Abstract
Free full text

Expression and nucleotide sequence of a plasmid-determined divalent cation efflux system from Alcaligenes eutrophus.
Abstract
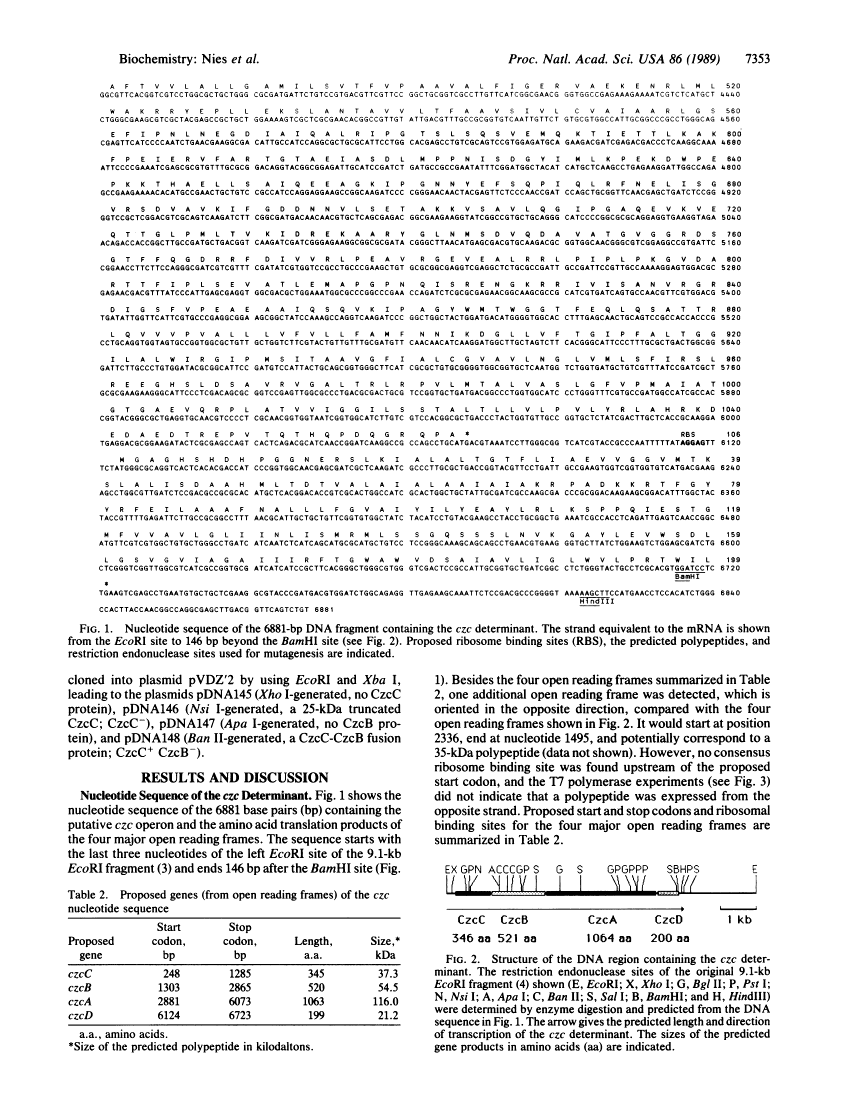
Resistance to cobalt, zinc, and cadmium specified by the czc determinant on plasmid pMOL30 in Alcaligenes eutrophus results from a cation efflux system. Five membrane-bound polypeptides that were expressed in Escherichia coli from this determinant under the control of a phage T7 promoter were assigned to four open reading frames identified in the nucleotide sequence of the 6881-base-pair fragment containing the czc putative operon. The contributions of the polypeptides to the cation efflux system were analyzed with deletion derivatives of the 6.9-kilobase fragment, constructed, and expressed in E. coli under the control of the phage T7 promoter and in A. eutrophus under the control of the lac promoter.
Full text
Full text is available as a scanned copy of the original print version. Get a printable copy (PDF file) of the complete article (1.1M), or click on a page image below to browse page by page. Links to PubMed are also available for Selected References.
Images in this article
Click on the image to see a larger version.
