Abstract
Free full text

Loss of BAP1 function leads to EZH2-dependent transformation
Introductory Paragraph
BAP1 and ASXL1 interact to form a polycomb deubiquitinase complex that removes monoubiquitin from histone H2A lysine 119 (H2AK119Ub). However, BAP1 and ASXL1 are mutated in distinct cancer types, consistent with independent roles in regulating epigenetic state and malignant transformation. Here we demonstrate that Bap1 loss results in increased trimethylated histone H3 lysine 27 (H3K27me3), elevated Ezh2 expression, and enhanced repression of Polycomb Repressive Complex 2 (PRC2) targets. These findings contrast with the reduction in H3K27me3 seen with Asxl1 loss. Conditional deletion of Bap1 and Ezh2 in vivo abrogates the myeloid progenitor expansion induced by Bap1 loss alone. Loss of Bap1 results in a marked decrease in H4K20 monomethylation (H4K20me1). Consistent with a role for H4K20me1 in EZH2 transcriptional regulation, expression of SETD8, the H4K20me1 methyltransferase, reduces EZH2 expression and abrogates the proliferation of BAP1-mutant cells. Further, mesothelioma cells that lack BAP1 are sensitive to EZH2 pharmacologic inhibition, suggesting a novel therapeutic approach for BAP1-mutant malignancies.
Genomic studies have identified somatic mutations in the tumor suppressors ASXL1 and BAP1 in different malignancies. Inactivating mutations in ASXL1 are most common in myeloid malignancies1-3, whereas recurrent BAP1 mutations are commonly observed in mesothelioma4, renal cell carcinoma5, and metastatic uveal melanoma6 suggesting BAP1 and ASXL1 have distinct roles in tumor suppression. These mutational profiles cannot be explained by differential tissue-specific BAP1 and ASXL1 expression (Supplementary Fig. 1a–c). The Drosophila ASXL1 homolog Asx and the BAP1 homolog Calypso form a complex which removes H2AK119Ub7. However, the BAP1-ASXL1 complex has not been shown to have a role in BAP1-mutant transformation. We sought to identify mechanisms by which BAP1 loss leads to transformation, independent of ASXL1, and to identify therapeutic vulnerabilities in BAP1-mutant cancer cells.
Recent studies have shown that somatic loss of Bap1 promotes hematopoietic transformation8. We investigated the impact of conditional Bap1 deletion on gene expression and chromatin state in hematopoietic cells (Supplementary Fig. 1d). Bap1 loss led to a fully penetrant myeloproliferative disease with splenomegaly, leukocytosis, anemia and progenitor expansion (Fig. 1a and Supplementary Fig. 1e–k). We observed increased proliferation and cell cycle progression in Bap1-deficient myeloid progenitors (Supplementary Fig. 1l). RNA sequencing analysis revealed the majority of differentially expressed genes in Bap1-deficient progenitors had reduced expression (P-adj < 0.001) (Supplementary Fig. 2a). Although we observed significant overlap between differentially expressed genes in Bap1 and Asxl1 KO progenitors9, in many cases we observed a paradoxical inverse effect on gene expression (n= 714 genes overlap, 247 inversely correlated, total n= 4246 Bap1 KO, n= 1761 Asxl1 KO) (Fig. 1b and Supplementary Fig. 2b). ASXL1 silencing leads to increased expression of HoxA cluster genes consistent with reduced PRC2 activity10. However, we observed reduced expression of HoxA gene members (Fig. 1c) and decreased expression of HoxA gene signatures in Bap1-deficient cells (Supplementary Fig. 2c). These data demonstrate that loss of Asxl1 and Bap1 have opposite effects on gene regulation.
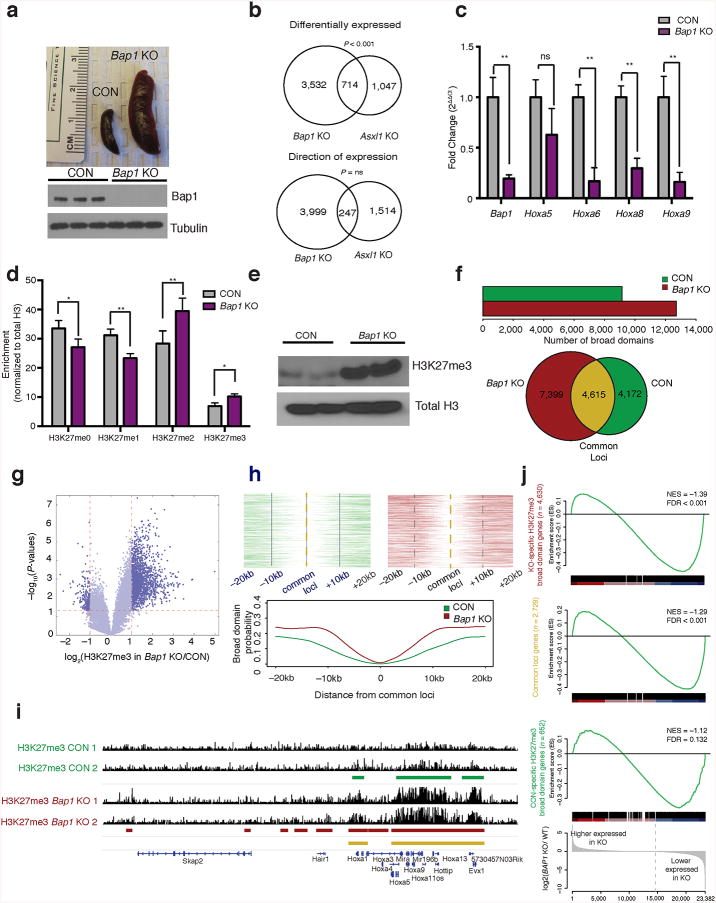
Bap1 deletion results in increased H3K27me3. (a) Spleen images three weeks after conditional deletion of Bap1 and verification of Bap1 deletion by Western blot of control (littermate Bap1f/f mice, CON) and Bap1 knockout (Mx1-Cre Bap1f/f mice, Bap1 KO) bone marrow. (b) Venn diagrams comparing myeloid progenitor gene expression in Bap1 and Asxl1 KO mice, P < 0.05; comparisons include gene overlap and genes changing in the same direction. (c)Quantitative real time PCR (qRT-PCR) of the HoxA cluster in sorted granulocyte-macrophage progenitors (GMPs; Lin–c-Kit+Sca1–CD34+ Fcγ+); from Bap1 KO and control mice (n = 3). (d) Mass spectrometric analysis of purified histones from c-Kit+ enriched bone marrow cells from Bap1 KO and controls normalized to total histone H3. (e) Western blot of H3K27me3 and total H3 in purified histones from Bap1 KO and control bone marrow. (f) Number of H3K27me3 broad domains that are called in CON and Bap1 KO samples. Venn diagram showing unique and overlapping broad domain (n= 2). (g) Peak calls from H3K27me3 ChIP-Seq in sorted GMP cells represented by a volcano plot displayed by ratio (KO/CON) vs. P-value. (h) Broad domain density as a function of distance from an H3K27me3 domain that was called in both CON and Bap1 KO samples. (i) H3K27me3 ChIP-seq at the HOXA locus. (j) GSEA demonstrating gene expression correlations to downregulated genes. Statistics were calculated with Student's t-test; *P < 0.05, **P < 0.005; ± s.d. values reported.
