Abstract
Free full text

Burkholderia pseudomallei Capsule Exacerbates Respiratory Melioidosis but Does Not Afford Protection against Antimicrobial Signaling or Bacterial Killing in Human Olfactory Ensheathing Cells
Abstract
Melioidosis, caused by the bacterium Burkholderia pseudomallei, is an often severe infection that regularly involves respiratory disease following inhalation exposure. Intranasal (i.n.) inoculation of mice represents an experimental approach used to study the contributions of bacterial capsular polysaccharide I (CPS I) to virulence during acute disease. We used aerosol delivery of B. pseudomallei to establish respiratory infection in mice and studied CPS I in the context of innate immune responses. CPS I improved B. pseudomallei survival in vivo and triggered multiple cytokine responses, neutrophil infiltration, and acute inflammatory histopathology in the spleen, liver, nasal-associated lymphoid tissue, and olfactory mucosa (OM). To further explore the role of the OM response to B. pseudomallei infection, we infected human olfactory ensheathing cells (OECs) in vitro and measured bacterial invasion and the cytokine responses induced following infection. Human OECs killed >90% of the B. pseudomallei in a CPS I-independent manner and exhibited an antibacterial cytokine response comprising granulocyte colony-stimulating factor, tumor necrosis factor alpha, and several regulatory cytokines. In-depth genome-wide transcriptomic profiling of the OEC response by RNA-Seq revealed a network of signaling pathways activated in OECs following infection involving a novel group of 378 genes that encode biological pathways controlling cellular movement, inflammation, immunological disease, and molecular transport. This represents the first antimicrobial program to be described in human OECs and establishes the extensive transcriptional defense network accessible in these cells. Collectively, these findings show a role for CPS I in B. pseudomallei survival in vivo following inhalation infection and the antibacterial signaling network that exists in human OM and OECs.
INTRODUCTION
Melioidosis, caused by the Gram-negative bacillus Burkholderia pseudomallei, is a disease endemic to Southeast Asia and Northern Australia (1). Melioidosis is associated with a range of clinical presentations, including subclinical, acute, and chronic forms. In acute melioidosis, almost any organ may be infected; however, the lungs are most frequently affected (2). In the Northern Territory of Australia, approximately 50% of melioidosis patients present with pneumonia as the primary diagnosis (1, 3). Interestingly, neurological melioidosis is also disproportionately prevalent in Northern Australia. The cells and molecular pathways that B. pseudomallei and other bacterial pathogens may utilize to enter the brain via nerves and olfactory mucosa (OM) after inhalation exposure and upper respiratory tract infection were recently reviewed (4). Risk factors that predispose individuals to melioidosis include diabetes mellitus, excessive alcohol consumption, renal impairment, and chronic lung disease (5). B. pseudomallei can be isolated from soil and standing water in regions where melioidosis is endemic, and infection can occur by either inhalation exposure or cutaneous inoculation (1). Following periods of heavy rainfall, inhalation of aerosolized bacteria from soil is predicted to be the most common mode of B. pseudomallei infection (6).
Mice have been used to study acute and chronic human melioidosis (7), and several groups have modeled acute inhalational melioidosis by intranasal (i.n.) inoculation (8,–13) because of the ease of this technique. However, i.n. infection as a challenge model of pneumonia has several limitations, including unpredictable delivery of microbes into the lungs, expulsion of inoculum from the nares, and a potential for diversion of the inoculum to the digestive tract. It has been suggested that i.n. inoculation may, in some cases, mimic some aspects of oropharyngeal aspiration (14, 15). In contrast, exposure to aerosolized B. pseudomallei closely models inhalational melioidosis and results in symmetrical delivery of infectious particles to the lungs (14). Previous studies using nose-only (16, 17) and whole-body (18) aerosol exposure systems have demonstrated that BALB/c mice are susceptible to exposure to aerosolized B. pseudomallei, which results in heavy bacterial loads in the lungs and systemic dissemination.
B. pseudomallei has several virulence factors that may contribute to pathogenesis. Sequencing of the B. pseudomallei K96243 genome revealed the presence of four operons encoding CPS I to CPS IV (19, 20). CPS I, encoded by the wcb operon on chromosome 1, is an unbranched, high-molecular-weight polymer with the structure 3-2-O-acetyl-6-deoxy-β-d-manno-heptopyranose-1 (21). CPS I was predicted to be a virulence determinant on the basis of subtractive hybridization with the closely related nonpathogenic species B. thailandensis (22,–24). Subsequent studies have also demonstrated that B. pseudomallei mutants deficient in CPS I had attenuated virulence in Syrian hamsters (22, 25) and BALB/c mice (9, 10, 24, 26, 27) following i.n. and intraperitoneal infections. However, the role of CPS I has not been assessed with a more physiologically relevant model of acute respiratory melioidosis based on inhalation exposure. In this study, we determined the role of CPS I in a mouse model of respiratory melioidosis following inhalation exposure to aerosolized B. pseudomallei and compared this with the i.n. model frequently used to study this disease. We also analyzed the spread of the bacteria and investigated the innate immune elements impacted by the presence of CPS I. Finally, human olfactory ensheathing cells (OECs), which are the glia of the primary olfactory system, were infected in vitro to identify parallels between the mouse infection model and human OEC responses that are relevant to neurological melioidosis. We demonstrate that CPS I is required for virulence in vivo, is involved in neutrophil recruitment, and stimulates key innate immune responses. Our findings highlight differences between the i.n. and inhalation routes of infection and define, for the first time, a comprehensive map of the anti-B. pseudomallei potential of human OECs based on RNA sequencing (RNA-Seq).
(This work was presented in part at the 7th World Melioidosis Congress, Bangkok, Thailand, 18 to 20 September 2013, and at the 2015 European Melioidosis Congress, Downing College, Cambridge, United Kingdom, 26 to 27 March 2015.)
MATERIALS AND METHODS
Bacterial strains and culture conditions.
B. pseudomallei strain MSHR520, previously designated isolate 08 (27, 28), is a clinical isolate originally cultured from a severe case of human melioidosis and was kindly provided by Bart Currie, Menzies School of Health Research. A CPS I mutant (MSHR520 Δcap mutant) was generated by allele replacement mutagenesis of the wcb operon in the wild-type (WT) MSHR520 strain by previously described methods (29, 30). Briefly, an 8,565-bp region containing the wcbD, wzm, wzt2, wcbE, wcbF, wcbG, wcbH, and wcbI genes on chromosome 1 was deleted and replaced with a selectable tetracycline resistance cassette. The deletion was confirmed by PCR and sequencing of the regions flanking the deleted genes and by phenotypic resistance to tetracycline. B. pseudomallei strains were grown aerobically at 37°C in Luria-Bertani (LB) broth with shaking. Cultures grown on LB agar were incubated at 37°C in 5% CO2. Streptomycin (100 μg/ml) was added to the medium when appropriate. There were no differences between the in vitro growth curve kinetics of the WT and Δcap mutant strains (data not shown).
Mouse infections.
Female BALB/c mice (5 weeks of age) were purchased from the Animal Resources Centre (Canning Vale, WA, Australia). Mice were housed in ventilated cages and given access to food and water ad libitum. All experimental procedures were performed in accordance with the Australian Code of Practice for the Care and Use of Animals for Scientific Purposes (National Health and Medical Research Council of Australia, 2004) and were approved by the Animal Ethics Committee of Griffith University (approval BDD/08/11/AEC). Stationary-phase B. pseudomallei cells were diluted 100-fold (for i.n. inoculation) or 1,000-fold (for aerosol generation) in phosphate-buffered saline (PBS). Numbers of bacteria were determined by plating on LB agar. For i.n. inoculation, 10 μl of a bacterial suspension (containing approximately 5.5 × 105 CFU) was equally distributed between the nasal nares of mice with a micropipette without anesthesia.
Inoculation by inhalation was performed with a whole-body Inhalation Exposure System (Glas-Col, Terre Haute, IN), which generates aerosol particles of 5 to 10 μm. Briefly, unanesthetized mice were placed in a stainless steel basket in the exposure chamber and 6.0 ml of a bacterial suspension (containing 2.7 × 107 to 3.6 × 107 CFU) was loaded into the nebulizer. The inoculum was aerosolized into a volume of 5 ft3 over a period of 20 min, followed by a 20-min cloud decay period during which airflow was maintained without introducing additional bacteria. During operation, the compressed air and vacuum flow meters were maintained at 10 and 60 ft3/h, respectively. At 24 or 48 h postinfection (p.i.), the mice (n = 10 per group) were euthanized by a lethal intraperitoneal injection of sodium pentobarbitone (Lethabarb) and tissues were collected for bacterial colony counts, cytokine assays, and tissue histopathology.
