Abstract
Background
Genomewide association studies can be used to identify disease-relevant genomic regions, but interpretation of the data is challenging. The FTO region harbors the strongest genetic association with obesity, yet the mechanistic basis of this association remains elusive.Methods
We examined epigenomic data, allelic activity, motif conservation, regulator expression, and gene coexpression patterns, with the aim of dissecting the regulatory circuitry and mechanistic basis of the association between the FTO region and obesity. We validated our predictions with the use of directed perturbations in samples from patients and from mice and with endogenous CRISPR-Cas9 genome editing in samples from patients.Results
Our data indicate that the FTO allele associated with obesity represses mitochondrial thermogenesis in adipocyte precursor cells in a tissue-autonomous manner. The rs1421085 T-to-C single-nucleotide variant disrupts a conserved motif for the ARID5B repressor, which leads to derepression of a potent preadipocyte enhancer and a doubling of IRX3 and IRX5 expression during early adipocyte differentiation. This results in a cell-autonomous developmental shift from energy-dissipating beige (brite) adipocytes to energy-storing white adipocytes, with a reduction in mitochondrial thermogenesis by a factor of 5, as well as an increase in lipid storage. Inhibition of Irx3 in adipose tissue in mice reduced body weight and increased energy dissipation without a change in physical activity or appetite. Knockdown of IRX3 or IRX5 in primary adipocytes from participants with the risk allele restored thermogenesis, increasing it by a factor of 7, and overexpression of these genes had the opposite effect in adipocytes from nonrisk-allele carriers. Repair of the ARID5B motif by CRISPR-Cas9 editing of rs1421085 in primary adipocytes from a patient with the risk allele restored IRX3 and IRX5 repression, activated browning expression programs, and restored thermogenesis, increasing it by a factor of 7.Conclusions
Our results point to a pathway for adipocyte thermogenesis regulation involving ARID5B, rs1421085, IRX3, and IRX5, which, when manipulated, had pronounced pro-obesity and anti-obesity effects. (Funded by the German Research Center for Environmental Health and others.).Free full text

FTO Obesity Variant Circuitry and Adipocyte Browning in Humans
Abstract
BACKGROUND
Genomewide association studies can be used to identify disease-relevant genomic regions, but interpretation of the data is challenging. The FTO region harbors the strongest genetic association with obesity, yet the mechanistic basis of this association remains elusive.
METHODS
We examined epigenomic data, allelic activity, motif conservation, regulator expression, and gene coexpression patterns, with the aim of dissecting the regulatory circuitry and mechanistic basis of the association between the FTO region and obesity. We validated our predictions with the use of directed perturbations in samples from patients and from mice and with endogenous CRISPR–Cas9 genome editing in samples from patients.
RESULTS
Our data indicate that the FTO allele associated with obesity represses mitochondrial thermogenesis in adipocyte precursor cells in a tissue-autonomous manner. The rs1421085 T-to-C single-nucleotide variant disrupts a conserved motif for the ARID5B repressor, which leads to derepression of a potent preadipocyte enhancer and a doubling of IRX3 and IRX5 expression during early adipocyte differentiation. This results in a cell-autonomous developmental shift from energy-dissipating beige (brite) adipocytes to energy-storing white adipocytes, with a reduction in mitochondrial thermogenesis by a factor of 5, as well as an increase in lipid storage. Inhibition of Irx3 in adipose tissue in mice reduced body weight and increased energy dissipation without a change in physical activity or appetite. Knockdown of IRX3 or IRX5 in primary adipocytes from participants with the risk allele restored thermogenesis, increasing it by a factor of 7, and overexpression of these genes had the opposite effect in adipocytes from nonrisk-allele carriers. Repair of the ARID5B motif by CRISPR–Cas9 editing of rs1421085 in primary adipocytes from a patient with the risk allele restored IRX3 and IRX5 repression, activated browning expression programs, and restored thermogenesis, increasing it by a factor of 7.
CONCLUSIONS
Our results point to a pathway for adipocyte thermogenesis regulation involving ARID5B, rs1421085, IRX3, and IRX5, which, when manipulated, had pronounced pro-obesity and anti-obesity effects. (Funded by the German Research Center for Environmental Health and others.)
Obesity affects more than 500 million people worldwide and contributes to type 2 diabetes, cardiovascular disorders, and cancer.1 Obesity is the result of a positive energy balance, whereby energy intake exceeds expenditure, resulting in the storage of energy, primarily as lipids in white adipocytes. Energy balance is modulated by food consumption and physical activity, as well as by the dissipation of energy as heat through constitutive thermogenesis in mitochondria-rich brown adipocytes in brown fat and through inducible thermogenesis in beige adipocytes in white fat.2–6 Thermogenesis is triggered by mechanisms within the cells themselves or by the sympathetic nervous system (e.g., through β-adrenergic receptor agonists), in response to exercise, diet, or exposure to cold. Regulators of mitochondrial thermogenesis (including IRF4, PGC1α, PRDM16, and TBX15) control the expression of the gene encoding uncoupling protein 1 (UCP1),4,7–9 which depolarizes the inner mitochondrial membrane, causing proton transfer and heat dissipation.
Body-mass index (BMI) has a strong genetic component (40 to 80% heritability) involving several genes that have expression in the hypothalamus and fulfill roles in appetite regulation.10,11 The strongest genomewide association signal lies in introns 1 and 2 of the gene FTO12,13; this region contains 89 common variants (Fig. 1A) that are in high linkage disequilibrium in Europeans (r2≥0.8)12,13 across approximately 47,000 nucleotides, which makes the identification of the likely causal variant challenging. The associated region lacks protein-altering variants, which has prompted numerous gene-regulatory studies17–21; these studies have predicted diverse and conflicting target genes and tissues, including FTO itself in a whole-body knockout,17 IRX3 in pancreas20 or brain,19 RBL2 in lymphocytes,21 and RPGRIP1L in brain.18 However, the identification of a mechanistic basis for the association between the FTO locus and obesity in humans has been elusive, the relevant cell types and target genes remain unresolved, and the causal variant remains uncharacterized. In this study, we sought to identify a causal variant with regulatory roles, its upstream regulator, and its downstream target gene in order to provide a candidate mechanistic basis for the association between FTO and obesity.
