Abstract
Free full text

Untangling autism
Abstract
A clever dissection of the roles of the Ptchd1 gene in the brains of mice demonstrates one way to untangle the complex relationships between the causes and symptoms of neurodevelopmental disorders.
Our current understanding of neurodevelopmental disorders can be thought of as a tangled mess of threads. The clinical presentations of such disorders are so variable that people can receive the same diagnosis despite not sharing a single symptom. Furthermore, the underlying risk factors — be they genetic or environmental — can be associated with not just one, but several disorders. Untangling these threads to link cause and outcome seems a Sisyphean task, but it is a crucial one if we are to develop treatment approaches that have a solid neurobiological basis. In this issue, Wells et al.1 (page 58) use a mouse model of a human genetic condition to grasp the loose end of a single thread and carefully extricate it from the tangled mess. In doing so, they map a precise set of genetically linked symptoms onto dysfunction of a neuronal structure that gates the flow of information across multiple brain circuits.
Wells and colleagues’ approach is based on clinical observations2,3 of a link between autism spectrum disorder and mutations in the PTCHD1 gene. Autism spectrum disorder involves a tremendously debilitating disruption of social and cognitive function, and is frequently associated with several neurodevelopmental diagnoses that have overlapping symptoms, including intellectual disability and attention deficit hyperactivity disorder.
As is typical for genes associated with neurodevelopmental conditions, PTCHD1 mutations are seen in only a small fraction of people with autism spectrum disorder4. Not everyone with the risk-associated gene variant develops the disorder, but the mutation does raise disease risk substantially: more than 40% of individuals with the mutation develop autism-like behaviours, compared with about 1% of the general population5. Nonetheless, how a single gene can alter brain function to produce particular symptoms remains unclear.
Wells et al. began to unravel these issues by generating mice that produce a truncated, non-functional form of the PTCHD1 protein. The authors report that Ptchd1 mutant mice exhibit behavioural abnormalities that are largely consistent with those seen in people with autism spectrum disorder, including sleep disruption, hyper-aggression and deficits in attention and learning.
In seeking to understand how the Ptchd1 mutation can produce such a broad set of symptoms, the authors were struck by the observation that, during early postnatal development, expression of the gene is highly enriched in a neuronal structure called the thalamic reticular nucleus (TRN) in the thalamus region of the brain. This structure sends inhibitory neuronal projections to all areas of the thalamus, and thus exerts powerful control over the flow of neuronal activity within and across brain circuits that process information about vision, movement, cognition and more. The TRN is known as the ‘guardian’ or ‘gatekeeper’ of activity in these thalamic brain circuits and has regulatory roles in sleep6 and attentional processes7,8. As such, dysfunction in the TRN could plausibly produce wide-ranging behavioural abnormalities.
Focusing on the TRN, Wells et al. traced a route from gene to behaviour in Ptchd1 mutant mice. The authors monitored the activity of TRN neurons, and found fewer bursts of activity in mutant animals than in controls. These changes were attributable to a drastic reduction in the activity of the SK-channel protein, which mediates passage of potassium ions across the cell membrane and normally promotes activity bursts. With reduced levels of channel activity, and reduced numbers of bursts, TRN neurons fail to properly inhibit activity in other regions of the thalamus, including the lateral geniculate nucleus — the main visual relay between the eyes and the brain’s visual cortex. This failure to properly inhibit thalamic relays seems to disrupt visual attention, among other behaviours.
Which of the wide-ranging abnormalities seen in the Ptchd1 mutant mice truly depend on this disruption of TRN function? To answer this question, Wells et al. generated mice that lacked the Ptchd1 gene exclusively in the TRN. In a remarkable dissociation of symptoms, they found that TRN-restricted Ptchd1 mutant mice display hyperactivity and deficits in sleep and attention, but do not exhibit the learning deficits and hyper-aggression observed in mice carrying the brain-wide mutation (Fig. 1). Moreover, using a pharmacological agent to restore SK-channel function in adult mice, Wells et al. were able to re-establish activity levels in the thalamic relays controlled by the TRN. This reversed abnormal hyperactivity and deficits in sleep and attention, but not impaired learning or hyper-aggression. Successful restoration of normal sleep and attention in adult animals raises hopes that treatments targeting the SK channel might benefit people with PTCHD1 mutations, and perhaps also those with autism spectrum disorder caused by other factors.