Tissue processing and quantitative culture of B. pseudomallei.
Heparinized blood was collected via cardiac puncture; the spleen, liver, and lungs were then collected. Nasal-associated lymphoid tissue (NALT) was obtained by previously described methods (31). Briefly, the mouse head was immobilized and the lower jaw was removed to expose the palate. The tip of the nose connecting the foreteeth was cut off, and the palate (containing NALT) was peeled back and separated from the rest of the nasal tissue. The OM was collected from both sides of the nasal cavity.
Tissues were homogenized in complete mini protease inhibitor cocktail (Roche, Castle Hill, NSW, Australia) with a Tissue Lyser II (Qiagen, Doncaster, VIC, Australia). Homogenates were then serially diluted in PBS, and 5-μl drops of each dilution were plated (in replicates of six) onto LB agar containing streptomycin. Inoculated agar plates were incubated for 24 h at 37°C, and colony counts were performed to determine the number of CFU per milliliter of blood and the number of CFU per gram of tissue. This assay had a limit of detection of approximately 33 CFU/ml. Tissue histopathology was analyzed by hematoxylin-and-eosin (H&E) staining and immunohistochemistry analysis for Ly6G (neutrophil-specific marker) as previously described (32, 33). Images were captured with an Aperio AT2 Scanner Console (Leica Biosystems, Inc., Vista, CA) and an AxioImager.M2 microscope (Carl Zeiss MicroImaging, Jena, Germany) fitted with a Plan-Apochromat X63/1.40 objective and an MRc 5 camera.
Cytokine and chemokine assays.
The levels of cytokines were determined in the lungs, NALT, and OM at 48 h p.i. with Bio-Plex Pro Mouse Cytokine 23-plex assays (Bio-Rad, Gladesville, NSW, Australia). Following the homogenization of lung, NALT, and OM tissues (as described above), an antibiotic cocktail consisting of trimethoprim (25 μg/ml), chloramphenicol (25 μg/ml), and imipenem (10 μg/ml) was added to each sample to inhibit the growth of B. pseudomallei. Samples were then stored at −80°C until analysis. Thawed samples were clarified by centrifugation at 1,000 × g for 20 min at 4°C, and the supernatant was collected and assayed according to the manufacturer's instructions. Plates were read with a Bio-Plex 200 dual-laser flow cytometer (Bio-Rad), and data were acquired with Bio-Plex Manager Software (version 5.0; Bio-Rad). We analyzed at 48 h p.i. in this experiment on the basis of previous studies that have shown that the concentrations of some cytokines peak at that time following B. pseudomallei infection (34, 35). For in vitro assays using human OECs, we measured cytokine levels at 24 h p.i. with Bio-Plex Pro Human Cytokine 27-plex assays (Bio-Rad).
In vitro human cell infections.
To analyze the interactions of B. pseudomallei WT MSHR520 and the CPS I-deficient mutant with human cells, we used human OECs (36, 37). These assays were used to define the adhesion and invasion of WT MSHR520 and the Δcap mutant and to determine the immune responses of these cells to B. pseudomallei. Methods for the routine culture and infection of human cells were performed essentially as previously described (38), with the following modifications. Human OECs were maintained in Dulbecco's modified Eagle's medium (DMEM)-F12 (1:1; Gibco, Grand Island, NY) supplemented with 10% (vol/vol) fetal bovine serum (Moregate BioTec, Bulimba, QLD, Australia), 2 mM GlutaMAX (Gibco), 20 μg/ml bovine pituitary extract (Life Technologies, Carlsbad, CA), 2 μM forskolin (Cell Signaling Technology, Danvers, MA), and 100 U/ml penicillin-streptomycin (Gibco) (36). OECs were used to seed 24-well plates and incubated at 37°C in 5% CO2 until approximately 80% confluence. Monolayers were then washed and infected with B. pseudomallei (six wells per strain) diluted in antibiotic-free medium (multiplicity of infection of 10) or medium alone (control). Adhesion of B. pseudomallei to OECs was determined at 30 min p.i. by washing monolayers with PBS to remove unattached bacteria and then enumerating adherent bacteria by colony counts on LB agar.
For antibiotic protection assays, bacteria were initially allowed to attach to OECs for 30 min, and then unattached bacteria were removed by washing in PBS. Medium containing kanamycin (250 μg/ml) was subsequently added to monolayers to kill extracellular B. pseudomallei, and the number of intracellular bacteria was determined at 2 and 24 h p.i. At these time points, supernatants were collected and an antibiotic cocktail of trimethoprim (25 µg/ml), chloramphenicol (25 µg/ml), and imipenem (10 µg/ml) was added before samples were stored at −80°C for cytokine assays.
RNA-Seq of the human OEC response to B. pseudomallei.
We undertook a comprehensive analysis of the response of OECs to B. pseudomallei by RNA-Seq and biological pathway mapping by using gene expression profiles. RNA was isolated from OECs at 2 and 24 h p.i. with RNeasy columns (Qiagen). mRNA sequencing was performed with an Illumina HiSeq2500 with the latest versions of the sequencing reagents and flow cells providing up to 300 Gb of sequence information per flow cell, essentially as previously described (39). Briefly, the quality of the total RNA isolated from OECs was assessed with the Agilent 2100 Bioanalyzer, followed by two rounds of poly(A)+ selection and conversion to cDNA. We used the TruSeq library generation kits in accordance with the manufacturer's instructions (Illumina, San Diego, CA). Library construction consisted of random fragmentation of the poly(A) mRNA, followed by cDNA synthesis with random primers. The ends of the cDNA were repaired and A tailed, and adaptors were ligated for indexing (up to 12 different barcodes per lane) during the sequencing runs. The cDNA libraries were quantitated by quantitative PCR in a Roche LightCycler 480 with the Kapa Biosystems kit for library quantitation (Kapa Biosystems, Woburn, MA) prior to cluster generation. Clusters were generated to yield approximately 725,000 to 825,000 clusters/mm2. Cluster density and quality were determined during the run after the first base addition parameters were assessed. We ran paired-end 2 × 5-bp sequencing runs to align the cDNA sequences with the reference genome as described below.
Data preprocessing and bioinformatic analysis.
TopHat version 2.0.11 was used to align the raw RNA-Seq fastq reads with the human hg19 genome from the University of California Santa Cruz with the short-read aligner Bowtie version 2.1.0 (40,–42). TopHat also analyzed the mapping results to identify splice junctions between exons. Cufflinks version 2.1.1 was used to align the reads from TopHat to assemble transcripts, estimate abundances, and test for differential expression and regulation (42, 43). Cuffmerge, as part of Cufflinks, was used to merge the assembled transcripts to a reference annotation and track Cufflinks transcripts across multiple experiments. Finally, Cuffdiff was used to identify significant changes in transcript expression, splicing, and promoter use. Genes that met certain criteria (i.e., changes of ±2.0-fold or more and q values of < 0.05) were further analyzed by using the Ingenuity Pathway Analysis (IPA; Qiagen, Redwood City, CA) and InnateDB Overrepresentation Analysis (ORA) (44) tools. Six biological replicates per treatment (each from an independent experiment) were used.
qRT-PCR analysis.
Quantitative reverse transcription (qRT)-PCR assays to compare the expression of selected target genes with mRNA sequencing data were performed with a LightCycler 480 Instrument Version II with RealTime ready Catalogue Assays consisting of forward and reverse primers with gene-specific hydrolysis probes (Universal ProbeLibrary; Roche Diagnostics GmbH, Penzberg, Germany). For a complete list of the primer sequences and Universal ProbeLibrary probe numbers used in this study, see Table S1 in the supplemental material. All reactions were set up with a robotic liquid sample handling epMotion 5075 LH (Eppendorf Australia) with epT.I.P.S. Motion Filtertips in 384-well plates. Reaction mixtures contained 50 pg to 50 ng of cDNA in a total reaction volume of 10 μl and were generated with the Transcriptor First Strand cDNA synthesis kit (Roche) and 1 μg of total RNA according to the manufacturer's instructions. The thermal cycling conditions were 95°C for 10 min, followed by repeated cycles of 95°C for 10 s, 60°C for 30 s, 72°C for 1 s, and 40°C for 30 s. Relative fold change values were calculated by using amplification efficiencies as described previously (45). All experiments conformed to the Minimum Information for Publication of Quantitative Real-Time PCR Experiments guidelines (46).