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Silver S, Misra TK. Plasmid-mediated heavy metal resistances. Annu Rev Microbiol. 1988;42:717–743. [Abstract] [Google Scholar]
- Mergeay M, Nies D, Schlegel HG, Gerits J, Charles P, Van Gijsegem F. Alcaligenes eutrophus CH34 is a facultative chemolithotroph with plasmid-bound resistance to heavy metals. J Bacteriol. 1985 Apr;162(1):328–334. [Europe PMC free article] [Abstract] [Google Scholar]
- Nies D, Mergeay M, Friedrich B, Schlegel HG. Cloning of plasmid genes encoding resistance to cadmium, zinc, and cobalt in Alcaligenes eutrophus CH34. J Bacteriol. 1987 Oct;169(10):4865–4868. [Europe PMC free article] [Abstract] [Google Scholar]
- Nies DH, Silver S. Plasmid-determined inducible efflux is responsible for resistance to cadmium, zinc, and cobalt in Alcaligenes eutrophus. J Bacteriol. 1989 Feb;171(2):896–900. [Europe PMC free article] [Abstract] [Google Scholar]
- Yanisch-Perron C, Vieira J, Messing J. Improved M13 phage cloning vectors and host strains: nucleotide sequences of the M13mp18 and pUC19 vectors. Gene. 1985;33(1):103–119. [Abstract] [Google Scholar]
- Deretic V, Chandrasekharappa S, Gill JF, Chatterjee DK, Chakrabarty AM. A set of cassettes and improved vectors for genetic and biochemical characterization of Pseudomonas genes. Gene. 1987;57(1):61–72. [Abstract] [Google Scholar]
- Misra TK. DNA sequencing: a new strategy to create ordered deletions, modified M13 vector, and improved reaction conditions for sequencing by dideoxy chain termination method. Methods Enzymol. 1987;155:119–139. [Abstract] [Google Scholar]
- Tabor S, Richardson CC. A bacteriophage T7 RNA polymerase/promoter system for controlled exclusive expression of specific genes. Proc Natl Acad Sci U S A. 1985 Feb;82(4):1074–1078. [Europe PMC free article] [Abstract] [Google Scholar]
- Nies A, Nies DH, Silver S. Cloning and expression of plasmid genes encoding resistances to chromate and cobalt in Alcaligenes eutrophus. J Bacteriol. 1989 Sep;171(9):5065–5070. [Europe PMC free article] [Abstract] [Google Scholar]
- Ames GF. Resolution of bacterial proteins by polyacrylamide gel electrophoresis on slabs. Membrane, soluble, and periplasmic fractions. J Biol Chem. 1974 Jan 25;249(2):634–644. [Abstract] [Google Scholar]
- Novick RP, Murphy E, Gryczan TJ, Baron E, Edelman I. Penicillinase plasmids of Staphylococcus aureus: restriction-deletion maps. Plasmid. 1979 Jan;2(1):109–129. [Abstract] [Google Scholar]
- Silver S, Nucifora G, Chu L, Misra TK. Bacterial resistance ATPases: primary pumps for exporting toxic cations and anions. Trends Biochem Sci. 1989 Feb;14(2):76–80. [Abstract] [Google Scholar]
- Nucifora G, Chu L, Misra TK, Silver S. Cadmium resistance from Staphylococcus aureus plasmid pI258 cadA gene results from a cadmium-efflux ATPase. Proc Natl Acad Sci U S A. 1989 May;86(10):3544–3548. [Europe PMC free article] [Abstract] [Google Scholar]
- Tynecka Z, Gos Z, Zajac J. Energy-dependent efflux of cadmium coded by a plasmid resistance determinant in Staphylococcus aureus. J Bacteriol. 1981 Aug;147(2):313–319. [Europe PMC free article] [Abstract] [Google Scholar]
- Weiss AA, Silver S, Kinscherf TG. Cation transport alteration associated with plasmid-determined resistance to cadmium in Staphylococcus aureus. Antimicrob Agents Chemother. 1978 Dec;14(6):856–865. [Europe PMC free article] [Abstract] [Google Scholar]
Associated Data
Articles from Proceedings of the National Academy of Sciences of the United States of America are provided here courtesy of National Academy of Sciences
Full text links
Read article at publisher's site: https://doi.org/10.1073/pnas.86.19.7351
Read article for free, from open access legal sources, via Unpaywall:
https://europepmc.org/articles/pmc298059?pdf=render
Citations & impact
Impact metrics
Citations of article over time
Article citations
A flow equilibrium of zinc in cells of Cupriavidus metallidurans.
J Bacteriol, 206(5):e0008024, 25 Apr 2024
Cited by: 2 articles | PMID: 38661374 | PMCID: PMC11112998
Diverse hydrocarbon degradation genes, heavy metal resistome, and microbiome of a fluorene-enriched animal-charcoal polluted soil.
Folia Microbiol (Praha), 69(1):59-80, 14 Jul 2023
Cited by: 1 article | PMID: 37450270
Interplay between Two-Component Regulatory Systems Is Involved in Control of Cupriavidus metallidurans Metal Resistance Genes.
J Bacteriol, 205(4):e0034322, 09 Mar 2023
Cited by: 3 articles | PMID: 36892288 | PMCID: PMC10127602
Taxonomic and genomic characterization of Sporosarcina cyprini sp. nov., moderately tolerant of Cr+6 and Cd+2 isolated from the gut of invasive fish Cyprinus carpio var. communis (Linn., 1758).
Antonie Van Leeuwenhoek, 116(3):193-206, 18 Nov 2022
Cited by: 0 articles | PMID: 36400900
Zinc Essentiality, Toxicity, and Its Bacterial Bioremediation: A Comprehensive Insight.
Front Microbiol, 13:900740, 31 May 2022
Cited by: 20 articles | PMID: 35711754 | PMCID: PMC9197589
Review Free full text in Europe PMC
Go to all (135) article citations
Data
Data behind the article
This data has been text mined from the article, or deposited into data resources.
BioStudies: supplemental material and supporting data
Similar Articles
To arrive at the top five similar articles we use a word-weighted algorithm to compare words from the Title and Abstract of each citation.
Nucleotide sequence and expression of a plasmid-encoded chromate resistance determinant from Alcaligenes eutrophus.
J Biol Chem, 265(10):5648-5653, 01 Apr 1990
Cited by: 82 articles | PMID: 2180932
Characterization of the inducible nickel and cobalt resistance determinant cnr from pMOL28 of Alcaligenes eutrophus CH34.
J Bacteriol, 175(3):767-778, 01 Feb 1993
Cited by: 125 articles | PMID: 8380802 | PMCID: PMC196216
CzcR and CzcD, gene products affecting regulation of resistance to cobalt, zinc, and cadmium (czc system) in Alcaligenes eutrophus.
J Bacteriol, 174(24):8102-8110, 01 Dec 1992
Cited by: 103 articles | PMID: 1459958 | PMCID: PMC207549
Plasmids for heavy metal resistance in Alcaligenes eutrophus CH34: mechanisms and applications.
FEMS Microbiol Rev, 14(4):405-414, 01 Aug 1994
Cited by: 49 articles | PMID: 7917428
Review