Abstract
Free full text

TRIM21 ubiquitylates SQSTM1/p62 and suppresses protein sequestration to regulate redox homeostasis
Associated Data
Summary
TRIM21 is a RING finger domain-containing ubiquitin E3 ligase whose expression is elevated in autoimmune disease. While TRIM21 plays an important role in immune activation during pathogen infection, little is known about its inherent cellular function. Here we show that TRIM21 plays an essential role in redox regulation by directly interacting with SQSTM1/p62 and ubiquitylating p62 at lysine(K)7 via K63-linkage. As p62 oligomerizes and sequesters client proteins in inclusions, the TRIM21-mediated p62 ubiquitylation abrogates p62 oligomerization and sequestration of proteins including Keap1, a negative regulator of antioxidant response. TRIM21-deficient cells display an enhanced antioxidant response and reduced cell death in response to oxidative stress. Genetic ablation of TRIM21 in mice confers protection from oxidative damages caused by arsenic-induced liver insult and pressure overload heart injury. Therefore, TRIM21 plays an essential role in p62-regulated redox homeostasis and may be a viable target for treating pathological conditions resulting from oxidative damage.
Introduction
Cellular redox regulation plays an essential role in organismal homeostasis. A major mechanism for cell redox homeostasis regulation is the Keap1 (Kelch-like ECH-associated protein 1)-Nrf2 (Nuclear factor erythroid 2-related factor 2) pathway (Harder et al., 2015), which can be regulated by SQSTM1/p62 (Katsuragi et al., 2015). p62 is a ubiquitin-binding protein that is enriched in protein aggregates (Shin, 1998) whose expression was found to be induced by oxidative stress (Ishii et al., 1996). One of Keap1’s functions is to act as a scaffold for the ubiquitin ligase Cullin 3, Ring-box 1 (Rbx1), and Nrf2 complex. Nrf2 is the transcriptional factor that, when in the nucleus, activates the expression of antioxidant enzymes by binding to the antioxidant response element (ARE) in their promoter region. Under normal conditions, Keap1 interacts with Nrf2 to retain the latter in the cytoplasm where Nrf2 is degraded via the ubiquitin-proteasome pathway (Kobayashi et al., 2004; Zhang et al., 2004). In response to proteotoxic and oxidative stress, p62 forms a homodimer which facilitates its oligomerization and interaction with ubiquitylated proteins that promotes their aggregation thereby sequestering the damaged or toxic proteins. A known mechanism for p62 dimerization is via the K7-D69 hydrogen bond in its PB1 domain (Wilson et al., 2003). The p62-mediated protein aggregate cargo can be subsequently delivered to autophagosomes for autophago-lysosomal degradation. As one of the protein inclusion client proteins is Keap1, the p62-mediated Keap1 sequestration and degradation frees Nrf2 from the Keap1 inhibition, allowing for its stabilization, nuclear translocation, transcriptional activation, and antioxidant response (Komatsu et al., 2010; Lau et al., 2010). In this study, we found that TRIM21 (Tripartite motif-containing protein 21), a ubiquitin E3 ligase, directly interacts with and ubiquitylates p62 to regulate its sequestration and cellular redox homeostasis.
Results
p62 is ubiquitylated at residue K7 via the K63-linkage
In our laboratory practice, we have constantly noticed that certain stressors such as the proteasome inhibitor MG132 enhances an upper shift of p62 in SDS-PAGE, suggesting a post-translational modification. Indeed, by transfecting cells with His-tagged ubiquitin and pull-down with nickel beads, p62 ubiquitylation was detected, which was further enhanced upon MG132 treatment (Fig. 1A). Serial deletion mutants of p62 revealed that deletion of the N-terminal 3–117 residues led to significant decrease of p62 ubiquitylation (Suppl. Fig. S1A). Lysine (K) to arginine (R) point mutations in the 3–117 region identified residue K7 as the critical residue for p62 ubiquitylation (Suppl. Fig. S1B and Fig. 1B). To determine whether p62(K7) ubiquitylation is via K48 or K63-linkage, K48-only or K63-only ubiquitin was expressed together with Flag and His-doubly tagged p62 (Fig. 1C). K48-only ubiquitin led to reduced levels of p62 ubiquitylation, possibly owing to the increased proteasomal degradation promoted by K48-linked ubiquitylation, whereas K63-only ubiquitin led to enhanced p62 ubiquitylation (Fig. 1C, p62wt lanes of Ni-NTA). This result suggests that p62 is ubiquitylated by both K48- and K63-linkages. Markedly, the K63-linked ubiquitylation was largely decreased in p62K7R mutant (Fig. 1C), indicating that K7 is ubiquitylated via K63-linkage.
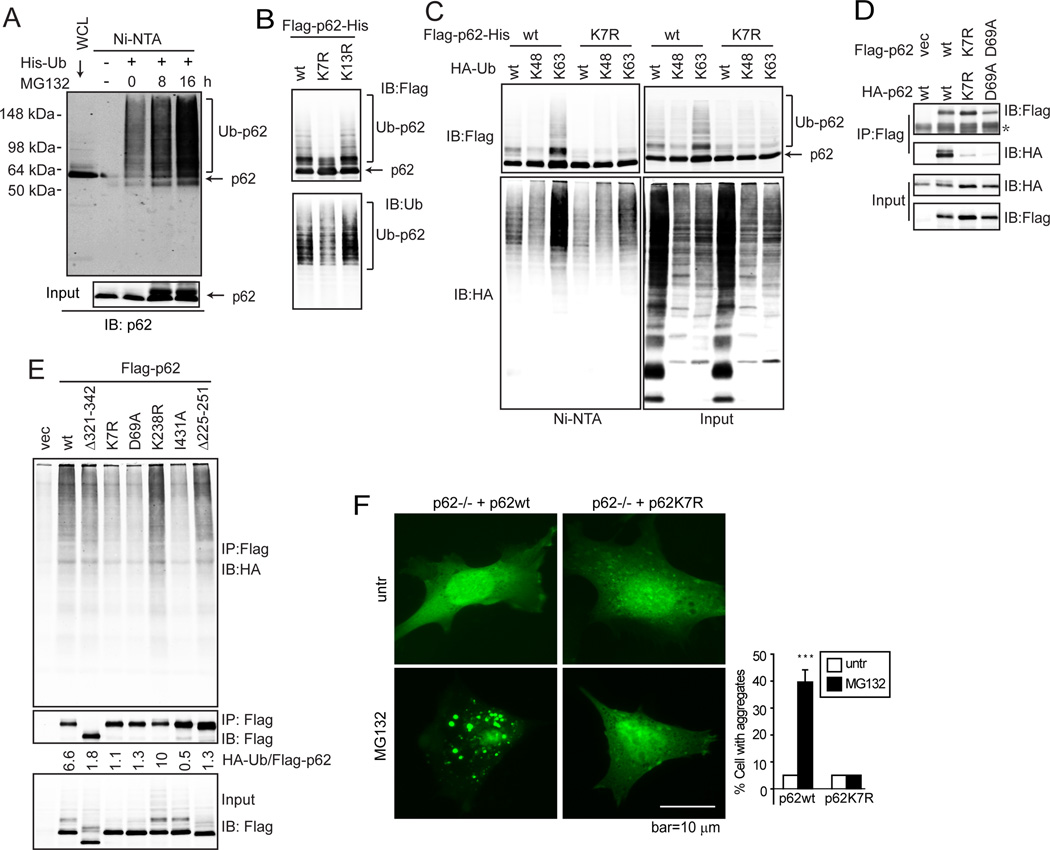
(A) Baby mouse kidney (BMK) cells were transfected with His-Ub for 24 h, then treated with MG132 (0.25 µM) and subjected to His pull-down (Ni-NTA) and immunoblotting (IB). (B) Flag- and His-doubly tagged p62wt, K7R, and K13R were transfected into HEK293T cells for 36 h. Cells were subjected to His-pull down and IB. (C) Flag and His-doubly tagged p62wt and K7R were co-transfected with HA-Ub wt, K48-only, and K63-only mutants in HEK293T cells. Cells were subjected to His pull-down and IB. (D) Indicated p62 mutants were co-expressed in HEK293T cells. Cells were subjected to His-pull down and IB. (E) Flag-tagged p62wt or indicated mutants were expressed in HEK293T cells. HA-Ub was expressed in another set of HEK293T cells. 24 h post transfection, p62 or mutant-expressing cell lysates were incubated with the same amount of the HA-Ub expressing cell lysates for 3 h. The mixtures were subjected to IP with FLAG antibody then IB with HA antibody. (F) p62−/− MEFs stably expressing GFP-LC3 were reconstituted with p62wt and p62K7R. Cells were treated with MG132 (0.25 µM) for 12 h, then observed under deconvolution microscope. The percentage of cells with GFP-LC3 aggregates was quantified by counting over 50 cells in each sample. Data shown are the mean plus S.D. of three countings. ***p<0.01.
Next we determined whether the K7 ubiquitylation affects p62 functions, one of which is to bind polyubiquitylated proteins and sequester them in aggregates (Moscat and Diaz-Meco, 2009). This is dependent on the ubiquitin-binding and dimerization properties of p62 (Bjorkoy et al., 2005). Using the p62 mutants tagged by either Flag or HA, we found that the K7R mutant lost its ability to form a homo-dimer (Fig. 1D), similar to the D69A dimerization deficient mutant (Bjorkoy et al., 2005). Importantly, the failure of dimerization led to the failure of p62 binding to polyubiquitylated proteins as seen for both the K7R and D69A mutants, similar to the ubiquitin-binding deficient mutant I431A (Seibenhener et al., 2004) (Fig. 1E). As ubiquitin-binding is essential for p62’s function to pack polyubiquitylated proteins into aggregates, we then tested whether K7R is able to mediate aggregate formation. Using MEFs that stably express GFP-LC3, we observed that while starvation-induced formation of LC3+-autophagosomes was not markedly affected, MG132-induced aggregate formation was abolished in p62−/− MEFs (Suppl. Fig. S1C). Reconstitution of p62−/− MEFs with wild-type p62 restored MG132-induced aggregate formation indicated by large-size LC3 puncta using either GFP-LC3 or endogenous LC3, whereas the K7R mutant failed to do so (Fig. 1F and Suppl. Fig. S1D). Taken together, these data indicate that p62 is ubiquitylated at K7 via K63-linkage, and that K7 is critical for p62 dimerization, ubiquitin binding, as well as aggregate formation.