Statistical analysis.
Data are presented as the mean and the standard error of the mean. Statistical analyses were performed with GraphPad Prism (version 5.0). Unpaired, two-tailed t tests were performed to compare colonization and cytokine data between the WT and Δcap mutant strains and between the routes of infection, as indicated in the figure legends. Cytokine data from human OECs were compared between experimental groups by one-way analysis of variance, followed by Tukey's multiple-comparison test. Statistical significance was accepted as a P value of <0.05.
Microarray data accession number.
Raw and processed data have been deposited in the Gene Expression Omnibus database under accession number GSE69312.
RESULTS
Acapsular B. pseudomallei colonizes respiratory tract and systemic tissues at lower levels than the WT strain.
BALB/c mice infected with the WT and Δcap mutant MSHR520 strains via inhalation and i.n. routes of infection exhibited heavy bacterial burdens in the lungs (Fig. 1A). At 24 h p.i., colonization of the lungs with the WT strain by the i.n. and inhalation routes of infection was similar. In contrast, the Δcap mutant strain colonized the lungs at that time point at significantly higher levels when delivered by inhalation (P < 0.05). At 48 h p.i., the Δcap mutant strain was attenuated in the lungs by both routes of infection, as evidenced by significantly fewer CFU per gram than the WT strain (P < 0.05). Significant attenuation of the Δcap mutant strain was also observed in the blood, spleen, and liver at 24 and 48 h p.i. compared to the WT (P < 0.05). The Δcap mutant strain was not detected in the blood at 24 h p.i. after infection by either route but was present at very low levels at 48 h following i.n. delivery. Inoculation with the WT strain by i.n. delivery was associated with significantly higher levels of B. pseudomallei in the blood, spleen, and liver at 48 h p.i. than was infection with that strain by inhalation (P < 0.05).
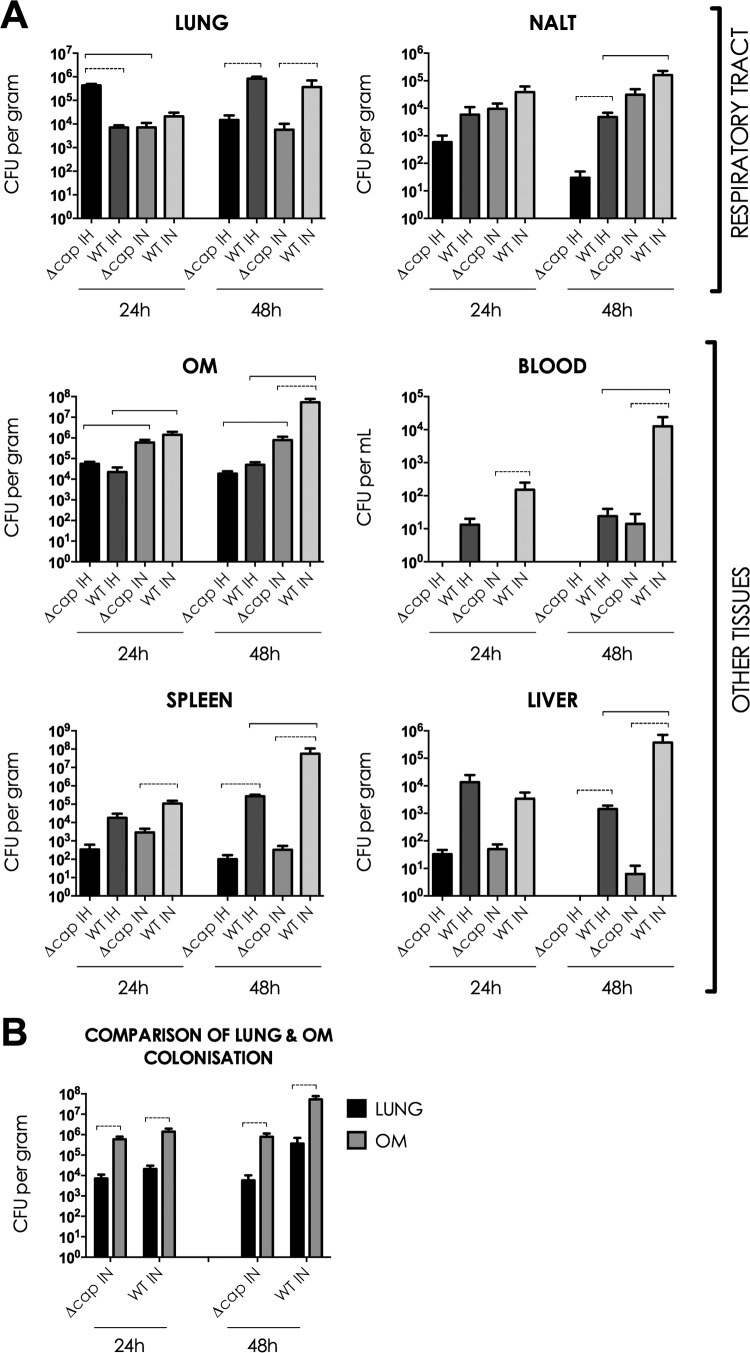
B. pseudomallei disseminates from the respiratory tract and colonizes systemic tissues following an inhalation or i.n. challenge at 24 or 48 h p.i. (A) The MSHR520 Δcap mutant strain was less virulent than the MSHR520 WT strain. (B) The OM was colonized at significantly higher levels than the lungs following i.n. inoculation. Unpaired, two-tailed t tests were used to compare colonization between (i) the WT and Δcap mutant strains (statistical significance [P < 0.05] is indicated by a dotted line), (ii) routes of infection (statistical significance [P < 0.05] is indicated by a solid line), and (iii) the OM and lung in i.n. infected mice. IH, inhalation; IN, intranasal.
B. pseudomallei was also isolated from NALT and the OM (Fig. 1A). There were no statistically significant differences in NALT and OM colonization by the WT and the Δcap mutant at 24 h p.i. (P > 0.05); however, at 48 h p.i., the Δcap mutant strain was attenuated in NALT following inhalation and in the OM following i.n. delivery (P < 0.05). Similar to colonization of the blood and liver, mice infected i.n. with the WT strain demonstrated greater bacterial burdens in NALT at 48 h p.i. than mice infected with that strain by inhalation. In this study, i.n. B. pseudomallei demonstrated a remarkable tropism for the OM. At both 24 and 48 h p.i., the numbers of WT and Δcap mutant bacteria recovered from the OM of mice infected by i.n. delivery were significantly higher than the bacterial loads in the lungs (P < 0.05; Fig. 1B). Loads were similar in the OM and lungs of mice infected by inhalation. Furthermore, colonization of the OM by both strains at 24 and 48 h p.i. was found to be significantly greater in mice infected by i.n. delivery than in those infected by inhalation (P < 0.05; Fig. 1A).
CPS I mediates innate immunity in the lungs, NALT, and OM.
Measurement of cytokines in the lungs, NALT, and OM at 48 h p.i. in the inhalation and i.n. models revealed a series of inflammatory response patterns that depend on the route of infection and the presence of CPS I. In the lungs, infection with the B. pseudomallei MSHR520 WT and Δcap mutant strains resulted in higher levels of proinflammatory cytokines, anti-inflammatory cytokines, and chemokines than in PBS controls (Fig. 2A). The WT strain induced significantly higher levels of cytokines and chemokines in the lungs than the Δcap mutant strain (P < 0.05). Numerous differences in cytokine concentrations were detected between the i.n. and inhalation routes of infection with the same strain. Specifically, the levels of interleukin-1α (IL-1α), IL-1β, IL-3, IL-12 (p40), IL-12 (p70), IL-17, granulocyte colony-stimulating factor (G-CSF), granulocyte-macrophage colony-stimulating factor (GM-CSF), gamma interferon (IFN-γ), tumor necrosis factor alpha (TNF-α), IL-6, IL-13, eotaxin, KC, monocyte chemoattractant protein 1 (MCP-1), and macrophage inflammatory protein 1α (MIP-1α) were significantly higher following inhalation of B. pseudomallei (when routes of infection with either one or both strains were compared) than after i.n. delivery (P < 0.05). In contrast, higher IL-10 levels were detected in the lungs of mice inoculated i.n. with the WT strain only.


