Abstract
Free full text

Avian retroviral long terminal repeats bind CCAAT/enhancer-binding protein.
Abstract
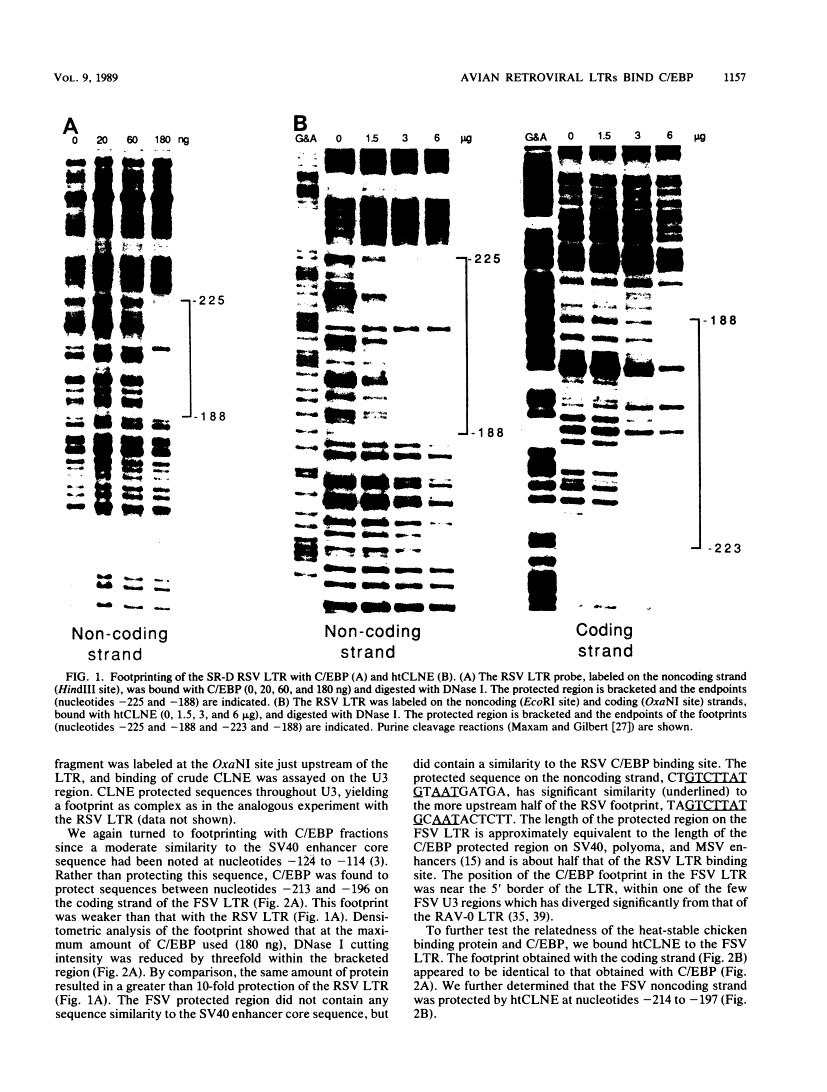
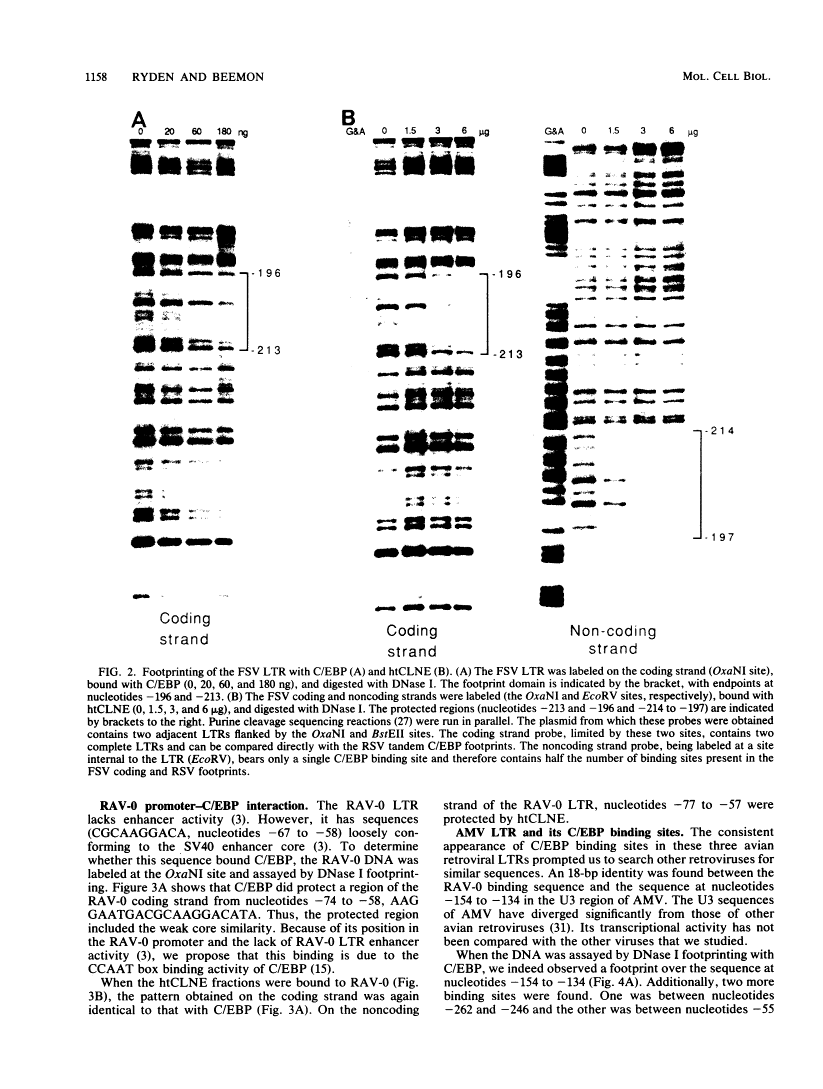
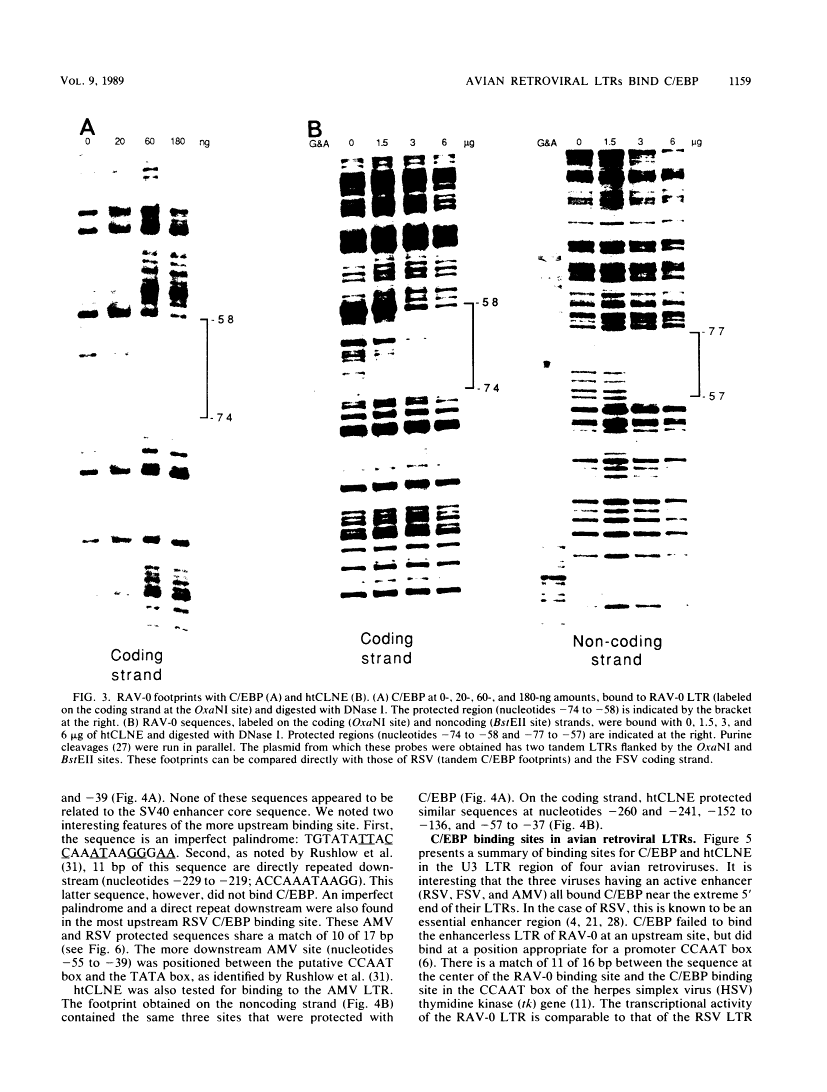
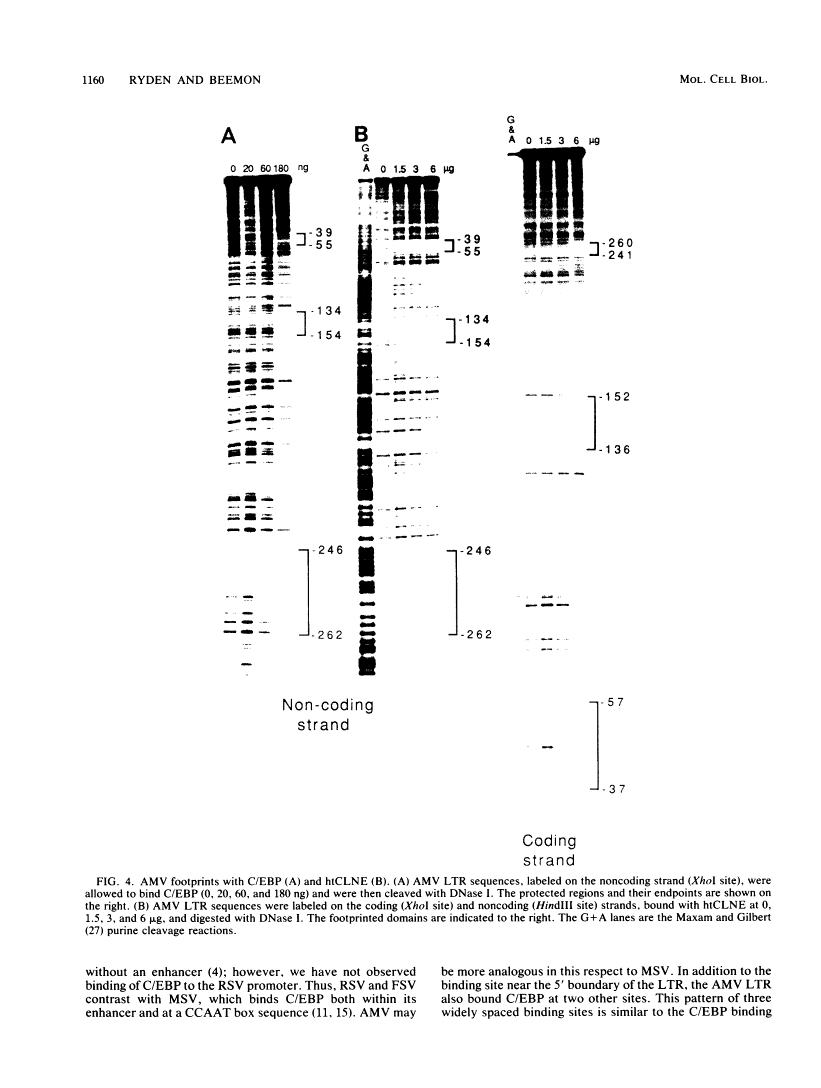
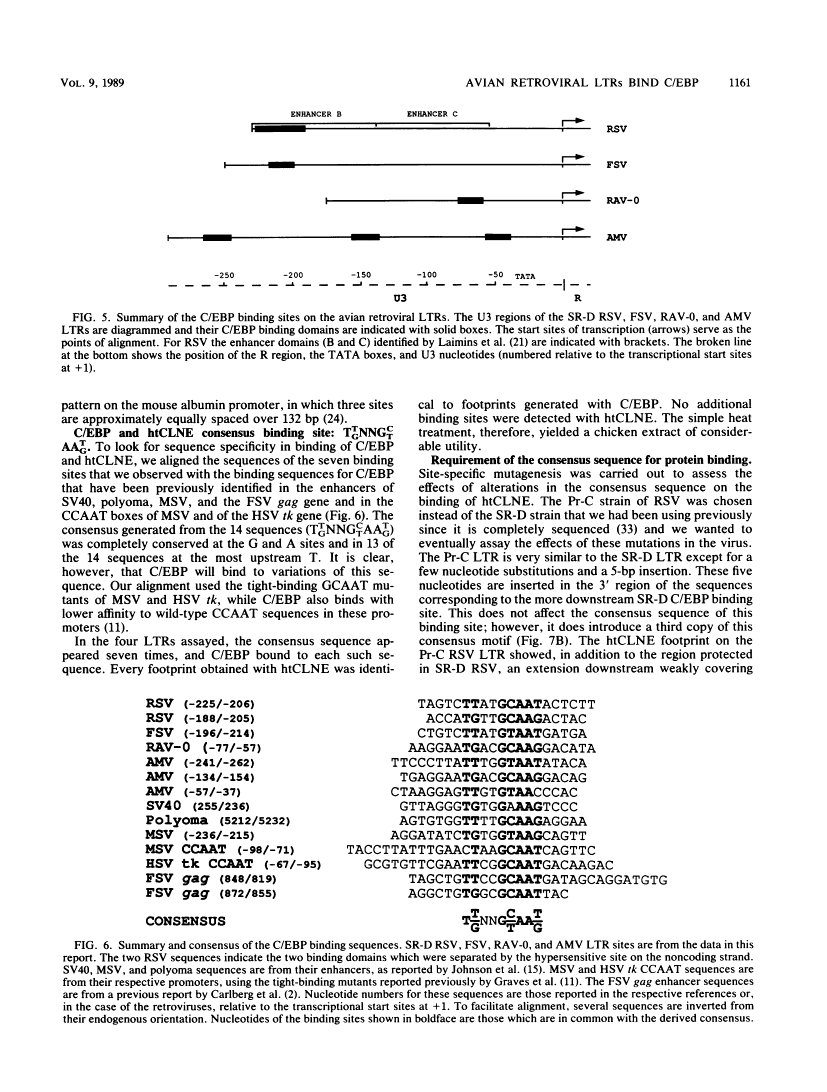
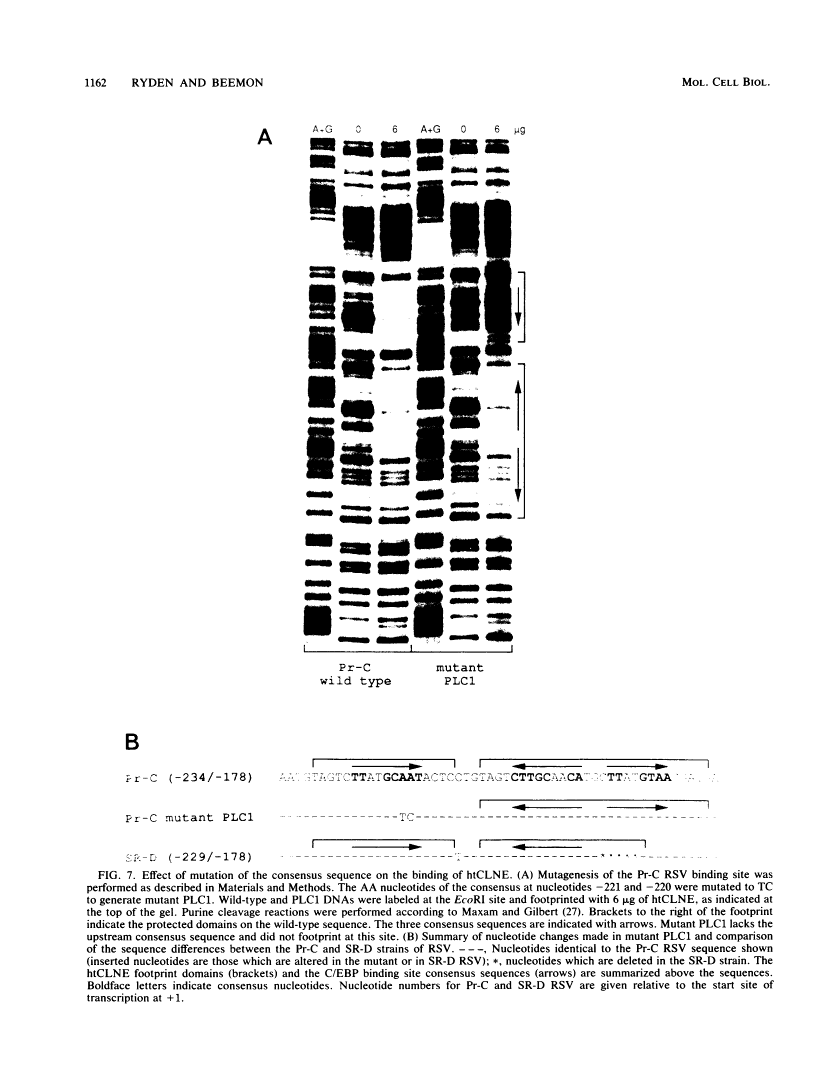
