Abstract
Free full text

A catalogue of 136 microbial draft genomes from Red Sea metagenomes
Abstract
Earth is expected to continue warming and the Red Sea is a model environment for understanding the effects of global warming on ocean microbiomes due to its unusually high temperature, salinity and solar irradiance. However, most microbial diversity analyses of the Red Sea have been limited to cultured representatives and single marker gene analyses, hence neglecting the substantial uncultured majority. Here, we report 136 microbial genomes (completion minus contamination is ≥50%) assembled from 45 metagenomes from eight stations spanning the Red Sea and taken from multiple depths between 10 to 500 m. Phylogenomic analysis showed that most of the retrieved genomes belong to seven different phyla of known marine microbes, but more than half representing currently uncultured species. The open-access data presented here is the largest number of Red Sea representative microbial genomes reported in a single study and will help facilitate future studies in understanding the physiology of these microorganisms and how they have adapted to the relatively harsh conditions of the Red Sea.
m. Phylogenomic analysis showed that most of the retrieved genomes belong to seven different phyla of known marine microbes, but more than half representing currently uncultured species. The open-access data presented here is the largest number of Red Sea representative microbial genomes reported in a single study and will help facilitate future studies in understanding the physiology of these microorganisms and how they have adapted to the relatively harsh conditions of the Red Sea.
Background & Summary
The Red Sea is an ideal marine environment to study microbial adaptation to physical conditions atypical of global oceans: high temperature, high salinity, and high irradiance. In late summer 2011, we undertook the King Abdullah University of Science and Technology (KAUST) Red Sea Expedition (KRSE2011) in the eastern Red Sea in order to map its diversity along environmental gradients that occur with changes in latitude, longitude, and depth1. This time of year is not only when temperatures and evaporation (and hence salinity) are highest, but also when a foreign water mass called the Gulf of Aden Intermediate Water (GAIW) intrudes into the Red Sea1,2 (Fig. 1). The GAIW brings nutrient-rich water to the Red Sea, providing nitrogen, phosphorus, and other elements to this otherwise oligotrophic sea, and is likely to introduce important microbial diversity.
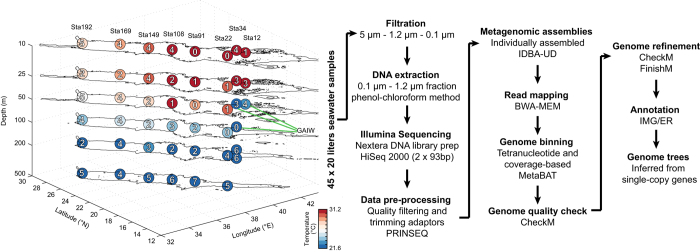
The circles superimposed on the Red Sea 3D map shows the sampling points during the King Abdullah University of Science and Technology Red Sea Expedition 2011. The green lines represent the three Gulf of Aden Intermediate Water (GAIW) sampling points. The numbers within the circles represent the number of genomes recovered from each of the sample. Colors represent the high (dark red) to low (dark blue) water temperature. A total of 45 samples of 20 l each were collected and filtered through a series of filters. For this study, DNA extraction was performed on the small microbial fractions (between 0.1 to 1.2 μm). Extracted DNA was sequenced on the Illumina HiSeq 2,000 generating paired-end reads (2×93
μm). Extracted DNA was sequenced on the Illumina HiSeq 2,000 generating paired-end reads (2×93 bp). Reads from each metagenome were cleaned and assembled individually. Genomes were binned based on tetranucleotide and coverage-based method, refined and quality checked. All 136 genomes were annotated by IMG/ER and taxonomically assigned based on genome trees inferred from single-copy genes.
bp). Reads from each metagenome were cleaned and assembled individually. Genomes were binned based on tetranucleotide and coverage-based method, refined and quality checked. All 136 genomes were annotated by IMG/ER and taxonomically assigned based on genome trees inferred from single-copy genes.
Insights into the taxonomic, evolutionary, and functional diversity of the Red Sea have largely been based on studies of pure cultures3–5 and single marker genes such as the 16S rRNA6,7, or internal transcribed spacer8. Recently, investigations of microbial ecology have steered towards whole genome-based culture-independent methods notably single-cell genomics and metagenomics9,10. Single-cell genomics is an exciting field that recovers complete and partial single cell genomes from complex environments, albeit the need of specialised equipment, high cost and relatively low throughput11–13. Metagenomics is paving the way forward by harnessing the recent wave of sequencing technology and bioinformatics advancements to recover genomes of individual populations or populations of closely related organisms14–16. Application of these methods has resulted in the recovery of numerous genomes of uncultivated microorganisms that have provided surprising insights into the diversity and function of microbial communities10,14,17–19.
During the KRSE2011, eight stations were sampled along a cruise track from south to north, capturing gradients in temperature, salinity, oxygen, and nutrients, including the unique GAIW water mass (Fig. 1 and Table 1 (available online only)). At each station, samples were collected from the surface to mesopelagic depths (10, 25, 50, 100, 200, and 500 m), except for stations 12 and 34, which had depths shallower than 500
m), except for stations 12 and 34, which had depths shallower than 500 m (Fig. 1 and Table 1 (available online only)), in order to capture a greater variation in environmental parameters and microbial diversity. Here, we successfully reconstructed 136 genomes from 45 individually assembled metagenomes (Figs 1 and and2,2, Tables 1 and and22 (available online only), Data Citation 1) by differential read coverage and tetranucleotide frequency methods. Of these, 43 were ‘near-complete’ with an estimated completion minus contamination of ≥90%, while the other 93 draft genomes had completion minus contamination of ≥50% (Table 2 (available online only)). To our knowledge, this is the largest number of microbial genomes from the Red Sea to be reported in a single study.
m (Fig. 1 and Table 1 (available online only)), in order to capture a greater variation in environmental parameters and microbial diversity. Here, we successfully reconstructed 136 genomes from 45 individually assembled metagenomes (Figs 1 and and2,2, Tables 1 and and22 (available online only), Data Citation 1) by differential read coverage and tetranucleotide frequency methods. Of these, 43 were ‘near-complete’ with an estimated completion minus contamination of ≥90%, while the other 93 draft genomes had completion minus contamination of ≥50% (Table 2 (available online only)). To our knowledge, this is the largest number of microbial genomes from the Red Sea to be reported in a single study.
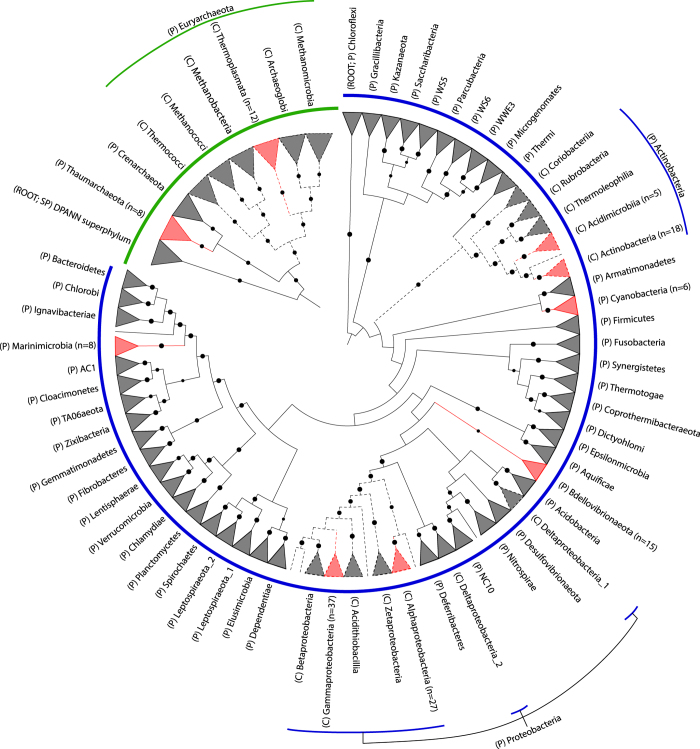
The clades represented by the triangles are collapsed at the phylum (P) level except for phyla containing genomes from this study which are expanded at the class (C) level and highlighted in red. Certain phyla have genome representatives only at the phylum level (Thaumarchaeota, Marinimicrobia, Cyanobacteria, and Bdellovibrionaeota). Numbers in parentheses indicate the count of recovered genomes from a particular taxonomic level. Dashed lines indicate nodes for class level. Robustness of the tree is indicated by black circles (size of circles scaled from 80 to 100% bootstrap support values). Trees were inferred independently. The archaeal tree was rooted with the DPANN superphylum9 while the bacterial tree was ‘arbitrarily’ rooted with the phylum Chloroflexi42 but should be treated as unrooted.