TRIM21 directly ubiquitylates p62 at residue K7 to inhibit its oligomerization and sequestration function
The failure of the K7R mutant to dimerize suggests two opposite possible scenarios: 1) Modification of K7 by ubiquitylation promotes p62 dimerization via the ubiquitin chain; or 2) Unmodified K7 is essential for p62 dimerization and its modification by ubiquitylation leads to the loss of function of p62. Importantly, an X-ray crystallography analysis has demonstrated that K7 is a basic cluster residue that would form a hydrogen bond with its corresponding acidic cluster residue D69 (Wilson et al., 2003). This supports the latter option that the non-ubiquitinated form of K7 is critical for p62 dimerization. To further understand the nature of p62 K7 ubiquitylation, and to unequivocally demonstrate that p62 K7 ubiquitylation plays a negatively regulates p62 dimerization and aggregation, we sought to identify the ubiquitin E3 ligase that catalyzes the p62 K7 ubiquitylation. In an in vitro ubiquitylation assay, we noticed that the immunoprecipitated Flag-p62 samples from HEK293T cells led to p62 ubiquitylation in the presence of purified E1, E2, and ubiquitin without the addition of an exogenous E3 (Fig. 2A). This suggests that an E3 enzyme was co-immunoprecipitated in the Flag-p62 IP samples. Indeed, mass spectrometry of Flag-p62 IP samples identified two putative E3 ligases: TRIM21 and RNF138. Expression of TRIM21 but not RNF138 led to p62 ubiquitylation (Fig. 2B and 2C), suggesting that TRIM21 is the E3 ligase for p62. A ligase-dead (LD) mutant TRIM21(C16A, C31A, and H33W) (Wada and Kamitani, 2006) revealed that the E3 ligase activity of TRIM21 was required for p62 ubiquitylation (Fig. 2D). An in vitro ubiquitylation assay further revealed that TRIM21 directly ubiquitylates p62 (Fig. 2E) via the K63-linkage (Fig. 2F), which was abolished in the p62K7R mutant (Fig. 2G). Silencing of TRIM21 abolished p62 ubiquitylation both at the basal level and upon MG132 treatment (Fig. 2H). Therefore, TRIM21 is a direct ubiquitin E3 ligase for the p62 residue K7.
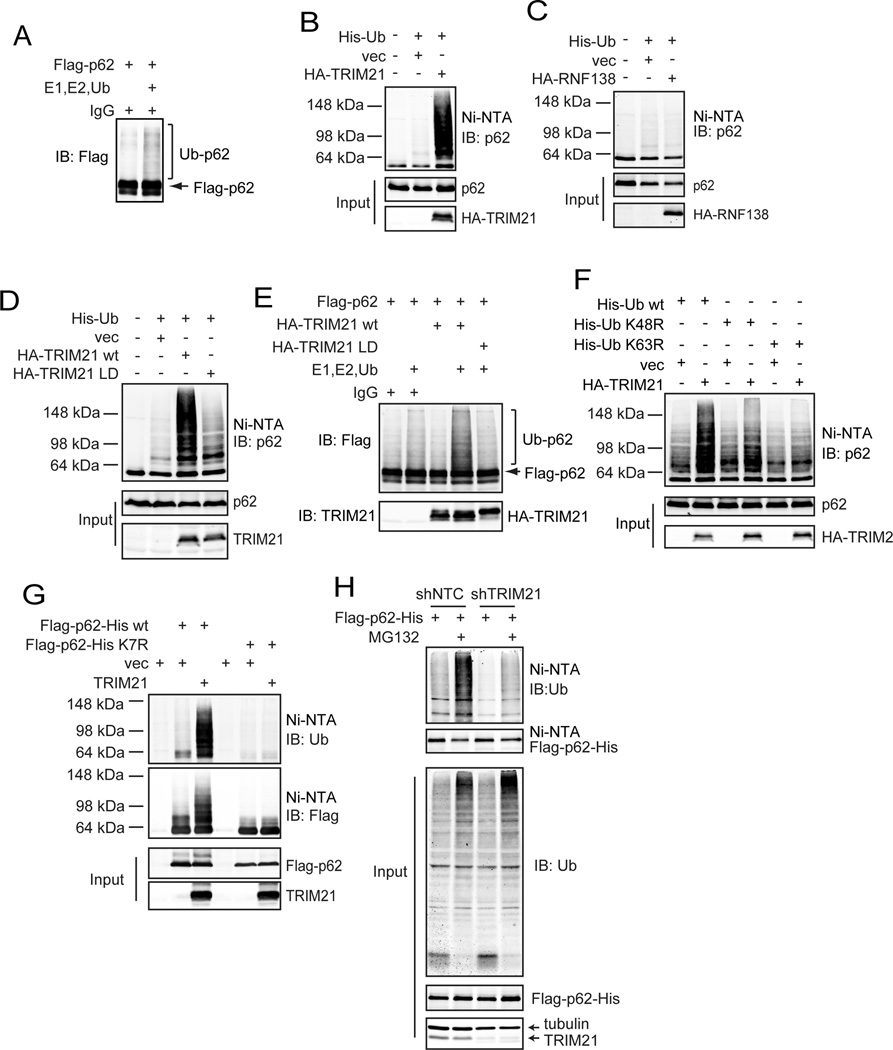
(A) Flag-p62 was expressed in HEK293T cells, and subjected to IP using the Flag antibody and eluted using Flag peptides. The eluates were incubated with in vitro ubiquitination assay system containing E1, E2, and ubiquitin, then subjected to IB. (B and C) His-Ub was transfected with vector and HA-TRIM21 (B) or HA-RNF138 (C) into HEK293T cells. 24 h later, cells were subjected to His pull-down followed by IB. (D) His-Ub was co-transfected with HA-TRIM21wt or HA-TRIM21-LD mutant in HEK293T cells. Cells were subjected to His pull-down and IB. (E) Purified Flag-p62 was incubated with IgG control, purified HA-TRIM21wt or LD mutant in the ubiquitination assaying mix, and probed for indicated proteins. (F) His-Ub (wt, K48R, and K63R) mutants were expressed with or without HA-TRIM21 in HEK293T cells. Cells were subjected to His pull-down and IB. (G) Flag- and His-doubly tagged p62wt or K7R mutant were co-expressed with vector or TRIM21 in HEK293T cells. Cells were subjected to His-pull down and IB. (H) MD-MBA-231 with stable expression of Flag-p62-His were infected with lentiviral non-targeting control (NTC) shRNA or shTRIM21. Cells were treated with MG132, then subjected to His pull-down and IB.
As the p62K7R mutant was unable to dimerize (Fig. 1D) and to promote protein aggregation (Fig. 1F), we tested whether TRIM21 plays a positive or negative role in p62 dimerization and aggregation using three approaches. First, we used the bimolecular fluorescence complementation (BiFC) assay (Huang et al., 2010). p62wt or the K7R mutant was fused to the split Venus proteins, Venus N(1–173) and Venus C(155–239), and expressed in HEK293T cells (Fig. 3A). BiFC was observed when p62wt-VN and p62wt-VC were co-expressed (Suppl. Fig. S2A and Fig. 3B), but not with the K7R mutants (Fig. 3B). Induction of wild-type TRIM21 using the estrogen receptor inducing system led to reduced BiFC, whereas the ubiquitin ligase-dead (LD) mutant TRIM21-LD failed to do so (Suppl. Fig. S2B and Fig. 3C), indicating that TRIM21 decreases p62 oligomerization in an E3 ligase activity-dependent manner.
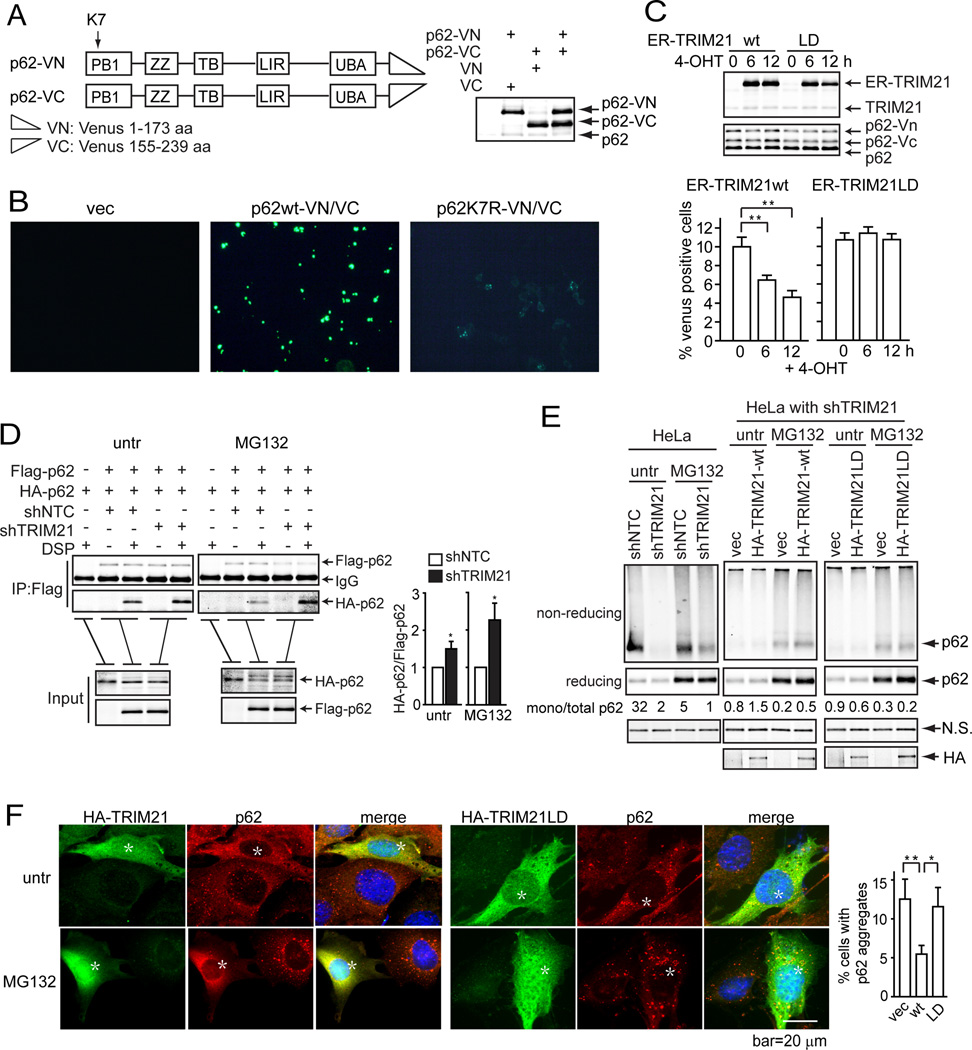
(A) Schematic for the strategy constructing p62 fused with Venus N-terminus (p62-VN) or C-terminus (p62-VC). p62-VN and p62-VC were expressed individually or simultaneously in HEK293T cells. The expression was determined by IB. (B) Wild-type or K7R p62-VN/p62-VC were expressed in HEK293T cells. Representative fluorescence microscopic images are shown. (C) p62-VN or p62-VC were expressed with estrogen receptor (ER)-fused TRIM21wt or LD mutant in HEK293T cells. 16 h later, cells were treated with 4-OHT for indicated times to induce TRIM21 expression (top panel). Venus fluorescence was determined by flow cytometry and quantified (bottom panel). (D) HeLa cells stably expressing both Flag- and HA-p62 were infected with lentivirus shNTC and shTRIM21. Cells were used for Flag IP, and IB for HA. The amount of HA-p62 was quantified by ImageJ and is shown in the right panel. *p<0.05. (E) HeLa with stable shTRIM21 were reconstituted with shRNA-resistant TRIM21wt or TRIM21 LD. Cells treated with MG132, then crosslinked with DSP, and lysed in IP lysis buffer with 1% SDS. The cells lysates mixed with loading buffer containing or not containing β-mercaptoethanol were subjected to IB. (F) TRIM21−/− MEFs reconstituted with TRIM21wt or LD were treated with MG132 (0.25 µM) for 12 h. Cells were used for IF using anti-TRIM21 (green) and anti-p62 (red) antibodies, and observed under deconvolution microscope. Cells with aggregates were quantified by blind counting. Shown on the right is the mean plus S.D. of three countings. *p<0.05, **p<0.01. Cells with high expression of TRIM21 wt or LD are marked with an asterisk.
