Abstract
Free full text

Tissue-of-origin Dictates Branched-Chain Amino Acid Metabolism in Mutant Kras-driven Cancers
Associated Data
Abstract
Tumor genetics guides patient selection for many new therapies, and cell culture studies have demonstrated that specific mutations can promote metabolic phenotypes. However, whether tissue context defines cancer dependence on specific metabolic pathways is unknown. Kras activation and Trp53 deletion in the pancreas or the lung result in pancreatic ductal adenocarinoma (PDAC) or non-small cell lung carcinoma (NSCLC) respectively, but despite the same initiating events, these tumors utilize branched-chain amino acids (BCAAs) differently. NSCLC tumors incorporate free BCAAs into tissue protein and use BCAAs as a nitrogen source while PDAC tumors have decreased BCAA uptake. These differences are reflected in expression levels of BCAA catabolic enzymes in both mice and humans. Loss of Bcat1 and Bcat2, the enzymes responsible for BCAA utilization, impairs NSCLC tumor formation, but these enzymes are not required for PDAC tumor formation, arguing that tissue-of-origin is an important determinant of how cancers satisfy their metabolic requirements.
Main Text
The development of new cancer therapeutics relies on underlying genetic features to identify sensitive patients (1). Mutations in both KRAS and TP53 are common genetic events found in tumors arising from many tissues and cancers with these mutations are often difficult to treat (2, 3). These genetic events, as well as others associated with cancer, contribute to the metabolic changes that support biomass accumulation and cancer cell proliferation (4). Oncogenic RAS signaling increases glucose and glutamine consumption to support anabolic processes including nucleotide, lipid and non-essential amino acid biosynthesis and can also drive extracellular protein and lipid scavenging (5). TP53 mutations increase glucose consumption and glycolytic flux, while inactivation of TP53 renders cancer cells more dependent on serine uptake and metabolism (6).
KRAS and TP53 mutations are found in most human pancreatic tumors (7) and are also common in lung adenocarcinoma (8). How mutant KRAS or disruption of TP53 affect cancer metabolism is based on cell culture studies in defined medium, although in vivo nutrient availability varies widely between tissues and vasculature changes can limit nutrient access within tumors (9, 10). The inability to model these differences in culture has therefore limited understanding of how tissue-of-origin influences tumor metabolism (11). Furthermore, environment can influence metabolic phenotypes in vitro (12–14), and metabolic dependencies in vivo can differ from those found in vitro (15). Metabolic differences between tumor types may also result from cell-autonomous effects, and tumor metabolic gene expression more closely resembles that of its tissue-of-origin than that of other tumors (16). The same oncogenic driver can also cause different metabolic phenotypes in lung and liver tumors (17). This raises the possibility that tumor type is a major determinant of some tumor metabolic dependencies in vivo.
Elevated plasma BCAA levels are found in early PDAC, but not in NSCLC, even when the tumors are initiated by the same genetic events (18). To confirm that tumor tissue-of-origin influences whole-body BCAA metabolism, we utilized LSL–KrasG12D/+; Trp53flox/flox (KP) mice. We crossed KP mice to mice harboring a Cre-recombinase allele driven by a Pdx-1 promoter (KP−/−C model) (19) or delivered viral Cre to the lungs of these mice (20) to generate models of PDAC and NSCLC respectively. Consistent with prior reports (18), mice with early PDAC have increased levels of plasma BCAAs while mice with early NSCLC exhibit decreased plasma BCAA levels (figs. S1, A–D). When cells derived from these tumors are implanted subcutaneously into syngeneic hosts, tumors derived from PDAC cells did not affect plasma BCAA levels (fig. S1E) (18), while tumors derived from NSCLC cells led to decreased plasma BCAAs (fig. S1F). These results suggest that tumor formation from NSCLC cells can cause depletion of circulating BCAAs.
To trace tissue-specific differences in BCAA metabolism in animals with pancreatic or lung tumors, mice were fed an amino acid defined diet in which 20% of leucine and valine were 13C-labeled. All groups of mice exhibited similar levels of plasma 13C-BCAA enrichment after one-week exposure to labeled diets (figs. S2, A and B). While PDAC tumors contained slightly decreased free BCAAs relative to normal pancreas, NSCLC tumors displayed a significant increase in labeled free BCAAs compared to normal lung (Fig. 1A and figs. S2, C and D). Importantly, these differences are not a reflection of different amino acid compositions of normal or tumor tissue in either the PDAC or NSCLC models (fig. S3). Because BCAAs are essential amino acids that animals cannot synthesize de novo (21), these results suggest that unlike PDAC tumors, NSCLC tumors display enhanced BCAA uptake.