DNA-protein interactions involving enhancer and promoter sequences within the U3 regions of several avian retroviral long terminal repeats (LTRs) were studied by DNase I footprinting. The rat CCAAT/enhancer-binding protein, C/EBP, bound to all four viral LTRs examined. The Rous sarcoma virus binding site corresponded closely to the 5' limit of the LTR enhancer; nucleotides -225 to -188 were protected as a pair of adjacent binding domains. The Fujinami sarcoma virus LTR bound C/EBP at a single site at nucleotides -213 to -195. C/EBP also bound to the promoter region of the enhancerless Rous-associated virus-0 LTR at nucleotides -77 to -57. The avian myeloblastosis virus LTR bound C/EBP at three sites: nucleotides -262 to -246, -154 to -134, and -55 to -39. We have previously observed binding of C/EBP to an enhancer in the gag gene of avian retroviruses. A heat-treated nuclear extract from chicken liver bound to all of the same retroviral sequences as did C/EBP. Alignment of the avian retroviral binding sequences with the published binding sites for C/EBP in two CCAAT boxes and in the simian virus 40, polyoma, and murine sarcoma virus enhancers suggested TTGNNGCTAATG as a consensus sequence for binding of C/EBP. When two bases of this consensus sequence were altered by site-specific mutagenesis of the Rous sarcoma virus LTR, binding of the heat-stable chicken protein was eliminated.
Full text
Full text is available as a scanned copy of the original print version. Get a printable copy (PDF file) of the complete article (3.2M), or click on a page image below to browse page by page. Links to PubMed are also available for Selected References.
Images in this article
Click on the image to see a larger version.