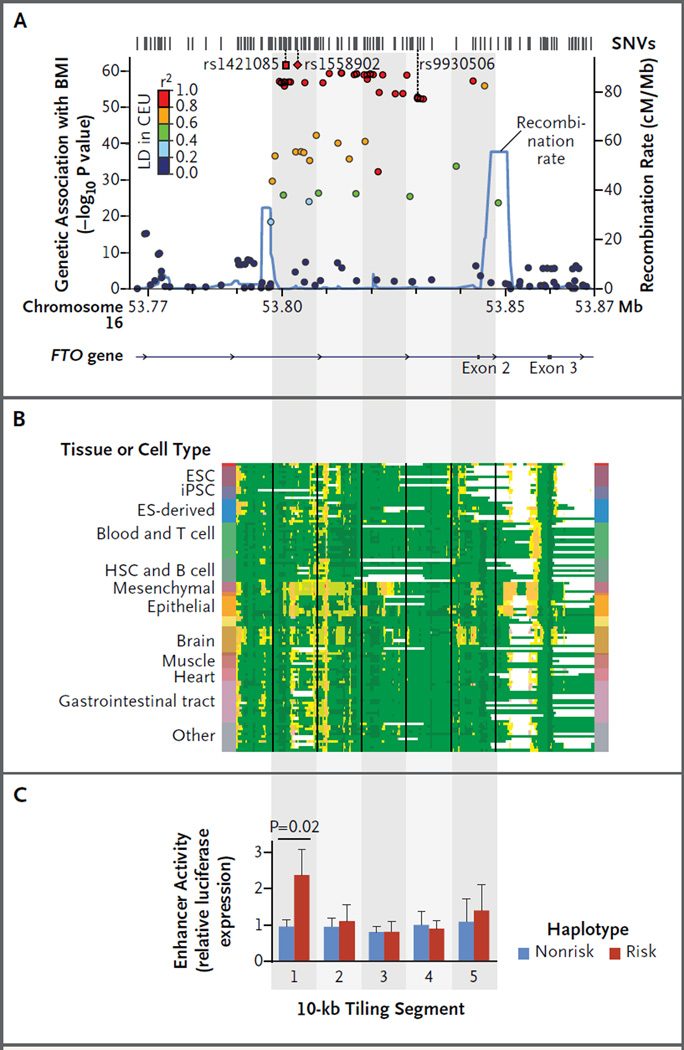
Panel A shows the genetic association with body-mass index (BMI) for all common FTO locus variants,14 including the reported single-nucleotide variant (SNV) rs1558902 (red diamond) and the predicted causal SNV rs1421085 (red square). Gray shading delineates consecutive 10-kb segments. CEU denotes a population of Utah residents with northern and western European ancestry, and LD linkage disequilibrium. Panel B shows chromatin state annotations for the locus across 127 reference epigenomes (rows) for cell and tissue types profiled by the Roadmap Epigenomics Project.15,16 For information on the colors used to denote chromatin states, see Figure S1A in the Supplementary Appendix. Vertical lines delineate the consecutive 10-kb segments shown in Panel A. ESC denotes embryonic stem cell, HSC hematopoietic stem cell, and iPSC induced pluripotent stem cell. Panel C shows human SGBS adipocyte enhancer activity, for 10-kb tiles, of the risk and nonrisk haplotypes with the use of relative luciferase expression. The boxes indicate means from seven triplicate experiments, and T bars indicate standard deviations.
METHODS
PARTICIPANTS
Primary human adipose–derived progenitor cell cultures were obtained from the subcutaneous adipose tissue of 100 healthy Europeans who were 20 to 50 years of age and had a BMI (the weight in kilograms divided by the square of the height in meters) of 20 to 24, which was in the normal range. The sample included 52 participants (hereafter referred to as risk-allele carriers) who were homozygous for the risk allele for the tag variant rs9930506 (which has been reported in genomewide association studies), as well as for the associated variants rs1421085 and rs1558902; together, these variants make up part of the risk haplotype, which in its entirety includes 89 variants in introns 1 and 2 of FTO that are common in European populations. The sample also included 48 participants who were homozygous for the nonrisk allele for all three of these variants (hereafter referred to as nonrisk-allele carriers). Primary cell cultures were used for the preparation of mitochondrial and nuclear messenger RNA, quantitative polymerase-chain-reaction (PCR) gene-expression analysis, assays of mitochondrial function and thermogenesis, determination of lipolysis rates, small interfering RNA–mediated knockdown, doxycycline-mediated overexpression, and CRISPR–Cas9 genome editing. In addition, whole adipose tissue and adipose-derived progenitor cells were obtained and RNA was isolated from a second European cohort of nongenotyped participants, including 12 severely obese patients (BMI, 35 to 52) who were undergoing bariatric surgery and 22 healthy nonobese participants (BMI, 18 to 28) who were undergoing elective surgery. We received approval from the local ethics committees in Germany, Norway, and Sweden. All participants gave written informed consent.
STUDY DESIGN
We used techniques of epigenomics, comparative genomics, human genetics, genome editing, and directed perturbations in samples from patients and from mice to dissect the regulatory circuitry and mechanistic basis of the FTO obesity-associated locus. Our methods are described in detail in the Methods section in the Supplementary Appendix, available with the full text of this article at NEJM.org.
On the basis of epigenomic annotations across 127 human cell types, we predicted the cell type in which the genetic variant was likely to act, and we validated the prediction with the use of haplotype-specific enhancer assays. We analyzed long-range chromatin interactions in the region surrounding FTO to define potential target genes, and to validate genetic targets, we conducted an expression quantitative-trait-locus (eQTL) analysis in primary human adipocytes from risk-allele carriers and nonrisk-allele carriers. We predicted the cellular processes affected by the obesity-associated variants on the basis of correlated expression with the target genes across participants, and we validated their genetic control with the use of a trans-eQTL analysis of energy-balance genes (i.e., an eQTL analysis of energy-balance genes at large genomic distances from the FTO locus) in adipocytes, as well as by measuring cellular phenotypes in risk-allele carriers and nonrisk-allele carriers. To examine the causal roles for the predicted target genes, we first used knockdown and overexpression of each target gene in primary human adipocytes from the subcutaneous fat of risk-allele carriers and nonrisk-allele carriers, followed by cellular phenotyping; second, we used generation of mice with a dominant negative allele of one of the target genes expressed in adipose tissue, followed by organism-level phenotyping, histologic measurements, and gene-expression profiling in major fat stores; and third, we used knockdown, overexpression, and knockout in three mouse adipocyte models.
We predicted the single-nucleotide variant (SNV) responsible for dysregulation of the target genes on the basis of a quantitative analysis of regulatory-motif conservation, and we used experimental alterations of the predicted causal nucleotide to validate its causality for enhancer activity and regulator binding. To predict the upstream regulator, we examined regulatory motif matches and regulator expression levels in adipocytes from obese and nonobese participants; we then experimentally validated its causality and epistasis with the risk haplotype by means of knockdown and overexpression in adipocytes from risk-allele carriers and nonrisk-allele carriers. Finally, we established the cell-autonomous causality of the predicted driver variant with the use of bidirectional CRISPR–Cas9 editing in primary adipocytes from risk-allele carriers and nonrisk-allele carriers, followed by measurement of target-gene expression, expression levels of energy-balance genes, and cellular phenotypic signatures of obesity in humans. Additional information on our methods is provided in the protocol (available at NEJM.org) and in the Supplementary Appendix.