ASXL1 directly interacts with the PRC2 complex and ASXL1 depletion reduces global and site-specific H3K27me310. Given the divergent effects of Asxl1 and Bap1 loss on gene expression, we investigated the impact of Bap1 deletion on H3K27me3. H3K27me3 levels were increased in Bap1 KO cells by mass spectrometry, Western blot, and ELISA (Fig. 1d,e and Supplementary Fig. 3a). Chromatin immunoprecipitation sequencing (ChIP-Seq) revealed a global increase in H3K27me3 broad domains11 and H3K27me3 “spreading” into nearby loci in Bap1 KO mice, as illustrated within the HoxA locus (Fig. 1f–i and Supplementary Fig. 3b). The sites marked with H3K27me3 in Bap1 KO cells primarily occurred in gene promoter regions (Supplementary Fig. 3c) that were enriched for enhanced repression (FDR < 0.001) (Fig. 1j). Genes dysregulated by Bap1 KO-associated H3K27me3 and gene repression were implicated inEZH2-dependent regulation, lineage commitment/differentiation and proliferation (Supplementary Fig. 3d,e). BAP1 silencing increased H3K27me3 and re-expression of BAP1 in Bap1-deficient cells, but not a deubiquitinase-deficient BAP1 allele, reduced H3K27me3 levels (Supplementary Fig. 4a–c).
We next assessed the role of PRC2-mediated H3K27me3 on BAP1-dependent transformation by investigating the impact of Ezh2 loss12 on transformation in vivo. Ezh2 deletion reduced H3K27me3 levels in Bap1/Ezh2-deficient mice compared to Bap1-knockout mice (Fig. 2a). Ezh2 deletion abrogated the myeloid malignancy induced by Bap1 loss, with reduced splenomegaly, leukocytosis and anemia (Fig. 2b–e, Supplementary Fig. 5a). Concomitant Bap1/Ezh2 loss reduced myeloid progenitor expansion, reduced the proportion of Mac1+Gr1+ myeloid cells, restored erythroid differentiation (CD71+Ter119+) and decreased proliferation of Bap1/Ezh2-deficient progenitors (Fig. 2f–g and Supplementary 5b,c). Consistent with the genetic data, treatment of Bap1 KO mice with the small molecule EZH2 inhibitor EPZ01198913decreased H3K27me3, splenomegaly, and white blood cell counts (Supplementary Fig. 5d and Fig. 2h,i). Ezh2 haploinsufficiency reduced, but did not abrogate, Bap1-deficient myeloproliferation (Supplementary Fig. 5e,f) consistent with a dose-dependent requirement for Ezh2. These data demonstrate that PRC2, and specifically Ezh2 activity, is required for Bap1-deficient myeloid transformation.
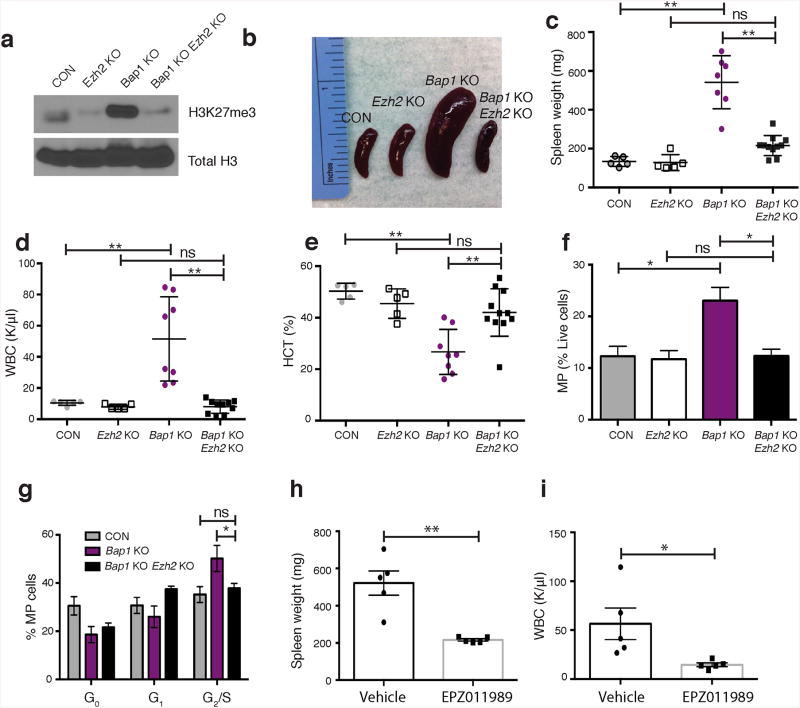
(a) Western blot of H3K27me3 levels in histones purified from bone marrow of Bap1 KO, Ezh2 KO, Bap1/Ezh2 KO and control mice. (b) Representative images of spleens and (c) enumeration of spleen weight from the indicated genotypes of mice, 3 weeks post pIpC. (d) Peripheral white blood cell counts and (e) hematocrit percentages as quantified by a Hemavet. (f) Flow cytometric enumeration of myeloid progenitors (Lin–c-Kit+Sca1–) (n= 3/group). (g) Cell cycle analyses in myeloid progenitors using Ki67 and DAPI stain (n = 3/group). (h) Spleen weights and (i)WBC counts of mice (n = 5/group) treated with EPZ011989 twice a day at 500 mg/kg for 16 days. Unless otherwise indicated, n = 5/CON, n = 5/Ezh2 KO, n = 8/Bap1 KO, and n = 11/ Bap1/Ezh2 KO. Statistics were calculated with Student's t-test; *P < 0.05, ** P< 0.005; ± s.d. values reported.