High concentrations of cytokines and chemokines were detected in B. pseudomallei-infected lung tissue, NALT, and the OM at 48 h p.i. In lung tissue (A), significant differences in cytokine/chemokine concentrations were observed between the WT and acapsular mutant B. pseudomallei strains and the routes of infection. In NALT (B), B. pseudomallei also induced increased concentrations of cytokines and chemokines. In the OM (C), infection with the WT strain was associated with greater levels of analytes than infection with the acapsular mutant. Higher concentrations of cytokines and chemokines were detected in the OM of mice infected by i.n. inoculation than in those infected by inhalation. Unpaired two-tailed t tests were used to compare the concentrations of cytokines/chemokines between the Δcap mutant and WT strains (statistical significance [P < 0.05] is indicated by a dotted line) and the routes of infection (statistical significance [P < 0.05] is indicated by a solid line). IH, inhalation; IN, intranasal.
In NALT, B. pseudomallei induced levels of cytokines and chemokines higher than those of PBS control mice (Fig. 2B); however, the concentrations of IL-1α, IL-12 (p70), and GM-CSF were not significantly different from those of the PBS control group (P > 0.05). The WT strain was generally associated with higher levels of cytokines and chemokines in NALT than was the Δcap mutant strain (P < 0.05), although the concentrations of IL-2, IL-9, IL-12 (p40), and IL-13 were not different between these strains (P > 0.05). Furthermore, the differences in cytokine concentrations observed between the Δcap mutant and WT strains were found to be dependent on the route of infection. When mice were infected by inhalation of B. pseudomallei, the Δcap mutant strain was associated with significantly lower concentrations of IL-17, TNF-α, IL-6, and IL-10 in NALT than was the WT strain (P < 0.05). However, when mice were infected by i.n. inoculation, the concentrations of these analytes were not different between the two strains (P > 0.05). In contrast to the lungs, the concentrations of a large number of cytokines and chemokines [IL-12 (p70), G-CSF, IFN-γ, TNF-α, IL-4, IL-10, IL-13, eotaxin, MIP-1α, MIP-1β, and RANTES] were significantly higher in NALT from mice infected i.n. than in that from mice infected by inhalation (P < 0.05).
In the OM at 48 h p.i., B. pseudomallei infection was associated with higher levels of all of the cytokines and chemokines tested than in mice treated with PBS (Fig. 2C). Infection with the WT strain generally induced higher concentrations of cytokines and chemokines in the OM than the acapsular mutant strain (P < 0.05). In the OM, there were striking differences between the inhalation and i.n. routes of infection in the levels of cytokines and chemokines detected when the strains were compared individually. The concentrations of IL-1α, IL-1β, IL-2, IL-3, IL-12 (p40), IL-12 (p70), G-CSF, GM-CSF, TNF-α, IL-4, IL-6, IL-10, IL-13, KC, MCP-1, MIP-1α, MIP-1β, and RANTES were significantly higher in the OM of mice infected i.n. (with either one, or both strains) than in that of mice infected by inhalation (P < 0.05).
The degree of the innate immune response reflects the bacterial load in NALT and the OM but not that in the lungs.
We compared tissue B. pseudomallei loads with cytokine levels at 48 h p.i. to determine if levels of cytokines in the lungs, NALT, and OM might simply reflect the local tissue bacterial burden (Table 1). Colonization of the lungs (Fig. 1A) demonstrated reduced numbers of CFU per gram of tissue infected with the Δcap mutant strain and no differences between the i.n. and inhalation models. When comparing lung colonization and cytokine data, most of the cytokines and chemokines tested (18 of 22; 82%) were inconsistent with B. pseudomallei colonization trends and did not reflect the bacterial burden in the tissue. The cytokines that reflected the lung colonization data included IL-2, IL-9, and MIP-1β.
TABLE 1
Comparison of cytokine response data with bacterial load colonization data on B. pseudomallei-infected mice at 48 h p.i.c
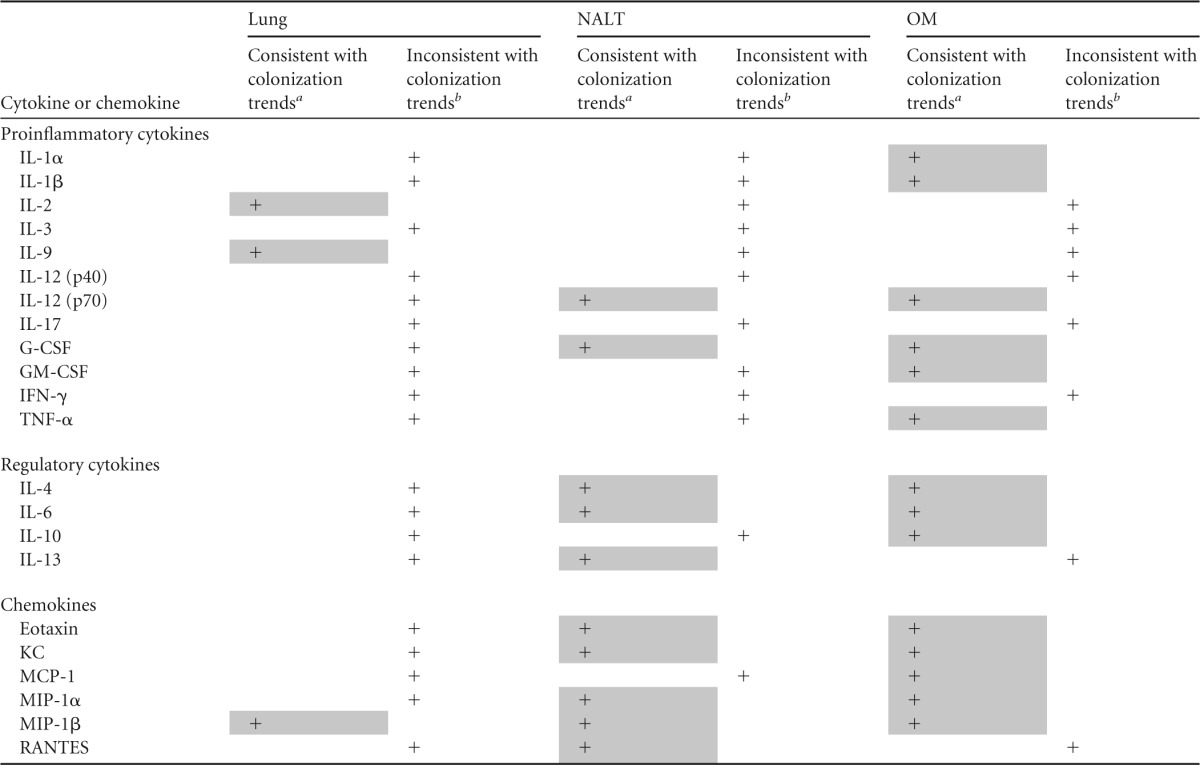
Colonization trends in NALT and the OM at 48 h p.i. were similar (Fig. 1A) and demonstrated lower numbers of CFU per gram in tissue infected with the Δcap mutant strain than tissue infected with the WT strain. Furthermore, higher numbers of B. pseudomallei bacteria were observed in tissues collected from mice infected by i.n. inoculation. The concentrations of 10 (of 12) proinflammatory cytokines (83%) in NALT were found to be inconsistent with these colonization trends; however, the concentrations of 75% (3 of 4) of the anti-inflammatory cytokines and 83% (5 of 6) of the chemokines did reflect bacterial burdens in NALT. In the OM, 50% of the proinflammatory cytokines tested were found to be consistent with bacterial numbers at that site. Similar to NALT, the concentrations of 75% of the anti-inflammatory cytokines and 83% of the chemokines in the OM were also reflective of B. pseudomallei colonization. MIP-1β was the only analyte that was consistent with colonization trends in the lungs, NALT, and OM. In contrast, IL-3, IL-12 (p40), IL-17, and IFN-γ were found to be inconsistent with B. pseudomallei colonization in each of these tissues.
Histopathology and neutrophil infiltration in B. pseudomallei-infected tissues.