(A and C–E) Mice were fed 13C-BCAA containing diet for seven days. (A) Relative ion counts by LC-MS analysis of fully-labeled, free BCAAs in tumors from PDAC and NSCLC mice and normal tissues from their respective control mice. Data are presented as mean ± SEM. N = 4 control and N = 4 PDAC; N = 4 control and N = 4 NSCLC. (B) Diagram of the leucine catabolic pathway. Red labels indicate metabolites measured by mass spectrometry. Blue circles indicate 13C-labeled carbons. KIC = α-ketoisocaproate. (C) Relative ion counts by GC-MS analysis of fully-labeled BCAAs from protein acid hydrolysates of tumors from PDAC and NSCLC mice and normal tissues from their respective control mice. Data are presented as mean ± SEM. N = 4 control and N = 4 PDAC; N = 4 control and N = 4 NSCLC. (D) Relative ion counts by LC-MS analysis of fully-labeled KIC in tumors from PDAC and NSCLC mice and normal tissues from their respective control mice. Data are presented as mean ± SEM. N = 4 control and N = 4 PDAC; N = 4 control and N = 4 NSCLC. (E) Citrate M+2 labeling (%) from [U-13C]-leucine by GC-MS analysis in tumors from PDAC and NSCLC mice and normal tissues from their respective control mice. Data are presented as mean ± SEM. N = 4 control and N = 4 PDAC; N = 4 control and N = 4 NSCLC. Two-tailed t test was used for all comparisons between two groups. * P<0.05, ** P<0.01, *** P<0.001
BCAAs have several potential metabolic fates in tissues (Fig. 1B). They can be directly incorporated into protein or reversibly transaminated by branched-chain amino acid transaminase (Bcat) to produce branched-chain α-ketoacids (BCKAs) and glutamate. BCKAs can regenerate BCAAs, be secreted, or be oxidatively decarboxylated by the branched-chain keto-acid dehydrogenase (Bckdh) complex to allow further oxidation of the carbon skeleton (21). In agreement with increased BCAA uptake in NSCLC tumors, lung tumors displayed increased labeled BCAA incorporation into protein compared to normal lung, while PDAC tumors incorporated less labeled BCAAs relative to normal pancreas (Fig. 1C and figs. S2, E and F). Analysis of metabolites derived from BCAA catabolism revealed that NSCLC tumors also had more labeled α-ketoisocaproate (KIC), the leucine-derived BCKA, while no change was observed in levels of this labeled metabolite in PDAC tumors (Fig. 1D and fig. S2G). No other differences in labeled BCAA catabolite levels were observed in NSCLC compared to normal tissues, but PDAC tumors showed decreased labeling of the tricarboxylic acid (TCA) cycle intermediate citrate relative to normal pancreas from labeled BCAAs (Fig. 1E and figs. S2, G–I). This is consistent with recent work demonstrating minimal catabolism of BCAAs to TCA intermediates in proliferating cells (22). We then explored whether excess KIC may be excreted by NSCLC tumors for further metabolism by other tissues such as liver, which has limited Bcat, but high Bckdh activity (21, 23). Consistent with this hypothesis, we observe increased labeling of downstream leucine metabolites in the livers of mice with lung tumors (fig. S4). Taken together, these data suggest that BCAA uptake and transamination, but not their subsequent catabolism, may provide a benefit to NSCLC tumors, potentially by acting as a source of nitrogen.
To examine whether NSCLC tumors, but not PDAC tumors, use BCAAs as a source of nitrogen, we fed mice a modified amino acid diet where 50% of leucine was labeled with 15N, allowing the fate of leucine-derived nitrogen to be traced (Fig. 2A). In agreement with 13C-tracing, mice with PDAC demonstrated no differences in free 15N-labeled leucine in tumors compared to control pancreas (fig. S5A), and had less 15N incorporation into other amino acids (fig. S5B). In contrast, increased levels of 15N-leucine were found in NSCLC tumors compared to normal lung (fig. S5C) with decreased plasma enrichment of 15N-leucine in mice with NSCLC tumors (Fig. 2B and fig. S5D). A relative increase in 15N-labeling of non-essential amino acids, as well as valine and isoleucine, was observed in both the free and tissue-protein amino acid pools of NSCLC compared to control lung (Fig. 2C and figs. S5, C and E). Given the reduced plasma enrichment transamination mediated by Bcat isoforms is active in NSCLC tumor tissue. Evidence for increased BCAA transamination in NSCLC compared to PDAC cells is also evident in vitro across a range of glutamine concentrations, however tissue culture does not recapitulate the same phenotypes observed in tumors (fig. S6). Downstream of non-essential amino acid biosynthesis, this nitrogen can also be used to generate nucleotides, primarily if aspartate is synthesized de novo in these tumors. Consistent with this possibility, we find increased incorporation of 15N-label in both aspartate and nucleotides (Figs. 2, C and D and fig. S5E). In some contexts, aspartate production is limiting for nucleotide biosynthesis and proliferation (24, 25), indicating that BCAA metabolism may be important for tumor growth.