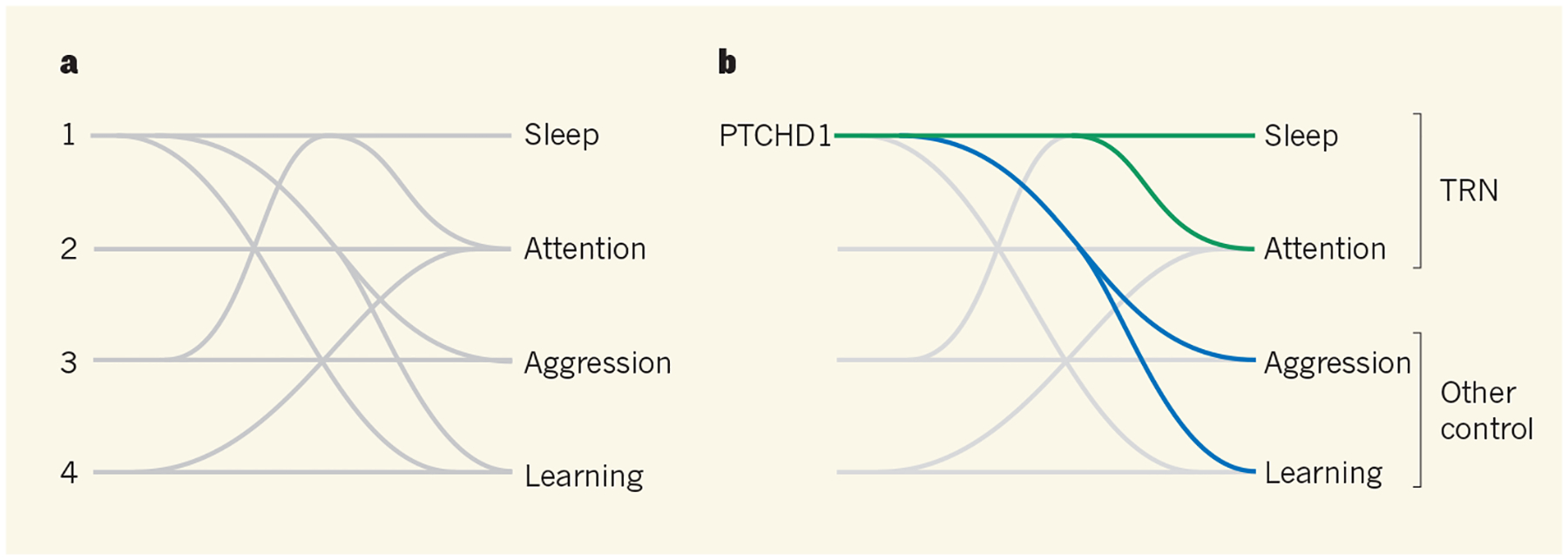
a, Autism spectrum disorder is caused by a range of environmental and genetic factors (symbolized here by numbers), each of which can lead to one or more of many symptoms. Untangling which causes lead to which symptoms is a major challenge. b, Wells et al.1 have done just that for the PTCHD1 protein, which is mutated in some people with autism spectrum disorder. They find that, in mice, PTCHD1 regulates sleep, attention, aggression and learning. Furthermore, sleep and attention, but not aggression and learning, are mediated by PTCHD1 expression in a neuronal structure called the thalamic reticular nucleus (TRN).
The authors’ study indicates that both TRN-specific deletion of PTCHD1 and systemic enhancements in SK-channel function affect the same subset of behavioural outcomes. This consistency strongly supports the idea that PTCHD1 exerts its effects on TRN function (and therefore on activity, sleep and attention) through the SK channel. However, the findings also argue that the effects of PTCHD1 on learning and aggression are caused by an altogether different mechanism that both acts outside the TRN and is independent of SK-channel function. Determining which brain regions and biochemical pathways might be responsible for these behavioural changes will probably be important for fully reversing the deficits caused by PTCHD1 mutations.
Wells et al. have placed their fingertips on the loose thread of PTCHD1 expression, following it to the TRN, through the SK channel and to its end in behaviours associated with neurodevelopmental disease. In doing so, they have identified the SK channel as a potential pharmacological target for treating autism spectrum disorder, while simultaneously highlighting the usefulness of animal models of human genetic conditions for studying neurodevelopmental and psychiatric diseases. Although the role of PTCHD1 in learning and aggression remains elusive, the current paper shows one way to navigate the daunting complexity of neuronal-circuit function to reveal the mechanisms by which gene dysfunction leads to changes in behaviour.
Contributor Information
SCOTT BOLKAN, Department of Neuroscience, Columbia University, New York, New York 10032, USA.
JOSHUA A. GORDON, Department of Psychiatry, Columbia University.
References
Full text links
Read article at publisher's site: https://doi.org/10.1038/nature17311
Read article for free, from open access legal sources, via Unpaywall:
https://www.nature.com/articles/nature17311.pdf
Citations & impact
Impact metrics
Article citations
Atypical co-development of the thalamus and cortex in autism: Evidence from age-related white-gray contrast change.
Hum Brain Mapp, 45(5):e26584, 01 Apr 2024
Cited by: 1 article | PMID: 38533724 | PMCID: PMC10966578
Chemogenetic activation of astrocytes promotes remyelination and restores cognitive deficits in visceral hypersensitive rats.
iScience, 26(1):105840, 20 Dec 2022
Cited by: 6 articles | PMID: 36619970 | PMCID: PMC9812719
Association of Race/Ethnicity and Social Disadvantage With Autism Prevalence in 7 Million School Children in England.
JAMA Pediatr, 175(6):e210054, 07 Jun 2021
Cited by: 73 articles | PMID: 33779707 | PMCID: PMC8008434
Similar Articles
To arrive at the top five similar articles we use a word-weighted algorithm to compare words from the Title and Abstract of each citation.
Thalamic reticular impairment underlies attention deficit in Ptchd1(Y/-) mice.
Nature, 532(7597):58-63, 23 Mar 2016
Cited by: 107 articles | PMID: 27007844 | PMCID: PMC4875756
Restlessness: the anatomy of a neuropsychiatric symptom.
Aust N Z J Psychiatry, 30(1):38-53, 01 Feb 1996
Cited by: 11 articles | PMID: 8724326
Review
Stimulant drug action in attention deficit hyperactivity disorder (ADHD): inference of neurophysiological mechanisms via quantitative modelling.
Clin Neurophysiol, 116(2):324-335, 01 Feb 2005
Cited by: 20 articles | PMID: 15661111
Combinatorial Targeting of Distributed Forebrain Networks Reverses Noise Hypersensitivity in a Model of Autism Spectrum Disorder.
Neuron, 104(3):488-500.e11, 21 Oct 2019
Cited by: 15 articles | PMID: 31648899 | PMCID: PMC7278896