Alternatively, we examined p62 oligomerization by co-expressing p62 tagged with either Flag or HA epitopes. Since a significant amount of p62 resides in protein aggregates, especially under the condition of proteotoxic stress such as treatment of MG132, we used the reversible crosslinking agent dithiobis succinimidyl propionate (DSP) to treat the cells then lysed the cells in strong detergent (1% SDS) to solubilize all proteins. The lysates were subjected to co-IP upon diluting the detergent to 0.1% SDS. Indeed, TRIM21 silencing led to increased p62 oligomerization at both basal condition and upon MG132 treatment (Fig. 3D). Moreover, since p62 oligomerization is important for its sequestration function, we determined whether p62-mediated aggregation is regulated by TRIM21. To this end, we detected the presence of p62 in aggregates by treating cells with the crosslinking agent DSP and running the cell lysates under either reducing or non-reducing condition (Suppl. Fig. 2C). Under the reducing condition, total p62 would be detected with a migration of 62 kDa, whereas under the non-reducing condition, only p62 monomers would be detected at 62 kDa and the oligomers or those conjugated with other proteins would display such a slow migration that it would not run through the gel (illustrated in Suppl. Fig. S2C). Therefore, higher ratio of the 62 kDa band in the non-reducing vs reducing conditions would indicate reduced p62 oligomerization or sequestration. MG132 treatment led to less p62 monomer in the non-reducing condition (Fig. 3E), indicating p62 aggregation and sequestration. TRIM21 silencing led to further decreased p62 monomer hence increased aggregation and sequestration (Fig. 3E). In TRIM21 knockdown cells, reconstitution of TRIM21wt but not the LD mutant led to increased p62 monomer indicating decreased p62 aggregation and sequestration (Fig. 3E).
The role of TRIM21 in negatively regulating p62 oligomerization was further supported using immunofluorescence analysis and fractionation assay, which shows that reconstitution of TRIM21wt but not the LD mutant led to diminishment of MG132-induced p62 aggregates (Fig. 3F and Suppl. Fig. S2D). Consistent with these findings, TRIM21 silencing led to increased p62 binding to ubiquitylated proteins (Suppl. Fig. S2E), and reconstitution of TRIM21wt but not the LD mutant led to diminished ubiquitin aggregates in TRIM21−/− cells (Suppl. Fig. S2F).
In addition to the proteasome inhibitor MG132, we also examined other conditions that are known to induce p62 aggregation including genetic ablation of autophagy-essential proteins Atg7 and Vps34. In Atg7flox/flox (Komatsu et al., 2005) and Vps34flox/flox (Jaber et al., 2012) MEFs, ablation of respective genes by Cre expression led to decreased p62 monomer in the non-reducing condition as a result of accumulated p62 aggregation due to the failure of autophagic clearance of p62 (Suppl. Fig. S3A and S3B). Overexpression of wild-type TRIM21, but not the LD mutant, led to increased amount of p62 monomer (Suppl. Fig. S3A and S3B), indicating that TRIM21 prevents p62 aggregation under the condition of genetic inhibition of autophagy. Consistently, the increased amount of insoluble polyubiquitylated proteins in Atg7−/− cells was reduced by the expression of TRIM21wt but not the LD mutant (Suppl. Fig. S3C). Taken together, these data indicate that residue K7 is critical for p62 dimerization, aggregation, and sequestration function, and that TRIM21 negatively regulates these functions by ubiquitylating p62 at residue K7.
TRIM21 interaction with p62 is via the PRYSPRY domain and is enhanced by proteotoxic stress
We next sought to characterize the molecular nature of the interaction between p62 and TRIM21. Two p62 deletion mutants Δ3–117 and Δ117–200 showed significantly decreased interaction with TRIM21 (Fig. 4A). Further deletion mutation revealed that the two domains a.a.3–61 and 163–200 had lost TRIM21 interaction (Fig. 4B). In TRIM21, deletion of a.a.252–476, which spans the PRYSPRY domain that is conserved among numerous TRIM family members and is important for their anti-viral function (Nisole et al., 2005; Song et al., 2005), led to a near complete loss of p62 interaction (Fig. 4C). It has been shown that residues D355, W381, W383, and F450 within the TRIM21 PRYSPRY domain are clustered at the center of the interface that interacts with IgG Fc (James et al., 2007). We reasoned that these residues may also be critical for TRIM21 interaction with other proteins. Indeed, the TRIM21W381/383A mutant abolished p62 interaction (Fig. 4D), p62 ubiquitylation (Fig. 4E), and the suppression of p62 aggregation (Fig. 4F). The importance of the TRIM21 ligase activity was also determined by the autophagic degradation of p62. Wild-type p62 was degraded by starvation-induced autophagy, but p62K7R, which is deficient in oligomerization (Fig. 1C), was not (Suppl. Fig. S4A). This is consistent with the notion that p62 autophagic degradation is dependent on p62 dimerization and aggregation (Itakura and Mizushima, 2011). Importantly, wild-type TRIM21 but not the W381/383A mutant suppressed starvation-induced p62 autophagic degradation (Suppl. Fig. S4B). These data indicate that W381 and W383 are required for TRIM21 interaction with p62 and for its function to ubiquitylate p62 and suppress p62 oligomerization.
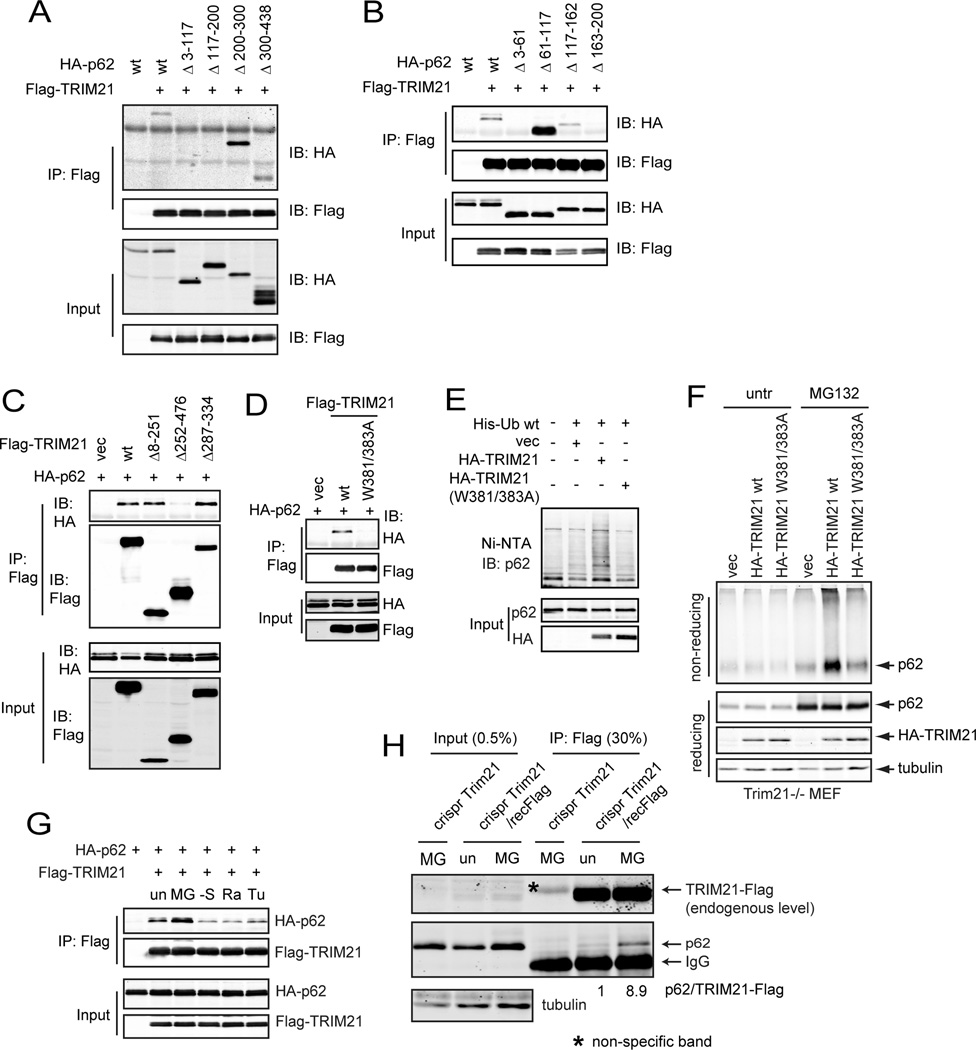
(A–C) Flag-tagged TRIM21 and HA-tagged p62wt or their truncation mutants were expressed in indicated combinations in HEK293T cells for 24 h. Cells were subjected to Flag IP and IB. p62 lost interaction with TRIM21 Δ213–117 and Δ117–200 (A). TRIM21 lost interaction with p62 Δ3–61, Δ117–162, and Δ163–200 (B). p62 lost interaction with TRIM21 Δ252–476 (C). (D) Flag-tagged TRIM21wt or W381/383A mutant and HA-tagged p62 were expressed in HEK293T cells for 24 h. Cells were subjected to Flag IP and IB. (E) HA-tagged TRIM21wt or W381/383A mutant and His-Ub were expressed in HEK293T cells. Cells were subjected to His pull-down and IB. (F) TRIM21−/− MEFs reconstituted with vector, TRIM21wt, and TRIM21 W381/383A mutant were treated with DSP and lysed in IP lysis buffer with 1% SDS. Lysates were mixed with loading buffer with/without β-mercaptoethanol and subjected to IB. (G) HA-tagged p62 and Flag-tagged TRIM21 were expressed in HEK293T cells for 24 h. Cells were treated with MG132 (0.5 µM) for 16 h, rapamycin (50 nM) for 16 h, tunicamycin (0.5 µg/ml) for 16 h, or cultured in serum-free medium for 6 h. Cells were subjected to Flag IP and IB. (H) A vector control or Flag tag was introduced into MEFs to fuse with the endogenous TRIM21 by CRISPR. Cells were treated with MG132 (0.25 µM) for 12 h, then subjected to Flag IP and IB.