Histopathological analysis of lung, NALT, OM, spleen, and liver samples from mice challenged with either WT or Δcap mutant B. pseudomallei MSHR520 revealed major differences in acute inflammation and neutrophil infiltration between the groups. Immunohistochemical analysis with a Ly6G-specific antibody to stain neutrophils demonstrated extensive cellular infiltration, particularly in NALT, the OM, the spleen, and the liver at 48 h following an i.n. challenge with WT MSHR520; however, an equivalent neutrophil infiltrate was absent from tissues collected from mice that were challenged with the MSHR520 Δcap mutant and uninfected control mice, as illustrated in Fig. 3 (shows low magnification) and Fig. 4 (high magnification). There was minimal inflammation (e.g., tissue neutrophilia, hemorrhage, loss of tissue architecture) in the NALT, OM, spleens, and livers of mice infected with the Δcap mutant strain, in contrast to those infected with the WT strain, but not in the lungs (which showed a moderate level of neutrophil infiltration). A moderately higher level of inflammation and neutrophil infiltration was observed in the lungs of WT-infected mice than in mice challenged with the Δcap mutant strain. The acute inflammatory pathology in NALT, the OM, the spleen, and the liver induced by infection with WT MSHR520 included tissue neutrophilia, hemorrhage, and karyorrhexis; histopathological changes were also evident at 24 h p.i., which is illustrated for OM in Fig. 5. Thus, massive neutrophil infiltration, acute inflammatory pathology, and loss of tissue architecture in several tissues, including NALT, OM, and the spleen, occur in mice following infection with the WT MSHR520 strain, but these pathological changes are not induced by the Δcap mutant.

Low-power magnification of histological sections prepared from tissues collected from BALB/c mice at 48 h p.i. and stained with anti-Ly6G antibody for neutrophils and with H&E. The columns show sections from mice challenged i.n. with WT MSHR520, the Δcap mutant, and PBS (uninfected controls) that were stained for Ly6G and with H&E. The scale bars indicate 600 μm for lung tissue, 300 μm for NALT and the OM, and 4 mm for spleen and liver tissues. The low magnifications chosen for each tissue illustrate the neutrophil infiltrate (brown stain) and the overall tissue architecture and acute inflammatory pathology induced following infection with WT MSHR520 but not the Δcap mutant (lung tissue, ×4; NALT and the OM, ×10; spleen and liver tissues, ×0.6).

High-power magnification of histological sections prepared from tissues collected from BALB/c mice at 48 h p.i. and stained with anti-Ly6G antibody for neutrophils and with H&E. The columns show tissue sections from mice challenged i.n. with WT MSHR520, the Δcap mutant, and PBS (controls) stained for Ly6G and with H&E. The scale bars indicate 50 μm for all of the tissue sections. The high magnification (×40) shows the neutrophil infiltrate (brown stain) and acute inflammatory pathology induced following infection with WT MSHR520 but not the Δcap mutant, especially for NALT, the OM, spleen tissue, and liver tissue.

Histological sections of OM prepared from tissues collected from BALB/c mice at 24 h p.i. and stained with anti-Ly6G antibody for neutrophils and with H&E. In uninfected control mice (A), fewer neutrophils are present, in contrast to mice challenged i.n. with WT MSHR520 (B), where a massive neutrophil infiltrate is present in the OM. The olfactory epithelium is indicated by the black arrowheads; the olfactory nerve fascicles, which are located in the lamina propria, are indicated by the white arrowheads. In a higher-magnification image of OM stained with H&E (C) (boxed area in panel B magnified from a sequential H&E-stained section), acute inflammatory pathology in the OM is shown, including severe tissue neutrophilia (white arrowheads), hemorrhage (black arrowheads), and B. pseudomallei cells (black arrows). The scale bars indicate 900 μm in panels A and B, which were taken at ×2.5 magnification, and 10 μm in panel C, which was taken at ×63 magnification.
Human OECs kill intracellular B. pseudomallei and express multiple inflammatory markers.
In vitro adhesion assays demonstrated that B. pseudomallei efficiently adhered to human OECs within 30 min and that this process occurred independently of CPS I (data not shown). Intracellular killing assays performed with OECs and Burkholderia under antibiotic protection conditions demonstrated efficient bacterial killing within 24 h p.i. in vitro. Comparisons of the numbers of the WT and Δcap mutant bacteria recovered from OECs showed no role for the capsule in protecting the bacteria from intracellular killing. Significantly fewer B. pseudomallei bacteria of both the WT and Δcap mutant strains were recovered from OECs at 24 h p.i. than at 2 h p.i. (Fig. 6). Thus, OECs efficiently kill >90% of the intracellular B. pseudomallei bacteria within 24 h and CPS I expression does not affect the intracellular survival of the bacteria. To gain insight into the antimicrobial response of OECs, we quantitated 27 cytokines at 2 and 24 h p.i. with multiplex protein assays. There were significant responses for several cytokines and chemokines in OEC supernatants in vitro at 24 h p.i., which included the IL-1 receptor antagonist, IL-2, TNF-α, G-CSF, IL-4, IL-15, basic fibroblast growth factor, IFN-γ-inducible protein 10, and MCP-1 (see Fig. S1 in the supplemental material). Consistent with the observation that CPS I did not affect intracellular survival, there were no significant differences in cytokine production between OECs infected with the WT and Δcap mutant strains (P > 0.05).

Intracellular killing of B. pseudomallei in human OECs in vitro occurs independently of bacterial CPS I. Equivalent numbers of B. pseudomallei bacteria were recovered from OEC cultures in antibiotic protection assays at 2 and 24 h p.i. with the WT and Δcap mutant strains. There were significantly fewer surviving B. pseudomallei bacteria at 24 h p.i. than at 2 h p.i., independently of capsule. Data represent three independent experiments combined and were analyzed by unpaired two-tailed t tests (***, P < 0.0001).
Human OECs exhibit an antimicrobial transcriptional program for defense against infection.
Initial RNA-Seq analysis of human OECs infected with Burkholderia demonstrated that these cells were capable of initiating an extensive defense response following a microbial challenge. RNA-Seq of infected and uninfected OECs revealed 24 and 378 genes with significantly altered expression at 2 and 24 h p.i., respectively (Fig. 7). Rapidly upregulated genes that were significantly expressed at 2 h p.i. and remained significantly elevated at 24 h p.i. included (fold changes at 2 and 24 h) CSF2 (2.0- and 6.5-fold), TNFAIP3 (1.9- and 4.5-fold), CXCL1 (1.7- and 16.1-fold), CXCL2 (1.7- and 5.3-fold), IL-8 (1.7- and 10.2-fold), CXCL3 (1.6- and 8.4-fold), REL (1.6- and 1.7-fold), PTX3 (1.5- and 3.2-fold), NFKBIA (1.4- and 2.0-fold), PTGS2 (1.4- and 1.8-fold), and IL-6 (1.3- and 6.2-fold). There were a few genes that exhibited significant downregulation at both time points, including KRT14/16/17 (−2.0- and −1.7-fold) and REXO1 (−2.4- and −2.2-fold), as well as two genes that was repressed and activated differentially at 2 and 24 h p.i., i.e., SNORA76/SNORD104 (3.0- and −14.1-fold) and HEATR6 (−31.4- and 13.4-fold). For the complete gene lists, see Table S2A in the supplemental material. A total of 15 genes were common to the two time points; 13 were expressed in the same direction in infected and control mice (i.e., upregulated or downregulated), and only two (SNORA76/SNORD104, HEATR6) exhibited contrasting expression patterns at the two time points. Together, these data define the first transcriptional signature of human OECs in response to a bacterial infection and reveal the capability of these cells to mount antimicrobial responses following in vitro infection.
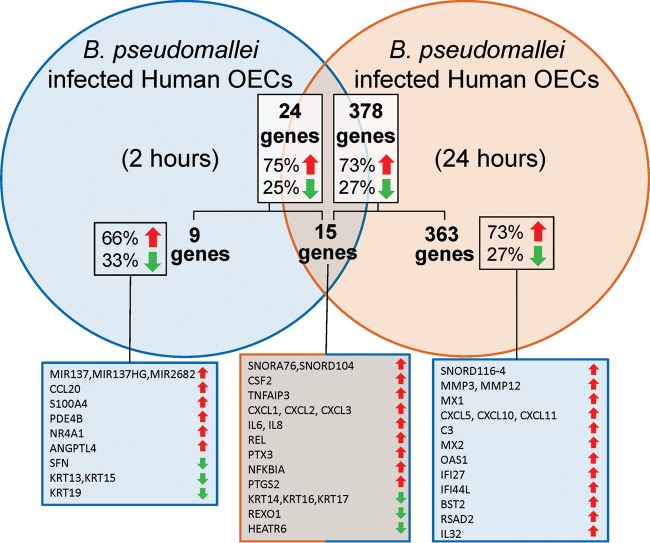
The antibacterial transcriptome of human OECs triggered in vitro following infection with B. pseudomallei. The Venn diagram shows the number of genes significantly activated or repressed at 2 h p.i. (24 genes) and 24 h p.i. (378 genes) and the numbers of genes common (15 genes) and unique to the time points (9 genes and 363 genes at 2 and 24 h, respectively). The percentages of genes in each group that are upregulated and downregulated are indicated by the red and green arrows, respectively. The genes up- and/or downregulated in the groups are listed in corresponding boxes at the bottom (only the top 15 genes upregulated at 24 h are shown).
