probe Annexin V.51 This is not consistent with rhodopsin being active as a scramblase in the plasma membrane. Similarly, PS is not found at the surface of COS-7 cells that over-express rhodopsin (S. Finnemann, A.K. Menon, unpublished observation). Most, if not all cells, express GPCRs and yet these cells also do not expose PS unless triggered to do so, for example during apoptosis. Thus the scramblase activity of rhodopsin and other GPCRs appears to be silent at the plasma membrane. A possible explanation is that the high cholesterol content of the plasma membrane suppresses the scramblase activity of these proteins. Cholesterol may promote GPCR dimerization or multimerization, with consequences for scrambling as discussed above. Alternatively, cholesterol may affect the transverse diffusion path for lipids by thickening and organizing the membrane.52 Rod photoreceptor discs have an unusual lipid composition that is known to influence rhodopsin’s function.53–56 New discs are rich in cholesterol (~30 mole % of total lipid, similar to the level of cholesterol in the plasma membrane), but become cholesterol-poor (~5 mole %) as they mature along the outer segment.57,58 Cholesterol inhibits rhodopsin activation and increases its stability54,56,59 : thus rhodopsin in the plasma membrane and in new rod discs is relatively inactive in terms of phototransduction, and similarly may be inactive as a scramblase in these membranes. An interesting point is that in cone outer segments, discs are continuous with the plasma membrane60 and therefore would be expected to be uniformly high in cholesterol. Thus, scrambling may not occur in the cone outer segment as it does in rod discs. An explicit test of the role of cholesterol in regulating scramblase activity requires measurement of scrambling after reconstitution of rhodopsin into cholesterol-containing lipid vesicles. Such efforts are underway (B. Ploier, I. Menon, M.A. Goren and A.K. Menon, unpublished work). Finally, disc phospholipids are rich in docosahexaenoic acid (DHA),61,62 a polyunsaturated fatty acid that increases rhodopsin’s signaling activity55 and that may, for the same reasons, also increase the rate of rhodopsin-mediated lipid scrambling.62,63 However, because DHA chains specifically associate with rhodopsin,64 the exchange and flipping of lipids at rhodopsin’s membrane surface could also be slowed. Clarification of these points is an objective for the future.
In contrast to the ideas suggested above for rhodopsin-mediated lipid scrambling, a relatively clearer picture has emerged for the recently discovered TMEM16 scramblases.19,65 The founding members of the TMEM16 family were shown to be Ca2+-activated Cl− channels, but other members of fungal origin appear to be Ca2+-dependent phospholipid scramblases that may or may not also transport ions. TMEM16 scramblases mediate rapid scrambling when reconstituted into large unilamellar vesicles in the presence of Ca2+ ions (similar to rhodopsin-mediated scrambling, Figure 3B) and slow, but detectable scrambling in the presence of a Ca2+ chelator.20,21 The crystal structure of a fungal TMEM16 homolog21 revealed the presence of a membrane-facing hydrophilic groove that could provide a path for phospholipids. Thus a phospholipid could reorient itself in the bilayer with its polar headgroup interacting with the hydrophilic grove while its acyl chains remain in the hydrophobic interior of the bilayer (Figure 5B). The Ca2+-dependence of scrambling by TMEM16 proteins can be explained by the effect of Ca2+ ions in controlling the conformation of the protein by dilating a partially open groove, or by enhancing the probability that the groove adopts an open, permissive conformation.
Biological function of rhodopsin-mediated lipid scrambling in discs
We speculate that rhodopsin-mediated lipid scrambling in discs is closely tied to the function of the ABC transporter ABCA4 (Figure 1), and in turn to one of the underlying causes of age-related macular degeneration (AMD). AMD is a major cause of visual impairment and blindness in older adults.66–68 A risk factor for dry AMD, exemplified in Stargardt’s and Best disease, is the accumulation of fluorescent lipofuscin in the retinal pigment epithelium (RPE). A major constituent of lipofuscin is the di-retinal conjugate A2E, a product of the visual cycle.69–71 The accumulation of A2E in RPE over a lifetime of light exposure causes progressive degeneration of RPE cells and their photoreceptor clients, destruction of the sensory retina and loss of central high acuity vision. In Stargardt’s and Best diseases, bis-retinoid accumulation results in blindness in the fourth decade of life.