We then studied how TRIM21-mediated p62 ubiquitylation might be regulated. In a co-overexpression system, treatment of MG132 but not serum starvation, rapamycin, or tunicamycin led to increased interaction between TRIM21 and p62 (Fig. 4G). Since the available TRIM21 and p62 antibodies were not suitable for immunoprecipitation, we introduced a Flag tag to the endogenous TRIM21 gene via CRISPR gene editing in MEFs (Suppl. Fig. S4C), so that Flag-tagged TRIM21 is expressed at the endogenous level. Co-IP analysis using a Flag antibody showed that MG132 induced the interaction between TRIM21 and p62 (Fig. 4H). These results are consistent with our observation that p62 ubiquitylation is enhanced upon MG132 treatment, and suggest that the TRIM21-mediated p62 ubiquitylation is a cell stress adaptation mechanism in response to proteasome inhibition in order to maintain a hemostatic level of functional p62.
TRIM21 negatively regulates p62-mediated sequestration of Keap1 and antioxidant response
An important function of p62 is to retain proteins in aggregates (sequestosomes) for subsequent autophagic degradation. One of the client proteins is Keap1, which negatively regulates the antioxidant response by interacting and preventing the nuclear accumulation of the transcriptional factor Nrf2, the master regulator to activate antioxidant gene transcription (Komatsu et al., 2010). As our results thus far indicate that TRIM21 negatively regulates p62-mediated protein sequestration by ubiquitylating p62 at K7, we sought to study whether this mechanism has an impact on cell’s antioxidant response. To this end, we used sodium arsenate (As(III)), which induces a detrimental level of intracellular reactive oxygen species (Barchowsky et al., 1996; Lynn et al., 1998). We first used p62−/− MEFs reconstituted with either p62wt or p62K7R. While As(III) treatment did not induce Keap1 aggregation in p62−/− MEFs, reconstitution of p62wt but not p62K7R led to aggregation of both p62 and Keap1 and their accumulation in the insoluble fraction (Fig. 5A and 5B). The ability of p62wt to sequester Keap1 was also indicated by increased Nrf2 and NQO1 expression (Fig. 5C and 5D). This As(III)-induced p62 sequestration is likely to be negatively regulated by TRIM21 since As(III) induced p62-TRIM21 interaction at the endogenous level in the TRIM21-Flag knock-in cells (Fig. 5E). This As(III)-induced p62 aggregation is suppressed by TRIM21wt but not the W381/383A mutant (Fig. 5F and Suppl. Fig. S5A). TRIM21 reconstitution led to decreased p62 and Keap1 in the insoluble fraction (Fig. 5G), decreased Nrf2 in the nucleus (Fig. 5H), and decreased Nrf2 stability (Fig. 5I and Suppl. Fig. S5B). Consistently, wild-type TRIM21 showed low level of NQO1 promoter activation (Fig. 5J) and increased oxidative stress (Fig. 5K). Together, these data indicate that p62 plays an antioxidant role by sequestrating Keap1 in response to As(III), and that TRIM21 negatively regulates this function by ubiquitylating p62.
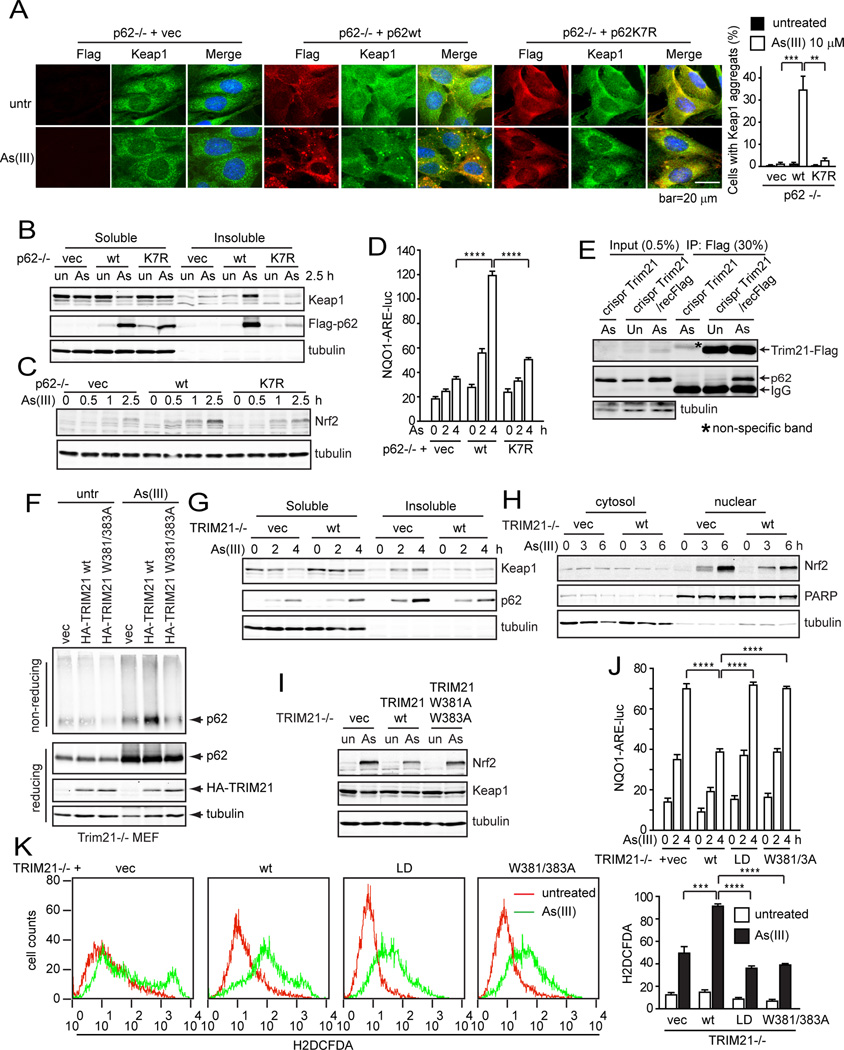
(A) p62−/− MEFs reconstituted with vector, Flag-p62wt, or Flag-p62K7R mutant were untreated or treated with sodium arsenate (As(III), 10 µM) for 6 h. Cells were subjected to IF with Flag and Keap1 antibodies, and observed under deconvolution microscope. Cells with Keap1 aggregates were counted blindly. Data shown are the averages plus S.D. of at least three countings with over 200 cells. **p<0.01, ***p<0.001. (B) p62−/− MEFs reconstituted with vector, Flag-p62wt, and Flag-p62K7R were treated with As(III) (10 µM) for 2.5 h. Cells were lysed in IP lysis buffer with 1% Triton X-100. The insoluble fraction was lysed in IP lysis buffer with 1% SDS. 30 µg soluble proteins and corresponding volume of insoluble proteins were used for IB. (C) p62−/− MEFs reconstituted with vector, Flag-p62wt, and Flag-p62K7R were left untreated or treated with As(III) (10 µM) for 4 h. Cells were treated with cycloheximide (CHX) for indicated times. Cells were lysed in RIPA (1% SDS) and probed for Nrf2 and tubulin. (D) p62−/− MEFs reconstituted with vector, Flag-p62wt, and Flag-p62K7R were transfected with NQO1-ARE-luc and pRL-TK (Renilla Luciferase Control Reporter Vector, Promega) plasmids. 16 h post transfection, cells were treated with As(III) and assayed for luciferase activity. (E) MEFs without/with CRISPR Flag fused into TRIM21 gene were treated with As(III) (10 µM) for 4 h, and subjected to Flag IP and IB. (F) TRIM21−/− MEFs reconstituted with vector, TRIM21wt, and TRIM21W381/383A mutant were treated with the crosslinking agent DSP and lyse in IP lysis buffer with 1% SDS. The lysates were mixed with loading buffer with/without β-mercaptoethanol and subjected to SDS-PAGE and IB. (G) TRIM21−/− MEFs reconstituted with vector and HA-TRIM21wt were treated with As(III). Cells were lysed in IP lysis buffer containing 1% Triton X-100. The insoluble fraction was dissolved in RIPA buffer containing 1% SDS. Both the Triton X-100 soluble and insoluble fractions were subjected to IB. (H) TRIM21−/− MEFs reconstituted with vector and HA-TRIM21wt were treated with As(III) (10 µM). Cell were fractionated to obtain the cytosol and nuclear fractions, and probed for indicated proteins. (I) TRIM21−/− MEFs reconstituted with vector, HA-TRIM21wt, or HATRIM21W381/ 383A mutants were treated with As(III) (10 µM) for 4 h. Cells were lysed in RIPA buffer containing 1% SDS and probed for indicated proteins. (J) TRIM21−/− MEFs reconstituted with vector, HA-TRIM21wt, HA-TRIM21-LD, or HA-TRIM21W381/383A were transfected with NQO1-ARE-luc and renilla luciferase pRL-TK constructs. 16 h later, cells were treated with As(III) (10 µM), and assayed for luciferase activity. (K) TRIM21−/− MEFs reconstituted with vector, HA-TRIM21wt, HA-TRIM21-LD, or HA-TRIM21W381/383A mutants were treated with As(III) (10 µM) for 4 h. Cells were stained with H2DCFDA and analyzed by flow cytometry. Quantification of relative H2DCFDA intensity (Geo mean plus S.D. of four repeats) is shown on the right. ***p<0.001, ****p<0.0001.