Table 1
| Isolation source | Water mass | Date and Time | Assembly size (Mbps) | No. of scaffolds | Largest scaffold size (Mbps) | N50 | Depth (m) | BioProject | BioSample | NCBI accession (assembled) | NCBI SRA accession (raw reads) |
|---|---|---|---|---|---|---|---|---|---|---|---|
| Red Sea water column Station 12 | Red Sea | 18/09/2011 10:26 | 44.11 | 29555 | 0.120 | 2072 | 10 | PRJNA289734 | SAMN03860258 | LUMR00000000 | SRR2102994 |
| Red Sea water column Station 12 | Red Sea | 75.83 | 46146 | 0.102 | 2508 | 25 | PRJNA289734 | SAMN03860259 | LUMQ00000000 | SRR2102995 | |
| Red Sea water column Station 12 | GAIW | 37.84 | 23158 | 0.157 | 2720 | 47 | PRJNA289734 | SAMN03860260 | LUMP00000000 | SRR2103006 | |
| Red Sea water column Station 22 | Red Sea | 19/09/2011 08:53 | 47.49 | 38508 | 0.060 | 1415 | 10 | PRJNA289734 | SAMN03860261 | LUMO00000000 | SRR2103017 |
| Red Sea water column Station 22 | Red Sea | 40.79 | 27718 | 0.072 | 1973 | 25 | PRJNA289734 | SAMN03860262 | LUMN00000000 | SRR2103028 | |
| Red Sea water column Station 22 | Red Sea | 41.68 | 29846 | 0.090 | 1775 | 50 | PRJNA289734 | SAMN03860263 | LUMM00000000 | SRR2103034 | |
| Red Sea water column Station 22 | Red Sea | 41.80 | 33190 | 0.129 | 1457 | 100 | PRJNA289734 | SAMN03860264 | LUML00000000 | SRR2103035 | |
| Red Sea water column Station 22 | Red Sea | 47.47 | 33409 | 0.133 | 1876 | 200 | PRJNA289734 | SAMN03860265 | LUMK00000000 | SRR2103036 | |
| Red Sea water column Station 22 | Red Sea | 80.71 | 45186 | 0.179 | 3201 | 500 | PRJNA289734 | SAMN03860266 | LUMJ00000000 | SRR2103037 | |
| Red Sea water column Station 34 | Red Sea | 20/09/2011 04:10 | 77.82 | 46638 | 0.221 | 2646 | 10 | PRJNA289734 | SAMN03860267 | LUMI00000000 | SRR2103038 |
| Red Sea water column Station 34 | Red Sea | 42.51 | 28685 | 0.221 | 2018 | 25 | PRJNA289734 | SAMN03860268 | LUMH00000000 | SRR2102996 | |
| Red Sea water column Station 34 | GAIW | 38.78 | 27506 | 0.253 | 1843 | 50 | PRJNA289734 | SAMN03860269 | LUMG00000000 | SRR2102997 | |
| Red Sea water column Station 34 | GAIW | 39.04 | 35615 | 0.037 | 1167 | 100 | PRJNA289734 | SAMN03860270 | LUMF00000000 | SRR2102998 | |
| Red Sea water column Station 34 | Red Sea | 63.41 | 42929 | 0.125 | 1962 | 200 | PRJNA289734 | SAMN03860271 | LUME00000000 | SRR2102999 | |
| Red Sea water column Station 34 | Red Sea | 64.65 | 37057 | 0.263 | 2832 | 258 | PRJNA289734 | SAMN03860272 | LUMD00000000 | SRR2103000 | |
| Red Sea water column Station 91 | Red Sea | 24/09/2011 20:54 | 35.42 | 26110 | 0.116 | 1716 | 10 | PRJNA289734 | SAMN03860273 | LUMC00000000 | SRR2103001 |
| Red Sea water column Station 91 | Red Sea | 28.80 | 21238 | 0.079 | 1783 | 25 | PRJNA289734 | SAMN03860274 | LUMB00000000 | SRR2103002 | |
| Red Sea water column Station 91 | Red Sea | 21.35 | 18107 | 0.070 | 1293 | 50 | PRJNA289734 | SAMN03860275 | LUMA00000000 | SRR2103003 | |
| Red Sea water column Station 91 | Red Sea | 51.35 | 45910 | 0.141 | 1194 | 100 | PRJNA289734 | SAMN03860276 | LULZ00000000 | SRR2103004 | |
| Red Sea water column Station 91 | Red Sea | 49.04 | 43484 | 0.129 | 1239 | 200 | PRJNA289734 | SAMN03860277 | LULY00000000 | SRR2103005 | |
| Red Sea water column Station 91 | Red Sea | 68.61 | 45543 | 0.194 | 2011 | 500 | PRJNA289734 | SAMN03860278 | LULX00000000 | SRR2103007 | |
| Red Sea water column Station 108 | Red Sea | 27/09/2011 21:03 | 68.31 | 39235 | 1.199 | 2885 | 10 | PRJNA289734 | SAMN03860279 | LULW00000000 | SRR2103008 |
| Red Sea water column Station 108 | Red Sea | 59.60 | 40690 | 0.160 | 1948 | 25 | PRJNA289734 | SAMN03860280 | LULV00000000 | SRR2103009 | |
| Red Sea water column Station 108 | Red Sea | 58.23 | 49013 | 0.058 | 1334 | 50 | PRJNA289734 | SAMN03860281 | LULU00000000 | SRR2103010 | |
| Red Sea water column Station 108 | Red Sea | 36.51 | 27461 | 0.142 | 1626 | 100 | PRJNA289734 | SAMN03860282 | LULT00000000 | SRR2103011 | |
| Red Sea water column Station 108 | Red Sea | 58.51 | 45536 | 0.079 | 1553 | 200 | PRJNA289734 | SAMN03860283 | LULS00000000 | SRR2103012 | |
| Red Sea water column Station 108 | Red Sea | 63.24 | 41604 | 0.139 | 2136 | 500 | PRJNA289734 | SAMN03860284 | LULR00000000 | SRR2103013 | |
| Red Sea water column Station 149 | Red Sea | 01/10/2011 05:00 | 56.10 | 31577 | 1.198 | 2984 | 10 | PRJNA289734 | SAMN03860285 | LULQ00000000 | SRR2103014 |
| Red Sea water column Station 149 | Red Sea | 62.95 | 34995 | 0.416 | 3178 | 25 | PRJNA289734 | SAMN03860286 | LULP00000000 | SRR2103015 | |
| Red Sea water column Station 149 | Red Sea | 79.82 | 47255 | 0.314 | 2763 | 50 | PRJNA289734 | SAMN03860287 | LULO00000000 | SRR2103016 | |
| Red Sea water column Station 149 | Red Sea | 38.33 | 25062 | 0.170 | 2222 | 100 | PRJNA289734 | SAMN03860288 | LULN00000000 | SRR2103018 | |
| Red Sea water column Station 149 | Red Sea | 66.80 | 52078 | 0.105 | 1503 | 200 | PRJNA289734 | SAMN03860289 | LULM00000000 | SRR2103019 | |
| Red Sea water column Station 149 | Red Sea | 85.38 | 54665 | 0.289 | 2146 | 500 | PRJNA289734 | SAMN03860290 | LULL00000000 | SRR2103020 | |
| Red Sea water column Station 169 | Red Sea | 03/10/2011 04:55 | 99.91 | 54636 | 1.199 | 3019 | 10 | PRJNA289734 | SAMN03860291 | LULK00000000 | SRR2103021 |
| Red Sea water column Station 169 | Red Sea | 83.57 | 45217 | 0.308 | 3232 | 25 | PRJNA289734 | SAMN03860292 | LULJ00000000 | SRR2103022 | |
| Red Sea water column Station 169 | Red Sea | 84.94 | 53578 | 0.158 | 2309 | 50 | PRJNA289734 | SAMN03860293 | LULI00000000 | SRR2103023 | |
| Red Sea water column Station 169 | Red Sea | 73.95 | 54373 | 0.149 | 1685 | 100 | PRJNA289734 | SAMN03860294 | LULH00000000 | SRR2103024 | |
| Red Sea water column Station 169 | Red Sea | 73.59 | 57258 | 0.155 | 1530 | 200 | PRJNA289734 | SAMN03860295 | LULG00000000 | SRR2103025 | |
| Red Sea water column Station 169 | Red Sea | 79.59 | 58308 | 0.400 | 1655 | 500 | PRJNA289734 | SAMN03860296 | LULF00000000 | SRR2103026 | |
| Red Sea water column Station 192 | Red Sea | 05/10/2011 10:56 | 98.63 | 57007 | 1.199 | 2789 | 10 | PRJNA289734 | SAMN03860297 | LULE00000000 | SRR2103027 |
| Red Sea water column Station 192 | Red Sea | 58.15 | 34483 | 0.321 | 2666 | 25 | PRJNA289734 | SAMN03860298 | LUMS00000000 | SRR2103029 | |
| Red Sea water column Station 192 | Red Sea | 87.61 | 47563 | 1.358 | 3315 | 50 | PRJNA289734 | SAMN03860299 | LUMT00000000 | SRR2103030 | |
| Red Sea water column Station 192 | Red Sea | 50.63 | 34015 | 0.680 | 2014 | 100 | PRJNA289734 | SAMN03860300 | LUMU00000000 | SRR2103031 | |
| Red Sea water column Station 192 | Red Sea | 45.78 | 30314 | 0.295 | 2337 | 200 | PRJNA289734 | SAMN03860301 | LUMV00000000 | SRR2103032 | |
| Red Sea water column Station 192 | Red Sea | 73.78 | 44069 | 0.278 | 2528 | 500 | PRJNA289734 | SAMN03860302 | LUMW00000000 | SRR2103033 |
Table 2
| Genome bins | Genome size (Mbps) | No. of scaffolds | IMG Gene count | GC (%) | Marker lineage for CheckM | Completeness (%) | Contamination (%) | Comp-Cont % | Isolation source | Depth (m) | Latitude/Longtitude | BioProject | BioSample | NCBI accession | IMG genome ID |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Acidimicrobiia bacterium REDSEA-S09_B7 | 2.02 | 226 | 1884 | 71.62 | k__Bacteria (UID1453) | 84.25 | 2.23 | 82.02 | Red Sea water column Station 22 | 500 | 17.996 N 39.799 E | PRJNA289734 | SAMN04534547 | LUMX00000000 | 2651870138 |
| Acidimicrobiia bacterium REDSEA-S14_B4 | 2.10 | 257 | 1654 | 71.6 | k__Bacteria (UID1453) | 85.86 | 2.56 | 83.3 | Red Sea water column Station 34 | 200 | 18.58 N 40.743 E | PRJNA289734 | SAMN04534548 | LUMY00000000 | 2651870139 |
| Acidimicrobiia bacterium REDSEA-S20_B6 | 1.76 | 338 | 2064 | 71.41 | k__Bacteria (UID1453) | 81.36 | 1.59 | 79.77 | Red Sea water column Station 91 | 200 | 20.525 N 38.781 E | PRJNA289734 | SAMN04534549 | LUMZ00000000 | 2651870140 |
| Acidimicrobiia bacterium REDSEA-S21_B10 | 1.73 | 328 | 2037 | 71.37 | k__Bacteria (UID1453) | 64.31 | 0.43 | 63.88 | Red Sea water column Station 91 | 500 | 20.525 N 38.781 E | PRJNA289734 | SAMN04534550 | LUNA00000000 | 2651870141 |
| Acidimicrobiia bacterium REDSEA-S33_B8N9 | 2.15 | 262 | 2380 | 71.46 | k__Bacteria (UID1453) | 80.48 | 5.56 | 74.92 | Red Sea water column Station 149 | 500 | 23.604 N 37.054 E | PRJNA289734 | SAMN04534551 | LUNB00000000 | 2651870142 |
| Acinetobacter sp. REDSEA-S21_B14 | 2.58 | 517 | 3085 | 39.1 | f__Moraxellaceae (UID4680) | 71.54 | 4.32 | 67.22 | Red Sea water column Station 91 | 500 | 20.525 N 38.781 E | PRJNA289734 | SAMN04534552 | LUNC00000000 | 2651870143 |
