Abstract
Background
Foodborne diseases are important worldwide, resulting in considerable morbidity and mortality. To our knowledge, we present the first global and regional estimates of the disease burden of the most important foodborne bacterial, protozoal, and viral diseases.Methods and findings
We synthesized data on the number of foodborne illnesses, sequelae, deaths, and Disability Adjusted Life Years (DALYs), for all diseases with sufficient data to support global and regional estimates, by age and region. The data sources included varied by pathogen and included systematic reviews, cohort studies, surveillance studies and other burden of disease assessments. We sought relevant data circa 2010, and included sources from 1990-2012. The number of studies per pathogen ranged from as few as 5 studies for bacterial intoxications through to 494 studies for diarrheal pathogens. To estimate mortality for Mycobacterium bovis infections and morbidity and mortality for invasive non-typhoidal Salmonella enterica infections, we excluded cases attributed to HIV infection. We excluded stillbirths in our estimates. We estimate that the 22 diseases included in our study resulted in two billion (95% uncertainty interval [UI] 1.5-2.9 billion) cases, over one million (95% UI 0.89-1.4 million) deaths, and 78.7 million (95% UI 65.0-97.7 million) DALYs in 2010. To estimate the burden due to contaminated food, we then applied proportions of infections that were estimated to be foodborne from a global expert elicitation. Waterborne transmission of disease was not included. We estimate that 29% (95% UI 23-36%) of cases caused by diseases in our study, or 582 million (95% UI 401-922 million), were transmitted by contaminated food, resulting in 25.2 million (95% UI 17.5-37.0 million) DALYs. Norovirus was the leading cause of foodborne illness causing 125 million (95% UI 70-251 million) cases, while Campylobacter spp. caused 96 million (95% UI 52-177 million) foodborne illnesses. Of all foodborne diseases, diarrheal and invasive infections due to non-typhoidal S. enterica infections resulted in the highest burden, causing 4.07 million (95% UI 2.49-6.27 million) DALYs. Regionally, DALYs per 100,000 population were highest in the African region followed by the South East Asian region. Considerable burden of foodborne disease is borne by children less than five years of age. Major limitations of our study include data gaps, particularly in middle- and high-mortality countries, and uncertainty around the proportion of diseases that were foodborne.Conclusions
Foodborne diseases result in a large disease burden, particularly in children. Although it is known that diarrheal diseases are a major burden in children, we have demonstrated for the first time the importance of contaminated food as a cause. There is a need to focus food safety interventions on preventing foodborne diseases, particularly in low- and middle-income settings.Free full text

World Health Organization Estimates of the Global and Regional Disease Burden of 22 Foodborne Bacterial, Protozoal, and Viral Diseases, 2010: A Data Synthesis
Abstract
Background
Foodborne diseases are important worldwide, resulting in considerable morbidity and mortality. To our knowledge, we present the first global and regional estimates of the disease burden of the most important foodborne bacterial, protozoal, and viral diseases.
Methods and Findings
We synthesized data on the number of foodborne illnesses, sequelae, deaths, and Disability Adjusted Life Years (DALYs), for all diseases with sufficient data to support global and regional estimates, by age and region. The data sources included varied by pathogen and included systematic reviews, cohort studies, surveillance studies and other burden of disease assessments. We sought relevant data circa 2010, and included sources from 1990–2012. The number of studies per pathogen ranged from as few as 5 studies for bacterial intoxications through to 494 studies for diarrheal pathogens. To estimate mortality for Mycobacterium bovis infections and morbidity and mortality for invasive non-typhoidal Salmonella enterica infections, we excluded cases attributed to HIV infection. We excluded stillbirths in our estimates. We estimate that the 22 diseases included in our study resulted in two billion (95% uncertainty interval [UI] 1.5–2.9 billion) cases, over one million (95% UI 0.89–1.4 million) deaths, and 78.7 million (95% UI 65.0–97.7 million) DALYs in 2010. To estimate the burden due to contaminated food, we then applied proportions of infections that were estimated to be foodborne from a global expert elicitation. Waterborne transmission of disease was not included. We estimate that 29% (95% UI 23–36%) of cases caused by diseases in our study, or 582 million (95% UI 401–922 million), were transmitted by contaminated food, resulting in 25.2 million (95% UI 17.5–37.0 million) DALYs. Norovirus was the leading cause of foodborne illness causing 125 million (95% UI 70–251 million) cases, while Campylobacter spp. caused 96 million (95% UI 52–177 million) foodborne illnesses. Of all foodborne diseases, diarrheal and invasive infections due to non-typhoidal S. enterica infections resulted in the highest burden, causing 4.07 million (95% UI 2.49–6.27 million) DALYs. Regionally, DALYs per 100,000 population were highest in the African region followed by the South East Asian region. Considerable burden of foodborne disease is borne by children less than five years of age. Major limitations of our study include data gaps, particularly in middle- and high-mortality countries, and uncertainty around the proportion of diseases that were foodborne.
Conclusions
Foodborne diseases result in a large disease burden, particularly in children. Although it is known that diarrheal diseases are a major burden in children, we have demonstrated for the first time the importance of contaminated food as a cause. There is a need to focus food safety interventions on preventing foodborne diseases, particularly in low- and middle-income settings.
Introduction
Foodborne diseases are globally important, as they result in considerable morbidity, mortality, and economic costs [1,2]. Many different diseases, including those due to bacteria, viruses, parasites, chemicals, and prions, may be transmitted to humans by contaminated food [3]. Outbreaks and sporadic cases of foodborne disease are regular occurrences in all countries of the world. In recent decades, globalization of the food supply has also meant that pathogens causing foodborne diseases are rapidly transported across international borders [4]. Foodborne disease outbreaks have led to adverse impacts on trade and food security [5,6]. In response to foodborne diseases, national governments and international bodies have established elaborate systems to control and improve food safety [7].
Recognizing that contaminated food is an important cause of human disease, estimates of disease burden of the various foodborne diseases has been sought to enable advocacy for improved food safety and to assist governments to prioritize efforts for enhancing food safety. Although several countries have estimated the number of cases, sequelae, deaths, and Disability Adjusted Life Years (DALYs) resulting from foodborne diseases at the national level, most have not [8]. Furthermore, global and regional estimates of the burden of foodborne diseases have not been available [1,2]. In 2007, the World Health Organization (WHO) established the Foodborne Disease Burden Epidemiology Reference Group (FERG) to estimate the global and regional burden of disease attributable to food from all causes [9]. FERG consists of thematic task forces to estimate the human health burden of (1) enteric bacterial and viral infections, (2) parasitic infections, and (3) illnesses due to chemicals and toxins. The Director General of WHO nominated members of the FERG following an open call for applications to governments, in the scientific press, and through global networks [1]. The Enteric Diseases Task Force (EDTF) of the FERG comprised 14 experts in the epidemiology of viral, bacterial and parasitic infections transmitted by food, and was supported by various resource advisors who were expert in various aspects of infectious disease transmission.
Methods
In this study, the EDTF estimates the global and regional disease health burden in 2010 resulting from the most common foodborne diseases. For a glossary of terms used in this paper see S1 Text.
FERG EDTF reviewed the epidemiology of all bacterial and viral diseases potentially transmitted by food and identified those for inclusion based on public health importance and data availability. We excluded enteroaggerative Escherichia coli, Vibrio parahaemolyticus, V. vulnificus, and Yersinia spp., which are infrequent causes of foodborne diseases, due to a lack of sufficient data for global estimation, ie–not commonly reported in systematic reviews of etiological agents of diarrhea [10]. After excluding these agents, we included 19 bacterial or viral diseases in our study. Of these 19 diseases, four are distinct manifestations of Salmonella enterica infection: invasive infections due to S. enterica serotype Typhi (Salmonella Typhi); invasive infections due to S. enterica serotype Paratyphi A (Salmonella Paratyphi A); invasive infections due to non-typhoidal S. enterica (iNTS); and diarrheal disease due to non-typhoidal S. enterica (NTS). We then determined that our approach for estimating the burden for diarrheal diseases could also be applied to protozoal diseases and included 3 protozoal diseases.
Diarrhea is a dominant feature for 14 of these diseases—ten caused by bacteria, three by protozoa, and one by a virus. One or more extra-intestinal manifestations including bacteremia, hepatitis, and meningitis are the dominant feature for the other eight diseases—seven caused by bacteria and one caused by a virus.
To identify data for estimation of incidence, mortality and sequelae for different agents, we conducted literature reviews to identify relevant studies, consulted with academics and expert committees, and evaluated systematic reviews that were conducted circa 2010. Where data sources were not available, the EDTF recommended that WHO commission systematic reviews into incidence and outcomes of specific agents, which were subsequently published in the peer-reviewed literature [11–22]. Where multiple data sources were available for a single agent, the EDTF made decisions to use the most contemporaneous and comprehensive sources, or incorporate both into the estimation process. In the following sections, we outline how we synthesized data in accordance with a documented framework reported elsewhere [23]. Table 1 summarizes our approach, while S2 Text provides a comprehensive description of the methods and distribution for all parameters used for estimating cases, sequelae, deaths and DALYs for each of the 22 diseases.