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Arrigo S, Yun M, Beemon K. cis-acting regulatory elements within gag genes of avian retroviruses. Mol Cell Biol. 1987 Jan;7(1):388–397. [Europe PMC free article] [Abstract] [Google Scholar]
- Carlberg K, Ryden TA, Beemon K. Localization and footprinting of an enhancer within the avian sarcoma virus gag gene. J Virol. 1988 May;62(5):1617–1624. [Europe PMC free article] [Abstract] [Google Scholar]
- Cullen BR, Raymond K, Ju G. Transcriptional activity of avian retroviral long terminal repeats directly correlates with enhancer activity. J Virol. 1985 Feb;53(2):515–521. [Europe PMC free article] [Abstract] [Google Scholar]
- Cullen BR, Raymond K, Ju G. Functional analysis of the transcription control region located within the avian retroviral long terminal repeat. Mol Cell Biol. 1985 Mar;5(3):438–447. [Europe PMC free article] [Abstract] [Google Scholar]
- Czernilofsky AP, DeLorbe W, Swanstrom R, Varmus HE, Bishop JM, Tischer E, Goodman HM. The nucleotide sequence of an untranslated but conserved domain at the 3' end of the avian sarcoma virus genome. Nucleic Acids Res. 1980 Jul 11;8(13):2967–2984. [Europe PMC free article] [Abstract] [Google Scholar]
- Efstratiadis A, Posakony JW, Maniatis T, Lawn RM, O'Connell C, Spritz RA, DeRiel JK, Forget BG, Weissman SM, Slightom JL, et al. The structure and evolution of the human beta-globin gene family. Cell. 1980 Oct;21(3):653–668. [Abstract] [Google Scholar]
- Flamant F, Le Guellec D, Verdier G, Nigon VM. Tissue specificity of retrovirus expression in inoculated avian embryos revealed by in situ hybridization to whole-body section. Virology. 1987 Sep;160(1):301–304. [Abstract] [Google Scholar]
- Goodwin GH. Identification of three sequence-specific DNA-binding proteins which interact with the Rous sarcoma virus enhancer and upstream promoter elements. J Virol. 1988 Jun;62(6):2186–2190. [Europe PMC free article] [Abstract] [Google Scholar]
- Gorman CM, Merlino GT, Willingham MC, Pastan I, Howard BH. The Rous sarcoma virus long terminal repeat is a strong promoter when introduced into a variety of eukaryotic cells by DNA-mediated transfection. Proc Natl Acad Sci U S A. 1982 Nov;79(22):6777–6781. [Europe PMC free article] [Abstract] [Google Scholar]
- Gowda S, Rao AS, Kim YW, Guntaka RV. Identification of sequences in the long terminal repeat of avian sarcoma virus required for efficient transcription. Virology. 1988 Jan;162(1):243–247. [Abstract] [Google Scholar]
- Graves BJ, Johnson PF, McKnight SL. Homologous recognition of a promoter domain common to the MSV LTR and the HSV tk gene. Cell. 1986 Feb 28;44(4):565–576. [Abstract] [Google Scholar]
- Hatamochi A, Golumbek PT, Van Schaftingen E, de Crombrugghe B. A CCAAT DNA binding factor consisting of two different components that are both required for DNA binding. J Biol Chem. 1988 Apr 25;263(12):5940–5947. [Abstract] [Google Scholar]
- Herman SA, Coffin JM. Differential transcription from the long terminal repeats of integrated avian leukosis virus DNA. J Virol. 1986 Nov;60(2):497–505. [Europe PMC free article] [Abstract] [Google Scholar]
- Herr W, Gluzman Y. Duplications of a mutated simian virus 40 enhancer restore its activity. Nature. 1985 Feb 21;313(6004):711–714. [Abstract] [Google Scholar]
- Johnson PF, Landschulz WH, Graves BJ, McKnight SL. Identification of a rat liver nuclear protein that binds to the enhancer core element of three animal viruses. Genes Dev. 1987 Apr;1(2):133–146. [Abstract] [Google Scholar]
- Jones NC, Rigby PW, Ziff EB. Trans-acting protein factors and the regulation of eukaryotic transcription: lessons from studies on DNA tumor viruses. Genes Dev. 1988 Mar;2(3):267–281. [Abstract] [Google Scholar]