RESULTS
EFFECT OF THE FTO LOCUS ON IRX3 AND IRX5 IN HUMAN ADIPOCYTE PROGENITOR CELLS
To identify the cell types in which the causal variant may act, we examined chromatin state maps15,16 of the FTO obesity region across 127 cell types. An unusually long enhancer (12.8 kb) in mesenchymal adipocyte progenitors indicated a major regulatory locus (Fig. 1B; and Fig. S1A, S1B, and S1C in the Supplementary Appendix). Haplotype-specific enhancer assays showed activity in association with the risk haplotype that was 2.4 times as high as that associated with the nonrisk haplotype in human SGBS adipocytes (i.e., adipocytes derived from a patient with the Simpson–Golabi–Behmel syndrome), which indicated genetic control of enhancer activity (Fig. 1C). Enhancers in brain cells and other cell types were considerably shorter than those in mesenchymal adipocyte progenitors and lacked allelic activity (Fig. S1C and S1D in the Supplementary Appendix).
To predict putative target genes, we examined large domains that had long-range three-dimensional chromatin interactions surrounding FTO and identified eight candidate genes (Fig. 2A and 2B). Among them, the developmental regulators IRX3 and IRX5 had genotype-associated expression, which indicated long-range (1.2-Mb) genetic control in primary preadipocytes (Fig. 2C). Genotype-associated expression was not observed in whole-adipose tissue, a finding consistent with previous reports23,24; this indicated that the effect was cell type–specific and restricted to preadipocytes, which represent a minority of cells in adipose tissue (Fig. S2A in the Supplementary Appendix).
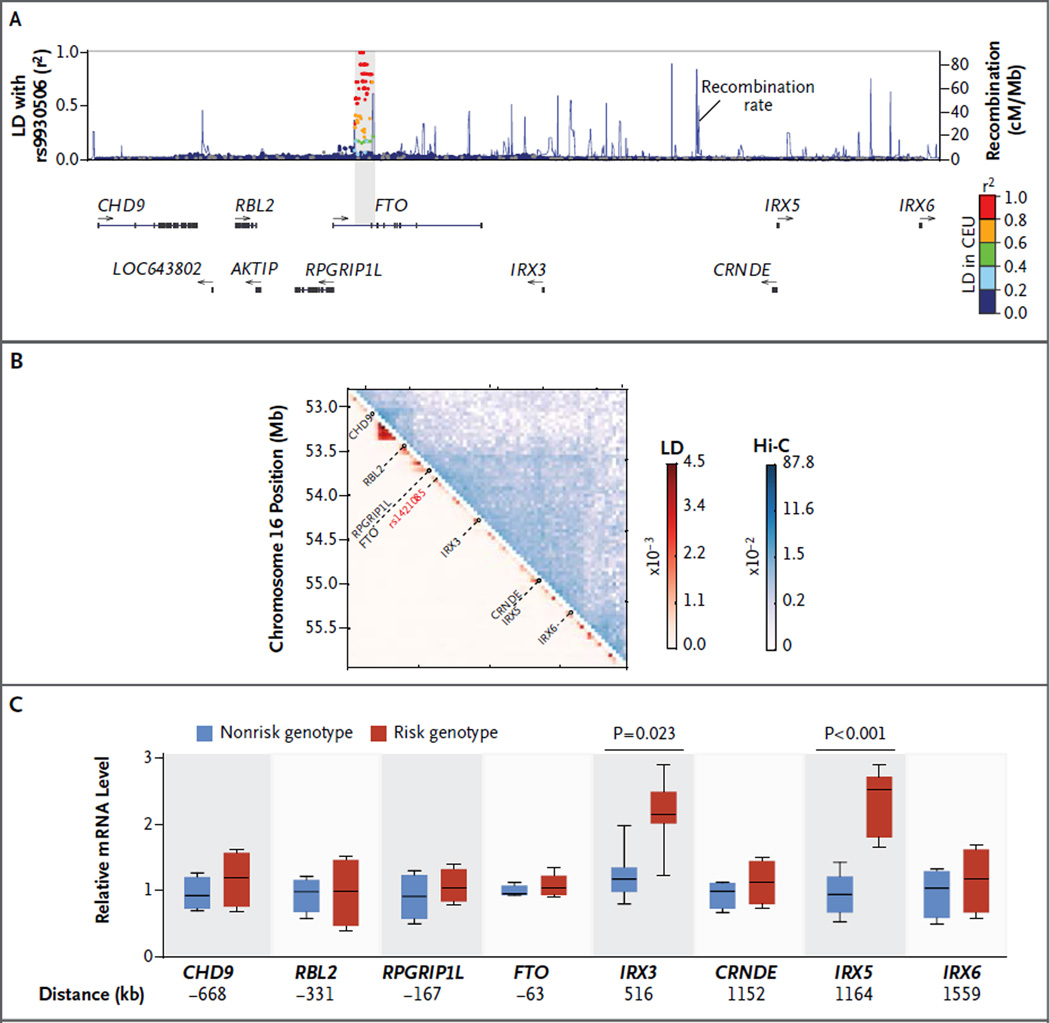
Panel A shows gene annotations and LD with array tag variant rs9930506 in a 2.5-Mb window; LD is expressed as r2 values in the CEU population. Arrows indicate the direction of transcription of annotated genes in the locus. Panel B shows chromosome conformation capture (Hi-C) interactions contact probabilities in human IMR90 myofibroblasts,22 revealing a 2-Mb topologically associating domain, and LD mean r2 statistics for all SNV pairs at 40-kb resolution. Panel C shows box plots for expression levels, after 2 days of differentiation, in human adipose progenitors isolated from 20 risk-allele carriers and 18 nonrisk-allele carriers, evaluated by means of a quantitative polymerase-chain-reaction analysis for all genes in the 2.5-Mb locus. The horizontal line within each box represents the median, the top and bottom of each box indicate the 75th and 25th percentile, and I bars indicate the range.
