Abstract
Free full text

Optimization and comparison of bottom-up proteomic sample preparation for early stage Xenopus laevis embryos
Associated Data
Abstract
Xenopus laevis is an important model organism in developmental biology. While there is a large literature on changes in the organism’s transcriptome during development, the study of its proteome is at an embryonic state. Several papers have been published recently that characterize the proteome of Xenopus laevis eggs and early stage embryos; however, proteomic sample preparation optimizations have not been reported. Sample preparation is challenging because a large fraction (~90% by weight) of the egg or early stage embryo is yolk. We compared three common protein extraction buffer systems, mammalian Cell-PE LB™ lysing buffer (NP40), sodium dodecyl sulfate (SDS), and 8M urea, in terms of protein extraction efficiency and protein identifications. SDS extracts contained the highest concentration of proteins, but this extract was dominated by a high concentration of yolk proteins. In contrast, NP40 extracts contained ~30% of the protein concentration as SDS extracts, but excelled in discriminating against yolk proteins, which resulted in more protein and peptide identifications. We then compared digestion methods using both SDS and NP40 extraction methods with one-dimensional reverse phase liquid chromatography tandem mass spectrometry (RPLC-MS/MS). NP40 coupled to a filter-aided sample preparation (FASP) procedure produced nearly twice the number of protein and peptide identifications compared to alternatives. When NP40-FASP samples were subjected to two-dimensional RPLC-ESI-MS/MS, a total of 5171 proteins and 38,885 peptides were identified from single stage of embryos (stage 2), increasing the number of protein identifications by 23% in comparison to other traditional protein extraction methods.
Introduction
Xenopus laevis is an important model organism for studying early vertebrate development. This organisms was used for the first nuclear transfer experiment that demonstrated totipotency of the nucleus from a differentiated cell [1], the first isolation of a gene from an organism [2], the generation of the first nucleotide sequence of a gene [3], and the purification of the first eukaryotic transcription factor [4]. Xenopus laevis has a number of useful properties for developmental studies. Fertilization and development occur outside of the mother, which allows direct observation of embryogenesis. Embryo development is reasonably fast to produce a fully developed nervous system within 4 days. For proteomic research, the relatively large size of the egg (~1.2 mm) simplifies a number of manipulations [5].
While there is rich literature that describes changes in the transcriptome during development [6], the analysis of the Xenopus laevis proteome is an early state. Western blotting and other traditional methods are commonly used to confirm protein identity during development [7,8]. However, these methods only characterize a handful of proteins and are limited to those targets for which affinity reagents are available.
Several quantitative bottom-up proteomics studies have appeared in the past two years for Xenopus embryos and eggs [9–12]. We reported the first proteomic study of early-stage vertebrate development [9]. That study used iTRAQ chemistry for precise quantification of changes in expression of 4000 proteins at six stages of early Xenopus development. Our study used NP40 to lyse the embryos, followed by protein purification with acetone precipitation and urea solubilization.
Three other papers dealing with Xenopus proteomics have appeared since our original report. Smits et al. employed label-free proteomics of single Xenopus eggs [10]. That paper also employed NP40 to lyse the eggs, following by purification with a (FASP) method [13]. Wũhr et al. described protein and transcript expression in unfertilized Xenopus eggs [12]. Recently, Peshkin et al. generated label-free quantification of slightly more than 5000 proteins across six stages of early development [11]. These last two studies used NP40 for lysing, followed by a methanol-chloroform protocol.
These early studies provide limited detail on sample preparation. Sample preparation is made challenging by the very large yolk protein concentration in early stage embryos; ~90% of proteins at stage 1 are yolk proteins [14]. With such a large portion of the protein being yolk, other, lower abundance proteins can be masked when subjected to traditional extraction and purification methods. In this manuscript, we systematically evaluate a number of protocols for sample preparation of early stage Xenopus embryos.