- Kane SE, Beemon K. Inhibition of methylation at two internal N6-methyladenosine sites caused by GAC to GAU mutations. J Biol Chem. 1987 Mar 5;262(7):3422–3427. [Abstract] [Google Scholar]
- Karnitz L, Faber S, Chalkley R. Specific nuclear proteins interact with the Rous sarcoma virus internal enhancer and share a common element with the enhancer located in the long terminal repeat of the virus. Nucleic Acids Res. 1987 Dec 10;15(23):9841–9859. [Europe PMC free article] [Abstract] [Google Scholar]
- Khoury G, Gruss P. Enhancer elements. Cell. 1983 Jun;33(2):313–314. [Abstract] [Google Scholar]
- Kunkel TA, Roberts JD, Zakour RA. Rapid and efficient site-specific mutagenesis without phenotypic selection. Methods Enzymol. 1987;154:367–382. [Abstract] [Google Scholar]
- Laimins LA, Tsichlis P, Khoury G. Multiple enhancer domains in the 3' terminus of the Prague strain of Rous sarcoma virus. Nucleic Acids Res. 1984 Aug 24;12(16):6427–6442. [Europe PMC free article] [Abstract] [Google Scholar]
- Landschulz WH, Johnson PF, Adashi EY, Graves BJ, McKnight SL. Isolation of a recombinant copy of the gene encoding C/EBP. Genes Dev. 1988 Jul;2(7):786–800. [Abstract] [Google Scholar]
- Landschulz WH, Johnson PF, McKnight SL. The leucine zipper: a hypothetical structure common to a new class of DNA binding proteins. Science. 1988 Jun 24;240(4860):1759–1764. [Abstract] [Google Scholar]
- Lichtsteiner S, Wuarin J, Schibler U. The interplay of DNA-binding proteins on the promoter of the mouse albumin gene. Cell. 1987 Dec 24;51(6):963–973. [Abstract] [Google Scholar]
- Luciw PA, Bishop JM, Varmus HE, Capecchi MR. Location and function of retroviral and SV40 sequences that enhance biochemical transformation after microinjection of DNA. Cell. 1983 Jul;33(3):705–716. [Abstract] [Google Scholar]
- Maniatis T, Goodbourn S, Fischer JA. Regulation of inducible and tissue-specific gene expression. Science. 1987 Jun 5;236(4806):1237–1245. [Abstract] [Google Scholar]
- Maxam AM, Gilbert W. Sequencing end-labeled DNA with base-specific chemical cleavages. Methods Enzymol. 1980;65(1):499–560. [Abstract] [Google Scholar]
- Norton PA, Coffin JM. Characterization of Rous sarcoma virus sequences essential for viral gene expression. J Virol. 1987 Apr;61(4):1171–1179. [Europe PMC free article] [Abstract] [Google Scholar]
- Overbeek PA, Lai SP, Van Quill KR, Westphal H. Tissue-specific expression in transgenic mice of a fused gene containing RSV terminal sequences. Science. 1986 Mar 28;231(4745):1574–1577. [Abstract] [Google Scholar]
- Ruddell A, Linial M, Schubach W, Groudine M. Lability of leukosis virus enhancer-binding proteins in avian hematopoeitic cells. J Virol. 1988 Aug;62(8):2728–2735. [Europe PMC free article] [Abstract] [Google Scholar]
- Rushlow KE, Lautenberger JA, Reddy EP, Souza LM, Baluda MA, Chirikjian JG, Papas TS. Nucleotide sequence analysis of the long terminal repeat of avian myeloblastosis virus and adjacent host sequences. J Virol. 1982 Jun;42(3):840–846. [Europe PMC free article] [Abstract] [Google Scholar]
- Sanger F, Nicklen S, Coulson AR. DNA sequencing with chain-terminating inhibitors. Proc Natl Acad Sci U S A. 1977 Dec;74(12):5463–5467. [Europe PMC free article] [Abstract] [Google Scholar]
- Schwartz DE, Tizard R, Gilbert W. Nucleotide sequence of Rous sarcoma virus. Cell. 1983 Mar;32(3):853–869. [Abstract] [Google Scholar]
- Sealey L, Chalkley R. At least two nuclear proteins bind specifically to the Rous sarcoma virus long terminal repeat enhancer. Mol Cell Biol. 1987 Feb;7(2):787–798. [Europe PMC free article] [Abstract] [Google Scholar]
- Shibuya M, Wang LH, Hanafusa H. Molecular cloning of the Fujinami sarcoma virus genome and its comparison with sequences of other related transforming viruses. J Virol. 1982 Jun;42(3):1007–1016. [Europe PMC free article] [Abstract] [Google Scholar]