EFFECT OF THE FTO LOCUS ON MITOCHONDRIAL THERMOGENESIS AND LIPID STORAGE
To identify the biologic processes affected by altered IRX3 and IRX5 expression in adipocytes, we used genomewide expression patterns in brown adipocyte–containing perirenal adipose tissue from a separate cohort of 10 nongenotyped, healthy kidney donors to identify genes with expression that was positively or negatively correlated with IRX3 and IRX5 expression. Genes that are associated with mitochondrial functions were found to have a negative correlation with IRX3 and IRX5, and genes with FXR and RXR lipid-metabolism functions were found to have a positive correlation, which suggests that IRX3 and IRX5 may play roles in energy dissipation and storage (Fig. 3A, and Table S1 in the Supplementary Appendix). IRX3 and IRX5 had consistently higher mean expression in white adipose tissue from nine participants, as well as negative correlation with PGC1A and UCP1 expression, as assessed with the use of interindividual expression patterns in perithyroid brown adipose tissue (Fig. S2B and S2C in the Supplementary Appendix); these findings indicated potential roles for IRX3 and IRX5 in the repression of thermogenesis.
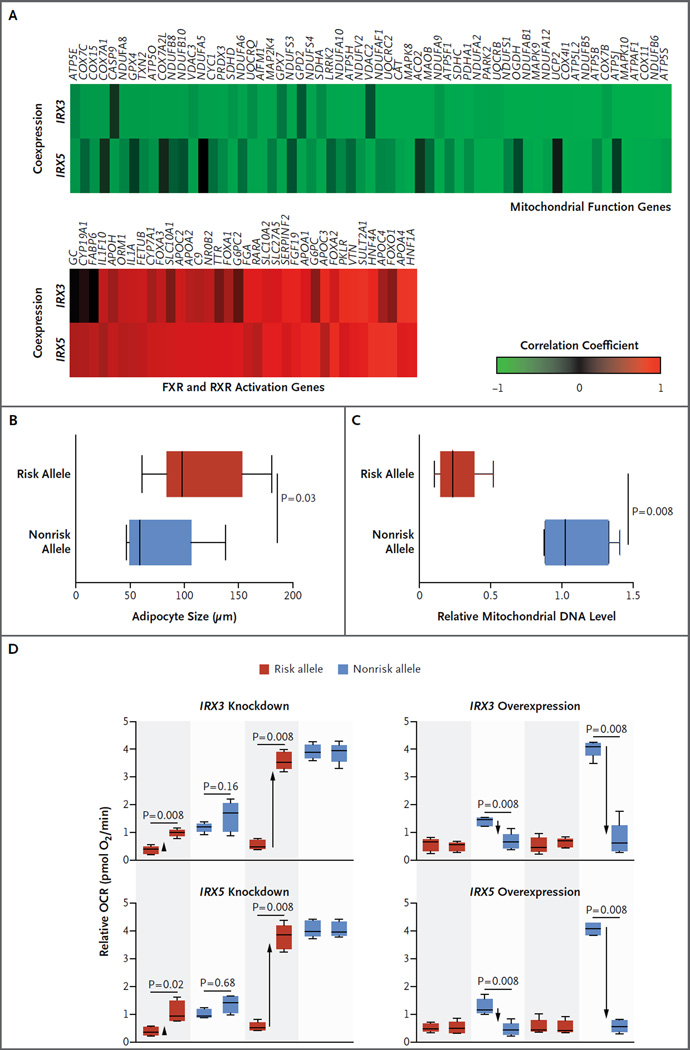
Panel A shows the mitochondrial and FXR and RXR activation genes with strongest positive (in red) or negative (in green) correlation with IRX3 and IRX5 in human perirenal adipose tissue from 10 participants. Panels B and C show box plots of the increased adipocyte diameter and decreased mitochondrial DNA content in isolated differentiated adipocytes from risk-allele carriers (16 and 8 participants, respectively) relative to nonrisk-allele carriers (26 and 8, respectively). The vertical line within each box represents the median, the left and right margins of each box indicate the interquartile range, and I bars indicate the range. Panel D shows box plots of the altered basal and isoproterenol-stimulated oxygen consumption rate (OCR) on small interfering RNA (siRNA) knockdown and doxycycline (DOX)–mediated overexpression of IRX3 and IRX5 in 8 risk-allele carriers and 10 nonrisk-allele carriers. The siRNA efficiency was 62% for IRX3 and 71% for IRX5. The horizontal line within each box represents the median, the top and bottom of each box indicate the interquartile range, and I bars indicate the range.
To examine the trans-eQTL genetic control of energy balance by the FTO obesity locus, we used primary preadipocytes from risk-allele carriers and nonrisk-allele carriers to evaluate the genes with mitochondrial and FXR and RXR functions that had expression patterns most closely correlated with those of IRX3 and IRX5, as well as several known markers of energy-balance regulation (Fig. S2D and S2E in the Supplementary Appendix). As compared with nonrisk-allele carriers, risk-allele carriers had lower expression of mitochondrial, browning, and respiration genes and higher expression of lipid-storage markers, which indicated a shift from energy dissipation to energy storage.
These differences in expression were also reflected in the cellular signatures of obesity. Risk-allele carriers had increased adipocyte size, reduced mitochondrial DNA content, and a loss of UCP1 response to β-adrenergic stimulus or cold exposure (Fig. 3B and 3C, and Fig. S2F in the Supplementary Appendix), as well as resistance to isoproterenol-mediated uncoupling, a decreased basal oxygen consumption rate, and a reduction in mitochondrial thermogenesis by a factor of 5 (Fig. S2G in the Supplementary Appendix); this indicated excessive accumulation of triglycerides, reduced mitochondrial oxidative capacity, reduced white adipocyte browning, and reduced thermogenesis.
ADIPOCYTE-AUTONOMOUS EFFECTS OF IRX3 AND IRX5 ON ENERGY BALANCE
We next quantified the effect that manipulation of IRX3 and IRX5 expression had on thermogenesis in primary preadipocytes that were isolated from both risk-allele carriers and nonrisk-allele carriers. In preadipocytes from risk-allele carriers, IRX3 and IRX5 knockdown restored oxygen consumption and thermogenesis response to nonrisk levels, increased thermogenesis by a factor of 7 (Fig. 3D), and restored UCP1 expression levels (Fig. S3A in the Supplementary Appendix). In preadipocytes from nonrisk-allele carriers, IRX3 and IRX5 overexpression reduced basal respiration and thermogenesis to risk-allele levels (with thermogenesis reduced by a factor of 8) (Fig. 3D) and decreased the expression of UCP1, other regulators of mitochondrial function and thermogenesis (PGC1A, PGC1B, and PRDM16), and the β-adrenergic receptor (ADRB3), which also regulates UCP1-independent thermogenesis programs (Fig. S3B and S3C in the Supplementary Appendix). These manipulations had no significant effect on preadipocytes from participants with the reciprocal genotypes, which indicated that IRX3 and IRX5 levels recapitulate the effect that the FTO genetic variant has on thermogenesis.