We next sought to delineate the mechanism by which Bap1 deletion increased H3K27me3 levels. We did not identify an interaction between BAP1 and EZH2 by co-immunoprecipitation (Fig. 3a). We observed increased mRNA and protein expression of Ezh2 and Suz12 (Fig. 3b and Supplementary Fig. 6a) consistent with a role for Bap1 in regulating PRC2 expression. Re-expression of BAP1 in Bap1 KO bone marrow cells reduced Ezh2 mRNA expression to normal levels (Supplementary Fig. 6b). We hypothesized that Bap1 loss might alter other histone marks, which would then alter chromatin state at key target loci, including EZH2. Mass spectrometry revealed a marked decrease in H4K20me1 in Bap1 KO cells (Fig. 3c) compared to other measured histone marks (Supplementary Fig. 6c). Expression of BAP1, but not ASXL1, increased H4K20me1 at the EZH2 locus (Supplementary Fig. 6d). We therefore hypothesized that loss of the H4K20me1 mark may have a role in BAP1-dependent gene expression. SETD8 is the only known methyltransferase that places H4K20me114. Expression of SETD8 in BAP1-mutant mesothelioma cells (H226, H2452) increased apoptosis and reduced proliferation, whereas wild-type (MSTO-211H and Meso10) cells were unaffected (Fig. 3d,e). SETD8 overexpression in mesothelioma cells decreased EZH2 mRNA and protein expression (Fig. 3f,g). Wild-type cell lines were more sensitive to the SETD8 inhibitor BVT59415 than BAP1-mutant cell lines (Supplementary Fig. 7a).
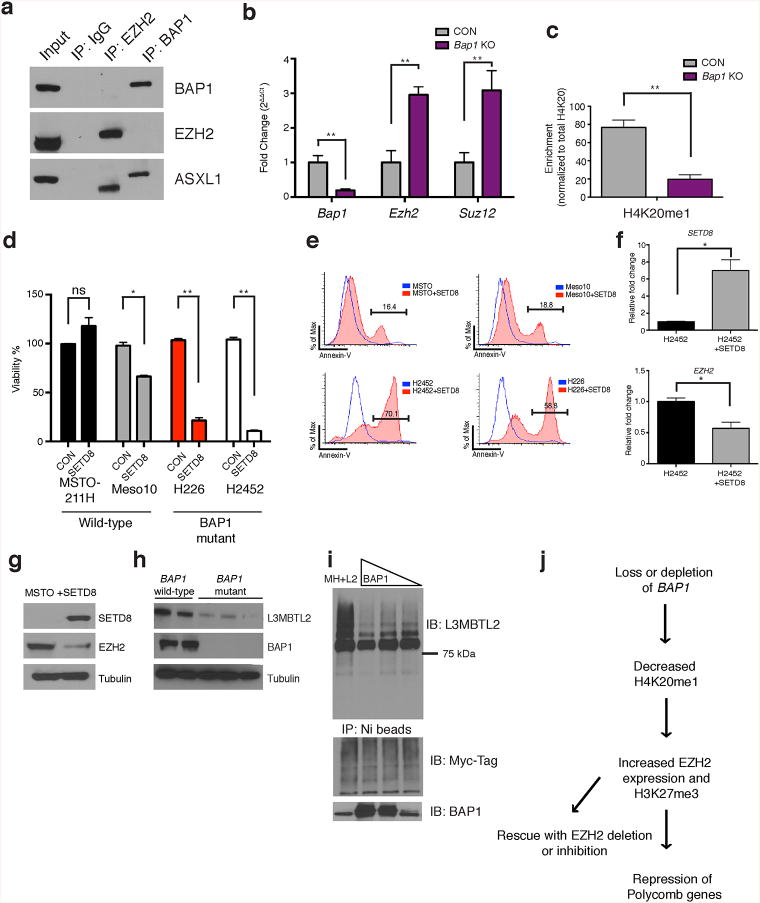
(a) Co-immunoprecipitation of endogenous EZH2 and BAP1 in SET2 cells followed by Western blot analysis (performed in presence of benzonase to inhibit interactions dependent on DNA). (b) Bap1,Ezh2 and Suz12 expression byqRT-PCR from sorted granulocyte-macrophage progenitor (GMP; Lin–c-Kit+Sca1–CD34+Fcγ+). (c) H4K20me1 quantification from histone mass spectrometry experiments. (d) Cell viability as assessed by Cell Titer Glo viability assay and (e) Annexin V assays for SETD8 overexpression experiments in BAP1 wild-type (MSTO-211H, Meso10) and ‘mutant’ cell lines (H226, homozygous deletion; H2452 catalytic mutation). (f) Quantitative qPCR for SETD8 and EZH2 in BAP1 mutant cells with SETD8 overexpression. (g) Western blot analysis for SETD8 and EZH2 in a BAP1 wild-type cell line. (h) L3MBTL2 and BAP1 expression in BAP1 wild-type (Met5a, JMN) and mutant mesothelioma cell lines (H226, H2452, H28). (i) 293T cells overexpressing Myc-His tagged ubiquitin and L3MBTL2 cDNA and varying levels of BAP1 (0, 5 μg, 2.5 μg, 1 μg). Co-immunoprecipitation experiments were conducted with Ni-beads and a series of stringent washes. (j) Model depicting the regulation of BAP1 leading to effects on chromatin and gene expression. Statistics were calculated with Student's t-test; *P < 0.05, **P < 0.005; ± s.d. values reported.