- Tsichlis PN, Donehower L, Hager G, Zeller N, Malavarca R, Astrin S, Skalka AM. Sequence comparison in the crossover region of an oncogenic avian retrovirus recombinant and its nononcogenic parent: genetic regions that control growth rate and oncogenic potential. Mol Cell Biol. 1982 Nov;2(11):1331–1338. [Europe PMC free article] [Abstract] [Google Scholar]
- Weber F, Schaffner W. Enhancer activity correlates with the oncogenic potential of avian retroviruses. EMBO J. 1985 Apr;4(4):949–956. [Europe PMC free article] [Abstract] [Google Scholar]
- Weiher H, König M, Gruss P. Multiple point mutations affecting the simian virus 40 enhancer. Science. 1983 Feb 11;219(4585):626–631. [Abstract] [Google Scholar]
- Zenke M, Grundström T, Matthes H, Wintzerith M, Schatz C, Wildeman A, Chambon P. Multiple sequence motifs are involved in SV40 enhancer function. EMBO J. 1986 Feb;5(2):387–397. [Europe PMC free article] [Abstract] [Google Scholar]
Associated Data
Articles from Molecular and Cellular Biology are provided here courtesy of Taylor & Francis
Full text links
Read article at publisher's site: https://doi.org/10.1128/mcb.9.3.1155-1164.1989
Free to read at mcb.asm.org
http://mcb.asm.org/cgi/content/abstract/9/3/1155
Free after 4 months at mcb.asm.org
http://mcb.asm.org/cgi/reprint/9/3/1155
Citations & impact
Impact metrics
Citations of article over time
Article citations
Phylogenetic Analysis and Pathogenicity Assessment of the Emerging Recombinant Subgroup K of Avian Leukosis Virus in South China.
Viruses, 10(4):E194, 13 Apr 2018
Cited by: 12 articles | PMID: 29652854 | PMCID: PMC5923488
A deep sequencing reveals significant diversity among dominant variants and evolutionary dynamics of avian leukosis viruses in two infectious ecosystems.
BMC Vet Res, 12(1):287, 19 Dec 2016
Cited by: 7 articles | PMID: 27993149 | PMCID: PMC5168851
Isolation, identification and evolution analysis of a novel subgroup of avian leukosis virus isolated from a local Chinese yellow broiler in South China.
Arch Virol, 161(10):2717-2725, 15 Jul 2016
Cited by: 40 articles | PMID: 27422398
A comprehensive analysis of the chorion locus in silkmoth.
Sci Rep, 5:16424, 10 Nov 2015
Cited by: 11 articles | PMID: 26553298 | PMCID: PMC4639761
Transcription factor CAAT/enhancer-binding protein is involved in regulation of expression of sterol carrier protein x in Spodoptera litura.
Insect Mol Biol, 24(5):551-560, 15 Jul 2015
Cited by: 4 articles | PMID: 26174044
Go to all (104) article citations
Similar Articles
To arrive at the top five similar articles we use a word-weighted algorithm to compare words from the Title and Abstract of each citation.
The VBP and a1/EBP leucine zipper factors bind overlapping subsets of avian retroviral long terminal repeat CCAAT/enhancer elements.
J Virol, 68(10):6232-6242, 01 Oct 1994
Cited by: 7 articles | PMID: 8083963 | PMCID: PMC237043
Mutation of the C/EBP binding sites in the Rous sarcoma virus long terminal repeat and gag enhancers.
J Virol, 67(5):2862-2870, 01 May 1993
Cited by: 27 articles | PMID: 8386280 | PMCID: PMC237611
Regulation of avian leukosis virus long terminal repeat-enhanced transcription by C/EBP-Rel interactions.
J Virol, 70(5):3051-3059, 01 May 1996
Cited by: 12 articles | PMID: 8627783 | PMCID: PMC190166
CArG, CCAAT, and CCAAT-like protein binding sites in avian retrovirus long terminal repeat enhancers.
J Virol, 66(4):1959-1970, 01 Apr 1992
Cited by: 20 articles | PMID: 1312613 | PMCID: PMC288984
Funding
Funders who supported this work.
NCI NIH HHS (1)
Grant ID: CA 33199
NCRR NIH HHS (1)
Grant ID: SO7 RR07041
OHS HRSA HHS (1)
Grant ID: ST32GM07231