To examine the organism-level effects of the repression of Irx3 in adipose tissue, we used adipose Irx3 dominant-negative (aP2-Irx3DN) mice. These mice had pronounced antiobesity characteristics, including reduced body size, body weight, fat mass, white and brown fat depots, and adipocyte size (Fig. S4A through S4G in the Supplementary Appendix). These aP2-Irx-3DN mice also had resistance to weight gain on a high-fat diet, increased energy expenditure both at night and during the day, and increased oxygen consumption both at room temperature (22°C) and in thermoneutral conditions (30°C), but they did not have significant differences from control mice in food intake or locomotor activity (Fig. S4A and S4H through S4L in the Supplementary Appendix). At the molecular and cellular levels, these mice had increased mitochondrial activity and thermogenesis marker expression, reduced lipid-storage marker expression in both white and brown fat compartments, and markedly smaller adipocytes than did control mice (Fig. S4M, S4N, and S4O in the Supplementary Appendix).
We next evaluated the tissue-autonomous versus brain-mediated roles of Irx3 by comparing the aP2-Irx3DN mice with hypothalamus dominant-negative Ins2-Irx3DN mice.19 The aP2-Irx3DN mice had a reduction in fat-mass ratio that was 3 times as great as that in Ins2-Irx3DN mice (a reduction of 57% vs. 19%), despite the fact that transgene expression in the hypothalamus was 3 times lower than that in Ins2-Irx3DN mice (Fig. S4P and S4Q in the Supplementary Appendix), which indicated that Irx3 has a hypothalamus-independent regulatory role in whole-body energy regulation. The phenotypic effects of Irx3 repression in aP2-Irx3DN mice were also stronger than those in whole-body Irx3 knockout mice, which suggested potential dominant repressor effects in adipocytes or other tissues, and were independent of Fto gene expression, which did not change (Fig. S4P and S4R in the Supplementary Appendix).
Our findings indicate that both Irx3 and Irx5 have cell-autonomous roles: manipulation of Irx3 and Irx5 led to energy-balance differences in three mouse cellular models, including mouse embryonic fibroblast–derived adipocytes, white 3T3-L1 preadipocytes, and β-adrenergic–stimulated beige ME3 preadipocytes (Fig. S5 in the Supplementary Appendix). In each case, our results indicated that Irx3 and Irx5 induced adipocyte lipid accumulation and repressed thermogenesis in a cell-autonomous way.
DETERMINATION OF THE CAUSAL VARIANT AND DISRUPTION OF REPRESSION BY ARID5B
To predict the causal variant, the disruption of which is necessary and sufficient to cause IRX3 and IRX5 dysregulation in human preadipocytes, we used phylogenetic module complexity analysis (PMCA)25 (Fig. 4A and 4B, and Fig. S6A and S6B in the Supplementary Appendix). The highest PMCA score was found for the rs1421085 T-to-C SNV, which is in perfect linkage disequilibrium with the most significant reported SNV, rs1558902, across multiple populations (1000 Genomes Phase 1 data), a finding that is consistent with a potentially causal role.
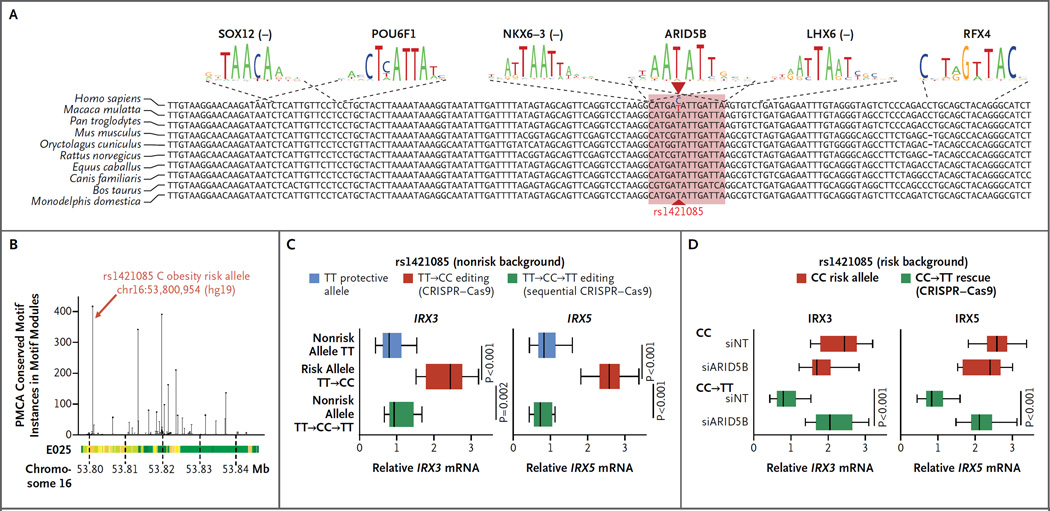
Panel A shows disruption of an ARID5B repressor motif in the evolutionarily conserved motif module surrounding rs1421085. The sequences shown at the top of the panel indicate the frequencies of each nucleotide, with the size scaled to indicate the information content (measured as entropy) at each position. Panel B shows adapted phylogenetic module complexity analysis (PMCA)25 scores in the FTO region for all 82 noncoding SNPs in LD (r2≥0.8) with tag SNV rs1558902, which was identified in a genomewide association study26; rs1421085 had the maximal score. Chromatin state annotation is shown for Roadmap Epigenomics reference genome E025, which corresponds to adipose-derived mesenchymal stem cells; for information on the colors used to denote chromatin states, see Figure S1A in the Supplementary Appendix. Panel C shows increased endogenous expression of IRX3 and IRX5 on single-nucleotide T-to-C editing of rs1421085 in the nonrisk haplotype of a nonrisk-allele carrier, using CRISPR–Cas9 (five clonal expansions). CRISPR–Cas9 reediting from the engineered C risk allele back to a T nonrisk allele with the use of an alternative single guide RNA restores low endogenous IRX3 and IRX5 gene expression. Panel D shows reduced expression of IRX3 and IRX5 on C-to-T editing of the risk allele in adipocyte progenitors from a risk-allele carrier. Knockdown of ARID5B increases IRX3 and IRX5 levels, as compared with the nontargeting control (siNT), only in the rescued allele, but not in the risk allele.
To evaluate whether rs1421085 plays a causal role in enhancer activity, we introduced the C allele into the nonrisk haplotype in our luciferase reporter assay. The T-to-C single-nucleotide alteration increased enhancer activity levels for 10-kb and 1-kb segments centered on the variant, in both orientations and both upstream and downstream of the transcription start, which indicated a gain of enhancer activity in association with the rs1421085 risk allele (Fig. S6C and S6D in the Supplementary Appendix).