Ablation of TRIM21 led to protection against arsenic-induced cell death and liver injury
We next examined the effect of TRIM21 on As(III)-induced cell death. Acute As(III) treatment induces ROS, DNA damage, and activation of poly(ADP-ribose) polymerase (PARP) as well as depletion of NAD and ATP, leading to bioenergetic catastrophe and cell death (Bernstam and Nriagu, 2000; Chou et al., 2004; Kang et al., 2004). Indeed, in wild-type cells, As(III) treatment led to DNA damage indicated by increased H2A.X phosphorylation (Fig. 6A), PARP activation indicated by increased poly(ADP) ribosylation (PAR) (Fig. 6B), and cell death in a manner that was inhibitable by ROS scavengers N-acetylcysteine (NAC) and 2,2,6,6-tetramethylpiperidine-N-oxyl (TEMPO) (Suppl. Fig. S6A) and by PARP inhibitors 3,4-Dihydro-5-[4-(1-piperidinyl)butoxyl]-1(2H)-isoquinolinone (DPQ) and 1,5-dihydroxyisoquinilone (DHIQ) (Suppl. Fig. S6B). Importantly, As(III)-induced DNA damage, PARP activation, and cell death were markedly suppressed in TRIM21−/− cells (Fig. 6A–D). Reconstitution of TRIM21wt but not the LD mutant re-sensitized cells to As(III) (Fig. 6E). Similarly, silencing of p62 led to increased sensitivity to As(III) (Suppl. Fig. S6C). Similar to TRIM21−/− MEFs, silencing of TRIM21 in MDA-MB-231 and MCF-7 cells also led to protection against As(III) toxicity (Suppl. Fig. S6D and S6E). The As(III)-induced cell death was not inhibited by necrostatin-1 (Nec-1) (Suppl. Fig. S6F). Interestingly, while TRIM21-deficient cells were refractory to As(III)-induced cell death, they were more susceptible to cell death induced by MNNG (Suppl. Fig. S6G) that was blocked by PARP inhibitor but not Nec-1 (Suppl. Fig. S6H), and to cell death induced by TNFα/cycloheximide/z-VAD (TCZ) (Suppl. Fig. S6I) that was blocked by Nec-1 (Suppl. Fig. S6J), suggesting that TRIM21 may play a protective role in response to harmful conditions other than oxidative stress. Nonetheless, the above data indicate that TRIM21 inhibition leads to protection against As(III)-induced oxidative stress.
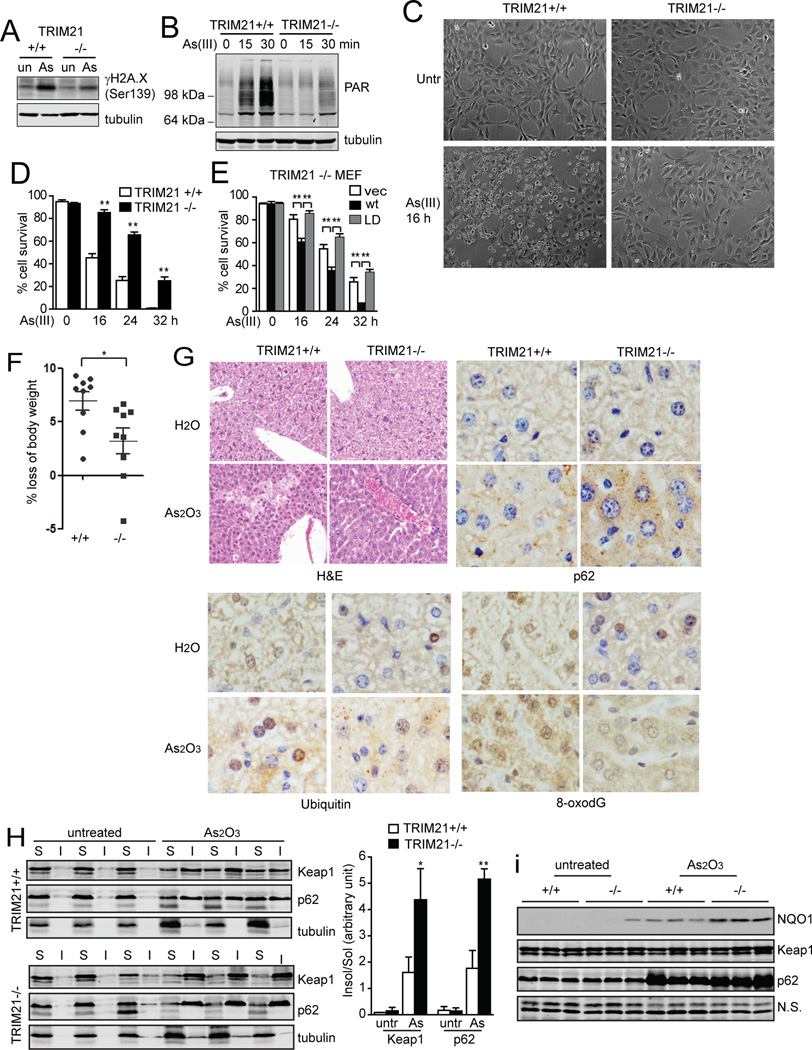
(A and B) TRIM21+/+ and −/− MEFs were treated with As(III) (10 µM) for 12 h (A), or for the indicated times (B). Cells were collected and the lysates were subjected to IB. (C and D) MEFs were treated with As(III) (10 µM). (C) Cells were observed under phase-contrast microscope 16 h after the treatment. Representative micrographs are shown. (D) Cell viability was determined by Trypan Blue. Data shown are the mean plus S.D. of three countings. **p<0.01. (E) TRIM21−/− MEFs reconstituted with vector, HA-TRIM21wt, or HA-TRIM21W381/383A mutant were treated with As(III) (10 µM). Cell viability was determined by Trypan Blue. Shown are the mean plus S.D. of three countings. **p<0.01. (F–I) Nine pairs of litter mates of TRIM21+/+ and −/− mice were gavaged with 30 mg/kg As2O3. (F) Body weight was measured before and after 24 h treatment, and loss of body weight was calculated and plotted. *p<0.05. (G) Livers from water or As2O3-treated mice were collected after 24 h. Tissues were processed for H&E staining and IHC. Representative images are shown. (H) TRIM21+/+ and TRIM21−/− mice were treated with As2O3 for 24 h. Liver tissues were homogenized and separated into detergent (1% Triton X-100)-soluble “S” and insoluble “I” fractions, and subjected to IB. The ratio of insoluble/soluble band intensities was normalized to the corresponding tubulin band, and is shown on the right. *p<0.05, **p<0.01. (I) Liver tissue was lysed in RIPA buffer containing 1% SDS and subjected to IB.
TRIM21 is ubiquitously expressed in all tissues (Suppl. Fig. S7A). To test the physiological relevance of TRIM21, we tested arsenic trioxide (As2O3)-induced liver injury using the whole body TRIM21 knockout mouse (Yoshimi et al., 2009). Sublethal dosage of As2O3 (30 mg/kg) was given to wild-type or TRIM21−/− littermates for 24 h. A significant body weight loss was observed in wild-type mice, which was significantly alleviated in TRIM21−/− mice (Fig. 6F). Hematoxylin/eosin (H&E) staining revealed moderate level of necrosis near the central vein in the wild-type mice, which was blocked in TRIM21−/− mice (Fig. 6G). Immunohistochemistry (IHC) analysis revealed that TRIM21−/− mice displayed more profound aggregation of p62 and ubiquitylated proteins, as well as reduced oxidative DNA damage indicated by 8-oxodG staining (Fig. 6G). Cell fractionation revealed that As2O3 treatment led to increased insoluble p62 and Keap1, which was markedly further enhanced and correlated with the higher NQO1 expression in TRIM21−/− livers (Fig. 6H and 6I). This was consistent with the increased p62-Keap1 colocalization (Suppl. Fig. S7B) as well as increased expression of a number of antioxidant genes upon As(III) treatment (Suppl. Fig. S7C). Together, these data indicate that ablation of TRIM21 confers protection against As2O3-induced oxidative burst, cell death, and liver injury.
TRIM21 knockout mice are protected from heart damage induced by transverse aortic constriction
Another in vivo condition where ROS plays an important role is cardiac dysfunction (Belch et al., 1991; von Harsdorf et al., 1999). A commonly used in vivo model in studying heart failure is pressure overload upon severe transverse thoracic aorta constriction (sTAC), which has been shown to induce ROS and can be alleviated by ROS scavengers (Date et al., 2002). We therefore examined sTAC-induced heart damage in TRIM21−/− mice. In the control mice, sTAC caused cardiac dilatation (Fig. 7A) associated with an acute drop in contractility and increased left ventricular diastolic and systolic volumes (Fig. 7B and 7C). In sharp contrast, the heart size and cardiac contractility was completely preserved in the banded TRIM21−/− mice with fractional shortening and ejection fraction levels similar to the sham-operated animals (Fig.7A–C). Close examination of the heart tissue revealed that sTAC induced disarrangement of cardiomyocytes with sarcoplasmic vacuoles in the wild-type mice. These abnormalities were absent in the hearts of banded TRIM21−/− mice (Fig. 7D, H&E). IHC analysis revealed that hearts from TRIM21−/− mice displayed more profound aggregation of p62 and ubiquitylated proteins compared with hearts from wild-type mice (Fig. 7D, IHC for p62 and ubiquitin). Consistently, while sTAC induced elevated H2A.X phosphorylation in wild-type hearts indicative of DNA damage, which was markedly suppressed and accompanied by increased antioxidant response indicated by increased NQO1 expression in TRIM21−/− hearts (Fig. 7E). These results suggest that loss of TRIM21 attenuated the ROS release from sTAC that leads to myocyte damage resulting in decreased cardiac contractility. Taken together with the protection from As2O3-induced liver damage in TRIM21−/− mice (Fig. 6), our results strongly indicate that ablation of TRIM21 can increase the antioxidant response and protect against pathological conditions where dysregulated ROS plays a major etiological role.