We hypothesized that BAP1 deubiquitinates a chromatin modulator that regulates H4K20me1. Analysis of ChIP-Seq data8,9 identified a cluster of genes with Bap1 occupancy, but not Asxl1 binding, and were enriched for an E-box motif (Supplementary Fig. 7b,c). Previous studies have shown the atypical polycomb proteins L3MBTL1 and L3MBTL2 bind E-box motifs, and maintain H4K20me116-19. L3mbtl1-deficient mice have no overt phenotype20, whereas L3mbtl2-deficient mice are embryonic lethal similar in timing to Bap1 loss8,17. We therefore investigated whether Bap1 loss led to alterations in L3mbtl2 expression. L3mbtl2 protein but not RNA expression was reduced in Bap1 KO hematopoietic cells (Supplementary Fig. 8a,b) and in BAP1-mutant mesothelioma cells compared to BAP1 wild-type mesothelioma cells (Fig. 3h). L3MBTL2 ubiquitination was reduced in cells expressing BAP1 (Fig. 3i) and proteasome inhibitor treatment increased L3MBTL2 stability in BAP1-mutant cells (Supplementary Fig. 8c). L3MBTL2 expression decreased EZH2 protein levels and EZH2 promoter activity (Supplementary Fig. 8d,e). Conversely, L3MBTL2 silencing increased expression of EZH2 (Supplementary Fig. 8f). We observed enrichment for L3MBTL2 and BAP1 at the EZH2 locus (Supplementary Fig. 8g,h). These data suggest that BAP1 and L3MBTL2 interact (Supplementary Fig. 8i) and co-occupy the EZH2 locus. BAP1 loss leads to reduced L3MBTL2 stability and increased EZH2 transcriptional output (Fig. 3j).
Analysis of TCGA data revealed EZH2 mRNA expression was increased in mesothelioma (Fig. 4a). EZH2 silencing induced apoptosis in BAP1-mutant mesothelioma cell lines, whereas wild-type cells continued to proliferate (Fig. 4b,c). EZH2 silencing abrogated in vivo tumor formation of BAP1-mutant but not wild-type cell lines (Fig. 4d). Overexpression of EZH2 in BAP1 wild-type cell lines increased proliferation and sensitivity to EZH2 inhibition (Supplementary Fig. 9a,b). BAP1-mutant cell lines were more sensitive to EZH2 inhibition (EPZ011989) in vitro in 2D and 3D culture (Fig. 4e,f). We next assessed the impact of EZH2 inhibition in vivo. EZH2 inhibition significantly reduced BAP1-mutant tumor size compared to vehicle treated mice, whereas wild-type tumors were less responsive to EZH2 inhibition (Fig. 4g,h). EZH2 inhibition abrogated pulmonary metastasis in a BAP1-mutant mesothelioma cell line with metastatic potential (Fig. 4g) consistent with a role for BAP1/EZH2 in metastasis6. EZH2 inhibition reduced invasion and increased E-Cadherin expression in vitro (Supplementary Fig. 9c,d). These data indicate that EZH2 represents a potential therapeutic target in BAP1-mutant cancer cells.
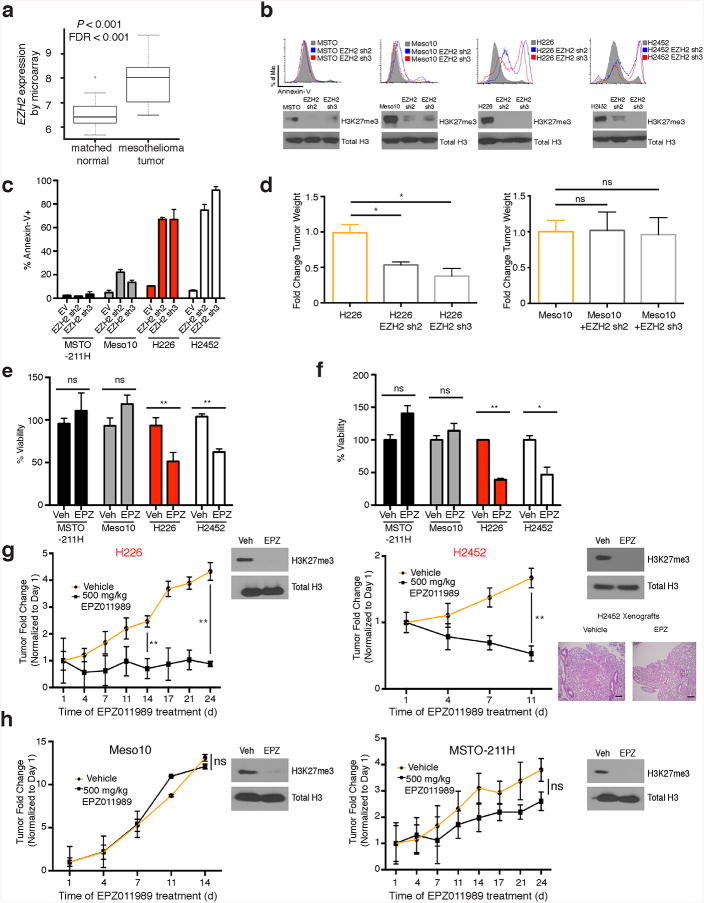
(a) Expression of EZH2 transcripts in TCGA mesothelioma patients compared to matched normals. (b) Annexin V assays in BAP1 wild-type and mutant cell lines expressing either empty vector or hairpins targeting EZH2. (c) Quantitation of Annexin V experiments in mesothelioma cell lines. (d) Tumor size of Meso10 and H226 cell lines expressing EZH2 hairpins implanted into NOD-SCID mice, n = 6/group (± s.e.m.). (e) 2D Cell Titer Glo viability assays after 2 week treatment with EPZ011989 at 1.25 μM. (f) 3D Cell Titer Glo viability assays after 3 weeks EPZ011989 treatment at 1.25 μM. (g) Tumor size formation from BAP1 mutant (MSTO and Meso10) or (h) wild-type cells (H226 and H2452) implanted into NOD-SCID mice and treated with either vehicle or 500 mg/kg BID EPZ011989 (± s.e.m). Tumors were measured 3X weekly, n = 6/group. Target inhibition was assessed by histone western blots in extracted tumors (shown in respective figures). Lung pathology of H2452 cells with vehicle and EPZ011989 treatment, magnification 100X, scale bars 400 μM. Arrow indicates bulk metastasized tumor. Statistics were calculated with Student's t-test; *P < 0.05, **P < 0.005; ± s.d. values reported.
Identification of oncogenic EZH2 mutations21-23 led to development of mutant-specific epigenetic therapies24. However, most mutations in epigenetic regulators result in loss-of-function, such that they do not represent tractable direct therapeutic targets. EZH2 inhibitors have recently entered clinical trials (NCT01897571, NCT01395601, NCT01082977) and our data suggest that BAP1 loss results in a mutation-specific dependency in PRC2 that should be further explored in preclinical and clinical studies. Our data resonate with recent studies suggesting a role for PRC2 inhibition in SWI/SNF-mutant rhabdoid tumors25,26 and analyses showing BAP1 mutations are mutually exclusive with mutations in the SWI/SNF complex27. Our data suggest that studies of mutations in epigenetic regulators inform the development of therapies that reverse mutant-specific effects on epigenetic state.