Materials and methods
Materials
Xenopus laevis animals were purchased from Nasco (Fort Atkinson, WI USA). Animals were housed in a pathogen-free environment on a 12 h light – 12 h dark schedule. NP40 was purchased from G-Biosciences (St. Louis, MO USA). 1,1,2 trichloro 1,1,2 trifluroethane (Freon) was purchased from AquaSolutions (Deer Park, TX USA). NuPage 4–12% Bis-Tris Gel, MOPS SDS Running Buffer, SimplyBlue SafeStain™, and LDS Samper Buffer were purchased from Novex (Carlsbad, CA USA). Complete, mini protease inhibitor cocktail, provided in EASYpacks, was purchased from Roche Diagnostics (Indianapolis, IN USA). Human Chorionic Gonadotropin (HCG), bovine pancreas TPCK-treated trypsin, urea, NH4HCO3, dithiothreitol (DTT), iodaoacetamide (IAA), cysteine, SDS, and acetone were purchased from Sigma-Aldrich (St. Louis, MO USA). Water was deionized by a Nano Pure system from Thermo scientific (Marietta, OH). Acetonitrile (ACN), formic acid (FA), Pierce C-18 Spin Columns, and Pierce BCA Protein Assays were purchased from Thermo Scientific (Marietta, OH USA). Sep-Pak Vac (1 cc, 100 mg) C-18 Cartridges were purchased from Waters (Ireland). Centrifugal Filter Units with a 30,000 NMWL cut-off were purchased from Millipore (Carrigtwohill, CO USA). Marc’s Modified Ringers (MMR) and MBS were made in-house (See Electronic Supplemental Material (ESM) page S-10).
Embryo culture and collection
The animal protocol was based on Reference 9. Briefly, all animal procedures were performed according to protocols approved by the University of Notre Dame Institutional Animal Care and Use Committee. Female Xenopus laevis were induced to spawn by injection with 600 units of HCG 12–15 hours prior to spawning. Prior to spawning, testes were isolated from male frogs that had been immersed in ice water for 15–20 minutes. Eggs and minced testes were combined in 2–5 mL 1/3 x MMR and incubated for 10 minutes. Eggs and testes were then flooded with 1/3 x MMR and incubated for another 20 minutes. After fertilization, embryos were dejellied using 1/3 x MMR plus two percent L-cysteine by incubating with swirling for 4 minutes; 20 embryos per Eppendorf tube were collected 60 minutes post fertilization (development stage 2). All procedures were carried out at room temperature.
Protein extraction
Each sample was suspended in 650 μL of 3% SDS, 8 M urea, or NP40 containing protease inhibitor. Once a sample was suspended, all further manipulations were done on ice. Each sample was homogenized for 60 seconds, followed by sonication (VWR Scientia, Batavia, IL USA) for 5 minutes twice to lyse embryos. Sample were centrifuged for 10 minutes at 12,000 g and supernatant was collected in a fresh tube and stored at −80 °C before use. In one experiment, equal volumes of Freon and protein mixture were combined, vortexed, and centrifuged at 16,000 g for 16 minutes to extract yolk protein from SDS. The bottom layer containing yolk-free protein was saved. Extraction with Freon was repeated three times and the products were pooled. In all cases, a BCA assay was performed to determine total protein concentration.
SDS-Page gel
20 μg of each extracted protein mixture was loaded onto the gel. The NuPage pre-loaded settings on the Invitrogen Life Technologies (Marietta, OH USA) were used. A current of 180–190 mA was applied to the gel for 35 minutes. Following separation, the gel was stained for 4.5 hours before being de-stained for 60 minutes with deionized water.
FASP digestion
The FASP digestion protocol was based on reference 13. 100 μg of sample lysate was obtained and denatured at 90 °C for 10 minutes. Lysate was reduced with 25 mM DTT and incubated at 60 °C for 30 minutes followed by alkalization with 50mM IAA of the sample for 30 minutes at room temperature. The 30 kDa membrane was moistened using 100 μL of 100 mM NH4HCO3 and centrifuged for 15 minutes at 13,000 g. Alkylated sample was placed on the membrane and centrifuged for 40 minutes at 14,000 g. Membrane was washed three times with 200 μL of 8 M urea and centrifuged at 18,000 g for 25 minutes each time. A final wash with 100 μL of 20 mM NH4HCO3 was centrifuged for 10 minutes at 16,000 g. A tryptic digest of 1:25 trypsin:protein was completed by vortexing the samples for 10 minutes, followed by incubating at 37°C for 12 hours. FA was used to quench the digest.