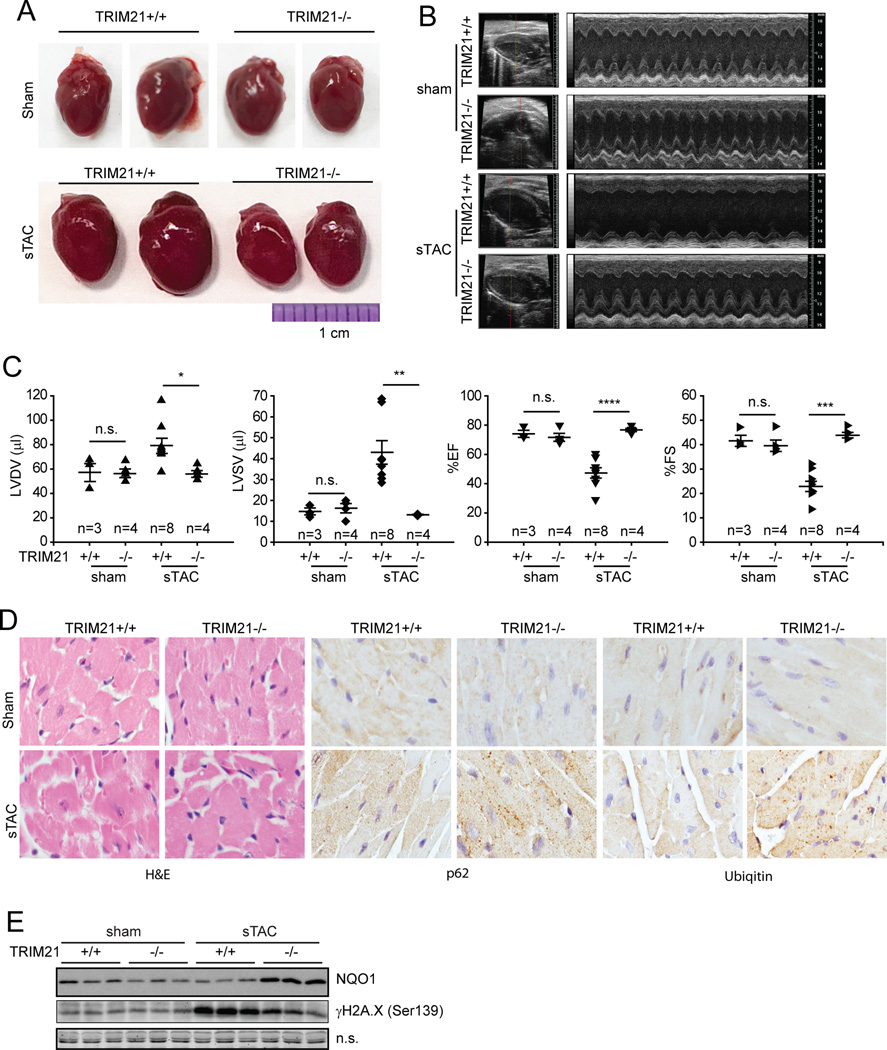
Sham or sTAC was performed on wild-type or TRIM21−/− mice. 4 d later, animals were subjected to echocardiography, then the hearts were isolated for further analyses. (A) Hearts from each group were photographed. (B) Representative B-mode (left ventricle labeled) and M-mode echocardiographic images are shown. (C) M-mode images from 9 cardiac cycles for each animal were used to calculate ventricular measurements: LVDV (left ventricular diastolic volume), LVSV (left ventricular systolic volume), fractional shortening, FS (fractional shortening) and EF (ejection fraction). Individual data points and the mean are shown. n.s.: nonsignificant; *p<0.05; **p<0.01; ***p<0.001; ****p<0.0001. (D) Hearts from sham or sTAC-treated mice were collected and examined with H&E and IHC staining. (E) Heart tissue was lysed in RIPA buffer containing 1% SDS and subjected to IB.
Discussion
In this study, we find that the ubiquitin E3 ligase TRIM21 directly interacts with and ubiquitylates p62 at residue K7, which inhibits p62 dimerization and sequestration function. K7 has been reported to form a hydrogen bond with D69 (Wilson et al., 2003), which is essential for p62 dimerization (Bjorkoy et al., 2005). Our findings demonstrate that there exists a naturally occurring post-translational modification of p62 K7. Our study also shows a molecular consequence is the failure of sequestering Keap1, leading to a reduced capacity of antioxidant response. TRIM21-deficient cells show enhanced p62 oligomerization and aggregation in response to proteotoxic and oxidative stresses (proteasome inhibition, arsenic), as well as upon ablation of autophagy-essential proteins Vps34 or Atg7. This is accompanied by enhanced antioxidant response and protection from superoxide-induced cell death and tissue damage.
On the other hand, although ROS is often considered harmful, it is not entirely surprising that a cell needs a negative regulator of the reductive response such as TRIM21 since certain levels of ROS may be beneficial by serving a critical role in cell signaling and anti-pathogenic response. Moreover, hyperactivating the reductive response may facilitate cancer cell survival and proliferation, hence promoting oncogenesis and therapy-resistance (Sayin et al., 2014; Schafer et al., 2009). Along this line, hyperactivation of p62 and Nrf2 was shown to promote tumorigenesis (DeNicola et al., 2011; Duran et al., 2008; Mathew et al., 2009), and genetic deletion of TRIM21 or reduced TRIM21 expression was associated with the development and poor prognosis in numerous cancers (Brauner et al., 2015; Ding et al., 2015; Zhao and Bepler, 2001). Therefore, by ubiquitylating p62 and suppressing cellular reductive response, TRIM21 may function as a pleiotropic antagonistic factor that is beneficial in maintaining developmental redox homeostasis yet detrimental under excessive oxidative insult.
TRIM21 is a RING finger domain-containing E3 ligase that belongs to the family of the tripartite motif (TRIM) family (Wada and Kamitani, 2006). It was initially discovered as a 52 kDa antigen of autoantibody to the ribonucleoprotein Ro/SSA in systemic lupus erythematosus (SLE) (Ben-Chetrit et al., 1988), whose increased expression was also found in Sjögren's syndrome (Ben-Chetrit et al., 1990). TRIM21 can interact with numerous proteins involved in both innate and adaptive immunity. While no obvious gross developmental defects was observed, TRIM21−/− mice showed increased dysregulation of NF-κB mediated proinflammatory cytokine production (Espinosa et al., 2009; Yoshimi et al., 2009). An important molecular feature of TRIM21 is its binding to immunoglobulin G (IgG) via its PRYSPRY domain (James et al., 2007), which plays an important role in innate immune activation during viral infection (Mallery et al., 2010; McEwan et al., 2013). While it was previously reported that p62 can modulate TRIM21 function and NF-κB activity (Kim and Ozato, 2009), we identified W381/383 in the TRIM21 PRYSPRY domain that are required for p62 interaction, implying a role of p62 in modulating TRIM21 IgG binding and immune response.
p62 is a major stress response regulator. Its expression and activity are critically regulated to maintain proper cell function and organismal homeostasis. p62 is regulated at multiple levels. At the transcription level, p62 expression is activated in a feed-forward loop by Nrf2 (Jain et al., 2010), by SP-1, AP-1, NF-κB and Ets-1(Vadlamudi and Shin, 1998), by the Ras/MEK/ERK1/2 pathway (Duran et al., 2008), and by the JNK/c-Jun pathway (Puissant et al., 2010). At the protein stability level, p62 is degraded along with the protein aggregates by autophagy (Pankiv et al., 2007) and is often served as an indicator for autophagy induction. At the post-translational level, p62 is phosphorylated at numerous sites, which enhances its activity (Ichimura et al., 2013; Lim et al., 2015; Linares et al., 2011; Matsumoto et al., 2011; Tanji et al., 2014). Here, we show that p62 activity can be negatively regulated by TRIM21-mediated K7 ubiquitylation, and that TRIM21-p62 interaction is induced by proteotoxic stress and oxidative stress. Our finding adds to the delicate system where p62 level needs to be critically regulated in order to maintain cellular homeostasis, and suggests a therapeutic potential by targeting TRIM21 in diseases involving proteotoxic and redox stress.
Experimental Procedures
Reagents
The following reagents were used: MG132 (Sigma-Aldrich, C2211), sodium arsenate (Sigma-Aldrich, 35000-1L-R), arsenic trioxide (Sigma-Aldrich, 311383), UBE2N/ UbcH13/Uev1a (BostonBiochem, E2-664), UBE1 (BostonBiochem, E-306), ubiquitin (BostonBiochem, U-100), UBE2D2 (BostonBiochem, E2-622), creatine phosphate (Amresco, 0271); creatine kinase (Affymetrix, 13915-100mg), Ni-NTA agarose (Qiagen, 133203974), Protein A (Roche, 11719408001), NAC (Sigma-Aldrich, A9165), Tempo (Sigma-Aldrich, 176141), DPQ (Sigma-Aldrich, D5314), DHIQ (Sigma-Aldrich, I138).
Lentiviral and retroviral infection
Lentiviruses and retroviruses were generated in HEK293T cells. The infection was performed as previously described (Pan et al., 2011).
Mouse experiments
All mouse experiments were done in compliance with the Institutional Animal Care and Use Committee guidelines at Stony Brook University and Rutgers University.
Statistics
Student’s t test was used to compare the differences between two groups. One-way ANOVA with Tukey’s post-hoc test was used for comparisons between more than two groups. Results were considered significant when p<0.05.
Image processing and densitometry measurements
Images captured by deconvolution and confocal microscopes were viewed and processed by AxioVision LE and Zeiss LSM image browser, respectively. Images were processed in Adobe Photoshop to enhance the brightness and contrast. Densitometry of immunoblot bands was determined by ImageJ or by the Odyssey Infrared Imaging System.
More detailed experimental procedures can be found in “Supplemental Information”.