Table 1
For more information, see S2 Text.
| Disease | Pathogens | Methods | Reference |
|---|---|---|---|
| Diarrheal Disease | Campylobacter spp., Cryptosporidium spp., Entamoeba histolytica, enterotoxigenic E. coli (ETEC), enteropathogenic E. coli (EPEC), Giardia lamblia, norovirus, nontyphoidal Salmonella enterica, Shigella spp | Synthesis of seven national studies into foodborne disease incidence for low-mortality countries in Region A combined with systematic reviews for middle- and high-mortality countries that was derived from 136 outpatient and community studies and 254 inpatient studies among children <5 years of age, and 97 inpatient, outpatient, and community studies among persons ≥5 years of age | [11] |
| Diarrheal Disease | Vibrio cholerae (cholera) | Data synthesis of WHO cholera reports and publicly available data sources. | [24] |
| Diarrheal Disease | Shiga-toxin producing E. coli | Systematic review of 16 published studies | [19] |
| Intoxications | Bacillus cereus, Clostridium perfringens, Staphylococcus aureus | Estimation from seven national studies in low-mortality sub-region A countries only | [11] |
| Intoxications | Clostridium botulinum | Estimation from five national studies in 55 EUR and AMR sub-region A countries only* | See S2 Text |
| Invasive enteric disease | Hepatitis A virus | Conversion of mortality data from Global Burden of Disease 2010 for hepatitis A to incidence | [25] |
| Invasive enteric disease | Typhoid, Paratyphoid fevers | Data from Global Burden of Disease 2010 | [26] |
| Invasive enteric disease | Invasive non-typhoidal Salmonella enterica (iNTS) | Systematic review of 10 published studies of incidence of invasive infections | [22] |
| Invasive enteric disease | Mycobacterium bovis | Systematic review of 140 data sources combined with incidence estimates for human TB infections. | [16,17] |
| Invasive enteric disease | Brucella spp. | Systematic review of 29 published studies of incidence combined with information from countries free of Brucella in livestock. | [15] |
| Invasive enteric disease | Listeria spp. | Systematic review of 87 published studies, including 43 containing data on incidence | [18] |
* EUR— European Region; AMR— Region of the Americas
Estimating Cases, Sequelae, and Deaths for Diarrheal Diseases
For diarrheal diseases caused by Campylobacter spp., Cryptosporidium spp., Entamoeba histolytica, enterotoxigenic E. coli (ETEC), enteropathogenic E. coli (EPEC), Giardia lamblia, norovirus, NTS, and Shigella spp., national estimates of foodborne diseases were only available from a limited number of countries. We therefore used two approaches, documented in Pires et al. [11], depending on the level of development of the country. The first approach, based on national estimates of the incidence of foodborne diseases from seven studies published between 2005–2014, was applied to the 61 countries in low-mortality (European Region (EUR) and other subregion A [27]) countries [3,28–35]. For countries with national estimates of incidence and mortality, we used these data. We used the median and associated uncertainty intervals for diarrheal diseases for the subregion to estimate incidence and mortality of diarrheal diseases for other countries within the subregion without national data [10].
The second approach was applied to the remaining 133 countries not in EUR or other subregion A categories (see S1 Text for a country list by subregion). For this approach, we modified the WHO Child Health Epidemiology Reference Group (CHERG) method to estimate diarrheal incidence and mortality for all age groups [10]. First, we estimated the overall incidence of diarrhea from all causes (i.e., “envelope” of diarrheal incidence) for 2010 by combining estimates of diarrheal incidence for children <5 years of age and persons ≥5 years of age [13,36]. We used the overall diarrheal mortality (i.e., envelope for diarrheal deaths) derived by WHO [37] for 2010. We derived an etiological proportion for each disease by region from systematic reviews of stool sample isolation or detection proportions from inpatient, outpatient, and community-based studies of persons with diarrhea. We followed the CHERG standard approach because there is limited information on pathogens among people who have died, and assumed that the distribution of pathogens observed among inpatients hospitalized with severe diarrhea represented the pathogen prevalence among diarrheal deaths [10]. To derive etiological proportions for children <5 years of age, we assumed that the distribution of pathogens in outpatient and community studies represented the pathogen prevalence among diarrheal episodes for those who did not die. We made the same assumption for persons ≥5 years of age but due to sparseness of data also included inpatient studies. For more details of methods, see Pires et al. [11]. For some pathogens we assumed that different etiological agents, such as Shigella spp., NTS and Campylobacter spp., had similar clinical profiles (S2 Text).
Estimates for cholera were based on the incidence among populations at risk for cholera in endemic and non-endemic countries [24]. The case fatality ratio (CFR) for cholera was 1% in Western Pacific Region (WPR) subregion B, 1% in South-East Asian Region (SEAR) subregion B (except 1.5% in Bangladesh), 1.3% in Eastern Mediterranean Region (EMR) subregion B, 3% in SEAR subregion D, 3.2% in EMR subregion D, and 3.8% in African Region (AFR) [24]. For all other countries, we assumed cholera occurred only among international travellers and did not result in deaths. In this instance, we applied the median incidence from non-endemic countries with available data for cholera.
Shiga-toxin producing E. coli (STEC) infection incidence and mortality were based on a systematic review described elsewhere [19]. Sequelae, more common among O157 infections, were hemolytic uremic syndrome (HUS) and end stage renal disease (ESRD). Based on review, we estimated 0.8% of O157 infections and 0.03% of infections caused by other serotypes result in HUS, and 3% of HUS cases result in ESRD. We estimated that the CFR for HUS was 3.7%; for ESRD the CFR was 20% in the 35 subregion A countries and 100% in other countries.
We used data on the incidence and mortality of foodborne intoxications caused by Bacillus cereus, Clostridium perfringens, and Staphylococcus aureus from national studies conducted in low-mortality countries. We applied the median incidence from national studies to the 61 countries in EUR and other subregion A countries. We did not attempt to estimate burden due to these three foodborne intoxications in high- and middle-mortality countries due to the absence of data on diseases caused by these pathogens in these countries. The median CFR from national studies was 0.003% for C. perfringens and 0.0025% for S. aureus; there were no B. cereus deaths. We considered that 31% of Guillain-Barré Syndrome (GBS) cases globally were associated with antecedent Campylobacter infection and that the CFR for GBS was 4.1% [38,39].
Estimating Cases, Sequelae, and Deaths of Extra-Intestinal Diseases
For diseases caused by hepatitis A virus, Brucella spp., Listeria monocytogenes, Mycobacterium bovis, iNTS, Salmonella Paratyphi A, and Salmonella Typhi we used a variety of approaches depending on availability of data.
We used Institute of Health Metrics and Evaluation (IHME) Global Burden of Disease (GBD) 2010 data to estimate the burden of disease for typhoid, paratyphoid, and hepatitis A [25]. IHME provided country-specific age-standardized prevalence data of typhoid and paratyphoid fever. These data were converted to incidence by dividing by duration, and partitioned into typhoid and paratyphoid assuming a 1.0 to 0.23 ratio [40]. Country-specific hepatitis A mortality data, stratified by age and sex, were converted to incidence assuming a CFR of 0.2%.
Rates of iNTS are highly correlated with HIV prevalence and malaria risk [22]. To estimate iNTS incidence globally, we used age-specific estimates of incidence from a systematic review [22] to construct a random effect log linear model using covariates of country specific HIV and malaria deaths, and the log of Gross Domestic Product. As data were sparse, we predicted incidence for all ages, which was converted to age-specific incidence based on age profiles for iNTS cases in low and high incidence settings [22]. From this, we predicted iNTS incidence among persons not infected with HIV [41,42]. To estimate deaths, we assumed that the CFR for iNTS in non-HIV infected individuals was a uniform distribution with a range 5–20% in sub-region B-E countries and range 3.9–6.6% in sub-region A countries [43].
Estimates for M. bovis infections were based on a systematic review where the proportion of human tuberculosis (TB) infections due to M. bovis ranged from 0.3% in Region of the Americas (AMR) to 2.8% in AFR [17]. We identified 51 countries that were free from M. bovis in cattle based on European Union certification and the World Organization for Animal Health [44]. All countries in a region except those free from M. bovis in cattle were assumed to have the same proportion of human TB infections due to M. bovis. To account for internationally acquired infections, all countries free of M. bovis in cattle were assigned the lowest observed proportion of human TB infections due to M. bovis (0.3%). To derive estimates of human M. bovis incidence, we multiplied WHO country-specific human TB incidence by the estimate of the proportion of human TB infections that were due to M. bovis [45]. To estimate mortality associated with M. bovis that accounted for HIV co-morbidity we used estimates of mortality due to human TB in HIV negative persons from WHO. We adjusted mortality data by assuming that the CFR for M. bovis was 20% lower than human TB, as M. bovis infections are more likely to be extrapulmonary [16].
To estimate the incidence and mortality for brucellosis we updated a systematic review and included additional data on 32 countries that were considered Brucella-free in livestock (free of B. arbortus in cattle and B. melitensis in sheep and goats) [15]. We imputed incidence data to countries without estimates using a Bayesian log-normal random effects model, except for countries that were Brucella-free in livestock [23]. To account for internationally acquired infections, all countries that were Brucella-free in livestock were assigned the median incidence of human brucellosis reported from these countries. The CFR for brucellosis was 0.05%, and 40% of cases resulted in chronic infections and 10% of cases in males resulted in orchitis [14].