Acetone precipitation (AP) with urea
The AP with urea protocol was based on reference 9. 50 μg of protein from each digest’s supernatant was precipitated with 300 μL of chilled acetone for 6 hours at −20 °C. The sample was then immediately centrifuged for 10 minutes at 10,000 g. Supernatant was removed and the protein pellet was further washed with an additional 300 μL of chilled acetone, centrifuged, supernatant was removed and the sample was allowed to air dry. 100 μL of 8 M urea was added to each sample to dissolve the pellet. Protein was denatured at 37 °C for 60 minutes before being reduced with 25 mM DTT for 30 minutes at 37°C. The protein was then alkylated with 50 mM IAA at room temperature in the dark for 30 minutes. Additional NH4HCO3 solution was added to dilute the urea concentration to approximately 2 M. A tryptic digest of 1:25 trypsin:protein was performed, followed by incubation at 37 °C for 12 hours. FA was used to quench the digest.
AP with NH4HCO3
The AP with NH4HCO3 protocol was based on reference 15. 100 μg of SDS and urea supernatant was denatured at 90 °C for 20 minutes followed by reduction of the protein with 25 mM DTT at 37 °C for 30 minutes. For alkylation, 50 mM IAA was added at room temperature for 30 minutes in the dark. Cold acetone equivalents were added five times and the protein mixture was vortexed vigorously. The sample was stored at −20 °C for 3 hours for protein precipitation before centrifugation at 4 °C for 30 minutes at 20,000 g. The supernatant was removed and the pellet was washed with an equivalent of 6:1, acetone:water and allowed to air dry. A tryptic digest of 1:25, trypsin:protein was performed, followed by incubation at 37 °C for 12 hours. FA was used to quench the digest.
Fractionation
Strong cation exchange fractionation was based on Reference 9. Samples were fractionated using a Water Alliance HPLC (Waters, Milford, MA, USA) at a flow rate of 0.8 mL/min. Approximately 250 μg of protein digest was loaded onto an Agilent Zorbax SB-C18 column (5 μm, 4.6 mm x 150 mm, Agilent Technologies, Wilmington, DE, USA). Sample was prepared in triplicate and pooled to obtain 300 μg of digest; 250 μg of the digest was injected onto the column. The mobile phase gradient was generated using buffer A (20 mM NH4HCO3 at a pH of 9) and buffer B (80% acetonitrile in 20 mM NH4HCO3 at a pH of 9). The samples were loaded onto the column with a 10-minute wash before collection, and then separated by a 60-minute gradient. Eluate from 10–70 minutes was collected in 60 fractions and each fraction was then vacuum concentrated. Fractions were lyophilized and re-suspended in 40 μL of 2% acetonitrile, 0.1% formic acid. Fifteen samples were then generated by pooling every 15th fraction (i.e. fractions 1, 16, 31, and 46 were pooled to generate sample 1; fractions 2, 17, 32, and 47 were pooled to generate sample 2; etc.). The pooled samples were then desalted.
Peptide desalting
Prior to 1D separation, all samples were desalted with C18 SPE columns. Prior to 2D separation, samples were desalted using a C18 spin column followed by vacuum concentration and lyophilization to a final concentration of 1 μg/μL protein:2% ACN and 0.1% FA solution.