The first step of A2E synthesis is the reversible formation of a Schiff-base adduct between a retinaldehyde and PE in disc membranes to form N-retinylidene-phosphatidylethanolamine (NRPE). Whereas it was originally thought that this reaction required all-trans-retinal, released from rhodopsin after light exposure,72–74 recent reports suggest instead that 11-cis-retinal is the critical molecule in this pathway.75,76 Thus, N-11-cis-retinylidene-PE (N-cis-R-PE) is formed from 11-cis-retinal that is delivered to the discs by the visual cycle, or after catabolism of rhodopsin. N-cis-R-PE isomerizes to N-all-trans-R-PE,76 before addition of a second retinaldehyde converts NRPE irreversibly to A2-PE in the disc; A2-PE is eventually hydrolyzed to A2E after discs are phagocytosed by the RPE. ABCA4 is predicted to play a key role in minimizing A2-PE production by flipping NRPE from the lumenal side to the cytoplasmic face of discs before it can be converted to A2-PE.75,76 On the cytoplasmic side of the disc, NRPE dissociates, releasing ATR to enter the visual cycle and leaving behind PE in the cytoplasmic leaflet of discs. In accordance with the bilayer couple hypothesis77 the deposition of PE in the cytoplasmic leaflet at the expense of the luminal leaflet would cause the disc membrane to bulge outwards, towards the cytoplasm. The associated bilayer stress would also prevent continued transport of NRPE by ABCA4. We believe (see also refs.78,79) that by restoring the balance of PE and other phospholipids across the membrane, rhodopsin acts as a release valve to normalize the disc membrane bilayer enabling ABCA4 to continue transporting NRPE. Thus, transbilayer lipid translocation by both ABCA4 and rhodopsin is needed to prevent A2E build-up.
What is the biological significance of lipid scrambling by GPCRs other than rhodopsin? These proteins are mainly located in the plasma membrane where their scramblase activity is likely to be suppressed by high levels of cholesterol as discussed above. However, GPCRs are integrated into the secretory pathway in the endoplasmic reticulum where cholesterol levels are low.52 Here they may provide the phospholipid scramblase activity that is necessary for the biogenic function of the ER.9,80
Conclusions and future directions
We have provided an overview of the discovery that rhodopsin is a phospholipid scramblase, accounting for the original observations of lipid scrambling in disc membranes. We speculate on why scrambling must occur in discs. Although we propose possible mechanisms by which rhodopsin and other GPCRs are able to scramble lipids, much more work needs to be done to arrive at a precise molecular understanding of how this transport process works. While it is possible to make comparisons with what has been learned about lipid scrambling by two other scramblases that have been recently discovered,12,20,21,81 leading to models of transbilayer lipid reorientation such as those depicted in Figure 5 and discussed extensively elsewhere,9,82 it is likely that rhodopsin’s mechanism of scrambling will be unique. Uncovering this mechanism is a key goal for the future.
Acknowledgments
We thank Lydia Caro, Birgit Ploier and Kalpana Pandey for comments on the manuscript, Indu Menon, Birgit Ploier and Silvia Finnemann for unpublished data, and Sam Canis for assistance. This work was supported by NIH grants EY024207 and GM106717 (A.K.M.), the Velux Stiftung (A.K.M.), the Qatar National Research Fund’s National Priority Research Program (5-669-1-112) (A.K.M.), and the Canada Excellence Research Chair program (O.P.E.). O.P.E. holds the Anne and Max Tanenbaum Chair in Neuroscience at the University of Toronto.