| Actinobacteria bacterium REDSEA-S36_B12 | 1.37 | 255 | 1734 | 62.57 | o__Actinomycetales (UID1663) | 60.63 | 0 | 60.63 | Red Sea water column Station 169 | 50 | 25.772 N 36.116 E | PRJNA289734 | SAMN04534553 | LUND00000000 | 2651870144 |
| Aeromicrobium sp. REDSEA-S32_B7 | 3.52 | 333 | 3794 | 71.86 | o__Actinomycetales (UID1697) | 95.73 | 6.23 | 89.5 | Red Sea water column Station 149 | 200 | 23.604 N 37.054 E | PRJNA289734 | SAMN04534555 | LUNF00000000 | 2651870146 |
| Aeromicrobium sp. REDSEA-S35_B1 | 3.56 | 83 | 3609 | 72.12 | o__Actinomycetales (UID1697) | 98.06 | 2.8 | 95.26 | Red Sea water column Station 169 | 25 | 25.772 N 36.116 E | PRJNA289734 | SAMN04534556 | LUNG00000000 | 2651870147 |
| Aeromicrobium sp. REDSEA-S38_B2 | 3.49 | 104 | 3608 | 72.12 | o__Actinomycetales (UID1697) | 98.53 | 1.9 | 96.63 | Red Sea water column Station 169 | 200 | 25.772 N 36.116 E | PRJNA289734 | SAMN04534557 | LUNH00000000 | 2651870148 |
| Aeromicrobium sp. REDSEA-S42_B4 | 3.47 | 54 | 3538 | 72.15 | o__Actinomycetales (UID1697) | 98.45 | 0.86 | 97.59 | Red Sea water column Station 192 | 50 | 27.897 N 34.507 E | PRJNA289734 | SAMN04534558 | LUNI00000000 | 2651870149 |
| Aeromicrobium sp. REDSEA-S44_B1 | 3.47 | 44 | 3560 | 72.09 | o__Actinomycetales (UID1697) | 98.91 | 0.95 | 97.96 | Red Sea water column Station 192 | 200 | 27.897 N 34.507 E | PRJNA289734 | SAMN04534559 | LUNJ00000000 | 2651870150 |
| Alteromonas macleodii str. REDSEA-S09_B2 | 4.34 | 102 | 3190 | 44.52 | c__Gammaproteobacteria (UID4761) | 98.83 | 0.51 | 98.32 | Red Sea water column Station 22 | 500 | 17.996 N 39.799 E | PRJNA289734 | SAMN04534560 | LUNK00000000 | 2651870151 |
| Alteromonas macleodii str. REDSEA-S10_B9 | 2.55 | 490 | 1239 | 44.64 | c__Gammaproteobacteria (UID4761) | 59.71 | 0.57 | 59.14 | Red Sea water column Station 34 | 10 | 18.58 N 40.743 E | PRJNA289734 | SAMN04534561 | LUNL00000000 | 2651870152 |
| Alteromonas macleodii str. REDSEA-S12_B5 | 2.64 | 514 | 2355 | 43.98 | c__Gammaproteobacteria (UID4761) | 59.42 | 2.39 | 57.03 | Red Sea water column Station 34 | 50 | 18.58 N 40.743 E | PRJNA289734 | SAMN04534562 | LUNM00000000 | 2651870153 |
| Alteromonas macleodii str. REDSEA-S14_B11 | 3.11 | 543 | 2135 | 44.58 | c__Gammaproteobacteria (UID4761) | 73.75 | 1.24 | 72.51 | Red Sea water column Station 34 | 200 | 18.58 N 40.743 E | PRJNA289734 | SAMN04534563 | LUNN00000000 | 2651870154 |
| Alteromonas macleodii str. REDSEA-S15_B11 | 3.79 | 457 | 3227 | 44.59 | c__Gammaproteobacteria (UID4761) | 89.9 | 1.25 | 88.65 | Red Sea water column Station 34 | 258 | 18.58 N 40.743 E | PRJNA289734 | SAMN04534564 | LUNO00000000 | 2651870155 |
| Candidatus Marinimicrobia (SAR406 cluster) bacterium REDSEA-S14_B6 | 1.43 | 234 | 2855 | 54.35 | k__Bacteria (UID2495) | 72.35 | 0.18 | 72.17 | Red Sea water column Station 34 | 200 | 18.58 N 40.743 E | PRJNA289734 | SAMN04534578 | LUOC00000000 | 2651870223 |
| Candidatus Marinimicrobia (SAR406 cluster) bacterium REDSEA-S15_B10 | 1.65 | 233 | 4044 | 54.08 | k__Bacteria (UID2495) | 71.03 | 0.1 | 70.93 | Red Sea water column Station 34 | 258 | 18.58 N 40.743 E | PRJNA289734 | SAMN04534579 | LUOD00000000 | 2651870224 |
| Candidatus Marinimicrobia (SAR406 cluster) bacterium REDSEA-S15_B13 | 1.48 | 201 | 3478 | 41.84 | k__Bacteria (UID2495) | 71.59 | 3.3 | 68.29 | Red Sea water column Station 34 | 258 | 18.58 N 40.743 E | PRJNA289734 | SAMN04534580 | LUOE00000000 | 2651870225 |
| Candidatus Marinimicrobia (SAR406 cluster) bacterium REDSEA-S27_B1N12 | 1.36 | 280 | 1643 | 51.44 | k__Bacteria (UID2495) | 60.19 | 2.2 | 57.99 | Red Sea water column Station 108 | 500 | 22.046 N 37.929 E | PRJNA289734 | SAMN04534581 | LUOF00000000 | 2651870226 |
| Candidatus Marinimicrobia (SAR406 cluster) bacterium REDSEA-S33_B13 | 1.27 | 264 | 1462 | 54.33 | k__Bacteria (UID2495) | 56.03 | 1.65 | 54.38 | Red Sea water column Station 149 | 500 | 23.604 N 37.054 E | PRJNA289734 | SAMN04534582 | LUOG00000000 | 2651870227 |
| Candidatus Marinimicrobia (SAR406 cluster) bacterium REDSEA-S38_B13 | 1.15 | 217 | 1313 | 41.56 | k__Bacteria (UID2495) | 58.34 | 1.2 | 57.14 | Red Sea water column Station 169 | 200 | 25.772 N 36.116 E | PRJNA289734 | SAMN04534583 | LUOH00000000 | 2651870228 |
| Candidatus Marinimicrobia (SAR406 cluster) bacterium REDSEA-S39_B11 | 1.09 | 225 | 1203 | 54.56 | k__Bacteria (UID2495) | 52.97 | 0.1 | 52.87 | Red Sea water column Station 169 | 500 | 25.772 N 36.116 E | PRJNA289734 | SAMN04534584 | LUOI00000000 | 2651870229 |
| Candidatus Marinimicrobia (SAR406 cluster) bacterium REDSEA-S39_B7 | 0.24 | 15 | 229 | 38.13 | k__Bacteria (UID2495) | 78.5 | 6.89 | 71.61 | Red Sea water column Station 169 | 500 | 25.772 N 36.116 E | PRJNA289734 | SAMN04534585 | LUOJ00000000 | 2651870230 |
| Candidatus Thioglobus (SUP05 cluster) sp. REDSEA-S03_B1 | 1.58 | 97 | 2264 | 38.39 | p__Proteobacteria (UID3880) | 87.92 | 3.86 | 84.06 | Red Sea water column Station 12 | 47 | 17.662 N 40.905 E | PRJNA289734 | SAMN04534667 | LURM00000000 | 2651870213 |
| Candidatus Thioglobus (SUP05 cluster) sp. REDSEA-S12_B1 | 1.61 | 77 | 1540 | 38.35 | p__Proteobacteria (UID3880) | 88.91 | 1.32 | 87.59 | Red Sea water column Station 34 | 50 | 18.58 N 40.743 E | PRJNA289734 | SAMN04534668 | LURN00000000 | 2651870214 |
| Candidatus Thioglobus (SUP05 cluster) sp. REDSEA-S14_B12 | 1.83 | 262 | 2336 | 39.85 | p__Proteobacteria (UID3880) | 67.29 | 7.86 | 59.43 | Red Sea water column Station 34 | 200 | 18.58 N 40.743 E | PRJNA289734 | SAMN04534669 | LURO00000000 | 2651870215 |
| Erythrobacter sp. REDSEA-S22_B4 | 2.69 | 10 | 2897 | 63.73 | o__Sphingomonadales (UID3310) | 99.93 | 0.43 | 99.5 | Red Sea water column Station 108 | 10 | 22.046 N 37.929 E | PRJNA289734 | SAMN04534565 | LUNP00000000 | 2651870156 |
| Erythrobacter sp. REDSEA-S28_B2 | 2.69 | 8 | 2894 | 63.71 | o__Sphingomonadales (UID3310) | 99.9 | 0.43 | 99.47 | Red Sea water column Station 149 | 10 | 23.604 N 37.054 E | PRJNA289734 | SAMN04534566 | LUNQ00000000 | 2654587888 |
| Erythrobacter sp. REDSEA-S34_B3 | 2.69 | 7 | 2893 | 63.73 | o__Sphingomonadales (UID3310) | 99.93 | 0.43 | 99.5 | Red Sea water column Station 169 | 10 | 25.772 N 36.116 E | PRJNA289734 | SAMN04534567 | LUNR00000000 | 2654587889 |
| Erythrobacter sp. REDSEA-S36_B6 | 2.93 | 99 | 3035 | 63.57 | o__Sphingomonadales (UID3310) | 97.09 | 2.53 | 94.56 | Red Sea water column Station 169 | 50 | 25.772 N 36.116 E | PRJNA289734 | SAMN04534568 | LUNS00000000 | 2654587890 |
| Erythrobacter sp. REDSEA-S37_B3 | 2.79 | 94 | 2968 | 63.54 | o__Sphingomonadales (UID3310) | 97.59 | 1 | 96.59 | Red Sea water column Station 169 | 100 | 25.772 N 36.116 E | PRJNA289734 | SAMN04534569 | LUNT00000000 | 2654587891 |
| Erythrobacter sp. REDSEA-S40_B1 | 2.69 | 7 | 2897 | 63.72 | o__Sphingomonadales (UID3310) | 99.93 | 0.43 | 99.5 | Red Sea water column Station 192 | 10 | 27.897 N 34.507 E | PRJNA289734 | SAMN04534570 | LUNU00000000 | 2654587892 |
| Erythrobacter sp. REDSEA-S41_B1 | 2.93 | 23 | 2933 | 63.59 | o__Sphingomonadales (UID3310) | 99.54 | 0.63 | 98.91 | Red Sea water column Station 192 | 25 | 27.897 N 34.507 E | PRJNA289734 | SAMN04534571 | LUNV00000000 | 2654587893 |
| Erythrobacter sp. REDSEA-S42_B5 | 2.68 | 12 | 2905 | 63.72 | o__Sphingomonadales (UID3310) | 99.83 | 0.43 | 99.4 | Red Sea water column Station 192 | 50 | 27.897 N 34.507 E | PRJNA289734 | SAMN04534572 | LUNW00000000 | 2654587894 |
| Erythrobacter sp. REDSEA-S43_B2 | 2.87 | 18 | 2918 | 63.64 | o__Sphingomonadales (UID3310) | 99.84 | 0.94 | 98.9 | Red Sea water column Station 192 | 100 | 27.897 N 34.507 E | PRJNA289734 | SAMN04534573 | LUNX00000000 | 2651870157 |
| Erythrobacter sp. REDSEA-S45_B7 | 2.89 | 285 | 3139 | 63.54 | o__Sphingomonadales (UID3310) | 89.24 | 1.25 | 87.99 | Red Sea water column Station 192 | 500 | 27.897 N 34.507 E | PRJNA289734 | SAMN04534574 | LUNY00000000 | 2651870158 |
| Idiomarina sp. REDSEA-S21_B4 | 2.26 | 60 | 2451 | 47.2 | c__Gammaproteobacteria (UID4761) | 95.29 | 0.42 | 94.87 | Red Sea water column Station 91 | 500 | 20.525 N 38.781 E | PRJNA289734 | SAMN04534575 | LUNZ00000000 | 2651870159 |
| Idiomarina sp. REDSEA-S27_B4 | 2.38 | 57 | 2550 | 47.27 | c__Gammaproteobacteria (UID4761) | 98.14 | 0.74 | 97.4 | Red Sea water column Station 108 | 500 | 22.046 N 37.929 E | PRJNA289734 | SAMN04534576 | LUOA00000000 | 2651870160 |