UPLC-ESI-MS/MS analysis
UPLC-ESI-MS/MS analysis was based on Reference 9. A nanoACQUITY UltraPerformance LC (UPLC©) system (Waters, Milford, MA USA) was used for peptide separation. Buffer A (0.1% FA in water) and buffer B (0.1% FA in ACN) were used as mobile phases for gradient separation. Peptides were automatically loaded onto a commercial C18 reverse phase column (Waters, 100 μm ID, 100 mm long, 1.7 μm particle, BEH130C18, column temperature 40 °C) with 2% buffer B for 10 minutes at a flow rate of 0.7 μL/min, followed by a 3-step gradient separation, 2 min from 2% to 8%, 114 minutes to 28% B, 3 minutes to 85% B, and maintained at 85% for the next 13 minutes. The column was equilibrated for 12 minutes with 2% B before analysis of the next sample. The eluted peptides from the C18 column were pumped through a capillary tip for electrospray, and analyzed by a Q-Exactive™ mass spectrometer (Thermo Fisher Scientific). For analysis using a Q-Exactive™ HF, all parameters were kept consistent except for the gradient, which was 2% buffer B for 15 minutes at a flow rate of 0.6 μL/min, followed by a 3-step gradient separation, 10 min from 2% to 12%, 120 minutes to 28% B, 10 minutes to 85% B, and maintained at 85% for the next 15 minutes. The column was equilibrated for 15 minutes with 2% B before analysis of the next sample. For each sample, 2 μg of peptide was used per run.
The electrospray voltage was 1.6 kV, and the ion transfer tube temperature was 280 °C. The S-Lens RF level was 50.00. Data acquisition was programmed in data-dependent acquisition (DDA) mode. For analysis using the Q-Exactive™, a top 12 method was used. Full MS scans were acquired in Orbitrap mass analyzer over 380–1800 m/z range with a resolution of 70,000 and the number of micro scans set to 1. The target value was 1.00E+06, the maximum injection time was 230 ms. For MS/MS scans, the twelve most intense peaks with charge state ≥2 were sequentially isolated and further fragmented in the higher-energy-collisional-dissociation (HCD) cell following one full MS scan. The normalized collision energy was 33%, and tandem mass spectra were acquired in the Orbitrap mass analyzer with resolution 35,000. The first fixed mass was 100.0. The number of micro scans was 1 and the ion selection threshold was 5.0E+04 counts. Peptide match and excludes isotopes were turned on. Dynamic exclusion was set at 30 seconds.
For analysis using the Q-Exactive™ HF, instrument settings included: a top 15 method, full MS scans were acquired in Orbitrap mass analyzer over 350–1500 m/z range with a resolution of 60,000, and the number of micro scans set to 1. The target value was 3.00 E+06, the maximum injection time was 30 ms. For MS/MS scans, the fifteen most intense peaks with charge state ≥2 were sequentially isolated and further fragmented in the higher-energy-collisional-dissociation (HCD) cell following one full MS scan. The normalized collision energy was 33%, and tandem mass spectra were acquired in the Orbitrap mass analyzer with resolution 30,000. The first fixed mass was 100.0. The number of micro scans was 1 and the ion selection threshold was 1.0 E+05 counts. Peptide match and excludes isotopes were turned on. Dynamic exclusion was set at 30 seconds.
Data analysis
Raw data files were analyzed using MaxQuant 1.5 with the default setting for deep proteome analysis [16]. Variable modifications were set to include: acetyl protein N-term, oxidation (M), acetyl (K), and deamination (NQ) using specific digestion with trypsin. Carbamylation was set as a fixed modification. A maximum of two-missed cleavages was allowed. The minimum charge of the peptide was set to 2 and the max charge was set to 7. The intensity threshold was set to 500 and the intensity determination was set to be the value at the maximum intensity. For label free quantification (LFQ), which employs delayed-normalization of the intensity, the minimum ratio count was set to 2 and fast LFQ searches were not allowed [17]. The maximum number of modifications per peptide was set to 5 and the maximum mass of a peptide was set to 4000 Da. A minimum of 1 unique peptide was required for protein identification. For searching and analyzing the data, the Uniprot database for Xenopus laevis from December 2014 was used. For comparison of protein identification numbers, the mRNA derived database was used [12]. When identifying and applying LFQ to proteins, the false discovery rate was set to 0.01 on both peptide and protein levels. Data are summarized in Excel spreadsheets included in the ESM.
Results and Discussion
We evaluated several experimental protocols to maximize the number of identified proteins from Xenopus laevis embryos at stage 2 of development.