There were a total of 277 and 101 genes that were upregulated and downregulated at 24 h p.i., respectively (see Table S2B in the supplemental material). The genes that exhibited significant upregulation only at 24 h p.i. (not at 2 h p.i.) included MMP12 (20.1-fold), MX1 (17.4-fold), CXCL5 (15.8-fold), CXCL10 (11.2-fold), C3 (10.4-fold), MX2 (8.2-fold), OAS1 (7.3-fold), IFI27 (6.6-fold), and IFI44L (6.6-fold). The list of strongly downregulated genes unique to the 24-h p.i. time point included MIR1204/PVT1 (−60.1-fold), ACTC1 (−17.9-fold), MYL3 (−7.0-fold), TNNI3 (−5.1-fold), TNNC1 (−5.0-fold), SCRIB (−4.1-fold), CRYAB (−3.1-fold), and CPSF1 (−2.4-fold).
The most highly activated canonical pathways in OECs infected with Burkholderia compared to PBS controls at 24 h p.i., identified by IPA, were agranulocyte and granulocyte adhesion and diapedesis, IL-17 signaling, role of pattern recognition receptors in recognizing microbes, TREM1 signaling, and role of cytokines in mediating communication between immune cells. The top canonical pathways, biological functions, and upstream mediators that define the OEC transcriptional signature in response to infection are summarized in Table 2. There were 10 canonical pathways identified with z-scores >1.8 standard deviations above the mean identified by IPA, including leukocyte extravasation signaling, IL-6 and IL-8 signaling, acute-phase response signaling, HMGB1 signaling, and IFN signaling (see Table S2C in the supplemental material). The top two networks of overexpressed genes, defined by IPA, are shown in Fig. S2 in the supplemental material. InnateDB, used to identify overrepresented pathways among those activated in OECs, identified many active pathways that were equivalent to those identified by IPA, such as immune-related and signaling pathways; the ORA-defined pathways are listed in Table S2D in the supplemental material.
TABLE 2
Top functional groupings of gene networks comprising significantly up- and downregulated genes in the response of human OECs to B. pseudomallei at 24 h p.i.a
| Functional grouping | Gene network |
|---|---|
| Canonical pathways | Role of pattern recognition receptors in recognizing bacteria and viruses |
| TREM1 | |
| HMGB1 | |
| Leukocyte extravasation | |
| Acute-phase response | |
| IFN, IL-6, IL-8 | |
| Biological functions | Cellular movement |
| Inflammatory response | |
| Immune cell trafficking | |
| Immunological disease | |
| Hematological system function | |
| Molecular transport | |
| Upstream mediators | Lipopolysaccharide |
| IL-1 receptor antagonist (IL1RN) | |
| TNF | |
| Poly(rI) · poly(rC) RNA | |
| IL-1β | |
| IFN-α and IFN-γ | |
| Pathways in InnateDB ORA | Cytokine-cytokine receptor interaction |
| Chemokine receptors bind chemokines | |
| NOD-like receptor signaling pathway | |
| IFN signaling | |
| Striated-muscle contraction | |
| Peptide ligand-binding receptors |
qRT-PCR analysis of OEC antimicrobial responses and validation of RNA-Seq data.
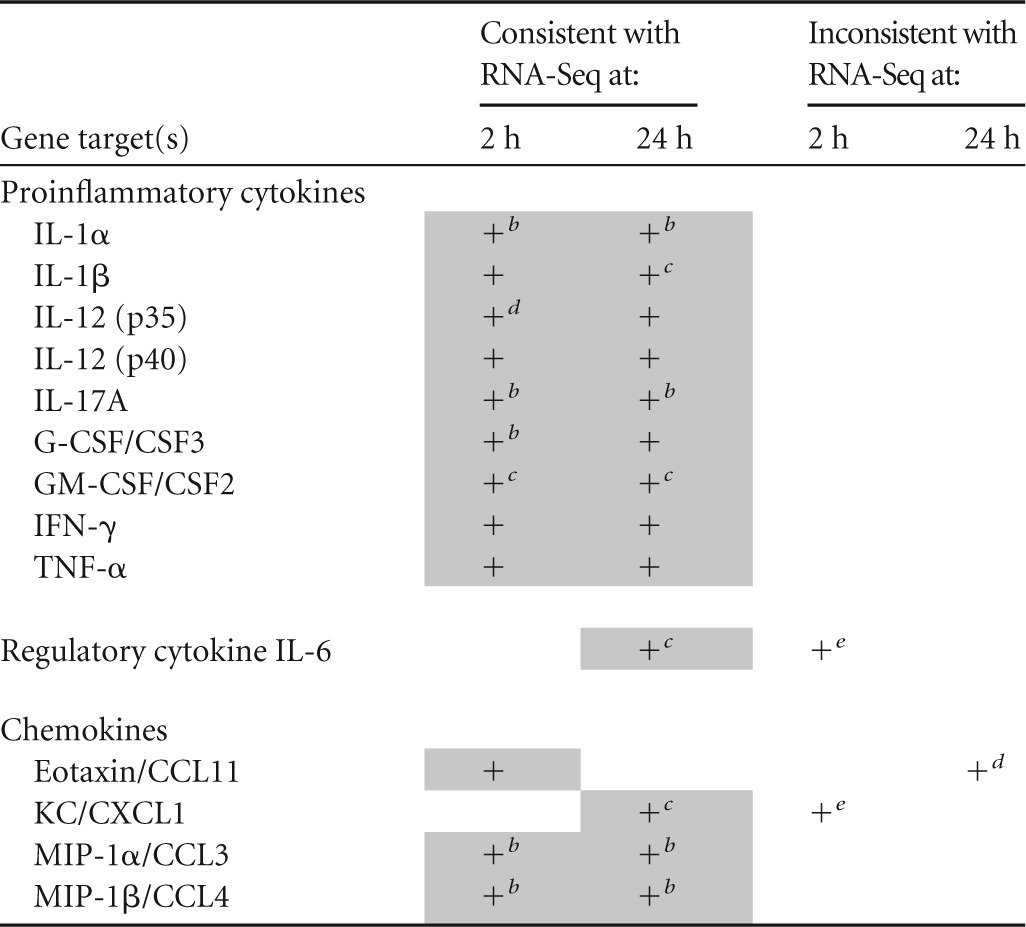
We quantitated mRNA for several genes chosen on the basis of RNA-Seq data by qRT-PCR; these results are summarized in Table 3. There was general agreement between the findings of qRT-PCR for compared to RNA-Seq. For example, we found that IL-1α, IL-1β, IL-12 (p35), IL-12 (p40), G-CSF/CSF3, GM-CSF/CSF2, IFN-γ, and TNF-α were significantly upregulated at 2 h p.i. according to qRT-PCR, which was reflected in the RNA-Seq data; IL-6, KC/CXCL1, and MIP-1α/CCL3 showed discordance between the two methods. All of the gene expression data at 24 h p.i. were consistent between the qRT-PCR and RNA-Seq results.
TABLE 3
Comparison of qRT-PCR and RNA-Seq data used to establish human OEC transcriptional responses to B. pseudomallei infectiona
DISCUSSION
Melioidosis is an emerging disease worldwide, with mortality rates as high as 42.6% in some regions where it is endemic (47). Treatment of the disease can be problematic (48), and there is no vaccine available. Various murine models have been used to characterize B. pseudomallei virulence factors and the host immune response. Here, we investigated the role of CPS I in a murine model of acute respiratory melioidosis. We have shown that CPS I affects colonization in this model and affects the innate immune response in the lungs, NALT, and OM. We have also demonstrated that these effects differ between the i.n. and inhalation routes of infection used here to model the pneumonic form of melioidosis. Analysis of histopathology in B. pseudomallei-infected mice showed that CPS I is a key factor in determining the severity of acute inflammation and neutrophil infiltration in various tissues, including NALT and the OM, which correlates with tissue cytokine responses and bacterial loads. Finally, we have elucidated, on a genome-wide scale, the antimicrobial transcriptional response of human OECs to B. pseudomallei.