We estimated the incidence and mortality for listeriosis using a systematic review that is described elsewhere [18]. In accordance with standard burden of disease practice, we excluded stillbirths, in our baseline burden estimates. The CFR was 14.9% for perinatal cases and 25.9% for other cases.
Incidence and mortality data for botulism were only available from countries in Europe and North America. We limited our estimation to the 55 countries in EUR and AMR subregion A, which was based on the median incidence derived from countries with national estimates of botulism. We estimated that 35% of botulism cases were severe and that the CFR of severe botulism was 15%.
Estimation of Foodborne Proportion
We assumed that all infections from L. monocytogenes, M. bovis, and from the four foodborne intoxications (B. cereus, C. botulinum, C. perfringens, and S. aureus) were foodborne. To estimate the proportion of the other enteric diseases that were transmitted by contaminated food, we relied upon results of a FERG structured expert elicitation [46]. In this expert elicitation, foodborne and waterborne were considered separate transmission pathways. Proportions of brucellosis and hepatitis A infections and intestinal protozoa acquired from contaminated food were estimated using global panels of experts. Foodborne proportions for the remaining enteric diseases were estimated using a panel of experts for each region separately, although several experts were serving on more than one panel.
Disability Weights
Disability weights for each clinical outcome for the enteric diseases were taken from the GBD 2010 study [47]. Disability weights for listeriosis meningitis and neurological sequelae were derived from GBD 2010 disability weights using a multiplicative methodology and expert judgment. Disability weights for individual conditions are specified in S2 Text.
Data Analysis
The FERG approach to computing DALYs from the estimated cases, sequelae, deaths, and other parameters described in this paper is described in more detail elsewhere, including the modelling of uncertainty intervals incorporating (where relevant) uncertainty of estimated foodborne proportions [23]. Estimates were produced using the 2012 revisions for United Nations country-level population data for 2010 [48]. Calculations followed disease-specific models defined by incidence and probability parameters, each with a distribution [23]. Uncertainty around input parameters were propagated using Monte Carlo simulations; 10,000 values were sampled from each input parameter to calculate 10,000 estimates of cases, deaths or DALYs. The 2.5th and 97.5th percentile of each set of the 10,000 estimates yielded a 95% Uncertainty Interval (UI). Further details of analyses and modelling of DALYs and components described in this paper are in S2 Text.
Results
Cases
We estimated that the 22 diseases in our study caused 2.0 billion (95% UI 1.5–3.0 billion) illnesses in 2010, 39% (95% UI 26–53%) in children <5 years of age. Among the 1.9 billion (95% UI 1.4–2.8 billion) cases of diarrheal diseases, norovirus was responsible for 684 million (95% UI 491–1,112 million) illnesses; the largest number of cases for any pathogen (Table 2). The pathogens resulting in the next largest number of cases were ETEC, Shigella spp., G. lamblia, Campylobacter spp. and NTS. Campylobacter spp. cases included 31,700 (95% UI 25,400–40,200) GBS cases. There were also 2.48 million (95% UI 1.69–5.38 million) STEC cases which included 3,330 (95% UI 2,160–6,550) with HUS and 200 (95% UI 15–760) with ESRD. Among the extra-intestinal diseases, the pathogens resulting in the most infections were hepatitis A virus, Salmonella Typhi and Salmonella Paratyphi A. Brucella spp. resulted in 0.83 million (95% UI 0.34–19.6 million) illnesses, which included almost 333,000 (95% UI 135,000–7,820,000) chronic infections and 83,300 (95% UI 33,800–1,960,000) episodes of orchitis. L. monocytogenes resulted in 14,200 (95% UI 6,100–91,200) illnesses which included 7830 (95% UI 3,400–50,500) cases of septicaemia, 3,920 (95% UI 1,650–24,900) cases of meningitis, and 666 (95% UI 207–4,710) cases with neurological sequelae.
Table 2
| PATHOGEN | ILLNESSES (95% UI) | DEATHS (95% UI) | DALYs (95% UI) | PROPORTION FOODBORNE (95% UI) | FOODBORNE ILLNESSES (95% UI) | FOODBORNE DEATHS (95% UI) | FOODBORNE DALYS (95% UI) |
|---|---|---|---|---|---|---|---|
| Diarrheal Disease | 1,912,159,038 (1,413,002,730–2,849,323,016) | 715,196 (603,325–846,397) | 55,139,959 (46,746,114–65,120,623) | 0.29 (0.22–0.36) | 548,285,159 (369,733,377–888,360,956) | 199,892 (136,903–286,616) | 15,780,400 (11,043,288–22,251,264) |
| Campylobacter spp.* | 166,175,078 (92,227,873–300,877,905) | 37,604 (27,738–55,101) | 3,733,822 (2,857,037–5,273,652) | 0.58 (0.44–0.69) | 95,613,970 (51,731,379–177,239,714) | 21,374 (14,604–32,584) | 2,141,926 (1,535,985–3,137,980) |
| Cryptosporidium spp. | 64,003,709 (43,049,455–104,679,951) | 27,553 (18,532–44,654) | 2,159,331 (1,392,438–3,686,925) | 0.13 (0.07–0.24) | 8,584,805 (3,897,252–18,531,196) | 3,759 (1,520–9,115) | 296,156 (119,456–724,660) |
| Entamoeba histolytica | 103,943,952 (47,018,659–210,632,459) | 5,450 (2,194–17,127) | 515,904 (222,446–1,552,466) | 0.28 (0.14–0.44) | 28,023,571 (10,261,254–68,567,590) | 1,470 (453–5,554) | 138,863 (47,339–503,775) |
| Enteropathogenic E. coli | 81,082,327 (40,716,656–171,419,480) | 122,760 (97,115–154,869) | 9,717,390 (7,602,047–12,387,029) | 0.30 (0.17–0.48) | 23,797,284 (10,750,919–62,931,604) | 37,077 (19,957–61,262) | 2,938,407 (1,587,757–4,865,590) |
| Enterotoxigenic E. coli | 240,886,759 (160,890,532–377,471,599) | 73,857 (53,851–103,026) | 5,887,541 (4,190,610–8,407,186) | 0.36 (0.24–0.50) | 86,502,735 (49,136,952–151,776,173) | 26,170 (14,887–43,523) | 2,084,229 (1,190,704–3,494,201) |
| Giardia spp. | 183,842,615 (130,018,020–262,838,002) | 0 (0–0) | 171,100 (115,777–257,315) | 0.15 (0.08–0.27) | 28,236,123 (12,945,655–56,996,454) | 0 (0–0) | 26,270 (11,462–53,577) |
| Norovirus | 684,850,131 (490,930,402–1,122,947,359) | 212,489 (160,595–278,420) | 15,105,714 (11,649,794–19,460,578) | 0.18 (0.11–0.30) | 124,803,946 (70,311,254–251,352,877) | 34,929 (15,916–79,620) | 2,496,078 (1,175,658–5,511,092) |
| Salmonella enterica, non-typhoidal | 153,097,991 (64,733,607–382,208,079) | 56,969 (43,272–88,129) | 4,377,930 (3,242,020–7,175,522) | 0.52 (0.35–0.67) | 78,439,785 (31,579,011–210,875,866) | 28,693 (17,070–49,768) | 2,183,146 (1,314,295–3,981,424) |