| Marine group II euryarchaeote REDSEA-S03_B6 | 1.32 | 210 | 1400 | 45.47 | p__Euryarchaeota (UID3) | 65.01 | 2.3 | 62.71 | Red Sea water column Station 12 | 47 | 17.662 N 40.905 E | PRJNA289734 | SAMN04534670 | LURP00000000 | 2651870292 |
| Marine group II euryarchaeote REDSEA-S10_B2 | 1.23 | 29 | 3698 | 50.45 | p__Euryarchaeota (UID3) | 76.13 | 1.2 | 74.93 | Red Sea water column Station 34 | 10 | 18.58 N 40.743 E | PRJNA289734 | SAMN04534671 | LURQ00000000 | 2651870293 |
| Marine group II euryarchaeote REDSEA-S11_B3N4 | 1.30 | 35 | 2197 | 50.98 | p__Euryarchaeota (UID3) | 81.96 | 0 | 81.96 | Red Sea water column Station 34 | 25 | 18.58 N 40.743 E | PRJNA289734 | SAMN04534672 | LURR00000000 | 2651870294 |
| Marine group II euryarchaeote REDSEA-S19_B7N8 | 1.17 | 155 | 1796 | 52.04 | p__Euryarchaeota (UID3) | 70.4 | 0.8 | 69.6 | Red Sea water column Station 91 | 100 | 20.525 N 38.781 E | PRJNA289734 | SAMN04534673 | LURS00000000 | 2651870295 |
| Marine group II euryarchaeote REDSEA-S25_B4N5 | 1.10 | 167 | 1272 | 52.14 | p__Euryarchaeota (UID3) | 58.43 | 0.27 | 58.16 | Red Sea water column Station 108 | 100 | 22.046 N 37.929 E | PRJNA289734 | SAMN04534674 | LURT00000000 | 2651870296 |
| Marine group II euryarchaeote REDSEA-S29_B8N9 | 1.10 | 131 | 1244 | 50.19 | p__Euryarchaeota (UID3) | 59.47 | 0 | 59.47 | Red Sea water column Station 149 | 25 | 23.604 N 37.054 E | PRJNA289734 | SAMN04534675 | LURU00000000 | 2651870297 |
| Marine group II euryarchaeote REDSEA-S30_B12 | 1.27 | 137 | 1374 | 36.9 | p__Euryarchaeota (UID3) | 67.79 | 0.8 | 66.99 | Red Sea water column Station 149 | 50 | 23.604 N 37.054 E | PRJNA289734 | SAMN04534676 | LURV00000000 | 2651870298 |
| Marine group II euryarchaeote REDSEA-S37_B2N9 | 1.21 | 6 | 155 | 49.08 | p__Euryarchaeota (UID3) | 76.47 | 0.06 | 76.41 | Red Sea water column Station 169 | 100 | 25.772 N 36.116 E | PRJNA289734 | SAMN04534677 | LURW00000000 | 2651870299 |
| Marine group II euryarchaeote REDSEA-S40_B11N13 | 1.24 | 118 | 1300 | 50.09 | p__Euryarchaeota (UID3) | 71.96 | 0 | 71.96 | Red Sea water column Station 192 | 10 | 27.897 N 34.507 E | PRJNA289734 | SAMN04534678 | LURX00000000 | 2651870300 |
| Marine group II euryarchaeote REDSEA-S41_B6 | 1.13 | 122 | 1216 | 49.97 | p__Euryarchaeota (UID3) | 71.35 | 1.92 | 69.43 | Red Sea water column Station 192 | 25 | 27.897 N 34.507 E | PRJNA289734 | SAMN04534679 | LURY00000000 | 2651870301 |
| Marine group II euryarchaeote REDSEA-S42_B7 | 1.19 | 43 | 1202 | 49.72 | p__Euryarchaeota (UID3) | 75.73 | 0 | 75.73 | Red Sea water column Station 192 | 50 | 27.897 N 34.507 E | PRJNA289734 | SAMN04534680 | LURZ00000000 | 2651870302 |
| Marine group II euryarchaeote REDSEA-S43_B8 | 1.10 | 105 | 1213 | 50.28 | p__Euryarchaeota (UID3) | 72.13 | 0 | 72.13 | Red Sea water column Station 192 | 100 | 27.897 N 34.507 E | PRJNA289734 | SAMN04534681 | LUSA00000000 | 2651870303 |
| Marinobacter sp. REDSEA-S15_B16 | 2.89 | 527 | 2873 | 57.64 | c__Gammaproteobacteria (UID4444) | 72.81 | 0.89 | 71.92 | Red Sea water column Station 34 | 258 | 18.58 N 40.743 E | PRJNA289734 | SAMN04534586 | LUOK00000000 | 2651870218 |
| Marinobacter sp. REDSEA-S21_B2N3 | 4.37 | 262 | 4343 | 57.01 | c__Gammaproteobacteria (UID4444) | 88.75 | 2.59 | 86.16 | Red Sea water column Station 91 | 500 | 20.525 N 38.781 E | PRJNA289734 | SAMN04534587 | LUOL00000000 | 2651870219 |
| Marinobacter sp. REDSEA-S27_B10 | 3.27 | 500 | 3781 | 57.24 | k__Bacteria (UID203) | 70 | 0 | 70 | Red Sea water column Station 108 | 500 | 22.046 N 37.929 E | PRJNA289734 | SAMN04534588 | LUOM00000000 | 2651870220 |
| Maritimibacter sp. REDSEA-S28_B5 | 4.25 | 122 | 4632 | 64.32 | f__Rhodobacteraceae (UID3356) | 97.93 | 0.68 | 97.25 | Red Sea water column Station 149 | 10 | 23.604 N 37.054 E | PRJNA289734 | SAMN04534590 | LUOO00000000 | 2651870216 |
| Maritimibacter sp. REDSEA-S40_B3 | 3.87 | 28 | 4511 | 64.35 | f__Rhodobacteraceae (UID3356) | 99.7 | 0.68 | 99.02 | Red Sea water column Station 192 | 10 | 27.897 N 34.507 E | PRJNA289734 | SAMN04534591 | LUOP00000000 | 2651870217 |
| Moraxellaceae bacterium REDSEA-S29_B6 | 2.23 | 154 | 2327 | 41.94 | c__Gammaproteobacteria (UID4201) | 91.59 | 0 | 91.59 | Red Sea water column Station 149 | 25 | 23.604 N 37.054 E | PRJNA289734 | SAMN04534592 | LUOQ00000000 | 2651870134 |
| Moraxellaceae bacterium REDSEA-S32_B1 | 2.38 | 80 | 2272 | 42.07 | c__Gammaproteobacteria (UID4201) | 98.13 | 0 | 98.13 | Red Sea water column Station 149 | 200 | 23.604 N 37.054 E | PRJNA289734 | SAMN04534593 | LUOR00000000 | 2654587887 |
| Moraxellaceae bacterium REDSEA-S35_B9 | 1.60 | 306 | 1900 | 41.91 | c__Gammaproteobacteria (UID4201) | 71.1 | 0.77 | 70.33 | Red Sea water column Station 169 | 25 | 25.772 N 36.116 E | PRJNA289734 | SAMN04534594 | LUOS00000000 | 2651870137 |
| Moraxellaceae bacterium REDSEA-S38_B3 | 2.41 | 100 | 2293 | 41.98 | c__Gammaproteobacteria (UID4201) | 97.33 | 0 | 97.33 | Red Sea water column Station 169 | 200 | 25.772 N 36.116 E | PRJNA289734 | SAMN04534595 | LUOT00000000 | 2651870132 |
| Moraxellaceae bacterium REDSEA-S42_B15 | 1.88 | 270 | 2124 | 41.99 | c__Gammaproteobacteria (UID4201) | 78.54 | 0.57 | 77.97 | Red Sea water column Station 192 | 50 | 27.897 N 34.507 E | PRJNA289734 | SAMN04534596 | LUOU00000000 | 2651870135 |
| Moraxellaceae bacterium REDSEA-S44_B2 | 2.35 | 95 | 2324 | 42.04 | c__Gammaproteobacteria (UID4201) | 97.13 | 1.15 | 95.98 | Red Sea water column Station 192 | 200 | 27.897 N 34.507 E | PRJNA289734 | SAMN04534597 | LUOV00000000 | 2651870133 |
| Moraxellaceae bacterium REDSEA-S45_B11 | 1.71 | 287 | 1938 | 42.04 | c__Gammaproteobacteria (UID4201) | 78.09 | 2.04 | 76.05 | Red Sea water column Station 192 | 500 | 27.897 N 34.507 E | PRJNA289734 | SAMN04534598 | LUOW00000000 | 2651870136 |
| Nitrosopelagicus sp. REDSEA-S08_B1 | 0.58 | 92 | 1823 | 35.43 | k__Archaea (UID2) | 58.9 | 5.83 | 53.07 | Red Sea water column Station 22 | 200 | 17.996 N 39.799 E | PRJNA289734 | SAMN04534599 | LUOX00000000 | 2651870205 |
| Nitrosopelagicus sp. REDSEA-S19_B12N3 | 1.51 | 307 | 1226 | 36.28 | k__Archaea (UID2) | 85.74 | 5.34 | 80.4 | Red Sea water column Station 91 | 100 | 20.525 N 38.781 E | PRJNA289734 | SAMN04534600 | LUOY00000000 | 2651870206 |
| Nitrosopelagicus sp. REDSEA-S25_B3 | 0.89 | 116 | 1294 | 34.1 | k__Archaea (UID2) | 74.76 | 1.94 | 72.82 | Red Sea water column Station 108 | 100 | 22.046 N 37.929 E | PRJNA289734 | SAMN04534601 | LUOZ00000000 | 2651870207 |
| Nitrosopelagicus sp. REDSEA-S27_B13N2 | 1.51 | 273 | 2009 | 37.26 | k__Archaea (UID2) | 69.84 | 4.85 | 64.99 | Red Sea water column Station 108 | 500 | 22.046 N 37.929 E | PRJNA289734 | SAMN04534602 | LUPA00000000 | 2651870208 |
| Nitrosopelagicus sp. REDSEA-S31_B2 | 1.01 | 86 | 1387 | 34.05 | k__Archaea (UID2) | 92.64 | 2.02 | 90.62 | Red Sea water column Station 149 | 100 | 23.604 N 37.054 E | PRJNA289734 | SAMN04534603 | LUPB00000000 | 2651870209 |
| Nitrosopelagicus sp. REDSEA-S32_B2 | 0.45 | 55 | 604 | 33.9 | k__Archaea (UID2) | 53.4 | 1.94 | 51.46 | Red Sea water column Station 149 | 200 | 23.604 N 37.054 E | PRJNA289734 | SAMN04534604 | LUPC00000000 | 2651870210 |
| Nitrosopelagicus sp. REDSEA-S37_B6 | 0.87 | 104 | 1233 | 34.05 | k__Archaea (UID2) | 89.81 | 1.94 | 87.87 | Red Sea water column Station 169 | 100 | 25.772 N 36.116 E | PRJNA289734 | SAMN04534605 | LUPD00000000 | 2651870211 |
| Nitrosopelagicus sp. REDSEA-S43_B1 | 0.98 | 115 | 1403 | 33.94 | k__Archaea (UID2) | 84.95 | 4.53 | 80.42 | Red Sea water column Station 192 | 100 | 27.897 N 34.507 E | PRJNA289734 | SAMN04534606 | LUPE00000000 | 2651870212 |
| Nocardioides sp. REDSEA-S22_B2 | 3.76 | 94 | 4050 | 71.84 | o__Actinomycetales (UID1697) | 94.43 | 1.38 | 93.05 | Red Sea water column Station 108 | 10 | 22.046 N 37.929 E | PRJNA289734 | SAMN04534607 | LUPF00000000 | 2651870231 |
| Nocardioides sp. REDSEA-S25_B9 | 2.25 | 485 | 2643 | 71.44 | o__Actinomycetales (UID1697) | 57.78 | 1.17 | 56.61 | Red Sea water column Station 108 | 100 | 22.046 N 37.929 E | PRJNA289734 | SAMN04534608 | LUPG00000000 | 2651870232 |
| Nocardioides sp. REDSEA-S28_B4 | 3.90 | 313 | 4194 | 71.71 | o__Actinomycetales (UID1697) | 94.91 | 3.54 | 91.37 | Red Sea water column Station 149 | 10 | 23.604 N 37.054 E | PRJNA289734 | SAMN04534609 | LUPH00000000 | 2651870233 |
| Nocardioides sp. REDSEA-S30_B4 | 3.68 | 104 | 3857 | 71.94 | o__Actinomycetales (UID1697) | 96.89 | 0.86 | 96.03 | Red Sea water column Station 149 | 50 | 23.604 N 37.054 E | PRJNA289734 | SAMN04534610 | LUPI00000000 | 2651870234 |