Online Methods
Animals
All animals were housed at Memorial Sloan Kettering Cancer Center. All animal procedures were completed in accordance with the Guidelines for the Care and Use of Laboratory Animals and were approved by the Institutional Animal Care and Use Committees at Memorial Sloan Kettering Cancer Center. The number of mice chosen in each experiment was chosen to give 90% statistical power with a 5% error level given the differences in standard deviation that was observed in the pilot study.
Generation of Bap1-deficient and Bap1/Ezh2-deficient mice
Embryonic stem cells targeting exons 6–12 of Bap1 were obtained from the European Conditional Mouse Consortium. A Frt-flanked premature stop cassette containing a lacZ and neomycin cassette was inserted upstream28. ES cell clones were expanded and injected into primary blastocysts. Generated mice were crossed to the germline Flp-deleter (The Jackson Laboratory) to excise the Frt-flanked cassette. These mice were subsequently crossed to the IFN-α-inducible Mx1-cre transgenic mice (The Jackson Laboratory) to assess the effects of inducible loss of Bap1 in the hematopoietic system. Bap1 fl/fl, Bap1 fl/+, and Bap1+/+ littermate mice were genotyped by PCR with the primers BAP1-up (actgcagcaatgtggatctg), BAP1-down (gaaaaggtctgacccagatca) using the following parameters: 95°C for 10min, followed by 40 cycles of 94°C for 10s, 65°C for 40s, and 72°C for 1min, and then 72°C for 5min. The WT allele was detected at 300 bp while the floxed allele was detected at 500 bp PCR. Excision after IFN-α-induction was confirmed by a PCR with primers to detect the floxed and excised band: BAP1-F (actgcagcaatgtggatctg), BAP1-F2 (gcgcaacgcaattaatgata), and BAP1-R (cagtgtccagaatggctcaa), using the same PCR parameters listed above.
Mx1-Cre-Bap1 f/f mice were crossed to Ezh2 f/f mice12
Mx-cre Bap1f/f conditional and Bap1f/f control mice received four intraperitoneal injections of polyI:polyC of 200 μL of a 1 mg/mL solution. Two weeks after excision, peripheral blood was collected via cheek bleeding using heparinized microhematocrit capillary tubes (Thermo Fisher Scientific). Excision was confirmed and peripheral blood counts were obtained using a HemaVet according to standard manufacturer's instruction. Formalin-fixed paraffin-embedded tissue sections were stained with hematoxylin and eosin (H&E). Deletion of Bap1 was confirmed by genomic excision PCR and Western blot analysis. Tails were submitted to the Transnetyx genotyping service (Cordova, TN) for qPCR-based genotyping for floxed and excised Ezh2 alleles. Excision was confirmed by Western blot. Animals were not excluded from genetic experiments.
Xenografts and in vivo EPZ011989 administration
Groups of 10 week old NOD-SCID mice were injected subcutaneously in the flank with6–10×106 mesothelioma cell lines (MSTO-211H, Meso10, H226 and H2452) in a 1:1 mixture of matrigel and media. When tumors reached a size of approximately 60–80mm3, we began treatment with either vehicle (0.5% NaCMC+0.1% Tween-80 in water) or EPZ011989. Either EPZ011989 or vehicle were given orally BID at a concentration of 500 mg/kg for the duration of the experiment. Tumor volumes were assessed in three dimensions using a caliper. Tumors or lung tissue were extracted following treatment and utilized for Western blotting to assess target inhibition. We pre-established criteria to exclude mice in xenograft experiments if tumors did not form after implantation (75% smaller than the mean of the implanted animals from the same group. Animals were not excluded from drug trials. For all xenograft drug studies, tumor size was followed for 10 days and mice were randomized at this point for tumor size before the trial. The genetic Bap1 KO EPZ011989 trial was conducted with randomization utilizing CBC analysis 3 weeks after polyI:polyC and confirming that WBC count averages were equivalent in both vehicle and treated groups. Five animals per group were treated orally with either vehicle (described above) or 500 mg/kg EPZ011989 BID for 16 days. Researchers were not blinded in these experiments.
Histological analyses
Mice were sacrificed and autopsied, and then dissected tissue samples were fixed for 24 hours in 4% paraformaldehyde, dehydrated, and embedded in paraffin. Paraffin blocks were sectioned at 4 μm and stained with H&E. Images were acquired using an Axio Observer A1 microscope (Carl Zeiss).
Cell culture
293T cells were cultured in Dulbecco's modified Eagle's medium (DMEM) supplemented with 10% fetal bovine serum (FBS) and nonessential amino acids. Human leukemia cell lines (SET2) and human mesothelioma cell lines (Met5a, MSTO-211H, Meso10, H2373, H226, H2452) were cultured in RPMI-1640 medium supplemented with 10% FBS.
RNA isolation, SMARTer amplification, Proton Transcriptome sequencing and analysis
Bone marrow cells were FACS sorted for GMPs (Lin–c-Kit+Sca1–CD34+Fcγ+) using the FACS Aria. Total RNA from 200–500K cells was extracted using TRIzol RNA Isolation Reagents (cat# 15596-026, Life Technologies). Quality of RNA was ensured before amplification by analyzing 20–50 pg of each sample using the RNA 6000 pico kit and a bioAnalyzer (Agilent). 10 ng of high quality (RIN>8) total RNA was subsequently amplified using the SMARTer® Universal Low Input RNA Kit for Sequencing (Clonetech Laboratory, cat# 634940) according to instructions provided by the manufacturer. Amplified material underwent whole transcriptome Library preparation according to the Ion Total RNA-Seq Kit v2 protocol (Life Technologies), with 16 cycles of PCR. Samples were barcoded and template-positive Ion PI™ Ion Sphere™ Particles (ISPs) were prepared using the ion one touch system II and Ion PI™ Template OT2 200kit v2 Kit (Life Technologies). Enriched particles were sequenced on a Proton sequencing system using 200 bp version 2 chemistry. An average of 70 to 80 million reads per sample were generated and 76 to 82% of the reads mapped to mRNA bases.