Acknowledgments
We thank Drs. Michael Frohman (Stony Brook University), Terje Johanson (University of Tromso, Norway), Huikuan Lin (M.D. Anderson Cancer Center), Noboru Mizushima (University of Tokyo), and Zhenyu Yue (Mt Sinai School of Medicine) for reagents. We thank Drs. Yanxiang Zhao (Hong Kong Polytechnic University) and David Goldfarb (University of Rochester) for insightful comments. This work was supported by grants from NIH (R01CA129536 and R01GM97355 to WXZ, R01CA172025 and R01DK108743 to JM, R01CA192642 to MTDM, 5P30CA030199 to MTDM and JM), Department of Veterans Affairs Merit Review Program (to RZL), and a key project (12KG107) from the Municipal Healthy Bureau of Tianjin, China (to BY).
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Author contributions
JAP and WXZ conceived the ideas and designed the experiments. JAP, YS, and WXZ wrote the paper. CD, JM, and KO provided critical reagents. All authors performed experiments or data analysis.References
- Barchowsky A, Dudek EJ, Treadwell MD, Wetterhahn KE. Arsenic induces oxidant stress and NF-kappa B activation in cultured aortic endothelial cells. Free Radic Biol Med. 1996;21:783–790. [Abstract] [Google Scholar]
- Belch JJ, Bridges AB, Scott N, Chopra M. Oxygen free radicals and congestive heart failure. British heart journal. 1991;65:245–248. [Europe PMC free article] [Abstract] [Google Scholar]
- Ben-Chetrit E, Chan EK, Sullivan KF, Tan EM. A 52-kD protein is a novel component of the SS-A/Ro antigenic particle. J Exp Med. 1988;167:1560–1571. [Europe PMC free article] [Abstract] [Google Scholar]
- Ben-Chetrit E, Fox RI, Tan EM. Dissociation of immune responses to the SS-A (Ro) 52-kd and 60-kd polypeptides in systemic lupus erythematosus and Sjogren's syndrome. Arthritis and rheumatism. 1990;33:349–355. [Abstract] [Google Scholar]
- Bernstam L, Nriagu J. Molecular aspects of arsenic stress. Journal of toxicology and environmental health Part B, Critical reviews. 2000;3:293–322. [Abstract] [Google Scholar]
- Bjorkoy G, Lamark T, Brech A, Outzen H, Perander M, Overvatn A, Stenmark H, Johansen T. p62/SQSTM1 forms protein aggregates degraded by autophagy and has a protective effect on huntingtin-induced cell death. J Cell Biol. 2005;171:603–614. [Europe PMC free article] [Abstract] [Google Scholar]
- Brauner S, Zhou W, Backlin C, Green TM, Folkersen L, Ivanchenko M, Lofstrom B, Xu-Monette ZY, Young KH, Moller Pedersen L, et al. Reduced expression of TRIM21/Ro52 predicts poor prognosis in diffuse large B-cell lymphoma patients with and without rheumatic disease. Journal of internal medicine. 2015 [Abstract] [Google Scholar]
- Chou WC, Jie C, Kenedy AA, Jones RJ, Trush MA, Dang CV. Role of NADPH oxidase in arsenic-induced reactive oxygen species formation and cytotoxicity in myeloid leukemia cells. Proc Natl Acad Sci U S A. 2004;101:4578–4583. [Europe PMC free article] [Abstract] [Google Scholar]
- Date MO, Morita T, Yamashita N, Nishida K, Yamaguchi O, Higuchi Y, Hirotani S, Matsumura Y, Hori M, Tada M, et al. The antioxidant N-2-mercaptopropionyl glycine attenuates left ventricular hypertrophy in in vivo murine pressure-overload model. Journal of the American College of Cardiology. 2002;39:907–912. [Abstract] [Google Scholar]
- DeNicola GM, Karreth FA, Humpton TJ, Gopinathan A, Wei C, Frese K, Mangal D, Yu KH, Yeo CJ, Calhoun ES, et al. Oncogene-induced Nrf2 transcription promotes ROS detoxification and tumorigenesis. Nature. 2011;475:106–109. [Europe PMC free article] [Abstract] [Google Scholar]
- Ding Q, He D, He K, Zhang Q, Tang M, Dai J, Lv H, Wang X, Xiang G, Yu H. Downregulation of TRIM21 contributes to hepatocellular carcinoma carcinogenesis and indicates poor prognosis of cancers. Tumour Biol. 2015 [Abstract] [Google Scholar]
- Duran A, Linares JF, Galvez AS, Wikenheiser K, Flores JM, Diaz-Meco MT, Moscat J. The signaling adaptor p62 is an important NF-kappaB mediator in tumorigenesis. Cancer Cell. 2008;13:343–354. [Abstract] [Google Scholar]
- Espinosa A, Dardalhon V, Brauner S, Ambrosi A, Higgs R, Quintana FJ, Sjostrand M, Eloranta ML, Ni Gabhann J, Winqvist O, et al. Loss of the lupus autoantigen Ro52/Trim21 induces tissue inflammation and systemic autoimmunity by disregulating the IL-23-Th17 pathway. J Exp Med. 2009;206:1661–1671. [Europe PMC free article] [Abstract] [Google Scholar]
- Harder B, Jiang T, Wu T, Tao S, de la Vega MR, Tian W, Chapman E, Zhang DD. Molecular mechanisms of Nrf2 regulation and how these influence chemical modulation for disease intervention. Biochem Soc Trans. 2015;43:680–686. [Europe PMC free article] [Abstract] [Google Scholar]
- Huang H, Choi SY, Frohman MA. A quantitative assay for mitochondrial fusion using Renilla luciferase complementation. Mitochondrion. 2010;10:559–566. [Europe PMC free article] [Abstract] [Google Scholar]
- Ichimura Y, Waguri S, Sou YS, Kageyama S, Hasegawa J, Ishimura R, Saito T, Yang Y, Kouno T, Fukutomi T, et al. Phosphorylation of p62 activates the Keap1-Nrf2 pathway during selective autophagy. Mol Cell. 2013;51:618–631. [Abstract] [Google Scholar]
- Ishii T, Yanagawa T, Kawane T, Yuki K, Seita J, Yoshida H, Bannai S. Murine peritoneal macrophages induce a novel 60-kDa protein with structural similarity to a tyrosine kinase p56lck-associated protein in response to oxidative stress. Biochem Biophys Res Commun. 1996;226:456–460. [Abstract] [Google Scholar]
- Itakura E, Mizushima N. p62 Targeting to the autophagosome formation site requires self-oligomerization but not LC3 binding. J Cell Biol. 2011;192:17–27. [Europe PMC free article] [Abstract] [Google Scholar]
- Jaber N, Dou Z, Chen JS, Catanzaro J, Jiang YP, Ballou LM, Selinger E, Ouyang X, Lin RZ, Zhang J, et al. Class III PI3K Vps34 plays an essential role in autophagy and in heart and liver function. Proc Natl Acad Sci U S A. 2012;109:2003–2008. [Europe PMC free article] [Abstract] [Google Scholar]
- Jain A, Lamark T, Sjottem E, Larsen KB, Awuh JA, Overvatn A, McMahon M, Hayes JD, Johansen T. p62/SQSTM1 is a target gene for transcription factor NRF2 and creates a positive feedback loop by inducing antioxidant response element-driven gene transcription. J Biol Chem. 2010;285:22576–22591. [Europe PMC free article] [Abstract] [Google Scholar]
- James LC, Keeble AH, Khan Z, Rhodes DA, Trowsdale J. Structural basis for PRYSPRY-mediated tripartite motif (TRIM) protein function. Proc Natl Acad Sci U S A. 2007;104:6200–6205. [Europe PMC free article] [Abstract] [Google Scholar]
- Kang YH, Yi MJ, Kim MJ, Park MT, Bae S, Kang CM, Cho CK, Park IC, Park MJ, Rhee CH, et al. Caspase-independent cell death by arsenic trioxide in human cervical cancer cells: reactive oxygen species-mediated poly(ADP-ribose) polymerase-1 activation signals apoptosis-inducing factor release from mitochondria. Cancer Res. 2004;64:8960–8967. [Abstract] [Google Scholar]
- Katsuragi Y, Ichimura Y, Komatsu M. p62/SQSTM1 functions as a signaling hub and an autophagy adaptor. The FEBS journal. 2015 [Abstract] [Google Scholar]
- Kim JY, Ozato K. The sequestosome 1/p62 attenuates cytokine gene expression in activated macrophages by inhibiting IFN regulatory factor 8 and TNF receptor-associated factor 6/NF-kappaB activity. J Immunol. 2009;182:2131–2140. [Europe PMC free article] [Abstract] [Google Scholar]
- Kobayashi A, Kang MI, Okawa H, Ohtsuji M, Zenke Y, Chiba T, Igarashi K, Yamamoto M. Oxidative stress sensor Keap1 functions as an adaptor for Cul3-based E3 ligase to regulate proteasomal degradation of Nrf2. Mol Cell Biol. 2004;24:7130–7139. [Europe PMC free article] [Abstract] [Google Scholar]
- Komatsu M, Kurokawa H, Waguri S, Taguchi K, Kobayashi A, Ichimura Y, Sou YS, Ueno I, Sakamoto A, Tong KI, et al. The selective autophagy substrate p62 activates the stress responsive transcription factor Nrf2 through inactivation of Keap1. Nat Cell Biol. 2010;12:213–223. [Abstract] [Google Scholar]
- Komatsu M, Waguri S, Ueno T, Iwata J, Murata S, Tanida I, Ezaki J, Mizushima N, Ohsumi Y, Uchiyama Y, et al. Impairment of starvation-induced and constitutive autophagy in Atg7-deficient mice. J Cell Biol. 2005;169:425–434. [Europe PMC free article] [Abstract] [Google Scholar]
- Lau A, Wang XJ, Zhao F, Villeneuve NF, Wu T, Jiang T, Sun Z, White E, Zhang DD. A noncanonical mechanism of Nrf2 activation by autophagy deficiency: direct interaction between Keap1 and p62. Mol Cell Biol. 2010;30:3275–3285. [Europe PMC free article] [Abstract] [Google Scholar]
- Lim J, Lachenmayer ML, Wu S, Liu W, Kundu M, Wang R, Komatsu M, Oh YJ, Zhao Y, Yue Z. Proteotoxic stress induces phosphorylation of p62/SQSTM1 by ULK1 to regulate selective autophagic clearance of protein aggregates. PLoS genetics. 2015;11:e1004987. [Europe PMC free article] [Abstract] [Google Scholar]