| Nocardioides sp. REDSEA-S31_B4 | 3.44 | 387 | 3841 | 71.68 | o__Actinomycetales (UID1697) | 83.54 | 1.21 | 82.33 | Red Sea water column Station 149 | 100 | 23.604 N 37.054 E | PRJNA289734 | SAMN04534611 | LUPJ00000000 | 2651870235 |
| Nocardioides sp. REDSEA-S33_B3 | 3.56 | 61 | 3796 | 72.09 | o__Actinomycetales (UID1697) | 98.19 | 0 | 98.19 | Red Sea water column Station 149 | 500 | 23.604 N 37.054 E | PRJNA289734 | SAMN04534612 | LUPK00000000 | 2651870236 |
| Nocardioides sp. REDSEA-S34_B5 | 4.17 | 291 | 4427 | 71.75 | o__Actinomycetales (UID1697) | 94.13 | 4.13 | 90 | Red Sea water column Station 169 | 10 | 25.772 N 36.116 E | PRJNA289734 | SAMN04534613 | LUPL00000000 | 2651870237 |
| Nocardioides sp. REDSEA-S36_B10 | 2.34 | 439 | 2744 | 71.08 | o__Actinomycetales (UID1697) | 59.33 | 1.26 | 58.07 | Red Sea water column Station 169 | 50 | 25.772 N 36.116 E | PRJNA289734 | SAMN04534614 | LUPM00000000 | 2651870238 |
| Nocardioides sp. REDSEA-S37_B12 | 2.85 | 503 | 3278 | 71.42 | o__Actinomycetales (UID1697) | 77.85 | 0.6 | 77.25 | Red Sea water column Station 169 | 100 | 25.772 N 36.116 E | PRJNA289734 | SAMN04534615 | LUPN00000000 | 2651870239 |
| Nocardioides sp. REDSEA-S39_B2 | 3.48 | 56 | 3796 | 72.16 | o__Actinomycetales (UID1697) | 97.67 | 0.05 | 97.62 | Red Sea water column Station 169 | 500 | 25.772 N 36.116 E | PRJNA289734 | SAMN04534616 | LUPO00000000 | 2651870240 |
| Nocardioides sp. REDSEA-S40_B4 | 3.70 | 116 | 3967 | 71.95 | o__Actinomycetales (UID1697) | 91.19 | 1.11 | 90.08 | Red Sea water column Station 192 | 10 | 27.897 N 34.507 E | PRJNA289734 | SAMN04534617 | LUPP00000000 | 2651870241 |
| Nocardioides sp. REDSEA-S43_B3 | 3.68 | 164 | 3948 | 71.9 | o__Actinomycetales (UID1697) | 96.07 | 0.35 | 95.72 | Red Sea water column Station 192 | 100 | 27.897 N 34.507 E | PRJNA289734 | SAMN04534618 | LUPQ00000000 | 2651870242 |
| Prochlorococcus sp. REDSEA-S17_B1 | 1.07 | 152 | 2041 | 30.99 | p__Cyanobacteria (UID2143) | 63.12 | 4.65 | 58.47 | Red Sea water column Station 91 | 25 | 20.525 N 38.781 E | PRJNA289734 | SAMN04534620 | LUPS00000000 | 2651870162 |
| Prochlorococcus sp. REDSEA-S22_B1 | 1.01 | 113 | 1338 | 31.11 | p__Cyanobacteria (UID2143) | 60.42 | 7.38 | 53.04 | Red Sea water column Station 108 | 10 | 22.046 N 37.929 E | PRJNA289734 | SAMN04534621 | LUPT00000000 | 2651870163 |
| Prochlorococcus sp. REDSEA-S23_B1 | 1.06 | 123 | 1381 | 30.87 | p__Cyanobacteria (UID2143) | 61.61 | 7.2 | 54.41 | Red Sea water column Station 108 | 25 | 22.046 N 37.929 E | PRJNA289734 | SAMN04534622 | LUPU00000000 | 2651870164 |
| Prochlorococcus sp. REDSEA-S28_B1 | 0.93 | 101 | 1181 | 31.35 | p__Cyanobacteria (UID2143) | 55.19 | 4.71 | 50.48 | Red Sea water column Station 149 | 10 | 23.604 N 37.054 E | PRJNA289734 | SAMN04534623 | LUPV00000000 | 2651870165 |
| Rhodobacteraceae bacterium REDSEA-S02_B3 | 2.07 | 113 | 2195 | 39.74 | f__Rhodobacteraceae (UID3340) | 80.46 | 1.04 | 79.42 | Red Sea water column Station 12 | 25 | 17.662 N 40.905 E | PRJNA289734 | SAMN04534625 | LUPX00000000 | 2651870273 |
| Rhodobacteraceae bacterium REDSEA-S03_B4 | 2.03 | 202 | 1380 | 39.69 | f__Rhodobacteraceae (UID3340) | 77.8 | 4.86 | 72.94 | Red Sea water column Station 12 | 47 | 17.662 N 40.905 E | PRJNA289734 | SAMN04534626 | LUPY00000000 | 2651870274 |
| Rhodobacteraceae bacterium REDSEA-S11_B6 | 1.89 | 192 | 2900 | 39.63 | k__Bacteria (UID203) | 77.43 | 8.62 | 68.81 | Red Sea water column Station 34 | 25 | 18.58 N 40.743 E | PRJNA289734 | SAMN04534628 | LUQA00000000 | 2651870277 |
| Rhodobacteraceae bacterium REDSEA-S29_B10 | 1.79 | 357 | 2262 | 40.1 | f__Rhodobacteraceae (UID3340) | 57.51 | 3.99 | 53.52 | Red Sea water column Station 149 | 25 | 23.604 N 37.054 E | PRJNA289734 | SAMN04534629 | LUQB00000000 | 2651870278 |
| Rhodobacteraceae bacterium REDSEA-S34_B6 | 2.41 | 111 | 2606 | 40.44 | f__Rhodobacteraceae (UID3340) | 89.51 | 1.57 | 87.94 | Red Sea water column Station 169 | 10 | 25.772 N 36.116 E | PRJNA289734 | SAMN04534630 | LUQC00000000 | 2651870280 |
| SAR116 cluster alphaproteobacterium REDSEA-S02_B12 | 1.51 | 247 | 2215 | 63.03 | c__Alphaproteobacteria (UID3305) | 74.63 | 0.44 | 74.19 | Red Sea water column Station 12 | 25 | 17.662 N 40.905 E | PRJNA289734 | SAMN04534631 | LUQD00000000 | 2654587886 |
| SAR116 cluster alphaproteobacterium REDSEA-S10_B10N8 | 1.58 | 247 | 2222 | 62.96 | c__Alphaproteobacteria (UID3305) | 78.7 | 0 | 78.7 | Red Sea water column Station 34 | 10 | 18.58 N 40.743 E | PRJNA289734 | SAMN04534632 | LUQE00000000 | 2651870131 |
| SAR324 cluster deltaproteobacterium REDSEA-S05_B4 | 1.75 | 357 | 2060 | 46.78 | k__Bacteria (UID3187) | 54.11 | 0.94 | 53.17 | Red Sea water column Station 22 | 25 | 17.996 N 39.799 E | PRJNA289734 | SAMN04534633 | LUQF00000000 | 2654587902 |
| SAR324 cluster deltaproteobacterium REDSEA-S06_B4 | 1.73 | 373 | 873 | 47.26 | k__Bacteria (UID3187) | 54.22 | 0.05 | 54.17 | Red Sea water column Station 22 | 50 | 17.996 N 39.799 E | PRJNA289734 | SAMN04534634 | LUQG00000000 | 2651870251 |
| SAR324 cluster deltaproteobacterium REDSEA-S08_B7 | 2.12 | 328 | 1504 | 43.26 | k__Bacteria (UID2495) | 55.09 | 4.09 | 51 | Red Sea water column Station 22 | 200 | 17.996 N 39.799 E | PRJNA289734 | SAMN04534635 | LUQH00000000 | 2651870252 |
| SAR324 cluster deltaproteobacterium REDSEA-S09_B3 | 3.35 | 83 | 1823 | 42.85 | k__Bacteria (UID3187) | 92.1 | 0 | 92.1 | Red Sea water column Station 22 | 500 | 17.996 N 39.799 E | PRJNA289734 | SAMN04534636 | LUQI00000000 | 2651870253 |
| SAR324 cluster deltaproteobacterium REDSEA-S10_B5 | 3.49 | 290 | 2962 | 47.12 | k__Bacteria (UID3187) | 90.84 | 0 | 90.84 | Red Sea water column Station 34 | 10 | 18.58 N 40.743 E | PRJNA289734 | SAMN04270322 | LNZD00000000 | 2651870254 |
| SAR324 cluster deltaproteobacterium REDSEA-S11_B7 | 2.54 | 416 | 1743 | 47.48 | k__Bacteria (UID3187) | 73.12 | 3.78 | 69.34 | Red Sea water column Station 34 | 25 | 18.58 N 40.743 E | PRJNA289734 | SAMN04534638 | LUQJ00000000 | 2651870255 |
| SAR324 cluster deltaproteobacterium REDSEA-S14_B10 | 1.95 | 455 | 3426 | 42.39 | k__Bacteria (UID3187) | 62 | 3.12 | 58.88 | Red Sea water column Station 34 | 200 | 18.58 N 40.743 E | PRJNA289734 | SAMN04534639 | LUQK00000000 | 2651870256 |
| SAR324 cluster deltaproteobacterium REDSEA-S15_B6 | 2.79 | 191 | 1471 | 42.4 | k__Bacteria (UID3187) | 86.18 | 1.73 | 84.45 | Red Sea water column Station 34 | 258 | 18.58 N 40.743 E | PRJNA289734 | SAMN04534640 | LUQL00000000 | 2651870257 |
| SAR324 cluster deltaproteobacterium REDSEA-S21_B5 | 3.05 | 128 | 3061 | 42.86 | k__Bacteria (UID3187) | 89.51 | 0.22 | 89.29 | Red Sea water column Station 91 | 500 | 20.525 N 38.781 E | PRJNA289734 | SAMN04534641 | LUQM00000000 | 2651870258 |
| SAR324 cluster deltaproteobacterium REDSEA-S26_B7 | 2.26 | 415 | 2573 | 42.53 | k__Bacteria (UID3187) | 69.56 | 1.05 | 68.51 | Red Sea water column Station 108 | 200 | 22.046 N 37.929 E | PRJNA289734 | SAMN04534642 | LUQN00000000 | 2651870259 |
| SAR324 cluster deltaproteobacterium REDSEA-S27_B3 | 3.30 | 80 | 3134 | 42.85 | k__Bacteria (UID3187) | 92.1 | 0 | 92.1 | Red Sea water column Station 108 | 500 | 22.046 N 37.929 E | PRJNA289734 | SAMN04534643 | LUQO00000000 | 2651870260 |
| SAR324 cluster deltaproteobacterium REDSEA-S33_B4 | 3.12 | 94 | 3069 | 42.82 | k__Bacteria (UID3187) | 92.1 | 0 | 92.1 | Red Sea water column Station 149 | 500 | 23.604 N 37.054 E | PRJNA289734 | SAMN04534644 | LUQP00000000 | 2651870261 |
| SAR324 cluster deltaproteobacterium REDSEA-S36_B13 | 1.37 | 323 | 1640 | 46.89 | k__Bacteria (UID3187) | 51.76 | 1.68 | 50.08 | Red Sea water column Station 169 | 50 | 25.772 N 36.116 E | PRJNA289734 | SAMN04534645 | LUQQ00000000 | 2651870262 |
| SAR324 cluster deltaproteobacterium REDSEA-S39_B5 | 3.04 | 266 | 3057 | 43 | k__Bacteria (UID3187) | 88.88 | 2.6 | 86.28 | Red Sea water column Station 169 | 500 | 25.772 N 36.116 E | PRJNA289734 | SAMN04534646 | LUQR00000000 | 2651870263 |
| SAR324 cluster deltaproteobacterium REDSEA-S45_B3 | 3.17 | 89 | 3038 | 42.89 | k__Bacteria (UID3187) | 92.1 | 0 | 92.1 | Red Sea water column Station 192 | 500 | 27.897 N 34.507 E | PRJNA289734 | SAMN04534647 | LUQS00000000 | 2651870264 |
| SAR86 cluster gammaproteobacterium REDSEA-S08_B3 | 1.56 | 177 | 2329 | 36.99 | c__Gammaproteobacteria (UID4443) | 61.92 | 4.16 | 57.76 | Red Sea water column Station 22 | 200 | 17.996 N 39.799 E | PRJNA289734 | SAMN04534648 | LUQT00000000 | 2651870265 |