RAW output BAMs were converted back to FASTQ using PICARD Sam2Fastq. The reads are first mapped to the mouse genome using rnaStar. The genome used was MM9 with junctions from ENSEMBL (Mus_musculus.NCBIM37.67) and a read overhang of 49. Then any unmapped reads were mapped to MM9 using BWA MEM (version 0.7.5a). The two mapped BAMs were then merged and sorted and gene level counts were computed using htseq-count (options -s y -m intersection-strict) and the same gene models (Mus_musculus.NCBIM37.67). Raw data was uploaded to the GEO database with the following accession number: GSE61360.
Analyses of TCGA data in AML and mesothelioma
Average gene expression from TCGA AML patients was expressed as mathematical mean and standard error of normalized read counts as provided by DESeq229. For TCGA mesothelioma patients, these same values were calculated from normalized read counts as provided by TCGA as level 3 data. To compare EZH2 expression of mesothelioma tumors to normal lung tissue30, GSE51024 was analyzed using Geo2R, which makes use of the Limma Bioconductor R package. Normalized expression values for EZH2 were extracted from this analysis, and used to generate the boxplots provided. Log2 Fold Change, nominal P-value, and Benjamini-Hochberg FDR were calculated by Geo2R/Limma.
Histone extraction, histone ELISAs, histone Western blots, and histone LC/MS
Histones were extracted by standard extraction techniques or overnight using the Active Motif Histone Extraction Minikit (40026). Histone ELISAs were conducted using the trimethyl K27 Elisa Kit (Active Motif, 53106) normalized to a H3K27me3 standard curve and total H3 protein. Histone Western blots were conducted with 3–5 μg of histones. For Histone LC/MS, 12 million control and Bap1 KO cells were lysed, nuclei were isolated and histones were extracted using 0.4N H2SO4 and chemically derivatized using propionic anhydride, as previously described31. Histones were then digested with trypsin and separated by nano-liquid chromatography (75 μm i.d., 15 cm long, packed with MagicC18aq media, dp 3 μ) coupled to a TSQ Quantum Ultra mass spectrometer. Data were analyzed with Skyline32 and relative quantification was performed by peak area.
Chromatin preparation and immunoprecipitation, ChIP Library preparation and sequencing, and analysis of ChIP-Seq data
Bone marrow cells were enriched for c-Kit+ cells (or sorted for GMPs) using the EasySep Mouse Hematopoietic cell Enrichment Kit (Stem Cell Technologies, 19756). 5×106 cells were fixed in a 1% methanol-free formaldehyde solution and then resuspended in SDS lysis buffer. Lysates were sonicated in an E220 focused-ultrasonicator (Covaris) to a desired fragment size distribution of 100–500 base pairs. IP reactions were performed using anti-trimethyl H3K27 (abcam, 6002), anti-monomethyl H4K20 (Abcam, 9051), and IgG (Santa Cruz, 2027) each on approximately 400,000 cells as previously described 33. ChIP assays were processed on an SX-8G IP-STAR Compact Automated System (Diagenode) using a Direct ChIP protocol as described elsewhere 34. Eluted chromatin fragments were then de-crosslinked and the DNA fragments purified using Agencourt AMPure XP beads (Beckman Coulter).
Barcoded libraries were prepared from the ChIP-enriched and input DNA using a NEBNext ChIP-Seq Library Prep Master Mix Set for Illumina (New England Biolabs) and TruSeq Adaptors (Illumina) according to manufacturer's instructions on an SX-8G IP-STAR Compact Automated System (Diagenode). Phusion High-Fidelity DNA Polymerase (New England Biolabs) and TruSeq PCR Primers (Illumina) were used to amplify the libraries, which were then purified to remove adaptor dimers using AMPure XP beads and multiplexed on the HiSeq 2000 (Illumina).
Reads were quality and adapter-trimmed using ‘trim_galore’ before aligning to mouse assembly mm9 with bowtie2 using the default parameters. Aligned reads with the same start position and orientation were collapsed to a single read before subsequent analysis. Density profiles were created by extending each read to the average library fragment size and then computing density using the BEDTools suite. For GMP sorted cells, enriched regions were discovered using MACS 1.4 with default parameters, and scored against matched input libraries. H3K27me3 broad domains were identified as described in Beguelin et al. Briefly, H3K27me3 ChIP-Seq reads were calculated in 1 kb bins genome-wide. Enriched regions were binned consecutively with read counts greater than one standard deviation of the genome-wide mean11. Enriched regions were mapped to specific genomic locations: promoters, introns, exons in addition to regions with specified distances from the transcription start site. All genome browser tracks and read density tables were normalized to sequencing depth. For comparison of ChIP-seq samples in control and KO conditions, the signals of three replicates per condition were tested using either the Mann-Whitney U test or the t-test. Cluster analysis was performed on normalized count data in Matlab with the kmeans clustering package. Motif analysis was performed in Homer using default parameters for the findMotifsGenome program.
Western blot and immunoprecipitation
Cells were lysed for Western blot and immunoprecipitation experiments in the following buffer: 150 mM NaCl, 20 mM Tris (pH 7.4), 5 mM EDTA, 1% Triton, protease arrest (EMD) and phosphatase inhibitors (Calbiochem). To perform immunoprecipitations in the presence of benzonase, cells were lysed in the BC-300 buffer: 20 mM Tris (pH 7.4), 10% glycerol, 300 mM KCl, 0.1% NP-40. The cleared lysate was treated with MgCl2 to 2.5 mM and benzonase was added at 1250 U/mL. The lysate was incubated for 1 hour with rotation and the reaction was terminated by adding 5 mM EDTA. DNA digestion was confirmed by running lysate on an ethidium bromide gel before setting up the immunoprecipitation experiment. Antibodies used included: BAP1 (C-4; Santa Cruz sc-28383; 1:1000), EZH2 (Active Motif, 39933, Active Motif, 39901, or Millipore, 07-689;1:10,000), SUZ12 (Abcam, Ab12073, 1:1000), ASXL1 (N-13; Santa Cruz sc-85283, 1:1000), L3MBTL2 (Active Motif, 39569, 1:1000), Myc-Tag (Cell Signaling, 2276; 1:2000), SETD8 (ab3798, 1:1000), Tubulin (Sigma, T9026, 1:10,000), H3K27me3 (Abcam, 6002 or Millipore, 07-449, 1:1000), H3 (Abcam, Ab1791, 1:10,000), and H4K20me1 (Abcam, Ab9051, 1:1000).