| Shigella spp. | 190,849,501 (97,832,995–363,915,689) | 65,796 (46,317–97,036) | 5,407,736 (3,771,300–8,107,456) | 0.27 (0.13–0.44) | 51,014,050 (20,405,214–118,927,631) | 15,156 (6,839–30,072) | 1,237,103 (554,204–2,520,126) |
| Shiga toxin-producing E. coli | 2,481,511 (1,594,572–5,376,503) | 269 (111–814) | 26,827 (12,089–72,204) | 0.48 (0.33–0.60) | 1,176,854 (754,108–2,523,007) | 128 (55–374) | 12,953 (5,951–33,664) |
| Vibrio cholerae | 3,183,394 (2,211,329–4,146,250) | 105,170 (78,671–126,058) | 7,347,635 (5,496,431–8,804,408) | 0.24 (0.10–0.46) | 763,451 (310,910–1,567,682) | 24,649 (10,304–50,042) | 1,722,312 (720,029–3,491,997) |
| Intoxications | 5,409,083 (2,187,762–12,929,293) | 175 (70–407) | 9,905 (3,993–23,527) | 1.00 | 5,409,083 (2,187,762–12,929,293) | 175 (70–407) | 9,905 (3,993–23,527) |
| Bacillus cereus ** | 256,775 (43,875–807,547) | 0 (0–0) | 45 (7–171) | 1.00 | 256,775 (43,875–807,547) | 0 (0–0) | 45 (7–171) |
| Clostridium botulinum *** | 475 (183–990) | 24 (7–65) | 1,036 (299–2,805) | 1.00 | 475 (183–990) | 24 (7–65) | 1,036 (299–2,805) |
| Clostridium perfringens ** | 3,998,164 (837,262–11,529,642) | 120 (25–351) | 6,963 (1,423–20,493) | 1.00 | 3,998,164 (837,262–11,529,642) | 120 (25–351) | 6,963 (1,423–20,493) |
| Staphylococcus aureus ** | 1,073,339 (658,463–1,639,524) | 25 (10–55) | 1,575 (702–3,244) | 1.00 | 1,073,339 (658,463–1,639,524) | 25 (10–55) | 1,575 (702–3,244) |
| Invasive enteric diseases | 77,929,723 (36,606,712–149,676,316) | 371,002 (218,593–631,271) | 23,070,841 (13,388,154–39,912,033) | 0.34 (0.17–0.52) | 25,569,838 (10,019,370–58,282,758) | 146,981 (81,052–274,835) | 9,107,557 (4,891,985–17,483,327) |
| Brucella spp. | 832,633 (337,929–19,560,440) | 4,145 (1,557–95,894) | 264,073 (100,540–6,187,148) | 0.47 (0.30–0.61) | 393,239 (143,815–9,099,394) | 1,957 (661–45,545) | 124,884 (43,153–2,910,416) |
| Hepatitis A | 46,864,406 (14,417,704–111,771,902) | 93,961 (29,602–221,677) | 4,580,758 (1,599,296–10,408,164) | 0.30 (0.14–0.49) | 13,709,836 (3,630,847–38,524,946) | 27,731 (7,169–77,320) | 1,353,767 (383,684–3,672,726) |
| Listeria monocytogenes | 14,169 (6,112–91,175) | 3,175 (1,339–20,428) | 118,340 (49,634–754,680) | 1.00 | 14,169 (6,112–91,175) | 3,175 (1,339–20,428) | 118,340 (49,634–754,680) |
| Mycobacterium bovis | 121,268 (99,852–150,239) | 10,545 (7,894–14,472) | 607,775 (458,364–826,115) | 1.00 | 121,268 (99,852–150,239) | 10,545 (7,894–14,472) | 607,775 (458,364–826,115) |
| Salmonella enterica, invasive non-typhoidal | 596,824 (596,824–596,824) | 63,312 (38,986–94,193) | 3,895,547 (2,401,034–5,790,874) | 0.48 (0.28–0.64) | 284,972 (167,455–384,321) | 29,391 (14,948–50,463) | 1,794,575 (886,443–3,107,172) |
| Salmonella enterica Paratyphi A | 4,826,477 (1,782,796–10,323,273) | 33,325 (12,309–71,278) | 2,367,164 (875,236–5,066,375) | 0.37 (0.19–0.58) | 1,741,120 (536,650–4,310,983) | 12,069 (3,784–29,521) | 855,730 (268,879–2,100,120) |
| Salmonella enterica Typhi | 20,984,683 (7,751,285–44,883,794) | 144,890 (53,519–309,903) | 10,292,017 (3,805,373–22,027,716) | 0.37 (0.19–0.58) | 7,570,087 (2,333,263–18,743,406) | 52,472 (16,454–128,350) | 3,720,565 (1,169,040–9,130,956) |
| TOTAL | 2,000,626,631 (1,494,986,030–2,942,534,533) | 1,092,584 (892,999–1,374,238) | 78,730,084 (64,963,913–97,740,062) | 0.29 (0.23–0.36) | 581,902,722 (400,741,151–922,031,380) | 350,686 (240,030–524,042) | 25,175,035 (17,547,264–37,021,003) |
* Includes Guillain-Barré Syndrome cases and deaths.
** 61 EUR and other subregion A (low mortality) countries only.
*** EUR and subregion A (low mortality) countries only, excluding WPR A countries.
Overall, 29% (95% UI 23–36%) of all 22 diseases were estimated to be transmitted by contaminated food equating to 582 million (95% UI 400–922 million) foodborne cases in 2010; 38% (95% UI 24–53%) in children <5 years of age. The pathogens resulting in the most foodborne cases were norovirus, Campylobacter spp., ETEC, NTS, and Shigella spp. A high proportion of foodborne infections caused by V. cholerae, Salmonella Typhi, and Salmonella Paratyphi A occurred in the African region (Table 3). A high proportion of foodborne infections caused by EPEC, Cryptosporidium spp., and Campylobacter spp. occurred among children <5 years of age (Table 4). Among the 11 diarrheal diseases, the rate ratio of foodborne cases occurring among children <5 years of age compared to those ≥5 years of age was 6.44 (95%UI 3.15–12.46).
Table 3
| PATHOGEN* | African Region (AFR) | Region of the Americas (AMR) | Eastern Mediterranean Region (EMR) | ||||||
|---|---|---|---|---|---|---|---|---|---|
| ILLNESSES (95% UI) | DEATHS (95% UI) | DALYs (95% UI) | ILLNESSES (95% UI) | DEATHS (95% UI) | DALYs (95% UI) | ILLNESSES (95% UI) | DEATHS (95% UI) | DALYs (95% UI) | |
| Diarrheal Disease | 9,830 (3,969–21,567) | 9 (5–14) | 687 (369–1,106) | 7,900 (4,497–13,850) | 0.5 (0.3–0.7) | 44 (30–63) | 16,387 (7,729–34,176) | 4 (2–6) | 354 (218–544) |
| Campylobacter spp.** | 2,221 (335–8,482) | 0.8 (0.4–1) | 70 (41–112) | 1,389 (490–3,207) | 0.07 (0.04–0.1) | 13 (8–18) | 1,873 (488–5,608) | 1 (0.6–1) | 90 (56–130) |
| Cryptosporidium spp. | 205 (35–813) | 0.2 (0.04–0.4) | 13 (3–37) | 114 (32–355) | 0.007 (0.002–0.02) | 0.6 (0.2–2) | 346 (52–1,287) | 0.04 (0.004–0.2) | 4 (0.4–20) |
| Entamoeba histolytica | 796 (98–3,868) | 0.05 (0.009–0.4) | 5 (0.9–39) | 212 (16–1,209) | 0.001 (0–0.009) | 0.3 (0.03–1) | 737 (79–3,110) | 0.02 (0.002–0.2) | 2 (0.3–14) |
| Enteropathogenic E. coli | 454 (125–1,215) | 2 (0.6–3) | 140 (50–282) | 189 (35–730) | 0.06 (0.01–0.1) | 5 (1–12) | 430 (116–1,222) | 0.7 (0.2–2) | 57 (18–131) |
| Enterotoxigenic E. coli | 982 (312–2,480) | 1 (0.6–3) | 109 (46–216) | 1,281 (299–3,295) | 0.05 (0.01–0.1) | 5 (1–12) | 4,971 (1,685–10,849) | 0.4 (0.1–1) | 35 (11–89) |
| Giardia spp. | 809 (172–2,574) | 0 (0–0) | 0.8 (0.2–3) | 309 (62–1,249) | 0 (0–0) | 0.3 (0.05–1) | 670 (133–2,193) | 0 (0–0) | 0.6 (0.1–2) |
| Norovirus | 1,749 (491–5,060) | 1 (0.3–3) | 81 (24–185) | 2,491 (898–6,186) | 0.1 (0.04–0.3) | 9 (3–23) | 2,796 (744–7,376) | 0.4 (0.1–1) | 33 (9–76) |
| Salmonella enterica, non-typhoidal | 896 (175–2,994) | 1 (0.5–2) | 89 (42–147) | 1,002 (378–1,990) | 0.1 (0.06–0.2) | 7 (4–12) | 1,610 (147–14,052) | 0.6 (0.3–1) | 54 (26–87) |
| Shigella spp. | 523 (45–2,865) | 0.5 (0.1–2) | 43 (8–124) | 278 (35–1,443) | 0.02 (0.003–0.05) | 1 (0.3–5) | 627 (55–4,648) | 0.4 (0.07–1) | 38 (6–117) |