Protein extraction efficiency of three lysis buffer systems
We first compared 3% SDS, NP40, and 8 M urea for lysis of the embryos. SDS is a harsh, denaturing, ionic detergent that is widely used for solubilizing membrane proteins [18]. SDS is a harsh detergents that has one of the highest efficiencies for protein extraction. Urea is a chaotrope that is commonly used for extracting soluble proteins [19]. NP40 is non-ionic detergent that has been reported to have high protein extraction efficiency [20–21].
A standard BCA assay, Table 1, was performed to determine the protein concentration after extraction. SDS generated a ~4-fold higher protein concentration than NP40 and ~2-fold higher protein concentration than 8M urea.
Table 1
Protein concentration estimated using standard BCA analysis
| Method | μg protein extracted (± standard deviation of triplicate measurements) |
|---|---|
| SDS | 126 ± 30 |
| SDS/Freon | 112 ± 12 |
| NP40 | 36 ± 5 |
| 8M Urea | 69 ± 26 |
Protein concentration estimated using standard BCA analysis
We also used a Freon to extract nonpolar yolk proteins from the SDS extract [22]. Surprisingly, Freon treatment did not result in a significant decrease in total protein concentration, Table 1. Only ~10–12% of the total protein was extracted from SDS samples, which included non-polar and yolk proteins (ESM Fig. S2). Nevertheless, it appears that lipovitellin 2B (31 kDa) and lipovitellin I (120 kDa) were partially removed by Freon extraction, Figure 1. Lipovitellin I protein has a noticeable decrease in the intensity of the 120 kDa band. The decrease in intensity is less obvious for lipovitellin 2B at 31 kDa [14]. In the following experiments, SDS extracts were subjected to Freon treatment before further preparation (ESM, Fig. S3).
In contrast, NP40 showed strong discrimination against yolk proteins compared with SDS, SDS/Freon, and 8 M urea, Fig. 1. However, other proteins appear to generate stronger signals in the 30–120 kDa range compared with other solubilization protocols.
Number of protein identifications using three lysis systems
The cell lysates from SDS/Freon, NP40, and urea extraction were further subjected to protein purification and digestion with the FASP method [13], followed by analysis using UPLC-ESI-MS/MS. NP40 produced 25% more protein identifications than 8 M urea and 50% more identifications than SDS/Freon, Table 2. Sequence coverage and the average number of peptides per protein were also elevated in NP40 samples, when compared to SDS/Freon and 8M urea samples, Table 2. This increases our confidence in the effectiveness of using NP40 as a preferred extraction buffer for early embryotic Xenopus laevis samples when yolk protein is in high abundance.
Table 2
Comparison of extraction methods
| Method | Peptides Identified | Proteins Identified | Average Peptides per Protein | Sequence Coverage (%) |
|---|---|---|---|---|
| NP40 | 14,000 ± 718 | 2270 ± 28 | 6.17 | 24.4 ± 0.4 |
| SDS/Freon | 3150 ± 801 | 1513 ± 155 | 2.08 | 6.6 ± 1.4 |
| 8 M Urea | 6660 ± 696 | 1820 ± 103 | 3.66 | 12.1 ± 1.2 |
Comparison of protein extraction methods. All methods using the FASP protocol for digestion of the proteins. The Q-Exactive HF mass spectrometer was used for analysis of these samples.
Although SDS/Freon produced extracts with the highest protein concentration, that extract generated a much smaller number of protein identifications than NP40 and 8M urea. The high concentration of yolk proteins in the SDS/Freon extract degraded the chromatographic separation of the tryptic digests, Fig. 2. Because of the high abundance of the yolk proteins, the migration times for the sample shifted, Fig. 2. Individual m/z values were extracted (ESM Fig. S7) to show the general shift of the retention times due to high abundance of a few proteins in the SDS/Freon samples.