The MSHR520 acapsular mutant strain colonized the lungs, blood, spleens, livers, NALT, and OM of mice at lower levels than the WT strain. This establishes that CPS I is a key virulence factor and is consistent with findings in hamster and mouse models that showed that acapsular B. pseudomallei strains have attenuated virulence (22, 24,–26, 49). CPS I aids the persistence of B. pseudomallei in blood by reducing complement C3b deposition on bacteria (25). In our study, the Δcap mutant strain was not detected in blood at 24 h p.i. (either route of infection) but was present at 48 h p.i. following i.n. infection (albeit at extremely low levels compared to the WT). In contrast, the WT strain was isolated from blood at both time points, irrespective of the infection route. These data show that CPS I enables the persistence of B. pseudomallei in blood and is consistent with a recent report that CPS I is required for dissemination from the lungs following intubation-mediated intratracheal instillation (50).
In the rodent nasal cavity, NALT (the equivalent of Waldeyer's ring in humans) exists as a pair of lymphoid cell aggregates and is the only organized mucosal lymphoid tissue in the murine upper respiratory tract (31). Previously, we reported that B. pseudomallei colonizes NALT following i.n. inoculation and hypothesized that NALT may be a portal to the lymphatic system, resulting in systemic infection (27). In the present study, we have demonstrated that B. pseudomallei colonizes NALT following infection via the i.n. and inhalation routes. Infection of the spleen and liver by the Δcap mutant strain at 24 h in the absence of detectable blood colonization, albeit without apparent replication by 48 h, supports the hypothesis that NALT may play a role in the dissemination of B. pseudomallei to systemic sites in the absence of hematogenous spread. The anatomy of the murine OM is very similar to that of the human OM, with the same cell types and anatomical arrangement (4, 28, 51, 52). We therefore examined the murine OM and showed that high levels of both the WT and Δcap mutant strains were recovered from the OM following inoculation by i.n. or inhalation delivery. We previously demonstrated that B. pseudomallei rapidly colonizes the OM and brain in the absence of detectable blood colonization (27), and B. pseudomallei rapidly penetrates the olfactory and trigeminal nerves to infect the central nervous system (28). In the primary olfactory nerve, OECs are the principal phagocytic cells and macrophages are largely excluded even after injury (53). OECs can phagocytose bacteria in vitro (54,–56). We did not investigate invasion of the central nervous system in the present study, but our data demonstrate a tropism of B. pseudomallei for the OM and a defensive reaction by OECs to the bacteria, which supports these previous findings.
We found that B. pseudomallei adhered to and invaded human OECs in a capsule-independent manner. CPS I does not protect against phagocytosis or reactive oxygen species-mediated killing (57); our findings are consistent with this previous report. Human OECs killed >90% of the intracellular B. pseudomallei within 24 h, suggesting that OECs have powerful antibacterial defense mechanisms. Previous studies showed that primary rodent OECs are capable of phagocytosing bacteria and respond to a bacterial challenge via the production of nitric oxide and induction of nuclear localization of NF-κB (54,–56, 58, 59). We used global gene expression analysis based on RNA-Seq to map the first antibacterial transcriptome of human OECs. Among the most interesting findings in these experiments were the infection-induced activation responses of 378 genes that mediate biological pathways controlling cellular movement, inflammation, immunological disease, and molecular transport. The majority of these genes found to be activated following B. pseudomallei infection have not previously been reported as part of the transcriptional response of OECs to any infection. Of particular interest are those genes identified at the intersection of the predicted upstream mediators and activated canonical pathways, such as IFN, IL-6, and TNF, which are known to be important in controlling melioidosis (60). Other pathways of gene activation uncovered in this study are novel in the context of both B. pseudomallei and OECs. Thus, the present study establishes the extent of antimicrobial responses in these cells on a genome-wide scale and, more importantly, highlights the complex and underrecognized gene networks and biological pathways that are present in human OECs and most likely contribute to the killing of microbes. The analysis of these gene networks is now important.
The innate immune response to B. pseudomallei has been characterized in BALB/c mice and is associated with the recruitment of inflammatory cells to infected organs and an overwhelming cytokine response (17, 34, 35, 60,–62). Toll-like receptor 2 recognizes B. pseudomallei lipopolysaccharide and mediates some responses in murine melioidosis (11) via myeloid differentiation primary-response gene 88 (MyD88) (63). We have demonstrated that B. pseudomallei induces high levels of cytokines in the lungs, NALT, and OM, findings consistent with these reports. This is the first report of the innate immune response to B. pseudomallei in NALT and the OM. We detected high levels of pro- and anti-inflammatory cytokines and chemokines in these tissues, suggesting a nonpolarized Th1 or Th2 response and activation of multiple inflammatory pathways. Our findings of elevated IL-6 levels in the OM and NALT, a response that was consistent with colonization trends in these tissues, are consistent with the production of IL-6 in OECs in a model of i.n. Staphylococcus aureus infection (64). Gan (65) suggested that the development of acute melioidosis is due not to a lack but rather an excess of inflammation and a failure of regulatory mechanisms. We observed increased levels of anti-inflammatory cytokines at 48 h p.i.; however, these were insufficient to inhibit the hyperproduction of proinflammatory responses in the lungs, NALT, and OM. Interestingly, the levels of many cytokines in the lungs, NALT, and OM were not proportionate to the B. pseudomallei colonization load in the tissue; similar results were reported for the spleen (66).
Infection with the Δcap mutant strain was associated with lower cytokine levels at 48 h p.i. than infection with the WT strain. This is consistent with findings that BALB/c mice i.n. infected with an acapsular B. pseudomallei strain produced lower levels of the monokine induced by IFN-γ, RANTES, and IFN-γ than the WT strain (9). In contrast to our findings, this previous study did not find differences in the concentrations of IL-1α, IL-1β, IL-6, eotaxin, G-CSF, GM-CSF, KC, MCP-1, MIP-1α, MIP-1β, or TNF between the acapsular mutant and the WT strain. In the present study, mice infected with the Δcap mutant or WT strain received approximately equal numbers of bacteria, whereas Warawa et al. (9) infected mice with a significantly larger dose of the acapsular mutant (3 × 106 CFU) than the WT strain (4 × 104 CFU). Thus, these differences may be attributable to dose-dependent effects in the different models.
Patients with severe melioidosis have been reported to have high serum IFN-γ, TNF-α, IL-6, IL-8, IL-10, IL-12, IL-15, IL-18, and IL-27 levels (67,–71). Of these, IL-6, IL-10, and IFN-γ may have prognostic value and are considered to be predictors of death (67,–69). In a mouse model of melioidosis, Santanirand et al. (72) demonstrated that the production of IFN-γ was essential for resistance to acute infection. In the present study, the levels of these cytokines were upregulated in response to B. pseudomallei infection compared to those in control mice. However, the levels of G-CSF, KC, and MCP-1 detected were highest in the lungs, NALT, and OM. KC and G-CSF are both potent chemoattractants and activators of neutrophils (73, 74), whereas MCP-1 specifically attracts macrophages and memory T cells (75). Whether the upregulation of cytokines and chemokines by OECs acts to recruit leukocytes in the olfactory nerves is uncertain, as previous studies have shown that cells of the immune system are largely excluded from the olfactory nerve and that OECs perform the role of innate immune cells instead (53, 76).
A number of studies have demonstrated that high numbers of neutrophils are recruited to B. pseudomallei-infected tissues (16, 60, 62, 77). Easton et al. (60) demonstrated that activated neutrophils are rapidly recruited to the lungs of C57BL/6 mice following i.n. B. pseudomallei infection. They also reported that depletion of Gr-1+ cells increased the susceptibility of mice to B. pseudomallei and resulted in a drastic reduction in TNF-α, IL-6, and IFN-γ levels. Neutrophil extracellular traps may also be involved in the response to B. pseudomallei infection (78). Collectively, these data suggest that neutrophils play a major role in the innate defense against melioidosis. Our finding that neutrophil recruitment, as well as inflammation, is severely compromised in response to infection with the acapsular mutant clearly indicates that the innate immune, neutrophil-mediated, response may be at least partly responsible for pathogenesis following infection by WT B. pseudomallei. The role of B. pseudomallei CPS I in mediating interactions with neutrophils is an area that merits further study.