| Shiga toxin-producing E. coli | 5 (2–9) | 0 (0–0.002) | 0.05 (0.02–0.1) | 16 (9–30) | 0.004 (0.001–0.01) | 0.3 (0.1–0.9) | 65 (37–97) | 0.002 (0–0.004) | 0.2 (0.1–0.5) |
| Vibrio cholerae | 43 (13–101) | 2 (0.5–4) | 1 12 (35–252) | 0.02 (0.008–0.05) | 0 (0–0) | 0 (0–0) | 9 (0.4–28) | 0.3 (0.01–1) | 20 (0.7–69) |
| Invasive enteric diseases | 425 (156–976) | 5 (3–8) | 307 (160–508) | 31 (11–81) | 0.3 (0.2–0.6) | 16 (8–35) | 394 (80–1,056) | 2 (0.7–4) | 108 (41–250) |
| Brucella spp. | 3 (0.4–110) | 0.02 (0.002–0.5) | 1 (0.1–34) | 3 (1–37) | 0.01 (0.005–0.2) | 0.9 (0.3–12) | 33 (10–187) | 0.2 (0.05–0.9) | 11 (3–60) |
| Hepatitis A | 232 (60–643) | 0.5 (0.1–1) | 23 (7–60) | 12 (3–33) | 0.02 (0.006–0.07) | 1 (0.3–3) | 237 (17–772) | 0.5 (0.04–2) | 23 (2–74) |
| Listeria monocytogenes | 0.1 (0–2) | 0.03 (0–0.6) | 1 (0–21) | 0.3 (0.1–1) | 0.07 (0.03–0.3) | 3 (1–11) | 0.1 (0–2) | 0.03 (0–0.6) | 1 (0–21) |
| Mycobacterium bovis | 7 (4–9) | 0.5 (0.3–0.7) | 30 (19–42) | 0.1 (0.05–0.2) | 0.007 (0.003–0.01) | 0.4 (0.2–0.8) | 1 (0.8–2) | 0.2 (0.08–0.3) | 9 (5–18) |
| Salmonella enterica., invasive non-typhoidal | 25 (12–37) | 3 (1–5) | 169 (71–306) | 0.7 (0.4–0.9) | 0.06 (0.03–0.1) | 3 (1–5) | 1 (0.7–2) | 0.1 (0.06–0.3) | 8 (3–14) |
| Salmonella enterica Paratyphi A | 25 (5–73) | 0.2 (0.04–0.5) | 12 (3–36) | 2 (0.4–7) | 0.02 (0.003–0.05) | 1 (0.2–4) | 17 (2–55) | 0.1 (0.01–0.4) | 9 (1–28) |
| Salmonella enterica Typhi | 108 (24–317) | 0.7 (0.2–2) | 53 (12–155) | 10 (2–32) | 0.07 (0.01–0.2) | 5 (0.9–16) | 73 (9–240) | 0.5 (0.06–2) | 37 (5–122) |
| TOTAL | 10,304 (4,279–22,108) | 14 (8–21) | 1,001 (562–1,543) | 7,937 (4,515–13,899) | 0.8 (0.5–1) | 61 (40–93) | 16,865 (8,051–34,712) | 6 (4–9) | 470 (286–728) |
| PATHOGEN* | European Region (EUR) | South-East Asian Region (SEAR) | Western Pacific Region (WPR) | GLOBAL | ||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| ILLNESSES (95% UI) | DEATHS (95% UI) | DALYs (95% UI) | ILLNESSES (95% UI) | DEATHS (95% UI) | DALYs (95% UI) | ILLNESSES (95% UI) | DEATHS (95% UI) | DALYs (95% UI) | ILLNESSES (95% UI) | DEATHS (95% UI) | DALYs (95% UI) | |
| Diarrheal Disease | 2,483 (1,439–4,136) | 0.3 (0.2–0.4) | 23 (16–30) | 7,074 (2,570–19,537) | 5 (2–9) | 363 (177–649) | 6,302 (2,501–17,289) | 0.3 (0.1–0.5) | 33 (17–54) | 7,968 (5,373–12,911) | 3 (2–4) | 229 (160–323) |
| Campylobacter spp.** | 522 (363–687) | 0.05 (0.03–0.09) | 9 (6–13) | 1,152 (200–3,372) | 0.4 (0.1–0.9) | 33 (9–83) | 876 (359–3,855) | 0.04 (0.02–0.1) | 10 (4–17) | 1,390 (752–2,576) | 0.3 (0.2–0.5) | 31 (22–46) |
| Cryptosporidium spp. | 21 (4–70) | 0.003 (0–0.009) | 0.2 (0.03–0.6) | 78 (10–474) | 0.09 (0.01–0.4) | 6 (0.9–29) | 32 (2–170) | 0.003 (0–0.03) | 0.3 (0.02–3) | 125 (57–269) | 0.05 (0.02–0.1) | 4 (2–11) |
| Entamoeba histolytica | 0 (0–0) | 0 (0–0) | 0 (0–0) | 256 (27–1,188) | 0.03 (0.004–0.2) | 3 (0.3–17) | 229 (0–1,598) | 0.001 (0–0.003) | 0.3 (0–1) | 407 (149–997) | 0.02 (0.007–0.08) | 2 (0.7–7) |
| Enteropathogenic E. coli | 8 (3–16) | 0 (0–0) | 0.005 (0.002–0.01) | 594 (62–2,775) | 0.9 (0.2–2) | 66 (15–146) | 166 (8–395) | 0.06 (0.003–0.1) | 5 (0.2–12) | 346 (156–915) | 0.5 (0.3–0.9) | 43 (23–71) |
| Enterotoxigenic E. coli | 6 (2–13) | 0 (0–0) | 0.004 (0.001–0.01) | 1,075 (229–3,521) | 0.6 (0.1–1) | 42 (10–104) | 555 (43–2,430) | 0.04 (0.003–0.1) | 4 (0.3–10) | 1,257 (714–2,206) | 0.4 (0.2–0.6) | 30 (17–51) |
| Giardia spp. | 54 (16–123) | 0 (0–0) | 0.03 (0.009–0.1) | 159 (16–903) | 0 (0–0) | 0.1 (0.01–0.9) | 354 (8–1,519) | 0 (0–0) | 0.3 (0.005–1) | 410 (188–828) | 0 (0–0) | 0.4 (0.2–0.8) |
| Norovirus | 1,652 (630–3,294) | 0.05 (0.02–0.1) | 4 (1–8) | 841 (113–5,631) | 1 (0.2–3) | 71 (15–230) | 1,305 (189–6,441) | 0.05 (0.004–0.2) | 4 (0.4–17) | 1,814 (1,022–3,653) | 0.5 (0.2–1) | 36 (17–80) |
| Salmonella enterica, non-typhoidal | 186 (118–275) | 0.1 (0.08–0.2) | 8 (5–14) | 908 (88–4,758) | 0.7 (0.2–2) | 49 (11–147) | 898 (170–6,428) | 0.02 (0.01–0.03) | 2 (1–7) | 1,140 (459–3,065) | 0.4 (0.2–0.7) | 32 (19–58) |
| Shigella spp. | 3 (0.9–8) | 0.003 (0–0.01) | 0.2 (0.03–0.8) | 1,084 (177–3,927) | 0.3 (0.07–1) | 25 (5–83) | 689 (19–2,549) | 0.04 (0.002–0.1) | 4 (0.1–10) | 741 (297–1,728) | 0.2 (0.1–0.4) | 18 (8–37) |
| Shiga toxin-producing E. coli | 18 (9–28) | 0.003 (0.001–0.006) | 0.3 (0.1–0.8) | 19 (2–95) | 0.002 (0–0.01) | 0.2 (0.02–1) | 3 (2–6) | 0 (0–0.001) | 0.05 (0.02–0.1) | 17 (11–37) | 0.002 (0–0.005) | 0.2 (0.09–0.5) |
| Vibrio cholerae | 0.03 (0.01–0.06) | 0 (0–0) | 0 (0–0) | 17 (0.7–52) | 0.4 (0.007–2) | 30 (0.5–109) | 0.2 (0.01–0.5) | 0.002 (0–0.005) | 0.1 (0.005–0.3) | 11 (5–23) | 0.4 (0.1–0.7) | 25 (10–51) |
| Invasive enteric diseases | 19 (9–51) | 0.2 (0.1–0.3) | 10 (7–19) | 872 (169–2,288) | 4 (1–9) | 250 (81–598) | 163 (30–391) | 0.9 (0.3–2) | 58 (19–134) | 372 (146–847) | 2 (1–4) | 132 (71–254) |
| Brucella spp. | 4 (1–33) | 0.02 (0.006–0.2) | 1 (0.4–11) | 2 (0.02–302) | 0.01 (0–1) | 0.8 (0.006–96) | 2 (0.5–61) | 0.01 (0.002–0.3) | 0.6 (0.1–19) | 6 (2–132) | 0.03 (0.01–0.7) | 2 (0.6–42) |
| Hepatitis A | 11 (3–28) | 0.02 (0.006–0.06) | 1 (0.3–3) | 494 (57–1,590) | 1 (0.1–3) | 48 (6–151) | 51 (3–164) | 0.1 (0.006–0.3) | 5 (0.3–15) | 199 (53–560) | 0.4 (0.1–1) | 20 (6–53) |
| Listeria monocytogenes | 0.2 (0.2–0.3) | 0.04 (0.03–0.06) | 2 (1–2) | 0.1 (0–2) | 0.03 (0–0.6) | 1 (0–21) | 0.2 (0.1–0.4) | 0.04 (0.03–0.09) | 1 (1–3) | 0.2 (0.09–1) | 0.05 (0.02–0.3) | 2 (0.7–11) |
| Mycobacterium bovis | 0.2 (0.1–0.3) | 0.02 (0.01–0.03) | 0.9 (0.7–1) | 2 (0.9–4) | 0.2 (0.1–0.5) | 13 (6–26) | 0.8 (0.4–1) | 0.04 (0.02–0.07) | 2 (1–4) | 2 (1–2) | 0.2 (0.1–0.2) | 9 (7–12) |
| Salmonella enterica., invasive non-typhoidal | 0.8 (0.6–1) | 0.07 (0.04–0.1) | 3 (2–5) | 2 (0.3–2) | 0.2 (0.03–0.3) | 9 (2–16) | 1 (0.5–2) | 0.1 (0.05–0.2) | 6 (2–10) | 4 (2–6) | 0.4 (0.2–0.7) | 26 (13–45) |
| Salmonella enterica Paratyphi A | 0.2 (0.03–1) | 0.002 (0–0.008) | 0.1 (0.01–0.6) | 58 (11–167) | 0.4 (0.1–1) | 29 (7–83) | 18 (3–47) | 0.1 (0.02–0.3) | 8 (1–20) | 25 (8–63) | 0.2 (0.05–0.4) | 12 (4–31) |
| Salmonella enterica Typhi | 1 (0.1–5) | 0.007 (0–0.03) | 0.5 (0.05–2) | 250 (50–725) | 2 (0.4–5) | 128 (29–361) | 77 (12–203) | 0.5 (0.07–1) | 33 (5–87) | 110 (34–272) | 0.8 (0.2–2) | 54 (17–133) |
| TOTAL | 2,506 (1,455–4,168) | 0.5 (0.3–0.6) | 32 (24–45) | 8,068 (3,294–20,663) | 9 (4–17) | 622 (306–1,145) | 6,491 (2,630–17,528) | 1 (0.6–2) | 93 (45–175) | 8,369 (5,723–13,318) | 5 (3–8) | 366 (255–538) |
* Table does not include four foodborne intoxications due to Clostridium botulinum, C. perfringens, S. aureus, and Bacillus cereus due to a lack of data for global estimation.