Flow cytometry analyses and antibodies
Surface marker staining of live bone marrow was conducted by first lysing cells with ammonium chloride-potassium bicarbonate lysis buffer and washing cells with phosphate-buffered saline (PBS). Cells were stained with antibodies in phosphate-buffered saline (PBS) for 20 minutes on ice. For hematopoietic stem and progenitor staining, cells were stained with a lineage cocktail including CD4 (RM4-5), CD3 (17A2), B220 (RA3-6B2), NK1.1 (PK136), Gr-1 (RB6-8C5), Cd11b (M1/70), and Ter119, allowing for mature lineage exclusion from the analysis. Cells were also stained with antibodies against c-Kit (2B8), Sca-1 (D7), FcγRII/III (2.4G2), and CD34 (RAM34). To assess the composition of the mature mononuclear cells we used Mac1, Gr-1, B220, and CD4/CD3. Cell cycle analysis was conducted by staining cells with the hematopoietic stem and progenitor mix described above. Cells were fixed using the FIX and PERM kit (Invitrogen cat# GAS-003). Cells were stained with Ki67 after fixation and then stained with DAPI before analysis on the LSR Fortessa. Sorting was conducted by staining as described in text and above and utilization of a FACSAria sorter.
Plasmids
The cDNA full-length clone of human FLAG-L3MBTL2 was obtained from Addgene (Plasmid 28232). The cDNA human full-length clone of HA-FLAG BAP1 was obtained from Addgene (Plasmid 22539). Deubiquitinase mutant constructs (C91A, C91S) were generated using Agilent site-directed mutagenesis kits and confirmed by full-length DNA sequencing. Short-hairpin RNAs were obtained from the RNAi Consortium (TRC) in a pLKO.1 puromycin vector. Sequences for the short-hairpins were as follows: human BAP1 (TRC Oligo IDs: TRCN0000078702 and TRCN0000078698), mouse BAP1 (TRCN0000030719 and TRCN0000030720), human L3MBTL2 (TRCN0000021724 and TRCN0000021726) and a control pLKO.1-puromycin vector encoding an shRNA for luciferase (shLUC).
Ubiquitin assays
HEK293T cells were seeded in a 10-cm dish and 24 hours later were transduced with 4 μg of a Myc-His-Ubi expression construct and control, 1 μg L3MBTL2 and/or 1–10 μg BAP1-GFP overexpression constructs. Forty-eight hours after the transfection, cells were lysed in a Guanidine HCl based lysis buffer: 6 M guanidine, 0.1 M NaH2PO4, 10 mM Tris, pH 8.0, and 10 mM BME. His-Ubi proteins were purified by incubation by 20 μL of Ni-NTA agarose (Qiagen) for 4 hours at room temperature. Beads were washed sequentially with 1 mL of 4 wash buffers: buffer A 6 M guanidine, 0.1 M NaH2PO4, 10 mM Tris, pH 8.0, 10 mM BME, and 0.2% Triton-X, buffer B 8 M urea, 0.1 M NaH2PO4, 10 mM Tris, pH 8.0, 10 mM BME, and 0.2% Triton-X, buffer C 0.1 M NaH2PO4, 10 mM Tris, pH 6.3, 10 mM BME, and 0.2% Triton-X, and buffer D 0.1 M NaH2PO4, 10 mM Tris, pH 6.3, 10 mM BME, and 0.1% Triton-X. All buffers were supplemented with 15 mM imidazole. His-tagged proteins were purified from the beads by boiling with 2x SDS Laemmli buffer supplemented with imidazole. Proteins were then analyzed by Western blot.
In vitro colony forming assays
Cells were sorted for Lin−c-Kit+Sca1+ cells using the FACSAria. 100 cells were plated in duplicate in methylcellulose (MethoCult GF M3434, Stem Cell Technologies). Colonies were counted 14 days after plating and colonies were collected by washing with PBS. Cells were then lysed for RNA and histone extraction.
Transient Transfection
293T cells were transfected with indicated constructs with X-treme gene transfection reagent (Roche). Protein and/or histones were extracted 48–72 hours after transfection.
Invasion Assays
Mesothelioma cells (MSTO-211H, H2373, H226 and H2452) were seeded in T75 flasks (100,000 cells). 12 hours later the plated cells were treated with GSK126 (0–2 μM) (Chemitek) and then left to proliferate for 7 days. 250,000 treated cells were then placed on the top of a Matrigel invasion chamber (BD Biosciences, cat no. 354480) in serum free media while the lower chamber contained media with serum. 22 hours later the cells on the bottom of the membrane were stained with crystal violet and quantitated with ImageJ.
Luciferase Assays
293T cells were transiently transfected with the pGL3 EZH2 promoter reporter construct and a Switchgear Renilla control construct in addition to EV, BAP1, and L3MBTL2 constructs. Cells were assessed for luciferase activity using the Dual Luciferase Reporter Assay System (Promega). Cells were seeded in 24 well plates and were cotransfected with 200 ng pGL3-EZH2-Luciferase, 200 ng of the Renilla luciferase control construct, and 500 ng of experimental constructs. Cells were incubated 48 hours after the transfection, lysed for 15 minutes at room temperature and luciferase activity was assessed on a luminometer. The Firefly luciferase readings were normalized to the Renilla transfection control.
Cell Titer Glo Viability Assays
Adherent mesothelioma cell lines were plated in 96 well dishes at about 100 cells per well to allow space for cells to proliferate. Cells were plated and given one day to adhere (an initial day one reading was taken at this time). ATP luminescence readings were taking at times specified in the manuscript. For assays in which we assessed response to EPZ001989, drug was added on day one and then replenished every four days.
Apoptosis Assays
For apoptosis analysis, 1×106 cells were stained using the Annexin V-FITC apoptosis detection kit (BD) according to the manufacturer's recommendations. DAP1 was used as a counterstain in these experiments and heat-shock controls were used as positive controls.
3D Growth Assays
To perform 3D assays in these cell lines, we utilized the technique using poly-HEMA as published for mesothelioma cell lines35. We prepared plates and spheroids as described with cells that had been pre-treated for 4 days with 500 nM EPZ011989. 5 days post-treatment we assessed viability with Cell Titer Glo assays.