To evaluate the effect of the variant on regulator binding, we used electrophoretic mobility-shift assays (EMSAs) of adipocyte nuclear extract with probes for the risk allele and the nonrisk allele of rs1421085. We found binding for the nonrisk allele, T, which lacked enhancer activity, but no binding for the risk allele, C; this indicated that the increased enhancer activity associated with the risk allele is probably due to a loss of repressor binding rather than to a gain of activator binding (Fig. S6E in the Supplementary Appendix).
We examined disrupted motifs and regulator expression to identify potential upstream regulators. The T-to-C substitution disrupted conserved motifs for NKX6-3, LHX6, and the ARID family of regulators (Fig. 4A). Among them, ARID5B had the highest expression in adipose tissue and adipocytes and was bound specifically to the nonrisk allele in EMSA competition experiments (Fig. S6E and S6F in the Supplementary Appendix). ARID5B is known to play both repressive and activating roles and was previously implicated in adipogenesis and lipid metabolism in mice.27,28. Among nonrisk-allele carriers, expression of ARID5B was negatively correlated with expression of IRX3 and IRX5, a finding consistent with ARID5B having a repressive role. No correlation was found in risk-allele carriers, which indicates a loss of ARID5B regulation (Fig. S6G in the Supplementary Appendix).
To evaluate the causal role of ARID5B, we next examined the effects of its knockdown and overexpression on IRX3 and IRX5. ARID5B knockdown increased IRX3 and IRX5 expression in primary preadipocytes from nonrisk-allele carriers to risk-allele levels, which indicates a loss of repression, but it had no effect on preadipocytes from risk-allele carriers, which indicates epistasis with the obesity-risk haplotype (Fig. S6H in the Supplementary Appendix). Consistent with this finding, in SGBS enhancer assays, ARID5B knockdown increased the activity of preadipocytes with the nonrisk allele to risk-allele levels, which indicates a loss of repression, but had no effect on risk-allele constructs, indicating epistasis with the rs1421085 risk allele (Fig. S6I in the Supplementary Appendix). ARID5B overexpression further reduced IRX3 and IRX5 levels in nonrisk-allele carriers, which indicated that repression was strengthened, but had no significant effect on risk-allele carriers, a finding consistent with impaired ARID5B repression in association with the risk haplotype (Fig. S6J in the Supplementary Appendix).
We also evaluated the cellular effects of ARID5B-directed perturbations in primary preadipocytes from risk-allele carriers and nonrisk-allele carriers. In preadipocytes from nonrisk-allele carriers, ARID5B knockdown reduced basal oxygen consumption and lipolysis (Fig. S6K and S6L in the Supplementary Appendix) and shifted expression patterns from mitochondrial to lipid markers (Fig. S2E in the Supplementary Appendix), which indicated that ARID5B plays causal roles in energy-balance regulation. In contrast, ARID5B knockdown had no effect on preadipocytes from risk-allele carriers, a finding consistent with a loss of ARID5B control.
These results suggest that the FTO obesity variant acts through disruption of ARID5B binding in the risk haplotype, leading to a loss of repression, a gain of enhancer activity, and increases in IRX3 and IRX5 expression (Fig. S6M in the Supplementary Appendix).
C-TO-T EDITING OF THE RS1421085 RISK VARIANT AND THE EFFECT ON THERMOGENESIS
Targeted genome editing technology involving CRISPR–Cas929 makes it possible to test the phenotypic effect of altering the predicted causal nucleotide rs1421085 in its endogenous genomic context, in isolation from the other obesity-associated genetic variants in the same haplotype. We used CRISPR–Cas9 in primary preadipocytes with two separate guide RNAs, one for rs1421085 C-to-T rescue of the ARID5B motif disruption in risk-allele carriers and one for rs1421085 T-to-C disruption of the ARID5B motif in nonrisk-allele carriers.
We first evaluated the effect of rs1421085 editing on IRX3 and IRX5 expression levels. Starting from preadipocytes of a nonrisk-allele carrier, T-to-C editing doubled endogenous IRX3 and IRX5 expression, to levels seen in risk-allele carriers; starting from the edited preadipocytes, C-to-T re-editing back to the nonrisk allele restored low expression levels (Fig. 4C). Starting from the risk haplotype, C-to-T editing reduced IRX3 and IRX5 to nonrisk-allele levels, but only in the presence of ARID5B (Fig. 4D); this established that disruption of ARID5B repression by rs1421085 is the mechanistic basis of the IRX3 and IRX5 dysregulatory event that mediates the effects of the FTO locus on obesity.
Next, we evaluated the role of rs1421085 editing during differentiation of white and beige adipocytes, by studying differences in expression between edited and unedited preadipocytes during differentiation. Unedited adipocytes from a risk-allele carrier had a peak in IRX3 and IRX5 expression during days 0 and 2 of preadipocyte differentiation into adipocytes; expression during early differentiation was reduced to nonrisk-allele levels by rs1421085 editing, which indicated a causal role of rs1421085 in developmental gene expression programs (Fig. 5A). The causal role of rs1421085 was further reflected in a significant increase in the expression of thermogenesis regulators (ADRB3, DIO2, PGC1A, and UCP1) and mitochondrial markers (NDUFA10, COX7A, and CPT1) in differentiating preadipocytes (Fig. 5B), which indicated that C-to-T editing of the risk allele rescued thermogenesis regulatory programs.