** Includes Guillain-Barré Syndrome cases and deaths.
Table 4
| PATHOGEN* | Age Group: <5 Years of Age | Age Group: ≥5 Years of Age | Ratio <5:≥5 | ||||||
|---|---|---|---|---|---|---|---|---|---|
| ILLNESSES | DEATHS | DALYs | ILLNESSES | DEATHS | DALYs | ILLNESSES | DEATHS | DALYs | |
| Number (95% UI) | Number (95% UI) | Number (95% UI) | Number (95% UI) | Number (95% UI) | Number (95% UI) | Ratio <5:≥5 (95% UI) | Ratio <5:≥5 (95% UI) | Ratio <5:≥5 (95% UI) | |
| Diarrheal Disease | 216,839,210 (148,937,428–309,926,253) | 91,621 (62,442–132,707) | 8,547,149 (5,903,945–12,254,175) | 327,209,075 (179,670,939–643,705,133) | 107,500 (69,907–163,979) | 7,205,002 (4,790,026–10,747,526) | 0.66 (0.32–1.28) | 0.86 (0.60–1.16) | 1.19 (0.86–1.60) |
| Campylobacter spp.** | 47,988,357 (22,436,891–102,663,926) | 13,861 (8,754–23,670) | 1,383,499 (911,878–2,279,897) | 42,883,268 (18,350,672–112,061,441) | 7,436 (4,930–9,974) | 750,578 (540,003–956,663) | 1.11 (0.34–3.47) | 1.91 (1.21–3.08) | 1.87 (1.26–2.92) |
| Cryptosporidium spp. | 5,986,213 (2,569,532–12,738,924) | 1,989 (678–5,683) | 185,057 (64,847–518,497) | 2,253,036 (774,628–8,639,265) | 1,673 (638–4,149) | 104,794 (40,408–256,055) | 2.61 (0.69–8.01) | 1.23 (0.42–2.72) | 1.83 (0.65–3.93) |
| Entamoeba histolytica | 8,480,759 (1,593,697–30,849,576) | 896 (90–4,852) | 92,213 (15,997–444,002) | 17,828,477 (5,378,578–50,963,825) | 524 (218–1,110) | 43,984 (20,149–85,551) | 0.48 (0.08–2.38) | 1.75 (0.18–8.71) | 2.14 (0.38–9.49) |
| Enteropathogenic E. coli | 17,312,780 (6,767,766–54,104,398) | 22,156 (11,944–37,473) | 2,004,543 (1,084,856–3,389,584) | 5,458,601 (2,145,370–16,561,005) | 14,647 (7,305–25,447) | 911,012 (457,215–1,575,768) | 3.20 (0.85–11.78) | 1.52 (1.03–2.29) | 2.21 (1.52–3.29) |
| Enterotoxigenic E. coli | 38,352,806 (21,144,875–64,795,160) | 14,056 (7,045–26,784) | 1,303,490 (668,837–2,446,758) | 46,811,878 (20,306,649–103,801,449) | 11,933 (6,382–18,887) | 767,975 (419,834–1,204,273) | 0.82 (0.35–1.96) | 1.21 (0.63–2.10) | 1.74 (0.95–2.93) |
| Giardia spp. | 18,773,028 (8,075,497–38,649,748) | 0 (0–0) | 20,677 (8,552–44,101) | 8,693,968 (3,337,657–24,195,602) | 0 (0–0) | 5,016 (1,945–13,791) | 2.11 (0.84–5.22) | N/A | 4.04 (1.57–10.28) |
| Norovirus | 34,582,700 (19,595,826–59,592,939) | 8,992 (4,251–19,347) | 844,376 (406,822–1,776,252) | 89,056,582 (46,054,795–206,532,318) | 25,807 (11,201–61,642) | 1,638,925 (730,924–3,844,771) | 0.38 (0.19–0.73) | 0.35 (0.22–0.54) | 0.52 (0.33–0.78) |
| Salmonella enterica, non-typhoidal | 15,274,234 (6,514,539–41,696,874) | 12,531 (6,562–30,779) | 1,149,675 (609,216–2,792,992) | 60,293,254 (18,488,275–189,066,838) | 15,807 (8,762–21,942) | 1,016,047 (576,408–1,405,079) | 0.26 (0.06–1.16) | 0.84 (0.44–1.83) | 1.19 (0.64–2.57) |
| Shigella spp. | 15,516,627 (5,416,319–38,620,351) | 8,863 (3,250–20,925) | 819,280 (309,576–1,909,450) | 34,049,173 (10,186,959–95,312,884) | 6,060 (2,734–11,511) | 404,144 (188,009–749,866) | 0.45 (0.13–1.70) | 1.49 (0.60–3.26) | 2.06 (0.87–4.43) |
| Shiga toxin-producing E. coli | 339,905 (217,805–728,708) | 63 (30–170) | 6,969 (3,278–17,751) | 836,948 (536,302–1,794,298) | 65 (24–204) | 5,989 (2,654–15,877) | 0.41 (0.41–0.41) | 0.96 (0.81–1.31) | 1.16 (1.09–1.29) |
| Vibrio cholerae | 114,518 (46,636–235,152) | 3,697 (1,546–7,506) | 331,395 (138,538–672,643) | 648,933 (264,273–1,332,530) | 20,952 (8,758–42,535) | 1,390,973 (581,491–2,820,499) | 0.18 (0.18–0.18) | 0.18 (0.18–0.18) | 0.24 (0.24–0.24) |
| Invasive enteric diseases | 4,336,215 (1,675,945–9,422,681) | 23,727 (11,866–45,950) | 2,180,916 (1,085,765–4,219,254) | 21,182,632 (8,375,340–49,059,198) | 123,026 (69,306–230,318) | 6,900,776 (3,799,471–13,355,093) | 0.21 (0.15–0.23) | 0.19 (0.14–0.22) | 0.32 (0.23–0.35) |
| Brucella spp. | 4,144 (1,527–93,225) | 21 (7–463) | 1,988 (687–44,999) | 389,106 (142,279–9,006,169) | 1,936 (654–45,081) | 122,904 (42,484–2,865,643) | 0.01 (0.01–0.01) | 0.01 (0.01–0.01) | 0.02 (0.02–0.02) |
| Hepatitis A | 2,165,243 (573,433–6,084,381) | 4,380 (1,132–12,211) | 411,592 (112,767–1,130,290) | 11,544,593 (3,057,415–32,440,565) | 23,351 (6,036–65,109) | 941,278 (269,448–2,538,627) | 0.19 (0.19–0.19) | 0.19 (0.19–0.19) | 0.44 (0.39–0.46) |
| Listeria monocytogenes | 1,240 (393–10,502) | 330 (126–2,138) | 30,750 (11,700–198,862) | 12,936 (5,716–80,766) | 2,851 (1,200–18,271) | 87,569 (36,830–561,221) | 0.10 (0.07–0.13) | 0.11 (0.08–0.16) | 0.34 (0.24–0.49) |
| Mycobacterium bovis | 869 (732–1,049) | 76 (58–101) | 7,134 (5,477–9,496) | 120,398 (99,119–149,188) | 10,470 (7,836–14,372) | 600,639 (452,917–816,737) | 0.01 (0.01–0.01) | 0.01 (0.01–0.01) | 0.01 (0.01–0.01) |
| Salmonella enterica, invasive non-typhoidal | 45,549 (25,019–62,638) | 4,700 (2,268–8,188) | 421,523 (203,340–733,940) | 239,467 (142,115–321,539) | 24,692 (12,655–42,246) | 1,373,635 (684,718–2,373,326) | 0.19 (0.17–0.20) | 0.19 (0.17–0.20) | 0.31 (0.29–0.32) |
| Salmonella enterica Paratyphi A | 357,814 (110,286–885,942) | 2,480 (778–6,067) | 227,507 (71,530–557,578) | 1,383,306 (426,364–3,425,041) | 9,588 (3,007–23,454) | 627,953 (197,302–1,541,909) | 0.26 (0.26–0.26) | 0.26 (0.26–0.26) | 0.36 (0.36–0.36) |
| Salmonella enterica Typhi | 1,555,715 (479,504–3,851,923) | 10,783 (3,381–26,377) | 989,159 (311,001–2,424,250) | 6,014,372 (1,853,758–14,891,483) | 41,689 (13,072–101,973) | 2,730,232 (857,835–6,703,954) | 0.26 (0.26–0.26) | 0.26 (0.26–0.26) | 0.36 (0.36–0.36) |
| TOTAL | 221,451,463 (153,244,508–315,075,166) | 116,613 (80,862–165,379) | 10,831,919 (7,587,557–15,271,603) | 350,711,509 (199,599,319–673,777,073) | 232,916 (152,283–368,498) | 14,250,088 (9,419,295–22,483,691) | 0.63 (0.32–1.18) | 0.50 (0.36–0.68) | 0.76 (0.56–1.01) |
* Table does not include four foodborne intoxications due to Clostridium botulinum, C. perfringens, S. aureus, and Bacillus cereus due to a lack of data for global estimation.