| SAR86 cluster gammaproteobacterium REDSEA-S09_B4 | 1.67 | 107 | 1534 | 37.05 | c__Gammaproteobacteria (UID4443) | 68.13 | 2.49 | 65.64 | Red Sea water column Station 22 | 500 | 17.996 N 39.799 E | PRJNA289734 | SAMN04534649 | LUQU00000000 | 2651870266 |
| SAR86 cluster gammaproteobacterium REDSEA-S20_B12N4 | 1.70 | 364 | 2043 | 38.32 | c__Gammaproteobacteria (UID4201) | 63.97 | 8.96 | 55.01 | Red Sea water column Station 91 | 200 | 20.525 N 38.781 E | PRJNA289734 | SAMN04534650 | LUQV00000000 | 2651870267 |
| SAR86 cluster gammaproteobacterium REDSEA-S21_B7 | 1.52 | 187 | 1781 | 37.04 | c__Gammaproteobacteria (UID4201) | 75.21 | 2.87 | 72.34 | Red Sea water column Station 91 | 500 | 20.525 N 38.781 E | PRJNA289734 | SAMN04534651 | LUQW00000000 | 2651870268 |
| SAR86 cluster gammaproteobacterium REDSEA-S45_B6 | 1.57 | 123 | 1781 | 37 | k__Bacteria (UID203) | 83.07 | 6.58 | 76.49 | Red Sea water column Station 192 | 500 | 27.897 N 34.507 E | PRJNA289734 | SAMN04534654 | LUQZ00000000 | 2651870271 |
| Sphingopyxis sp. REDSEA-S22_B5 | 3.06 | 103 | 3606 | 65.03 | o__Sphingomonadales (UID3310) | 98.03 | 0.73 | 97.3 | Red Sea water column Station 108 | 10 | 22.046 N 37.929 E | PRJNA289734 | SAMN04534655 | LURA00000000 | 2651870196 |
| Sphingopyxis sp. REDSEA-S23_B6 | 3.24 | 118 | 3560 | 65.08 | o__Sphingomonadales (UID3310) | 96.02 | 0.34 | 95.68 | Red Sea water column Station 108 | 25 | 22.046 N 37.929 E | PRJNA289734 | SAMN04534656 | LURB00000000 | 2651870197 |
| Sphingopyxis sp. REDSEA-S24_B7 | 1.75 | 439 | 2156 | 65.3 | o__Sphingomonadales (UID3310) | 52.05 | 1.47 | 50.58 | Red Sea water column Station 108 | 50 | 22.046 N 37.929 E | PRJNA289734 | SAMN04534657 | LURC00000000 | 2651870198 |
| Sphingopyxis sp. REDSEA-S29_B3 | 3.46 | 43 | 3540 | 65.19 | o__Sphingomonadales (UID3310) | 98.64 | 0.68 | 97.96 | Red Sea water column Station 149 | 25 | 23.604 N 37.054 E | PRJNA289734 | SAMN04534659 | LURE00000000 | 2651870200 |
| Sphingopyxis sp. REDSEA-S34_B10 | 1.75 | 399 | 2178 | 65.03 | o__Sphingomonadales (UID3310) | 50.93 | 0.76 | 50.17 | Red Sea water column Station 169 | 10 | 25.772 N 36.116 E | PRJNA289734 | SAMN04534660 | LURF00000000 | 2651870201 |
| Sphingopyxis sp. REDSEA-S38_B16 | 2.14 | 337 | 2485 | 64.84 | o__Sphingomonadales (UID3310) | 68.27 | 3.72 | 64.55 | Red Sea water column Station 169 | 200 | 25.772 N 36.116 E | PRJNA289734 | SAMN04534661 | LURG00000000 | 2651870202 |
| Sphingopyxis sp. REDSEA-S40_B6 | 3.45 | 73 | 3558 | 65.18 | o__Sphingomonadales (UID3310) | 97.42 | 0.51 | 96.91 | Red Sea water column Station 192 | 10 | 27.897 N 34.507 E | PRJNA289734 | SAMN04534662 | LURH00000000 | 2651870203 |
| Sphingopyxis sp. REDSEA-S42_B3 | 2.91 | 11 | 3455 | 65.26 | o__Sphingomonadales (UID3310) | 99.64 | 0.34 | 99.3 | Red Sea water column Station 192 | 50 | 27.897 N 34.507 E | PRJNA289734 | SAMN04534663 | LURI00000000 | 2651870204 |
| Synechococcus sp. REDSEA-S01_B1 | 1.80 | 78 | 2216 | 62.76 | p__Cyanobacteria (UID2143) | 95.92 | 0.82 | 95.1 | Red Sea water column Station 12 | 10 | 17.662 N 40.905 E | PRJNA289734 | SAMN04534664 | LURJ00000000 | 2651870193 |
| Synechococcus sp. REDSEA-S02_B4 | 1.78 | 81 | 1724 | 62.76 | p__Cyanobacteria (UID2143) | 95.02 | 0.27 | 94.75 | Red Sea water column Station 12 | 25 | 17.662 N 40.905 E | PRJNA289734 | SAMN04534665 | LURK00000000 | 2651870194 |
| Unclassified gammaproteobacterium REDSEA-S03_B5 | 1.17 | 129 | 1411 | 38.75 | p__Proteobacteria (UID3880) | 67.34 | 1.22 | 66.12 | Red Sea water column Station 12 | 47 | 17.662 N 40.905 E | PRJNA289734 | SAMN04534682 | LUSB00000000 | 2651870282 |
| Unclassified gammaproteobacterium REDSEA-S08_B8 | 1.22 | 235 | 2798 | 51.01 | p__Proteobacteria (UID3882) | 55.59 | 0.61 | 54.98 | Red Sea water column Station 22 | 200 | 17.996 N 39.799 E | PRJNA289734 | SAMN04534683 | LUSC00000000 | 2651870283 |
| Unclassified gammaproteobacterium REDSEA-S09_B13 | 2.28 | 371 | 4019 | 51.81 | p__Proteobacteria (UID3880) | 73.03 | 1.22 | 71.81 | Red Sea water column Station 22 | 500 | 17.996 N 39.799 E | PRJNA289734 | SAMN04534684 | LUSD00000000 | 2651870284 |
| Unclassified gammaproteobacterium REDSEA-S12_B4 | 1.30 | 146 | 3073 | 38.79 | p__Proteobacteria (UID3880) | 71.75 | 4.27 | 67.48 | Red Sea water column Station 34 | 50 | 18.58 N 40.743 E | PRJNA289734 | SAMN04534685 | LUSE00000000 | 2651870285 |
| Unclassified gammaproteobacterium REDSEA-S14_B7 | 2.48 | 348 | 1860 | 52.09 | p__Proteobacteria (UID3880) | 83.9 | 1.37 | 82.53 | Red Sea water column Station 34 | 200 | 18.58 N 40.743 E | PRJNA289734 | SAMN04534686 | LUSF00000000 | 2651870286 |
| Unclassified gammaproteobacterium REDSEA-S15_B12 | 2.90 | 309 | 1677 | 52.01 | p__Proteobacteria (UID3880) | 89.74 | 1.93 | 87.81 | Red Sea water column Station 34 | 258 | 18.58 N 40.743 E | PRJNA289734 | SAMN04534687 | LUSG00000000 | 2651870287 |
| Unclassified gammaproteobacterium REDSEA-S21_B8 | 2.72 | 374 | 3048 | 52.02 | p__Proteobacteria (UID3880) | 89.65 | 2.44 | 87.21 | Red Sea water column Station 91 | 500 | 20.525 N 38.781 E | PRJNA289734 | SAMN04534688 | LUSH00000000 | 2651870288 |
| Unclassified gammaproteobacterium REDSEA-S26_B10 | 1.41 | 345 | 1799 | 52.17 | p__Proteobacteria (UID3882) | 54.2 | 1.88 | 52.32 | Red Sea water column Station 108 | 200 | 22.046 N 37.929 E | PRJNA289734 | SAMN04534689 | LUSI00000000 | 2651870289 |
| Unclassified gammaproteobacterium REDSEA-S27_B14 | 1.43 | 327 | 1742 | 52.05 | p__Proteobacteria (UID3880) | 57.13 | 2.44 | 54.69 | Red Sea water column Station 108 | 500 | 22.046 N 37.929 E | PRJNA289734 | SAMN04534690 | LUSJ00000000 | 2654587903 |
| Unclassified gammaproteobacterium REDSEA-S33_B15 | 1.53 | 372 | 1853 | 52.31 | p__Proteobacteria (UID3882) | 56 | 1.02 | 54.98 | Red Sea water column Station 149 | 500 | 23.604 N 37.054 E | PRJNA289734 | SAMN04534691 | LUSK00000000 | 2651870290 |
| Unclassified gammaproteobacterium REDSEA-S45_B9 | 2.31 | 340 | 2626 | 51.86 | p__Proteobacteria (UID3880) | 70.43 | 2.03 | 68.4 | Red Sea water column Station 192 | 500 | 27.897 N 34.507 E | PRJNA289734 | SAMN04534692 | LUSL00000000 | 2651870291 |
Phylogenomic analysis based on sets of single-copy marker genes universal to either the bacterial or archaeal domain showed that the 136 genomes encompassed seven phyla across these domains: Thaumarchaeota, Euryarchaeota, Actinobacteria, Cyanobacteria, Bdellovibrionaeota, Proteobacteria, and Marinimicrobia (Fig. 2 and Table 2 (available online only)). As expected, most of the recovered genomes were affiliated with known marine microorganisms such as phototrophic Prochlorococcus20,21 and Synechococcus22,23; representative of clades first discovered in the Sargasso Sea (SAR86, SAR116, SAR324 and SAR406)24–26; common marine bacteria in tropical biomes such as Alteromonas macleodii27; an ammonia oxidizing thaumarchaeon from the genus Nitrosopelagicus28; euryarchaeotal Marine Group II organisms reported to be abundant in surface waters29; members of the Alpha- and Gamma-proteobacteria such as Aeromicrobium, Erythrobacter, Maritimibacter, Idiomarina, Marinobacter, Candidatus Thioglobus (SUP05 cluster) and several unclassified Gammaproteobacteria, consistent with the high relative abundance of these two groups in the recent Tara Oceans survey30. Additionally, actinobacterial Acidiimicrobia and Nocardioides genomes thought to be responsible for secondary metabolite production in marine ecosystems31 were recovered from the metagenomes. An important strength of this dataset is the recovery of multiple, closely-related genomes from different stations or depths in the Red Sea (Data Citation 2). When complemented with physicochemical data1, genome plasticity between these organisms to confer fitness under varying conditions can be investigated in future studies.
To allow easy access to the genomes, all 136 genomes were functionally annotated and deposited into the National Centre for Biotechnology Information (NCBI) and Integrated Microbial Genomes (IMG) databases32. The wealth of metagenomic and genomic data described here greatly expands the repertoire of microbial genomic information from the Red Sea which might help to better understand the effects of global warming to ocean microbiomes. These datasets will also strengthen studies to better understand the drivers of marine nutrient cycling, help approaches for bioprospecting for novel thermo- and halo-philic enzymes, and allow for a better understanding of microbial adaptation strategies against high temperature, salinity and solar irradiance.