- Linares JF, Amanchy R, Greis K, Diaz-Meco MT, Moscat J. Phosphorylation of p62 by cdk1 controls the timely transit of cells through mitosis and tumor cell proliferation. Mol Cell Biol. 2011;31:105–117. [Europe PMC free article] [Abstract] [Google Scholar]
- Lynn S, Shiung JN, Gurr JR, Jan KY. Arsenite stimulates poly(ADP-ribosylation) by generation of nitric oxide. Free Radic Biol Med. 1998;24:442–449. [Abstract] [Google Scholar]
- Mallery DL, McEwan WA, Bidgood SR, Towers GJ, Johnson CM, James LC. Antibodies mediate intracellular immunity through tripartite motif-containing 21 (TRIM21) Proc Natl Acad Sci U S A. 2010;107:19985–19990. [Europe PMC free article] [Abstract] [Google Scholar]
- Mathew R, Karp CM, Beaudoin B, Vuong N, Chen G, Chen HY, Bray K, Reddy A, Bhanot G, Gelinas C, et al. Autophagy suppresses tumorigenesis through elimination of p62. Cell. 2009;137:1062–1075. [Europe PMC free article] [Abstract] [Google Scholar]
- Matsumoto G, Wada K, Okuno M, Kurosawa M, Nukina N. Serine 403 Phosphorylation of p62/SQSTM1 Regulates Selective Autophagic Clearance of Ubiquitinated Proteins. Mol Cell. 2011;44:279–289. [Abstract] [Google Scholar]
- McEwan WA, Tam JC, Watkinson RE, Bidgood SR, Mallery DL, James LC. Intracellular antibody-bound pathogens stimulate immune signaling via the Fc receptor TRIM21. Nat Immunol. 2013;14:327–336. [Europe PMC free article] [Abstract] [Google Scholar]
- Moscat J, Diaz-Meco MT. p62 at the crossroads of autophagy, apoptosis, and cancer. Cell. 2009;137:1001–1004. [Europe PMC free article] [Abstract] [Google Scholar]
- Nisole S, Stoye JP, Saib A. TRIM family proteins: retroviral restriction and antiviral defence. Nature reviews Microbiology. 2005;3:799–808. [Abstract] [Google Scholar]
- Pan JA, Ullman E, Dou Z, Zong WX. Inhibition of Protein Degradation Induces Apoptosis through a Microtubule-Associated Protein 1 Light Chain 3-Mediated Activation of Caspase-8 at Intracellular Membranes. Mol Cell Biol. 2011;31:3158–3170. [Europe PMC free article] [Abstract] [Google Scholar]
- Pankiv S, Clausen TH, Lamark T, Brech A, Bruun JA, Outzen H, Overvatn A, Bjorkoy G, Johansen T. p62/SQSTM1 binds directly to Atg8/LC3 to facilitate degradation of ubiquitinated protein aggregates by autophagy. J Biol Chem. 2007;282:24131–24145. [Abstract] [Google Scholar]
- Puissant A, Robert G, Fenouille N, Luciano F, Cassuto JP, Raynaud S, Auberger P. Resveratrol promotes autophagic cell death in chronic myelogenous leukemia cells via JNK-mediated p62/SQSTM1 expression and AMPK activation. Cancer Res. 2010;70:1042–1052. [Abstract] [Google Scholar]
- Sayin VI, Ibrahim MX, Larsson E, Nilsson JA, Lindahl P, Bergo MO. Antioxidants accelerate lung cancer progression in mice. Science translational medicine. 2014;6:221ra215. [Abstract] [Google Scholar]
- Schafer ZT, Grassian AR, Song L, Jiang Z, Gerhart-Hines Z, Irie HY, Gao S, Puigserver P, Brugge JS. Antioxidant and oncogene rescue of metabolic defects caused by loss of matrix attachment. Nature. 2009;461:109–113. [Europe PMC free article] [Abstract] [Google Scholar]
- Seibenhener ML, Babu JR, Geetha T, Wong HC, Krishna NR, Wooten MW. Sequestosome 1/p62 is a polyubiquitin chain binding protein involved in ubiquitin proteasome degradation. Mol Cell Biol. 2004;24:8055–8068. [Europe PMC free article] [Abstract] [Google Scholar]
- Shin J. P62 and the sequestosome, a novel mechanism for protein metabolism. Archives of pharmacal research. 1998;21:629–633. [Abstract] [Google Scholar]
- Song B, Gold B, O'Huigin C, Javanbakht H, Li X, Stremlau M, Winkler C, Dean M, Sodroski J. The B30.2(SPRY) domain of the retroviral restriction factor TRIM5alpha exhibits lineage-specific length and sequence variation in primates. J Virol. 2005;79:6111–6121. [Europe PMC free article] [Abstract] [Google Scholar]
- Tanji K, Miki Y, Ozaki T, Maruyama A, Yoshida H, Mimura J, Matsumiya T, Mori F, Imaizumi T, Itoh K, et al. Phosphorylation of serine 349 of p62 in Alzheimer's disease brain. Acta neuropathologica communications. 2014;2:50. [Europe PMC free article] [Abstract] [Google Scholar]
- Vadlamudi RK, Shin J. Genomic structure and promoter analysis of the p62 gene encoding a non-proteasomal multiubiquitin chain binding protein. FEBS Lett. 1998;435:138–142. [Abstract] [Google Scholar]
- von Harsdorf R, Li PF, Dietz R. Signaling pathways in reactive oxygen species-induced cardiomyocyte apoptosis. Circulation. 1999;99:2934–2941. [Abstract] [Google Scholar]
- Wada K, Kamitani T. Autoantigen Ro52 is an E3 ubiquitin ligase. Biochem Biophys Res Commun. 2006;339:415–421. [Abstract] [Google Scholar]
- Wilson MI, Gill DJ, Perisic O, Quinn MT, Williams RL. PB1 domain-mediated heterodimerization in NADPH oxidase and signaling complexes of atypical protein kinase C with Par6 and p62. Mol Cell. 2003;12:39–50. [Abstract] [Google Scholar]
- Yoshimi R, Chang TH, Wang H, Atsumi T, Morse HC, 3rd, Ozato K. Gene disruption study reveals a nonredundant role for TRIM21/Ro52 in NF-kappaB-dependent cytokine expression in fibroblasts. J Immunol. 2009;182:7527–7538. [Europe PMC free article] [Abstract] [Google Scholar]
- Zhang DD, Lo SC, Cross JV, Templeton DJ, Hannink M. Keap1 is a redox-regulated substrate adaptor protein for a Cul3-dependent ubiquitin ligase complex. Mol Cell Biol. 2004;24:10941–10953. [Europe PMC free article] [Abstract] [Google Scholar]
- Zhao B, Bepler G. Transcript map and complete genomic sequence for the 310 kb region of minimal allele loss on chromosome segment 11p15.5 in non-small-cell lung cancer. Oncogene. 2001;20:8154–8164. [Abstract] [Google Scholar]
Full text links
Read article at publisher's site: https://doi.org/10.1016/j.molcel.2016.02.007
Read article for free, from open access legal sources, via Unpaywall:
http://www.cell.com/article/S1097276516000915/pdf
Citations & impact
Impact metrics
Citations of article over time
Alternative metrics
Article citations
Ubiquitination regulates autophagy in cancer: simple modifications, promising targets.
J Transl Med, 22(1):985, 31 Oct 2024
Cited by: 0 articles | PMID: 39482684 | PMCID: PMC11526641
Review Free full text in Europe PMC
ER-phagy restrains inflammatory responses through its receptor UBAC2.
EMBO J, 43(21):5057-5084, 16 Sep 2024
Cited by: 0 articles | PMID: 39284914 | PMCID: PMC11535055
Cardiomyocyte-derived USP28 negatively regulates antioxidant response and promotes cardiac hypertrophy via deubiquitinating TRIM21.
Theranostics, 14(16):6236-6248, 30 Sep 2024
Cited by: 0 articles | PMID: 39431010 | PMCID: PMC11488095
Nuclear p62 condensates stabilize the promyelocytic leukemia nuclear bodies by sequestering their ubiquitin ligase RNF4.
Proc Natl Acad Sci U S A, 121(43):e2414377121, 17 Oct 2024
Cited by: 0 articles | PMID: 39418304
TRIM44 enhances autophagy via SQSTM1 oligomerization in response to oxidative stress.
Sci Rep, 14(1):18974, 16 Aug 2024
Cited by: 0 articles | PMID: 39152142 | PMCID: PMC11329658
Go to all (116) article citations
Data
Data behind the article
This data has been text mined from the article, or deposited into data resources.
BioStudies: supplemental material and supporting data
Similar Articles
To arrive at the top five similar articles we use a word-weighted algorithm to compare words from the Title and Abstract of each citation.
Severe Fever with Thrombocytopenia Syndrome Virus NSs Interacts with TRIM21 To Activate the p62-Keap1-Nrf2 Pathway.
J Virol, 94(6):e01684-19, 28 Feb 2020
Cited by: 26 articles | PMID: 31852783 | PMCID: PMC7158713
The Ubiquitin E3 Ligase TRIM21 Promotes Hepatocarcinogenesis by Suppressing the p62-Keap1-Nrf2 Antioxidant Pathway.
Cell Mol Gastroenterol Hepatol, 11(5):1369-1385, 19 Jan 2021
Cited by: 29 articles | PMID: 33482392 | PMCID: PMC8024979
The selective autophagy substrate p62 activates the stress responsive transcription factor Nrf2 through inactivation of Keap1.
Nat Cell Biol, 12(3):213-223, 21 Feb 2010
Cited by: 1411 articles | PMID: 20173742
p62/SQSTM1 functions as a signaling hub and an autophagy adaptor.
FEBS J, 282(24):4672-4678, 16 Oct 2015
Cited by: 447 articles | PMID: 26432171
Review
Funding
Funders who supported this work.
NCI NIH HHS (9)
Grant ID: R01 CA172025
Grant ID: 5P30CA030199
Grant ID: R01 CA129536
Grant ID: R01CA192642
Grant ID: R01CA129536
Grant ID: R01CA172025
Grant ID: P30 CA030199
Grant ID: P30 CA072720
Grant ID: R01 CA192642
NIDDK NIH HHS (2)
Grant ID: R01 DK108743
Grant ID: R01DK108743
NIGMS NIH HHS (2)
Grant ID: R01GM97355
Grant ID: R01 GM097355