** Includes Guillain-Barré Syndrome cases and deaths.
Deaths
We estimated that the 22 diseases in our study caused 1.09 million (95% UI 0.89–1.37 million) deaths in 2010, 34% (95% UI 29–38%) in children <5 years of age. Among the diarrheal diseases, norovirus was responsible for the most deaths. Other diarrheal pathogens responsible for large numbers of deaths were EPEC, V. cholerae, and Shigella spp. The 37,600 (95% UI 27,700–55,100) deaths attributed to Campylobacter spp. included 1,310 (95% UI 887–1,880) deaths from GBS. Among the extra-intestinal enteric diseases, the pathogens resulting in the most deaths were Salmonella Typhi, hepatitis A virus, iNTS and Salmonella Paratyphi A.
Overall, the 22 diseases in our study resulted in 351,000 (95% UI 240,000–524,000) deaths due to contaminated food in 2010; 33% (95% UI 27–40%) in children <5 years of age. The enteric pathogens resulting in the most foodborne deaths were Salmonella Typhi, EPEC, norovirus, iNTS, NTS, and hepatitis A. The mortality rates of foodborne diseases were consistently highest in the African region followed by the South Eastern Asian region (Table 3). Among the 11 diarrheal diseases due to contaminated food, the rate ratio of deaths in children <5 years of age compared to those ≥5 years of age was 8.37 (95%UI 5.90–11.4). For all 22 diseases, the rate ratio of deaths in children <5 years of age compared to those ≥5 years of age was 4.85 (95%UI 3.54–6.59).
DALYs
We estimated that the 22 diseases in our study caused 78.7 million (95% UI 65.0–97.7 million) DALYS in 2010, 43% (95% UI 38–48%) in children <5 years of age. The pathogens resulting in the most DALYs were norovirus, Salmonella Typhi, EPEC, V. cholerae, ETEC, and hepatitis A (Table 2).
We estimate that the 22 diseases in our study resulted in 25.2 million (95% UI 17.5–37.0 million) DALYs due to contaminated food; 43% (95% UI 36–50%) in children <5 years of age. Fig 1 shows the relative burden of foodborne enteric infections, if iNTS and NTS were grouped together. The pathogen resulting in the most foodborne DALYs was nontyphoidal S. enterica, if iNTS were included (4.07 million DALYs; 95% UI 2.49–6.27 million DALYs). Other pathogens resulting in substantial foodborne DALYs included: Salmonella Typhi, EPEC, norovirus, and Campylobacter spp. The rates of DALYs for foodborne diseases were highest in the African region. Overall, the 22 diseases transmitted by contaminated food resulted in 10.8 million (95% UI 7.59–15.3 million) DALYs in children <5 years of age compared to 14.3 million (95% UI 9.42–22.5 million) DALYs in those ≥5 years of age.
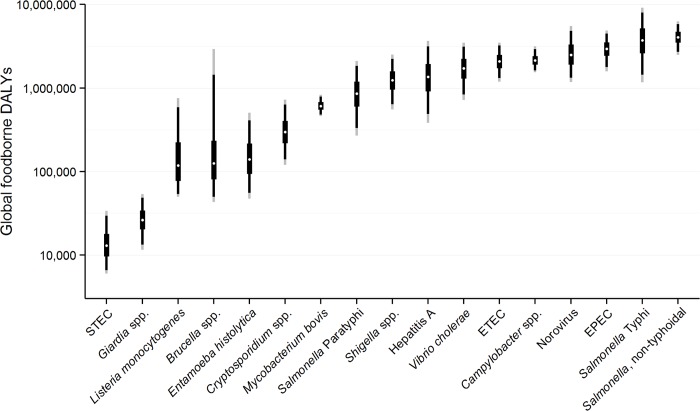
Note figure is on a logarithmic scale. The figure shows the median (white dot); Inter-Quartile Range = 50%UI = 25%/75% percentiles (thick black line); 90% UI = 5%/95% percentiles (thin black line); 95% UI = 2.5%/97.5% percentiles (thin grey line). Note, figure does not include four foodborne intoxications due to Clostridium botulinum, C. perfringens, S. aureus, and Bacillus cereus due to a lack of data for global estimation. In addition, data for non-typhoidal Salmonella enterica infections and invasive non-typhoidal S. enterica have been combined.
Discussion
To our knowledge, we estimate for the first time the substantial worldwide burden of foodborne bacterial, viral, and protozoal diseases in humans, particularly among children. Although children <5 years of age represent only 9% of the global population, we found that 43% of the disease burden from contaminated food occurred in this group. Foodborne illnesses from diarrheal and invasive non-typhoidal S. enterica resulted in the largest disease burden reflecting the ubiquitous nature of Salmonella, the severe nature of illness, and the fact that young children are commonly infected [19]. Large human disease burdens are also imposed by foodborne infections due to norovirus and typhoid. It is important to recognize that diseases with lower burden may still warrant intervention. In particular, certain foodborne diseases may represent a larger problem in some regions. For example, the most substantial burden due to foodborne cholera occurs in African and Asian regions. Similarly, the burden of brucellosis and M. bovis infections were highest in the Middle Eastern and African regions.
To develop these comprehensive estimates of the disease burden of foodborne diseases, we adopted an innovative approach to incorporate the highest quality data available for each foodborne disease [11]. Due to their quality, we gave highest priority to studies with national estimates of foodborne diseases. Since studies with national estimates were only available in a few countries, we adapted the CHERG approach for estimating the disease burden of diarrheal diseases [10,36]. This approach was facilitated by the availability of estimates of the envelope of diarrheal deaths, along with recent advances in diarrheal disease diagnosis, such as widespread application of polymerase chain reaction (PCR) for norovirus detection [20,49].
In our study, norovirus resulted in the largest number of cases of foodborne diseases and overall burden, highlighting the global importance of this agent [20]. However, the disease model we used in the 135 middle- and high-mortality counties included only norovirus infections that resulted in a diarrheal illness. If we also included estimates for norovirus infections that resulted in vomiting without diarrhea, there would be an estimated additional 163 million norovirus cases in these countries [50]. We also found, similar to what has been reported in national studies, that in the countries where we applied the modified CHERG approach, the etiological cause of almost half of diarrheal cases and deaths remained unknown. This was likely due, in large part, to pathogens that are possibly foodborne but with insufficient data for estimation, and unknown agents not yet discovered. In this study, we focused our attention on the burden due to pathogens that were known to be transmitted by contaminated food.
When we examined the human health impact of different pathogens, various serotypes of Salmonella resulted in the greatest foodborne burden. If we consider the combined burden attributable to S. enterica from all invasive (including iNTS, Salmonella Typhi and Salmonella Paratyphi A) and diarrheal infections, there were an estimated 8.76 million (95% UI 5.01–15.6 million) DALYs from all transmission sources and 6.43 million (95% UI 3.08–13.2 million) DALYs from contaminated food. This highlights the significant public health importance of Salmonella infections and the urgency of control, particularly for invasive infections in low- and middle-income settings where most of the mortality occurs [51–53].
Twelve of the diseases included in our study were also included in the GBD 2010 study [25,26,54]. For three diseases (typhoid, paratyphoid and hepatitis A) we used GBD 2010 data to derive estimates of incidence. For the other nine diarrheal diseases, we elected to conduct our own analysis or used updated data from commissioned systematic reviews to derive estimates. Our study builds upon the GBD 2010 study by providing estimates of the proportion of each disease acquired from food; we also provide, in addition to estimates of deaths, estimates of the number of illnesses for each of the diseases [26]. Before we applied our estimate of foodborne proportions to each pathogen, our estimates of the disease burden for a few pathogens, in terms of the estimated number of deaths and DALYs were relatively similar for diseases in common with GBD 2010 and FERG.
However, there were important differences in other estimates. The GBD 2010 study estimates of deaths due to E. histolytica, Cryptosporidium spp., and Campylobacter spp. were 10 times, 4 times and 3 times greater, respectively, than the FERG estimates [25]. The FERG estimate for DALYs for nontyphoidal S. enterica combining diarrheal and invasive infections was 4 times greater than GBD 2010 [54]. The FERG estimates are relatively similar to previous global estimates of cholera, typhoid fever, salmonellosis, and shigellosis [19,24,40,55]. The GBD 2010 study estimates included ‘cysts and liver abscesses’ as a complication of typhoid fever, which has been questioned [56]. However, we understood this categorization to be a proxy for serious typhoid fever and incorporated these data into our estimates. The CFR for each of the diseases included in our estimate are comparable to those reported in national studies. There is a continuing need for high quality studies assessing the foodborne disease burden at the national level. Our methodology for estimating the disease burden attributable to foodborne transmission can be used in future studies.