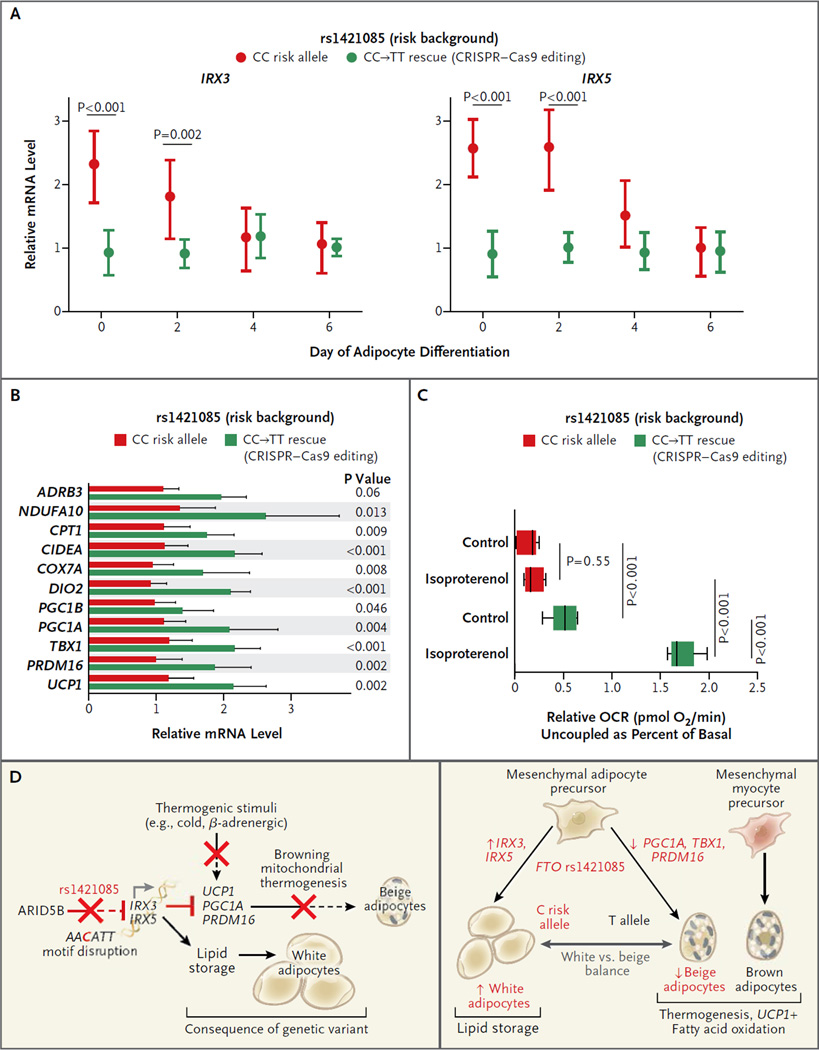
Panel A shows increased mean expression of IRX3 and IRX5 during early adipocyte differentiation specifically for the risk allele; increased expression levels are rescued by C-to-T genome editing. I bars indicate standard deviations. Panel B shows increased expression of thermogenic and mitochondrial genes on C-to-T endogenous single-nucleotide editing of rs1421085 in adipocyte progenitors from a patient with the risk allele. Panel C shows increased basal and isoproterenol-stimulated OCR on C-to-T single-nucleotide endogenous rescue of rs1421085 in adipocytes from a risk-allele carrier. Panel D shows a summary of our mechanistic model of the FTO locus association with obesity, implicating a developmental shift favoring lipid-storing white adipocytes over energy-burning beige adipocytes. At its core lies a single-nucleotide T-to-C variant, rs1421085, which disrupts a conserved ARID5B repressor motif and activates a mesenchymal superenhancer and its targets (IRX3 and IRX5), leading to reduced heat dissipation by mitochondrial thermogenesis (a process that is regulated by UCP1, PGC1α, and PRDM16) and to increased lipid storage in white adipocytes.
Last, we evaluated the role of rs1421085 editing in cellular signatures of obesity by quantifying phenotypic differences between edited and unedited adipocytes. A causal role in the regulation of energy balance was indicated by the fact that C-to-T rescue of rs1421085 in edited adipocytes resulted in a reduction in gene expression for lipid storage and lipolytic markers (Fig. S2E and S8A in the Supplementary Appendix), an increase by a factor of 4 in basal metabolic rate and β-adrenergic oxygen consumption, and an increase by a factor of 7 in thermogenesis (Fig. 5C, and Fig. S7B in the Supplementary Appendix).
In particular, rescue of the ARID5B motif in C-to-T edited preadipocytes restored the strong dependence of mitochondrial respiration on ARID5B that is seen in nonrisk-allele carriers (Fig. S7C in the Supplementary Appendix). These results indicate that the rs1421085 T-to-C single-nucleotide alteration underlies the association between FTO and obesity by disrupting ARID5B-mediated repression of IRX3 and IRX5. This disruption leads to a developmental shift from browning to whitening programs and loss of mitochondrial thermogenesis (Fig. 5D).
DISCUSSION
Our work elucidates a potential mechanistic basis for the genetic association between FTO and obesity and indicates that the causal variant rs1421085 can disrupt ARID5B repressor binding; this disruption results in derepression of IRX3 and IRX5 during early adipocyte differentiation. This process could lead to a cell-autonomous shift from white adipocyte browning and thermogenesis to lipid storage, increased fat stores, and body-weight gain.
To translate the results of genomewide association studies into mechanistic insights, we combined public resources (epigenomic annotations, chromosome conformation, and regulatory motif conservation), targeted experiments for risk and nonrisk haplotypes (enhancer tiling, gene expression, and cellular profiling), and directed perturbations in human primary cells and mouse models (regulator–target knockdown and overexpression and CRISPR–Cas9 genome editing). These methods are specific to the elucidation of noncoding variants, which constitute the majority of signals in genomewide association studies; 80% of the trait-associated loci identified in such studies lack protein-altering variants, and 93% of the top hits are noncoding.30
The FTO association with obesity is unusual in many ways. First, rs1421085 has both a high frequency and a strong effect size,31 which suggests positive selection or bottlenecks (e.g., 44% frequency in European populations vs. 5% in African populations). Second, rs1421085 has switchlike behavior in enhancer activity, target-gene expression, and cellular phenotypes, possibly because of selective pressures on energy-balance control for rapid adaptation. Third, rs1421085 acts specifically in the early differentiation of preadipocytes, which emphasizes the importance of profiling diverse tissues, cell types, and developmental stages. Fourth, enhancer activity is found only for the risk allele, which emphasizes the importance of profiling both alleles. Finally, rs1421085 leads to a gain of function (increased enhancer, IRX3, and IRX5 activity); this is a rare property in protein-coding variants but may be common in noncoding variants.
The apparent genetic link between obesity and cell-autonomous adipocyte browning suggests a central role of beige adipocyte thermogenesis in whole-body energy metabolism in humans, a role that is consistent with that suggested in recent reports on PRDM16 in mice.9 IRX3 and IRX5 have evolutionarily conserved roles, and the ARID5B motif lies in a module that is functionally conserved across multiple mammalian species; this indicates that adaptive thermogenesis circuits are conserved, and IRX3 and IRX5 probably play both UCP1-dependent and UCP1-independent roles. Even though IRX3 and IRX5 dysregulation by rs1421085 was restricted to early differentiation, their effects persisted in mature adipocytes, and the targeting of these genes can have broader effects.
Last, we found that direct manipulation of the ARID5B–rs1421085–IRX3/IRX5 regulatory axis in primary cell cultures of adipocytes from patients reversed the signatures of obesity. This indicates that in addition to changes in physical activity and nutrition, manipulation of mitochondrial thermogenesis26 offers a potential third pathway for shifting between energy storage and expenditure in a brain-independent and tissue-autonomous way in humans.