Methods
Metagenomic sequencing and assembly
Seawater samples were collected from eight stations and from different depths (10, 25, 50, 100, 200, and 500 m; locations are shown in Fig. 1) during summer as part of KRSE2011 (ref. 1). Genomic DNA was extracted from the 0.1–1.2
m; locations are shown in Fig. 1) during summer as part of KRSE2011 (ref. 1). Genomic DNA was extracted from the 0.1–1.2 μm size fraction using an established phenol-chloroform extraction protocol1,33. Paired-end libraries (2×100
μm size fraction using an established phenol-chloroform extraction protocol1,33. Paired-end libraries (2×100 bp) were prepared using Nextera DNA Library Prep Kit (Illumina) and sequenced on a HiSeq 2000 (Illumina). Reads were quality checked and trimmed using PRINSEQ v0.20.4 (ref. 34) generating read lengths of ~93
bp) were prepared using Nextera DNA Library Prep Kit (Illumina) and sequenced on a HiSeq 2000 (Illumina). Reads were quality checked and trimmed using PRINSEQ v0.20.4 (ref. 34) generating read lengths of ~93 bp and a total of ~10 million reads per sample with median insert sizes ranging from 183–366 bp1 (Data Citation 1). Trimmed metagenome reads were individually assembled (Table 1 (available online only)) using IDBA-UD v1.1.1 (ref. 35) using the ‘--pre-correction’ option. To obtain coverage profile of contigs from each metagenomic assembly, the trimmed reads were mapped back to contigs using BWA v0.7.12 (ref. 36) with the bwa-mem algorithm.
bp and a total of ~10 million reads per sample with median insert sizes ranging from 183–366 bp1 (Data Citation 1). Trimmed metagenome reads were individually assembled (Table 1 (available online only)) using IDBA-UD v1.1.1 (ref. 35) using the ‘--pre-correction’ option. To obtain coverage profile of contigs from each metagenomic assembly, the trimmed reads were mapped back to contigs using BWA v0.7.12 (ref. 36) with the bwa-mem algorithm.
Genome binning, refinement, and annotation
For each metagenome, genome bins were recovered based on tetranucleotide frequencies and read coverage using MetaBAT v0.26.1 (ref. 37) with default parameters. The completeness and contamination of the bins were assessed using CheckM v1.0.3 (ref. 38) using the lineage-specific workflow (Table 2 (available online only)). Bins were further refined using the CheckM ‘merge’ and ‘outliers’ commands which merge bins with complementary sets of marker genes to improve completeness and remove contigs from bins which appear to be outliers relative to reference GC and tetranucleotide distributions in order to reduce contamination38. The FinishM v0.0.7 (https://github.com/wwood/finishm) ‘roundup’ workflow which comprise of ‘wander’ and ‘gapfill’ modes was used to scaffold contigs together and fill gaps within individual bins. The ‘wander’ mode uses a de Bruijn graph (kmer length of 51 bp and coverage cutoff of 5) to determine contig ends which are connected while the ‘gapfill’ mode align the reads to regions of ambiguous nucleotides and replaces them with the appropriate nucleotides. Genome bins that passed the quality filter of completion minus contamination of ≥50% were submitted to IMG/ER32 for gene calling and functional annotation.
bp and coverage cutoff of 5) to determine contig ends which are connected while the ‘gapfill’ mode align the reads to regions of ambiguous nucleotides and replaces them with the appropriate nucleotides. Genome bins that passed the quality filter of completion minus contamination of ≥50% were submitted to IMG/ER32 for gene calling and functional annotation.
Genome tree construction
The archaeal and bacterial genome trees (Fig. 2) were inferred from the concatenation of 122 and 120 proteins, respectively, identified as being present in ≥90% of the genomes in their respective domains and, when present, single-copy in ≥95% of genomes (Supplementary Tables 1 and 2). These marker genes were aligned using HMMER v3.1b1 (ref. 39) and the tree inference from the concatenated alignment with FastTree v2.1.7 (ref. 40) under the WAG+GAMMA models (Data Citation 2). Support values were determined using 100 non-parametric bootstrap replicates41. The archaeal tree was rooted with the DPANN (Diapherotrites, Parvarchaeota, Aenigmarchaeota, Nanohaloarchaeota, and Nanoarchaeota) superphylum in concordance with a recent large-scale phylogenomic study9 while the bacterial tree was ‘arbitrarily’ rooted with the phylum Chloroflexi42 but should be treated as unrooted. The trees were visualized in ARB43, annotated by iTOL44 and edited in Illustrator CC 2014 (Adobe).
Code availability
All versions of third-party software and scripts used in this study are described and referenced accordingly in the Methods sub-sections for ease of access and reproducibility.
Data Records
The raw Illumina sequencing paired-end reads (Table 1 (available online only)), 45 assembled metagenome sequences (Table 1 (available online only)) and 136 assembled genome sequences (Table 2 (available online only)), generated from the KAUST Red Sea Expedition 2011, are available from NCBI databases (Data Citation 1). The genome trees and associated fasta amino acid alignment files are available from Figshare (Data Citation 2).
Technical Validation
To validate the completeness and contamination of the genomes, we accessed the number of marker genes present in all bacterial and archaeal genomes using CheckM38. The genomes were also manually cleaned from vector contamination by comparing against the UniVec core database (ftp://ftp.ncbi.nlm.nih.gov/pub/UniVec/).
Usage Notes
The annotated genome assemblies can be downloaded and accessed via the Integrated Microbial Genomes (IMG) system (https://img.jgi.doe.gov/cgi-bin/m/main.cgi). The IMG genome IDs are provided in Table 2 (available online only).
Additional Information
How to cite this article: Haroon, M. F. et al. A catalogue of 136 microbial draft genomes from Red Sea metagenomes. Sci. Data 3:160050 10.1038/sdata.2016.50 (2016).
Supplementary Material
Acknowledgments
We acknowledge the people who were involved in the KAUST Red Sea Expedition 2011 and those that helped to generate the data, include, but are not limited to, those named here: Matt Cahill, Mamoon Rashid, Vinu Manikandan, David Ngugi and Ahmed Shibl. This work was supported by King Abdullah University of Science and Technology (KAUST), Saudi Basic Industries Corporation (SABIC) fellowship to L.R.T., and SABIC presidential chair to U.S.
Footnotes
The authors declare no competing financial interests.
Data Citations
- Haroon M. F. 2015. National Center for Biotechnology Information (NCBI) BioProject database. PRJNA289734
- Haroon M. F. 2016. Figshare. https://dx.doi.org/10.6084/m9.figshare.3362899.v1
References
- Thompson L. R. et al. Metagenomic covariation along densely sampled environmental gradients in the Red Sea. bioRxiv 10.1101/055012 (2016). [Europe PMC free article] [Abstract] [Google Scholar]
- Churchill J. H., Bower A. S., McCorkle D. C. & Abualnaja Y. The transport of nutrient-rich Indian Ocean water through the Red Sea and into coastal reef systems. Journal of Marine Research 72, 165–181 (2014). [Google Scholar]
- Sagar S. et al. Cytotoxic and apoptotic evaluations of marine bacteria isolated from brine-seawater interface of the Red Sea. BMC Complementary and Alternative Medicine 13, 1–8 (2013). [Europe PMC free article] [Abstract] [Google Scholar]
- Jimenez-Infante F. et al. Genomic differentiation among two strains of the PS1 clade isolated from geographically separated marine habitats. FEMS microbiology ecology 89, 181–197 (2014). [Abstract] [Google Scholar]
- Zhang G., Haroon M. F., Zhang R., Hikmawan T. & Stingl U. Draft Genome Sequence of Pseudoalteromonas sp. Strain XI10 Isolated from the Brine-Seawater Interface of Erba Deep in the Red Sea. Genome Announcements 4, e00109–16 (2016). [Europe PMC free article] [Abstract] [Google Scholar]
- Fuller N. J. et al. Clade-specific 16S ribosomal DNA oligonucleotides reveal the predominance of a single marine Synechococcus clade throughout a stratified water column in the Red Sea. Applied and Environmental Microbiology 69, 2430–2443 (2003). [Europe PMC free article] [Abstract] [Google Scholar]
- Qian P.-Y. et al. Vertical stratification of microbial communities in the Red Sea revealed by 16S rDNA pyrosequencing. The ISME journal 5, 507–518 (2011). [Europe PMC free article] [Abstract] [Google Scholar]
- Ngugi D. K. & Stingl U. Combined analyses of the ITS loci and the corresponding 16S rRNA genes reveal high micro-and macrodiversity of SAR11 populations in the Red Sea. PLoS ONE 7, e50274 (2012). [Europe PMC free article] [Abstract] [Google Scholar]
- Rinke C. et al. Insights into the phylogeny and coding potential of microbial dark matter. Nature 499, 431–437 (2013). [Abstract] [Google Scholar]
- Brown C. T. et al. Unusual biology across a group comprising more than 15% of domain Bacteria. Nature 523, 208–211 (2015). [Abstract] [Google Scholar]
- Grötzinger S. W. et al. Mining a database of single amplified genomes from Red Sea brine pool extremophiles—improving reliability of gene function prediction using a profile and pattern matching algorithm (PPMA). Frontiers in Microbiology 5, 134 (2014). [Europe PMC free article] [Abstract] [Google Scholar]
- Clingenpeel S., Clum A., Schwientek P., Rinke C. & Woyke T. Reconstructing each cell’s genome within complex microbial communities - dream or reality? Frontiers in Microbiology 5 (2015). [Europe PMC free article] [Abstract] [Google Scholar]
- Gawad C., Koh W. & Quake S. R. Single-cell genome sequencing: current state of the science. Nat. Rev. Genet. 17, 175–188 (2016). [Abstract] [Google Scholar]
- Albertsen M. et al. Genome sequences of rare, uncultured bacteria obtained by differential coverage binning of multiple metagenomes. Nature Biotechnology 31, 533–538 (2013). [Abstract] [Google Scholar]
- Nielsen H. B. et al. Identification and assembly of genomes and genetic elements in complex metagenomic samples without using reference genomes. Nat. Biotech. 32, 822–828 (2014). [Abstract] [Google Scholar]
- Sangwan N., Xia F. & Gilbert J. A. Recovering complete and draft population genomes from metagenome datasets. Microbiome 4, 1–11 (2016). [Europe PMC free article] [Abstract] [Google Scholar]
- Haroon M. F. et al. Anaerobic oxidation of methane coupled to nitrate reduction in a novel archaeal lineage. Nature 500, 567–570 (2013). [Abstract] [Google Scholar]
- Soo R. M. et al. An expanded genomic representation of the phylum Cyanobacteria. Genome biology and evolution 6, 1031–1045 (2014). [Europe PMC free article] [Abstract] [Google Scholar]
- Evans P. N. et al. Methane metabolism in the archaeal phylum Bathyarchaeota revealed by genome-centric metagenomics. Science 350, 434–438 (2015). [Abstract] [Google Scholar]
- Moore L. R., Rocap G. & Chisholm S. W. Physiology and molecular phylogeny of coexisting Prochlorococcus ecotypes. Nature 393, 464–467 (1998). [Abstract] [Google Scholar]