In comparing the overall burden of our findings, the diseases we included in our study resulted in 79 million DALYs in 2010. This represent approximately 3% of the 2.49 billion DALYs reported in the GBD 2010 study [54]. The GBD 2010 study estimated that approximately 25% of DALYs globally were due to deaths and disability in children younger than 5 years of age, while we estimated that 43% of the DALYs in our study were among children <5 years of age.
There are obvious policy implications of our findings. Countries and international agencies must prioritize food safety to prevent foodborne illness, particularly among young children. The highest burden of disease due to contaminated food was in the African region, largely due to iNTS in children. The considerable regional difference in the burden of foodborne diseases suggests that current control methods exist to avoid an important proportion of the current burden. Our results should stimulate research into the epidemiology of foodborne diseases, with a view to informing prevention efforts. There is an urgent need to identify and implement effective food hygiene interventions in low- and middle-income countries.
A limitation of our estimates of the consequences for human health of foodborne diseases is that, due to data gaps particularly in middle- and high-mortality countries, we included only a few sequelae in our estimates. We did not include data on post-infectious sequelae, such as reactive arthritis and irritable bowel syndrome following foodborne infections. Studies of the burden of enteric pathogens in low-mortality countries highlight that excluding these sequelae under-represents the true burden of disease, but reliable data were not available from middle- and high-mortality countries [57]. Where we did include sequelae, there were insufficient data to account for age-specific effects. We also excluded stillbirths; this exclusion only affected disease burden estimates for L. monocytogenes. A recent review estimated that listeriosis, which we assume is all foodborne, causes 273 stillbirths globally annually [18].
We were unable to distinguish between the effects on health from the condition under study and that of co-morbid conditions, which is common to many studies of human health. For example, salmonellosis and M. bovis infections may occur as HIV co-infections. If FERG included deaths among HIV-infected persons, there would have substantial additional deaths due to invasive nontyphoidal S. enterica deaths, some of which could presumably be averted by improvements in food safety. For some other pathogens in our estimates, such as L. monocytogenes and Cryptosporidium spp., we were unable to account for the excess mortality due to HIV infection due to a lack of reliable data.
Another important limitation of our attempt to quantify the disease burden due of foodborne disease is the inherent difficulty in estimating the proportion of illness acquired from food [58]. We relied on a structured expert elicitation study for these estimates. We were unable to estimate differences in mode of transmission by age, despite this potentially being important. Expert elicitation studies can result in highly variable proportions attributed to food, depending on the nature of experts included in elicitation studies [59,60]. Without specific studies attributing sources of infection, it is difficult to obtain accurate estimates of foodborne transmission, but this finding regarding the need for more attribution studies is an important outcome of our study [61]. For example, the FERG expert elicitation study estimated that 18% of norovirus was foodborne compared with 14% estimated from a recent study based on outbreak genotyping [21].
The major limitation of our study was the lack of reliable data from many regions of the world. In particular, for the most populous regions of the world we had the least data for some pathogens [10]. We tried to use the best data available and attempted to make reasoned assumptions wherever possible [8]. For some agents, such as toxin-mediated illnesses, we elected to limit our estimates to countries where diseases were endemic or where there was sufficient data. Further data on burden of enteric diseases from low- and middle-income settings, particularly high quality epidemiological data, are needed to improve our understanding of foodborne diseases [8,11].
Despite these limitations, our estimates of the disease burden due to 22 foodborne diseases should provide policy makers with information for advocacy for improved regulation and control of foodborne diseases. Of particular importance, we estimate that almost half of the burden of foodborne disease occurs in children under 5 years of age. This previously underappreciated disease burden requires greater attention from governments and resourcing to improve food safety. In our study we highlight regions where diseases, such as tuberculosis and brucellosis are still transmitted by contaminated food. Our results highlight the benefits of countries conducting national studies estimating and attributing the incidence, hospitalizations and deaths due to foodborne diseases to improve understanding of burden and improve control measures. Our regional estimates could be used to fill data gaps for countries attempting to estimate the burden of disease due to contaminated food.
Supporting Information
S2 Text
Information sheets for foodborne agents.(DOCX)
Acknowledgments
We would like to acknowledge the assistance and leadership of the WHO Secretariat over the life of the FERG initiative, particularly Amy Cawthorne, Natsumi Chiba, Tim Corrigan, Tanja Kuchenmüller, Yuki Minato, and Claudia Stein. We acknowledge the Institute for Health Metrics and Evaluation (Seattle, WA, USA) for providing data on the global burden of selected diseases. Our work is based on the excellent contribution of commissioned scientists in providing data for these estimates, including: Marion Koopmans, Benjamin Lopman, Shannon Majowicz, Charline Maertens de Noordhout, Elaine Scallan, Linda Verhoef, along with many others. Anita McGrogan provided an update of her GBS review for FERG. Daniel Graciaa, Danielle Dang, Gregory Mak, and Joshua Kolbert provided exceptional research assistance. We thank other members of the EDTF who contributed to the work of the taskforce, including George Nasinyama, but not to the preparation of this manuscript.
Funding Statement
This study was commissioned and paid for by the World Health Organization (WHO). Copyright in the original work on which this article is based belongs to WHO. The authors have been given permission to publish this article. We acknowledge the support from the Bill & Melinda Gates Foundation that funded CFL, CFW, and REB through the Child Health Epidemiology Reference Group (CHERG). The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
Data Availability
All relevant data are within the paper and its Supporting Information files.
References
Articles from PLOS Medicine are provided here courtesy of PLOS
Full text links
Read article at publisher's site: https://doi.org/10.1371/journal.pmed.1001921
Read article for free, from open access legal sources, via Unpaywall:
https://journals.plos.org/plosmedicine/article/file?id=10.1371/journal.pmed.1001921&type=printable
Citations & impact
Impact metrics
Citations of article over time
Alternative metrics
Smart citations by scite.ai
Explore citation contexts and check if this article has been
supported or disputed.
https://scite.ai/reports/10.1371/journal.pmed.1001921
Article citations
Prevalence of Shigella species and antimicrobial resistance patterns in Africa: systematic review and meta-analysis.
BMC Infect Dis, 24(1):1217, 29 Oct 2024
Cited by: 0 articles | PMID: 39472797 | PMCID: PMC11520789
Review Free full text in Europe PMC
Microbiological contamination of lettuce (Lactuca sativa) reared with tilapia in aquaponic systems and use of bacillus strains as probiotics to prevent diseases: A systematic review.
PLoS One, 19(11):e0313022, 11 Nov 2024
Cited by: 0 articles | PMID: 39527521 | PMCID: PMC11554229
Review Free full text in Europe PMC
The Efficacy of a Ferric Sillen Core-Linked Polymer in Suppressing the Pathogenicity of <i>Campylobacter jejuni</i>.
Animals (Basel), 14(21):3150, 02 Nov 2024
Cited by: 0 articles | PMID: 39518873 | PMCID: PMC11545373
Occurrence of Extended-spectrum β-lactamase (ESBL) and Carbapenemase-producing Escherichia coli isolated from Childhood Diarrhoea in Yaoundé, Cameroon.
BMC Microbiol, 24(1):401, 10 Oct 2024
Cited by: 0 articles | PMID: 39385062 | PMCID: PMC11465673
Characterization of the <i>Bacillus cereus</i> Group Isolated from Ready-to-Eat Foods in Poland by Whole-Genome Sequencing.
Foods, 13(20):3266, 14 Oct 2024
Cited by: 0 articles | PMID: 39456328 | PMCID: PMC11506886
Go to all (635) article citations
Data
Data behind the article
This data has been text mined from the article, or deposited into data resources.
BioStudies: supplemental material and supporting data
Similar Articles
To arrive at the top five similar articles we use a word-weighted algorithm to compare words from the Title and Abstract of each citation.
World Health Organization Estimates of the Global and Regional Disease Burden of 11 Foodborne Parasitic Diseases, 2010: A Data Synthesis.
PLoS Med, 12(12):e1001920, 03 Dec 2015
Cited by: 317 articles | PMID: 26633705 | PMCID: PMC4668834
Review Free full text in Europe PMC
World Health Organization Global Estimates and Regional Comparisons of the Burden of Foodborne Disease in 2010.
PLoS Med, 12(12):e1001923, 03 Dec 2015
Cited by: 631 articles | PMID: 26633896 | PMCID: PMC4668832
Review Free full text in Europe PMC
Aetiology-Specific Estimates of the Global and Regional Incidence and Mortality of Diarrhoeal Diseases Commonly Transmitted through Food.
PLoS One, 10(12):e0142927, 03 Dec 2015
Cited by: 200 articles | PMID: 26632843 | PMCID: PMC4668836
Burden of Disease Estimates of Seven Pathogens Commonly Transmitted Through Foods in Denmark, 2017.
Foodborne Pathog Dis, 17(5):322-339, 22 Nov 2019
Cited by: 12 articles | PMID: 31755845