Inoculation by i.n. challenge has been used to model acute respiratory melioidosis and may mimic some aspects of an inhalation challenge (79); however, there have been no direct comparisons of pathogenesis between these routes of infection. In a prior aerosol challenge study, Massey et al. (80) compared disease progression in mice with histological analysis and cytokine responses. That study showed that the levels of cytokines and chemokines in serum, including KC, G-CSF, and MCP-1, correlated with tissue inflammation, leukocyte infiltration, and pathology at sites of infection. We did not measure serum cytokine levels, but we observed heavy loads of WT MSHR520 B. pseudomallei bacteria in tissues, including OM, lung, and NALT, that correlated with highly elevated levels of cytokines (e.g., G-CSF, KC, MCP-1) and tissue inflammatory pathology, including massive neutrophil infiltration; in mice infected with the acapsular mutant strain we observed lighter bacterial loads, lower levels of cytokines, and comparatively minimal tissue pathology. Overall, these findings are largely consistent with those reported by Massey et al. (80) and suggest a correlation among the bacterial load, the level of cytokine production, histopathology, and B. pseudomallei virulence. Variables such as B. pseudomallei strain virulence can dramatically influence the severity of experimental infection in mice (81). In addition to being infection route dependent (i.n. versus inhalation), phenotypes such as the ability of B. pseudomallei to enter the brain via the OM and nerve fascicles may also depend on the genotype of the isolate used. In addition to inflammatory responses being bacterial strain dependent, there may be bacterial tropism for different target organs.
This study establishes that significant differences in colonization and cytokine responses exist between the i.n. and inhalation routes of infection, a fact that has major implications for the modeling of this form of disease. It has been suggested that inhalation of B. pseudomallei results in the most severe form of melioidosis (17, 50); however, our data suggest that the number of bacteria recovered from blood, the spleen, the liver, NALT, and the OM is higher in mice infected i.n. and leads to proportionately strong cytokine responses in NALT and the OM. In contrast, Gutierrez et al. (50) recently demonstrated that i.n. delivery of B. pseudomallei resulted in significantly less systemic bacterial dissemination than inoculation via intubation-mediated intratracheal instillation; however, those studies were performed with C57BL/6 mice, which demonstrate responses to B. pseudomallei that are markedly different from those of BALB/c mice (7, 34). It is possible that the increased bacterial burden associated with i.n. inoculation, relative to inhalation, is due to delivery of the bacteria directly to the nasal cavity, which may facilitate dissemination via NALT and/or blood. Surprisingly, in the lungs, there were minimal differences in B. pseudomallei colonization between the i.n. and inhalation routes of infection. Despite this, the concentrations of most of the cytokines and chemokines tested were higher in the lungs of mice exposed to aerosolized B. pseudomallei. Thus, the immunopathogenesis of acute respiratory melioidosis, in terms of innate cytokine responses, depends on the route of infection. In addition, the inflammatory responses could also be dependent on the bacterial strain used and the bacterial tropism for different target organs, as discussed above. Future studies could address strain comparisons.
In summary, we show that CPS I facilitates bacterial colonization following i.n. and inhalation exposure in a BALB/c mouse model of acute respiratory melioidosis. CPS I stimulated key innate immune responses in the lungs, NALT, and OM and acute inflammatory pathology, including severe tissue neutrophilia in NALT, OM, the spleen, and the liver, demonstrating that CPS I acts a major virulence determinant. Mice inoculated with the Δcap mutant strain exhibited greater resistance to infection than those inoculated with WT B. pseudomallei, most likely because of lighter bacterial loads in their tissues, less acute inflammatory histopathology, and a controlled immune response. This study establishes, for the first time, that the i.n. and inhalation routes of infection lead to distinct disease outcomes in mice, which has implications for future experimental modeling of human respiratory melioidosis. Finally, this study defines the antimicrobial transcriptional program that is activated in human OECs for defense against B. pseudomallei.
ACKNOWLEDGMENTS
We thank Saeed M. Hashimi for excellent technical assistance and expert advice on the epMotion 5075 LH (Eppendorf Australia), which was used to perform robotic liquid handling as part of the qRT-PCR assays in the study.
Funding Statement
SYL is supported by a Fellowship from Conselho Nacional de Desenvolvimento Científico e Tecnológico (CNPq)-Brazil. GCU is supported by an Australian Research Council Future Fellowship. The funders had no role in study design, data collection and interpretation, or the decision to submit the work for publication.
Footnotes
Supplemental material for this article may be found at http://dx.doi.org/10.1128/IAI.01546-15.
REFERENCES
Articles from Infection and Immunity are provided here courtesy of American Society for Microbiology (ASM)
Full text links
Read article at publisher's site: https://doi.org/10.1128/iai.01546-15
Read article for free, from open access legal sources, via Unpaywall:
https://iai.asm.org/content/iai/84/7/1941.full.pdf
Citations & impact
Impact metrics
Citations of article over time
Alternative metrics
Article citations
The Immunological Roles of Olfactory Ensheathing Cells in the Treatment of Spinal Cord Injury.
Front Immunol, 13:881162, 20 May 2022
Cited by: 10 articles | PMID: 35669779 | PMCID: PMC9163387
Review Free full text in Europe PMC
Streptococcus agalactiae Infects Glial Cells and Invades the Central Nervous System via the Olfactory and Trigeminal Nerves.
Front Cell Infect Microbiol, 12:793416, 24 Feb 2022
Cited by: 4 articles | PMID: 35281448 | PMCID: PMC8907725
Chlamydia pneumoniae can infect the central nervous system via the olfactory and trigeminal nerves and contributes to Alzheimer's disease risk.
Sci Rep, 12(1):2759, 17 Feb 2022
Cited by: 25 articles | PMID: 35177758 | PMCID: PMC8854390
Burkholderia pseudomallei pathogenesis in human skin fibroblasts: A Bsa type III secretion system is involved in the invasion, multinucleated giant cell formation, and cellular damage.
PLoS One, 17(2):e0261961, 03 Feb 2022
Cited by: 4 articles | PMID: 35113856 | PMCID: PMC8812868
Novel factor in olfactory ensheathing cell-astrocyte crosstalk: Anti-inflammatory protein α-crystallin B.
Glia, 69(4):1022-1036, 12 Dec 2020
Cited by: 11 articles | PMID: 33314354 | PMCID: PMC9469687
Go to all (10) article citations
Data
Data behind the article
This data has been text mined from the article, or deposited into data resources.
BioStudies: supplemental material and supporting data
GEO - Gene Expression Omnibus
- (1 citation) GEO - GSE69312
Similar Articles
To arrive at the top five similar articles we use a word-weighted algorithm to compare words from the Title and Abstract of each citation.
Role for the Burkholderia pseudomallei capsular polysaccharide encoded by the wcb operon in acute disseminated melioidosis.
Infect Immun, 77(12):5252-5261, 14 Sep 2009
Cited by: 26 articles | PMID: 19752033 | PMCID: PMC2786491
Genome wide transcriptome profiling of a murine acute melioidosis model reveals new insights into how Burkholderia pseudomallei overcomes host innate immunity.
BMC Genomics, 11:672, 27 Nov 2010
Cited by: 33 articles | PMID: 21110886 | PMCID: PMC3017868
Comprehensive identification of virulence factors required for respiratory melioidosis using Tn-seq mutagenesis.
Front Cell Infect Microbiol, 5:78, 04 Nov 2015
Cited by: 28 articles | PMID: 26583079 | PMCID: PMC4631991
Melioidosis: molecular aspects of pathogenesis.
Expert Rev Anti Infect Ther, 12(12):1487-1499, 14 Oct 2014
Cited by: 42 articles | PMID: 25312349 | PMCID: PMC4409121
Review Free full text in Europe PMC
Funding
Funders who supported this work.
Department of Health | National Health and Medical Research Council (1)
Grant ID: APP1020394
Department of Industry, Innovation, Science, Research and Tertiary Education, Australian Government | Australian Research Council (1)
Grant ID: FT110101048
 a and
a and 