Biographies

Anant K. Menon is a Professor of Biochemistry at Weill Cornell Medical College. He received his undergraduate education at the Indian Institute of Technology, Kanpur and his doctorate in Chemistry at Cornell University. As a postdoctoral fellow at The Rockefeller University he elucidated the pathway for the biosynthesis of glycosylphosphatidylinositol (GPI)-anchored proteins. His recent work focuses on problems of membrane biogenesis, specifically how lipids are transported across and between membranes. His laboratory currently studies the molecular mechanisms of intracellular sterol transport, and the scramblase-mediated transbilayer movement of phospholipids. This review highlights the Menon laboratory’s discovery of rhodopsin’s scramblase activity.

Oliver P. Ernst
Oliver P. Ernst obtained his Dr. rer. nat. (chemistry/biochemistry) from the University of Freiburg, Germany, in 1994. After research training at Rockefeller University, he joined the Charité–Universitätsmedizin Berlin in 1995, where he became a group leader and made his habilitation in biophysics in 2003. Since 2011, he has been a full professor in the Departments of Biochemistry and Molecular Genetics at the University of Toronto. He holds the Canada Excellence Research Chair in Structural Neurobiology and the Max and Anne Tanenbaum Chair in Neuroscience. His research focuses on rhodopsin and visual signal transduction as well as other GPCRs.
References
Full text links
Read article at publisher's site: https://doi.org/10.1039/c5pp00195a
Read article for free, from open access legal sources, via Unpaywall:
https://europepmc.org/articles/pmc4764046?pdf=render
Citations & impact
Impact metrics
Citations of article over time
Article citations
Optimizing properties of translocation-enhancing transmembrane proteins.
Biophys J, 123(10):1240-1252, 13 Apr 2024
Cited by: 0 articles | PMID: 38615194 | PMCID: PMC11140465
A cholesterol switch controls phospholipid scrambling by G protein-coupled receptors.
J Biol Chem, 300(2):105649, 16 Jan 2024
Cited by: 4 articles | PMID: 38237683 | PMCID: PMC10874734
Scramblase activity of proteorhodopsin confers physiological advantages to Escherichia coli in the absence of light.
iScience, 26(12):108551, 22 Nov 2023
Cited by: 0 articles | PMID: 38125024 | PMCID: PMC10730872
Lactose Permease Scrambles Phospholipids.
Biology (Basel), 12(11):1367, 25 Oct 2023
Cited by: 2 articles | PMID: 37997967 | PMCID: PMC10669175
Retinal-phospholipid Schiff-base conjugates and their interaction with ABCA4, the ABC transporter associated with Stargardt disease.
J Biol Chem, 299(5):104614, 16 Mar 2023
Cited by: 6 articles | PMID: 36931393 | PMCID: PMC10127136
Go to all (30) article citations
Similar Articles
To arrive at the top five similar articles we use a word-weighted algorithm to compare words from the Title and Abstract of each citation.
A Fluorescence-based Assay of Phospholipid Scramblase Activity.
J Vis Exp, (115), 20 Sep 2016
Cited by: 29 articles | PMID: 27684510 | PMCID: PMC5092049
Constitutive phospholipid scramblase activity of a G protein-coupled receptor.
Nat Commun, 5:5115, 08 Oct 2014
Cited by: 70 articles | PMID: 25296113 | PMCID: PMC4198942
Phospholipid Scrambling by G Protein-Coupled Receptors.
Annu Rev Biophys, 51:39-61, 21 Dec 2021
Cited by: 19 articles | PMID: 34932914 | PMCID: PMC9521775
Review Free full text in Europe PMC
Kinetics of rhodopsin's chromophore monitored in a single photoreceptor.
Methods Mol Biol, 1271:327-343, 01 Jan 2015
Cited by: 3 articles | PMID: 25697533
Funding
Funders who supported this work.
Canada Excellence Research Chairs, Government of Canada
NEI NIH HHS (2)
Grant ID: R21 EY024207
Grant ID: EY024207
NIGMS NIH HHS (2)
Grant ID: GM106717
Grant ID: R01 GM106717
National Institutes of Health (2)
Grant ID: EY024207
Grant ID: GM106717