- Partensky F., Hess W. & Vaulot D. Prochlorococcus, a marine photosynthetic prokaryote of global significance. Microbiology and molecular biology reviews 63, 106–127 (1999). [Europe PMC free article] [Abstract] [Google Scholar]
- Moore L. R., Goericke R. & Chisholm S. W. Comparative physiology of Synechococcus and Prochlorococcus: influence of light and temperature on growth, pigments, fluorescence and absorptive properties. Marine ecology progress series. Oldendorf 116, 259–275 (1995). [Google Scholar]
- Palenik B. et al. The genome of a motile marine Synechococcus. Nature 424, 1037–1042 (2003). [Abstract] [Google Scholar]
- Giovannoni S. J., Britschgi T. B., Moyer C. L. & Field K. G. Genetic diversity in Sargasso Sea bacterioplankton. Nature 345, 60–63 (1990). [Abstract] [Google Scholar]
- Britschgi T. B. & Giovannoni S. J. Phylogenetic analysis of a natural marine bacterioplankton population by rRNA gene cloning and sequencing. Applied and Environmental Microbiology 57, 1707–1713 (1991). [Europe PMC free article] [Abstract] [Google Scholar]
- Haroon M. F., Thompson L. R. & Stingl U. Draft genome sequence of uncultured SAR324 bacterium lautmerah10, binned from a Red Sea metagenome. Genome Announcements 4, e01711–e01715 (2016). [Europe PMC free article] [Abstract] [Google Scholar]
- Ivars-Martinez E. et al. Comparative genomics of two ecotypes of the marine planktonic copiotroph Alteromonas macleodii suggests alternative lifestyles associated with different kinds of particulate organic matter. The ISME journal 2, 1194–1212 (2008). [Abstract] [Google Scholar]
- Santoro A. E. et al. Genomic and proteomic characterization of ‘Candidatus Nitrosopelagicus brevis’: An ammonia-oxidizing archaeon from the open ocean. Proceedings of the National Academy of Sciences 112, 1173–1178 (2015). [Europe PMC free article] [Abstract] [Google Scholar]
- DeLong E. F. Archaea in coastal marine environments. Proceedings of the National Academy of Sciences 89, 5685–5689 (1992). [Europe PMC free article] [Abstract] [Google Scholar]
- Sunagawa S. et al. Structure and function of the global ocean microbiome. Science 348, 1261359 (2015). [Abstract] [Google Scholar]
- Bull A. T. & Stach J. E. Marine actinobacteria: new opportunities for natural product search and discovery. Trends in microbiology 15, 491–499 (2007). [Abstract] [Google Scholar]
- Markowitz V. M. et al. IMG: the integrated microbial genomes database and comparative analysis system. Nucleic Acids Research 40, D115–D122 (2012). [Europe PMC free article] [Abstract] [Google Scholar]
- Rusch D. et al. The Sorcerer II Global Ocean Sampling expedition: Northwest Atlantic through eastern tropical Pacific. PLoS Biology 5, e77 (2007). [Europe PMC free article] [Abstract] [Google Scholar]
- Schmieder R. & Edwards R. Quality control and preprocessing of metagenomic datasets. Bioinformatics 27, 863–864 (2011). [Europe PMC free article] [Abstract] [Google Scholar]
- Peng Y., Leung H. C., Yiu S.-M. & Chin F. Y. IDBA-UD: a de novo assembler for single-cell and metagenomic sequencing data with highly uneven depth. Bioinformatics 28, 1420–1428 (2012). [Abstract] [Google Scholar]
- Li H. & Durbin R. Fast and accurate long-read alignment with Burrows–Wheeler transform. Bioinformatics 26, 589–595 (2010). [Europe PMC free article] [Abstract] [Google Scholar]
- Kang D. D., Froula J., Egan R. & Wang Z. MetaBAT, an efficient tool for accurately reconstructing single genomes from complex microbial communities. PeerJ 3, e1165 (2015). [Europe PMC free article] [Abstract] [Google Scholar]
- Parks D. H., Imelfort M., Skennerton C. T., Hugenholtz P. & Tyson G. W. CheckM: assessing the quality of microbial genomes recovered from isolates, single cells, and metagenomes. Genome Research 25, 1043–1055 (2015). [Europe PMC free article] [Abstract] [Google Scholar]
- Eddy S. R. Accelerated Profile HMM Searches. PLoS Computational Biology 7, e1002195 (2011). [Europe PMC free article] [Abstract] [Google Scholar]
- Price M. N., Dehal P. S. & Arkin A. P. FastTree 2—Approximately maximum-likelihood trees for large alignments. PLoS ONE 5, e9490 (2010). [Europe PMC free article] [Abstract] [Google Scholar]
- Felsenstein J. Confidence limits on phylogenies: an approach using the bootstrap. Evolution 39, 783–791 (1985). [Abstract] [Google Scholar]
- Dagan T., Roettger M., Bryant D. & Martin W. Genome Networks Root the Tree of Life between Prokaryotic Domains. Genome Biology and Evolution 2, 379–392 (2010). [Europe PMC free article] [Abstract] [Google Scholar]
- Ludwig W. et al. ARB: a software environment for sequence data. Nucleic Acids Research 32, 1363–1371 (2004). [Europe PMC free article] [Abstract] [Google Scholar]
- Letunic I. & Bork P. Interactive Tree Of Life v2: online annotation and display of phylogenetic trees made easy. Nucleic Acids Research 39, W475–W478 (2011). [Europe PMC free article] [Abstract] [Google Scholar]
Articles from Scientific Data are provided here courtesy of Nature Publishing Group
Full text links
Read article at publisher's site: https://doi.org/10.1038/sdata.2016.50
Read article for free, from open access legal sources, via Unpaywall:
https://www.nature.com/articles/sdata201650.pdf
Citations & impact
Impact metrics
Citations of article over time
Alternative metrics
Article citations
Novel, active, and uncultured hydrocarbon-degrading microbes in the ocean.
Appl Environ Microbiol, 90(9):e0122424, 23 Aug 2024
Cited by: 0 articles | PMID: 39177328 | PMCID: PMC11409719
Metagenome sequencing and 107 microbial genomes from seamount sediments along the Yap and Mariana trenches.
Sci Data, 11(1):887, 15 Aug 2024
Cited by: 0 articles | PMID: 39147792 | PMCID: PMC11327340
Metagenome sequencing and 103 microbial genomes from ballast water and sediments.
Sci Data, 10(1):536, 10 Aug 2023
Cited by: 1 article | PMID: 37563185 | PMCID: PMC10415351
Characterization of the Environmental Plasmidome of the Red Sea.
Microbiol Spectr, 11(4):e0040023, 03 Jul 2023
Cited by: 2 articles | PMID: 37395658 | PMCID: PMC10434023
Evolutionary pathways for deep-sea adaptation in marine planktonic Actinobacteriota.
Front Microbiol, 14:1159270, 10 May 2023
Cited by: 1 article | PMID: 37234526 | PMCID: PMC10205998
Go to all (35) article citations
Data
Data behind the article
This data has been text mined from the article, or deposited into data resources.
BioStudies: supplemental material and supporting data
Data Citations
- (1 citation) DOI - 10.6084/m9.figshare.3362899.v1
BioProject
- (50 citations) BioProject - PRJNA289734
BioSamples (Showing 45 of 45)
- (1 citation) BioSamples - SAMN03860274
- (1 citation) BioSamples - SAMN03860296
- (1 citation) BioSamples - SAMN03860273
- (1 citation) BioSamples - SAMN03860295
- (1 citation) BioSamples - SAMN03860272
- (1 citation) BioSamples - SAMN03860294
- (1 citation) BioSamples - SAMN03860271
- (1 citation) BioSamples - SAMN03860293
- (1 citation) BioSamples - SAMN03860270
- (1 citation) BioSamples - SAMN03860292
- (1 citation) BioSamples - SAMN03860291
- (1 citation) BioSamples - SAMN03860290
- (1 citation) BioSamples - SAMN03860259
- (1 citation) BioSamples - SAMN03860258
- (1 citation) BioSamples - SAMN03860279
- (1 citation) BioSamples - SAMN03860278
- (1 citation) BioSamples - SAMN03860277
- (1 citation) BioSamples - SAMN03860299
- (1 citation) BioSamples - SAMN03860276
- (1 citation) BioSamples - SAMN03860298
- (1 citation) BioSamples - SAMN03860275
- (1 citation) BioSamples - SAMN03860297
- (1 citation) BioSamples - SAMN03860263
- (1 citation) BioSamples - SAMN03860285
- (1 citation) BioSamples - SAMN03860262
- (1 citation) BioSamples - SAMN03860284
- (1 citation) BioSamples - SAMN03860261
- (1 citation) BioSamples - SAMN03860283
- (1 citation) BioSamples - SAMN03860260
- (1 citation) BioSamples - SAMN03860282
- (1 citation) BioSamples - SAMN03860300
- (1 citation) BioSamples - SAMN03860281
- (1 citation) BioSamples - SAMN03860301
- (1 citation) BioSamples - SAMN03860280
- (1 citation) BioSamples - SAMN03860302
- (1 citation) BioSamples - SAMN03860269
- (1 citation) BioSamples - SAMN03860268
- (1 citation) BioSamples - SAMN03860267
- (1 citation) BioSamples - SAMN03860289
- (1 citation) BioSamples - SAMN03860266
- (1 citation) BioSamples - SAMN03860288
- (1 citation) BioSamples - SAMN03860265
- (1 citation) BioSamples - SAMN03860287
- (1 citation) BioSamples - SAMN03860264
- (1 citation) BioSamples - SAMN03860286
Show less
Nucleotide Sequences (Showing 45 of 45)
- (1 citation) ENA - SRR2103038
- (1 citation) ENA - SRR2103016
- (1 citation) ENA - SRR2103017
- (1 citation) ENA - SRR2103018
- (1 citation) ENA - SRR2103019
- (1 citation) ENA - SRR2103001
- (1 citation) ENA - SRR2103023
- (1 citation) ENA - SRR2103002
- (1 citation) ENA - SRR2103024
- (1 citation) ENA - SRR2103003
- (1 citation) ENA - SRR2103025
- (1 citation) ENA - SRR2103004
- (1 citation) ENA - SRR2103026
- (1 citation) ENA - SRR2103020
- (1 citation) ENA - SRR2103021
- (1 citation) ENA - SRR2103000
- (1 citation) ENA - SRR2103022
- (1 citation) ENA - SRR2102998
- (1 citation) ENA - SRR2103009
- (1 citation) ENA - SRR2102999
- (1 citation) ENA - SRR2102994
- (1 citation) ENA - SRR2103005
- (1 citation) ENA - SRR2103027
- (1 citation) ENA - SRR2102995
- (1 citation) ENA - SRR2103006
- (1 citation) ENA - SRR2103028
- (1 citation) ENA - SRR2102996
- (1 citation) ENA - SRR2103007
- (1 citation) ENA - SRR2103029
- (1 citation) ENA - SRR2102997
- (1 citation) ENA - SRR2103008
- (1 citation) ENA - SRR2103034
- (1 citation) ENA - SRR2103012
- (1 citation) ENA - SRR2103035
- (1 citation) ENA - SRR2103013
- (1 citation) ENA - SRR2103036
- (1 citation) ENA - SRR2103014
- (1 citation) ENA - SRR2103037
- (1 citation) ENA - SRR2103015
- (1 citation) ENA - SRR2103030
- (1 citation) ENA - SRR2103031
- (1 citation) ENA - SRR2103010
- (1 citation) ENA - SRR2103032
- (1 citation) ENA - SRR2103011
- (1 citation) ENA - SRR2103033
Show less
EBI Metagenomics/MGnify
Similar Articles
To arrive at the top five similar articles we use a word-weighted algorithm to compare words from the Title and Abstract of each citation.
Red Sea SAR11 and Prochlorococcus Single-Cell Genomes Reflect Globally Distributed Pangenomes.
Appl Environ Microbiol, 85(13):e00369-19, 17 Jun 2019
Cited by: 5 articles | PMID: 31028022 | PMCID: PMC6581160
Diversification and niche adaptations of Nitrospina-like bacteria in the polyextreme interfaces of Red Sea brines.
ISME J, 10(6):1383-1399, 11 Dec 2015
Cited by: 40 articles | PMID: 26657763 | PMCID: PMC5029188
Metagenomic covariation along densely sampled environmental gradients in the Red Sea.
ISME J, 11(1):138-151, 15 Jul 2016
Cited by: 27 articles | PMID: 27420030 | PMCID: PMC5315489
Metagenomic studies of the Red Sea.
Gene, 576(2 pt 1):717-723, 23 Oct 2015
Cited by: 20 articles | PMID: 26526132
Review