In summary, our work elucidates a mechanistic basis for the strongest genetic association with obesity. Our results indicate that the SNV rs1421085 underlies the genetic association between the FTO locus and obesity. The SNV disrupts an evolutionarily conserved motif for the ARID5B repressor, which leads to loss of binding, derepression of a potent preadipocyte superenhancer, and activation of downstream targets IRX3 and IRX5 during early differentiation of mesenchymal progenitors into adipocyte subtypes. This results in a cell-autonomous shift from white adipocyte browning to lipid-storage gene expression programs and to repression of basal mitochondrial respiration, a decrease in thermogenesis in response to stimulus, and an increase in adipocyte size. Manipulation of the uncovered pathway, including knockdown or overexpression of the upstream regulator ARID5B, genome editing of the predicted causal variant rs1421085, and knockdown or overexpression of target genes IRX3 and IRX5, had a significant effect on obesity phenotypes.
Acknowledgments
Supported by grants from the Clinical Cooperation Group “Nutrigenomics and Type 2 Diabetes” of the Helmholtz Zentrum München, German Research Center for Environmental Health, and Technische Universität München, Freising-Weihenstephan (to Dr. Claussnitzer); fellowships from the Heart and Stroke Foundation of Canada and the Hospital for Sick Children (to Dr. Kim); grants from the Canadian Institute of Health Research (to Drs. Hui and Drucker); and grants from Else Kröner-Fresenius Foundation, Virtual Institute “Molecular Basis of Glucose Regulation and Type 2 Diabetes” received from the Helmholtz Zentrum München, Western Norway Regional Health Authority, the Swedish federal government under the LUA/ALF agreement, and the National Institutes of Health (R01HG004037, R01GM113708, R01HG008155, and RC1HG005334).
Appendix
The authors’ affiliations are as follows: Beth Israel Deaconess Medical Center and Hebrew SeniorLife, Gerontology Division, Harvard Medical School, Boston (M.C., Y.-H.H.); Massachusetts Institute of Technology (MIT) Computer Science and Artificial Intelligence Laboratory (M.C., G.Q., W.M., N.A.A., M.K.), and Broad Institute of MIT and Harvard, Cambridge (M.C., G.Q., W.M., M.K.); Clinical Cooperation Group “Nutrigenomics and Type 2 Diabetes,” Helmholtz Center Munich (M.C., H.H.), and Else Kröner-Fresenius Center for Nutritional Medicine, Klinikum rechts der Isar, ZIEL–Institute for Food and Health, Technische Universität München (M.C., V.G., I.S.S., H.H.), Munich, Germany; KG Jebsen Center for Diabetes Research, Department of Clinical Science, University of Bergen, and Hormone Laboratory, Haukeland University Hospital, Bergen, Norway (S.N.D., C.H., G.M.); Program in Developmental and Stem Cell Biology, Hospital for Sick Children, and Department of Molecular Genetics, University of Toronto (K.-H.K., V.P., J.L., C.-C.H.), and Department of Medicine, Lunenfeld-Tanenbaum Research Institute, Mt. Sinai Hospital (J.L.B., D.J.D.), Toronto; and the Department of Molecular and Clinical Medicine, Institute of Medicine, University of Gothenburg, Gothenburg, Sweden (P.-A.S.).
Footnotes
Disclosure forms provided by the authors are available with the full text of this article at NEJM.org.
REFERENCES
Full text links
Read article at publisher's site: https://doi.org/10.1056/nejmoa1502214
Read article for free, from open access legal sources, via Unpaywall:
https://www.nejm.org/doi/pdf/10.1056/NEJMoa1502214?articleTools=true
Citations & impact
Impact metrics
Citations of article over time
Alternative metrics
Smart citations by scite.ai
Explore citation contexts and check if this article has been
supported or disputed.
https://scite.ai/reports/10.1056/nejmoa1502214
Article citations
A guide to studying 3D genome structure and dynamics in the kidney.
Nat Rev Nephrol, 15 Oct 2024
Cited by: 0 articles | PMID: 39406927
Review
Depot-specific mRNA expression programs in human adipocytes suggest physiological specialization via distinct developmental programs.
PLoS One, 19(10):e0311751, 14 Oct 2024
Cited by: 0 articles | PMID: 39401200 | PMCID: PMC11472956
Translation of genome-wide association study: from genomic signals to biological insights.
Front Genet, 15:1375481, 03 Oct 2024
Cited by: 0 articles | PMID: 39421299 | PMCID: PMC11484060
Review Free full text in Europe PMC
Melanoma-specific mutation hotspots in distal, non-coding, promoter-interacting regions implicate novel candidate driver genes.
Br J Cancer, 131(10):1644-1655, 04 Oct 2024
Cited by: 0 articles | PMID: 39367275 | PMCID: PMC11555344
Dealkylation of Macromolecules by Eukaryotic α-Ketoglutarate-Dependent Dioxygenases from the AlkB-like Family.
Curr Issues Mol Biol, 46(9):10462-10491, 20 Sep 2024
Cited by: 0 articles | PMID: 39329974 | PMCID: PMC11431407
Review Free full text in Europe PMC
Go to all (736) article citations
Other citations
Data
Data behind the article
This data has been text mined from the article, or deposited into data resources.
BioStudies: supplemental material and supporting data
SNPs (3)
- (14 citations) dbSNP - rs1421085
- (3 citations) dbSNP - rs1558902
- (2 citations) dbSNP - rs9930506
Similar Articles
To arrive at the top five similar articles we use a word-weighted algorithm to compare words from the Title and Abstract of each citation.
FTO Obesity Risk Variants Are Linked to Adipocyte IRX3 Expression and BMI of Children - Relevance of FTO Variants to Defend Body Weight in Lean Children?
PLoS One, 11(8):e0161739, 25 Aug 2016
Cited by: 16 articles | PMID: 27560134 | PMCID: PMC4999231
FTO Intronic SNP Strongly Influences Human Neck Adipocyte Browning Determined by Tissue and PPARγ Specific Regulation: A Transcriptome Analysis.
Cells, 9(4):E987, 16 Apr 2020
Cited by: 15 articles | PMID: 32316277 | PMCID: PMC7227023
Human abdominal subcutaneous-derived active beige adipocytes carrying FTO rs1421085 obesity-risk alleles exert lower thermogenic capacity.
Front Cell Dev Biol, 11:1155673, 14 Jun 2023
Cited by: 2 articles | PMID: 37416800 | PMCID: PMC10321670
Understanding the Biology of Thermogenic Fat: Is Browning A New Approach to the Treatment of Obesity?
Arch Med Res, 48(5):401-413, 01 Jul 2017
Cited by: 49 articles | PMID: 29102386
Review
Funding
Funders who supported this work.
CIHR
NHGRI NIH HHS (6)
Grant ID: R01 HG008155
Grant ID: RC1HG005334
Grant ID: RC1 HG005334
Grant ID: R01 HG004037
Grant ID: R01HG004037
Grant ID: R01HG008155
NIGMS NIH HHS (2)
Grant ID: R01GM113708
Grant ID: R01 GM113708