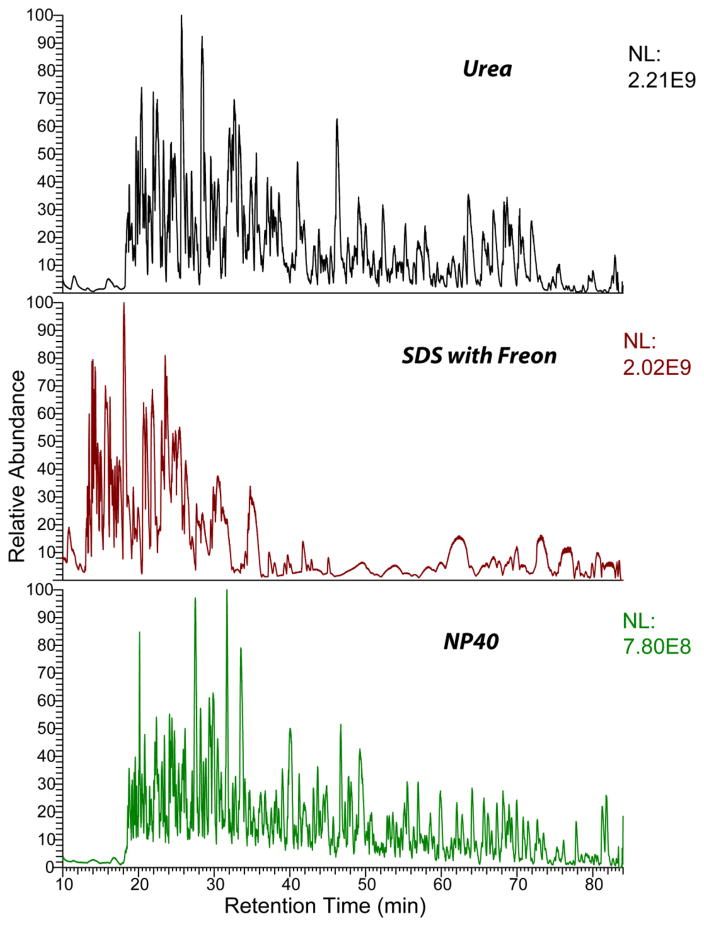
Base peak chromatograms for tryptic digests prepared from the three extraction methods. Chromatograms were extracted from data obtained from the Q-Exactive HF mass spectrometer.
We analyzed the overlap of identified proteins from the three lysis buffer systems. Nearly 95% of the proteins from SDS method were included in the NP40 method, and 96% of the proteins from 8 M urea method were included in the NP40 method. Combination of three lysis methods generated 2355 protein IDs, which is ~4% more than the NP40 method alone (ESM Fig. S4). Care was also taken to ensure sample preparation was not a factor in protein identification. Few differences were found between runs on the mass spectrometer and between sample preparation times (ESM Fig. S5). We also used the MaxLFQ function in MaxQuant to perform label free quantitation (LFQ) for proteins from the three lysis buffers. A cluster analysis was performed based on the LFQ intensity of proteins, Fig. S1 in the ESM [17]. Triplicate preparations of NP40, SDS, and 8 M urea samples grouped together, and the three lysis buffer data can be clearly distinguished. As expect, yolk proteins have significantly higher protein intensity in SDS and 8M urea data compared with NP40 data. Interestingly, spindle microtubule, ATP binding, zinc ion binding, and carbohydrate binding proteins have higher intensity in the NP40 data. ESM Fig. S6 provides the distribution of proteins according to cellular components, cellular part, and organelle. NP40 outperformed SDS frequently, as would be expected due to the increased number of proteins extracted by NP40.
Comparisons of different protein purification and digestion protocols
Based on the results of Table 2, NP40 was identified as the optimal lysis system, and NP40 extraction was used to evaluate a set of three protein purification and digestion methods. FASP, AP-Urea, and AP-NH4HCO3 were compared in terms of the protein and peptide IDs, Table 3. NP40 using the FASP method generated by far the highest number of protein and peptide identifications, with nearly a factor of two higher numbers of identifications compared with the alternatives.