Statistical Analyses
The Student's t-test was used to analyze statistical significance unless described in the text. Normality tests were used to test the assumption of a normal distribution. Prizm GraphPad Software was used for statistical calculations. Error was calculated using s.d. unless otherwise noted, *P < 0.05, **P < 0.005.
Replicates
c-Kit enrichment ChIP-Seq experiments were conducted with four biological replicates, with two replicates combined for sequencing. GMP ChIP-Seq experiments were conducted with three replicates for each group. c-Kit and GMP RNA-Seq was conducted with three biological replicates from each group. Histone mass spec experiments were conducted with three biological and three technical replicates. Genetic phenotyping experiments were replicated three times independently. Flow cytometric experiments were replicated independently two-three times. Pilot studies were conducted with drug studies and results were replicated in a larger study to achieve enough statistical power. In vitro experiments were replicated two-three times, with viability experiments being completed in triplicate.
Acknowledgments
This work was supported by the Pershing Square Sohn Prize (R.L.L.), by grant 2R01GM096056 (M.L.), by grant CA172636 (R.L.L. and A.M.), and by grant F31 CA180642-02 (L.M.L.). Work in the MSKCC Core facilities which supported these studies is supported by P30 CA008748. R.L.L. is a Leukemia and Lymphoma Society Scholar. We would like to thank V. Rotter (Weizmann Institute of Science, Israel), X. Jiang (Memorial Sloan Kettering Cancer Center), and M. Ladanyi (Memorial Sloan Kettering Cancer Center) for generously providing plasmids for this work. We would like to thank D. Schienberg (Memorial Sloan Kettering Cancer Center) for generously sharing the mesothelioma cell lines used in this work.
Footnotes
Author Contributions: L.M.L., O.A., and R.L.L. designed the study. L.M.L., W.B., A.C., E.P., M.K., K.K., J.M., I.K., E.H.D., X.S., Y.R.C., and O.A. performed the experiments. L.M.L., W.B., R.K., M.T., B.S., T.H., A.C., and A.K. performed ChIP-and RNA-Seq, sequencing, and subsequent downstream analyses. B.D., S.K.K., J.B.C, G.B, E.D., O.O., P.A., P.M.T., N.L.K, M.L., H.K., A.M., S.A.A., and R.L.L. participated in data analysis and discussions. L.M.L. and R.L.L. prepared the manuscript with input from all authors.
Accession codes: RNA-Seq and ChIP-Seq data are available via the GEO database, accession numbers GSE61577 and GSE61360, respectively.
References
Full text links
Read article at publisher's site: https://doi.org/10.1038/nm.3947
Read article for free, from open access legal sources, via Unpaywall:
https://www.ncbi.nlm.nih.gov/pmc/articles/PMC4636469
Citations & impact
Impact metrics
Citations of article over time
Alternative metrics
Smart citations by scite.ai
Explore citation contexts and check if this article has been
supported or disputed.
https://scite.ai/reports/10.1038/nm.3947
Article citations
Primary Intrahepatic Mesothelioma: Case Series and Systematic Review of Literature.
J Gastrointest Cancer, 55(4):1520-1529, 14 Aug 2024
Cited by: 0 articles | PMID: 39141212
Review
Generation and Characterization of Induced Pluripotent Stem Cells Carrying An ASXL1 Mutation.
Stem Cell Rev Rep, 20(7):1889-1901, 17 Jun 2024
Cited by: 0 articles | PMID: 38884929
Contemporary management of mesothelioma.
Breathe (Sheff), 20(2):230175, 01 Jun 2024
Cited by: 1 article | PMID: 39015660 | PMCID: PMC11250169
Review Free full text in Europe PMC
Potential roles of UCH family deubiquitinases in tumorigenesis and chemical inhibitors developed against them.
Am J Cancer Res, 14(6):2666-2694, 15 Jun 2024
Cited by: 0 articles | PMID: 39005671
Review
Targeted Therapy in Mesotheliomas: Uphill All the Way.
Cancers (Basel), 16(11):1971, 22 May 2024
Cited by: 0 articles | PMID: 38893092 | PMCID: PMC11171080
Review Free full text in Europe PMC
Go to all (215) article citations
Data
Data behind the article
This data has been text mined from the article, or deposited into data resources.
BioStudies: supplemental material and supporting data
Clinical Trials (3)
- (1 citation) ClinicalTrials.gov - NCT01395601
- (1 citation) ClinicalTrials.gov - NCT01082977
- (1 citation) ClinicalTrials.gov - NCT01897571
GEO - Gene Expression Omnibus (2)
- (1 citation) GEO - GSE61577
- (1 citation) GEO - GSE61360
Similar Articles
To arrive at the top five similar articles we use a word-weighted algorithm to compare words from the Title and Abstract of each citation.
Structural basis of histone H2A lysine 119 deubiquitination by Polycomb repressive deubiquitinase BAP1/ASXL1.
Sci Adv, 9(32):eadg9832, 09 Aug 2023
Cited by: 10 articles | PMID: 37556531 | PMCID: PMC10411902
Tumor-derived neomorphic mutations in ASXL1 impairs the BAP1-ASXL1-FOXK1/K2 transcription network.
Protein Cell, 12(7):557-577, 18 Jul 2020
Cited by: 16 articles | PMID: 32683582 | PMCID: PMC8225741
Targeting Excessive EZH1 and EZH2 Activities for Abnormal Histone Methylation and Transcription Network in Malignant Lymphomas.
Cell Rep, 29(8):2321-2337.e7, 01 Nov 2019
Cited by: 84 articles | PMID: 31747604
EZH2 methyltransferase and H3K27 methylation in breast cancer.
Int J Biol Sci, 8(1):59-65, 18 Nov 2011
Cited by: 151 articles | PMID: 22211105 | PMCID: PMC3226033
Review Free full text in Europe PMC
Funding
Funders who supported this work.
NCATS NIH HHS (1)
Grant ID: UL1 TR000457
NCI NIH HHS (7)
Grant ID: P30 CA060553
Grant ID: R01 CA187109
Grant ID: P30 CA008748
Grant ID: R01 CA173636
Grant ID: F31 CA180642
Grant ID: CA172636
Grant ID: R01 CA169784
NIGMS NIH HHS (3)
Grant ID: 2R01GM096056
Grant ID: R01 GM096056
Grant ID: R01 GM120570

 1,4,14,†
1,4,14,†