Table 3
Comparison of digestion methods
| Method | Peptide Identifications | Protein Identifications | Peptides per Protein | Sequence Coverage (%) |
|---|---|---|---|---|
| NP40, FASP | 15,065 | 2201 | 6.85 | 23.6 ± .1 |
| NP40, AP-Urea | 8924 | 1210 | 7.38 | 17.8 ± 0.1 |
| NP40, AP-NH4HC03 | 6968 | 1080 | 6.45 | 13.9 ± 0.4 |
| SDS/Freon, FASP | 1961 | 907 | 2.16 | 6.5 ± 0.3 |
| SDS/Freon, AP-Urea | 3235 | 739 | 4.37 | 1.8 ± 0.2 |
| SDS/Freon, AP-NH4HC03 | 2111 | 782 | 2.69 | 8.1 ± 0.1 |
Comparison of digestion methods using either SDS/Freon or NP40. All samples for this comparison were run on the Q-Exactive mass spectrometer.
The better performance of the FASP method is likely due two reasons. First, the FASP method produces a much cleaner proteome digest than other two methods. Lipids and other metabolites are easily removed with FASP method since they flow through the filter membrane. In contrast, they are not completely removed by AP. Second, FASP appears to minimize sample loss, which is commonly unavoidable during AP.
Large-scale proteome analysis of Xenopus stage 2 embryo lysate
We used the optimized sample preparation protocol (NP40-FASP) to generate a tryptic digest from a lysate from stage 2 Xenopus embryos. The digest was fractionated using high-pH RPLC, followed by low-pH RPLC separation and ESI-MS/MS analysis.
Because SDS extracted more proteins than NP40, Table 1, SDS may have extracted some proteins that were masked in the RPLC-MS/MS data of Table 2 due to the limited separation capacity of one-dimensional RPLC. We also used the SDS-FASP method to compare the digest efficiency, which was also fractionated with high-pH RPLC before low-pH RPLC-ESI-MS/MS analysis.
Using the two-dimensional RPLC strategy, 4957 proteins and 32,620 peptides (average 6.58 peptides/protein) were identified with NP40-FASP method; in contrast, 4010 proteins and 16,464 peptides (4.11 peptides/protein) were obtained with SDS/Freon-FASP method. SDS/Freon-FASP generated 80% of the number of protein IDs as NP40-FASP. However, over 98% of the proteins identified in the SDS/Freon-FASP analysis were also contained in the NP40-FASP results; combination of the two techniques produced a negligible increase in the number of protein identifications compared to the use of NP40-FASP alone (ESM Fig. S4B). Sequence coverage for the NP40-FASP samples averaged 25.2%, while sequence coverage for SDS/Freon-FASP samples averaged 13.6%. This gives us greater confidence in the NP40 analysis as it not only digs deeper into the proteome and also provides higher sequence coverage.
Finally, we also searched our raw fractionated data for NP40 and SDS/Freon against a Xenopus reference database derived from mRNA generated by Wühr et al. [12]. The new database search generated 5171 protein and 38,885 peptide identifications (7.51 peptides/protein identified) for the NP40-FASP data.
As noted in the introduction, there have been four publications addressing early stage Xenopus developmental proteomics [9–12]. Those studies focused on the changes in protein expression during several stages of development, and provided no information on sample preparation optimization. This set of papers employed different loading amounts, different chromatographic instruments and mass spectrometers, and different searching algorithms and databases; comparison of the number of protein identifications between these experiments is inappropriate. In this paper, we focus on a comparison of sample preparation protocols, with the goal of optimizing the number of protein identifications. Our comparison of protein identifications considered a number of parameters, and employed the same instrumentation for each of those comparisons, although different mass spectrometers were used for different parameters.
We have recently compared several sample preparation protocols for generation of the secretome from murine islets of Langerhans [23]. That sample is much simpler than Xenopus embryos, and generated far fewer protein identifications. Nevertheless, the secretome is dominated by one highly abundant protein, insulin, which is analogous to the highly abundant yolk protein present in the Xenopus embryos. In both cases, a FASP protocol provided the most protein identifications.
Supplementary Material
216_2016_9564_MOESM1_ESM
216_2016_9564_MOESM2_ESM
Acknowledgments
We give a special thanks to Olivia F. Cox and Dr. Paul Huber for providing the embryos used in this study. We thank Dr. William Boggess and Matthew Champion in the Notre Dame Mass Spectrometry and Proteomics Facility for their help. This work was supported by grants from the National Institutes of Health (R01GM096767 and R01HD084399). Elizabeth H. Peuchen acknowledges support from a National Science Foundation Graduate Research Fellowship under Grant No. DGE-1313583.
Footnotes
Conflict of Interest
The authors declare that they have no conflict of interest.
Compliance with Ethical Standards
All animal procedures were performed according to protocols approved by the University of Notre Dame Institutional Animal Care and Use Committee.
References
Full text links
Read article at publisher's site: https://doi.org/10.1007/s00216-016-9564-2
Read article for free, from open access legal sources, via Unpaywall:
https://www.ncbi.nlm.nih.gov/pmc/articles/PMC4926613
Citations & impact
Impact metrics
Citations of article over time
Alternative metrics
Smart citations by scite.ai
Explore citation contexts and check if this article has been
supported or disputed.
https://scite.ai/reports/10.1007/s00216-016-9564-2
Article citations
A Novel Rat Model of Venous Hypertensive Myelopathy Produced by Arteriovenous Bypass Plus Venous Stenosis.
Neurosurgery, 95(3):709-721, 15 Apr 2024
Cited by: 0 articles | PMID: 38619238
ASAP─Automated Sonication-Free Acid-Assisted Proteomes─from Cells and FFPE Tissues.
Anal Chem, 95(6):3291-3299, 01 Feb 2023
Cited by: 3 articles | PMID: 36724070 | PMCID: PMC9933881
Capillary Electrophoresis Mass Spectrometry for Scalable Single-Cell Proteomics.
Front Chem, 10:863979, 08 Apr 2022
Cited by: 15 articles | PMID: 35464213 | PMCID: PMC9024316
Mass spectrometry based proteomics for developmental neurobiology in the amphibian Xenopus laevis.
Curr Top Dev Biol, 145:205-231, 25 May 2021
Cited by: 2 articles | PMID: 34074530 | PMCID: PMC8314003
Review Free full text in Europe PMC
Quantitative capillary zone electrophoresis-mass spectrometry reveals the N-glycome developmental plan during vertebrate embryogenesis.
Mol Omics, 16(3):210-220, 01 Jun 2020
Cited by: 2 articles | PMID: 32149324 | PMCID: PMC7299754
Go to all (18) article citations
Data
Data behind the article
This data has been text mined from the article, or deposited into data resources.
BioStudies: supplemental material and supporting data
Similar Articles
To arrive at the top five similar articles we use a word-weighted algorithm to compare words from the Title and Abstract of each citation.
Over 4100 protein identifications from a Xenopus laevis fertilized egg digest using reversed-phase chromatographic prefractionation followed by capillary zone electrophoresis-electrospray ionization-tandem mass spectrometry analysis.
Proteomics, 16(23):2945-2952, 01 Dec 2016
Cited by: 19 articles | PMID: 27723263 | PMCID: PMC5140748
Single Cell Proteomics Using Frog (Xenopus laevis) Blastomeres Isolated from Early Stage Embryos, Which Form a Geometric Progression in Protein Content.
Anal Chem, 88(13):6653-6657, 22 Jun 2016
Cited by: 49 articles | PMID: 27314579 | PMCID: PMC4940028
Shotgun analysis of membrane proteomes by an improved SDS-assisted sample preparation method coupled with liquid chromatography-tandem mass spectrometry.
J Chromatogr B Analyt Technol Biomed Life Sci, 911:6-14, 23 Oct 2012
Cited by: 5 articles | PMID: 23217299
Sample Clean-up Strategies for ESI Mass Spectrometry Applications in Bottom-up Proteomics: Trends from 2012 to 2016.
Proteomics, 17(20), 12 Apr 2017
Cited by: 22 articles | PMID: 28271630
Review
Funding
Funders who supported this work.
NICHD NIH HHS (1)
Grant ID: R01 HD084399
NIGMS NIH HHS (2)
Grant ID: R01 GM071666
Grant ID: R01 GM096767
National Institutes of Health (1)
Grant ID: R01HD084399
National Science Foundation (1)
Grant ID: DGE-1313583