(A) Diagram of leucine transamination by Branched-chain amino acid transferase (Bcat) and nitrogen (green circles) fate after transamination. (B–D) NSCLC mice were fed 15N-leucine containing diet for six days. (B) Relative ion counts by GC-MS analysis of M+1 labeled amino acids in plasma of control and NSCLC mice. Data are presented as mean ± SEM. N = 5 control and N = 6 NSCLC. (C) Relative ion counts by GC-MS analysis of M+1 labeled amino acids from protein acid hydrolysates of control mouse lung tissue and NSCLC mouse tumors. Data are presented as mean ± SEM. N = 6 control and N = 6 NSCLC. (D) M+1 labeling (%) from 15N-leucine of deoxynucleic acids from nucleic acid digest of control mouse lung tissue and NSCLC mouse tumors. Data are presented as mean ± SEM. N = 6 control and N = 6 NSCLC. Two-tailed t test was used for all comparisons between two groups. * P<0.05, ** P<0.01, *** P<0.001
To test whether gene expression differences might contribute to differential BCAA metabolism, we used quantitative RT-PCR to analyze mRNA levels in NSCLC and PDAC tumors compared to their respective normal tissues. Consistent with increased BCAA uptake and KIC generation in NSCLC tumors, these tumors displayed increased expression of the primary BCAA transporter Slc7a5 (also called the neutral amino acid transporter Lat1) and increased levels of Bcat2 and Bckdh (Figs. 3, A, C and D). In contrast, PDAC exhibited decreased expression of these genes relative to normal pancreas (Figs. 3, B–D). Importantly, we also observed increased inhibitory phosphorylation of the Bckdh complex in lung tumors (Figs. 3, C and D). Bcat expression enables utilization of BCAAs as a source of nitrogen by lung tumors, and inhibition of Bckdh prevents further catabolism of these amino acids.
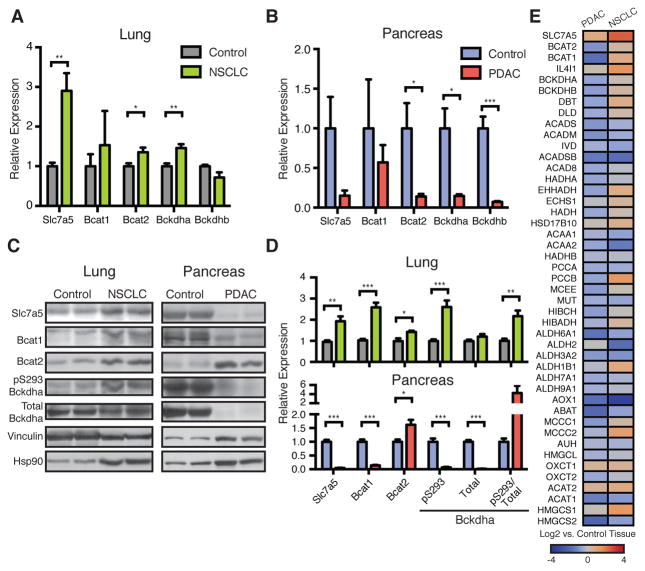
(A) Relative Expression of BCAA metabolic pathway genes in normal lung and NSCLC tumors from KP mice. Data are presented as mean ± SEM. N = 6 control and N = 6 NSCLC. (B) Relative expression of BCAA metabolic pathway genes in normal pancreas and PDAC tumors from KP mice. Data are presented as mean ± SEM. N = 7 control and N = 5 PDAC. (C) Immunoblots of proteins involved in BCAA metabolism in representative normal lung and NSCLC tumors (left) and representative normal pancreas and PDAC tumors (right) from KP mice. (D) Quantification of (C). Data are presented as mean ± SEM. N = 6 control and N = 6 NSCLC; N = 4 control and N = 4 PDAC. (E) Comparison of BCAA metabolic pathway gene expression in human NSCLC and PDAC tumors relative to their adjacent paired normal tissues. Overall expression of BCAA metabolism genes is significantly decreased in PDAC (P<0.0001). Two-tailed t test was used for all comparisons between two groups unless otherwise stated. * P<0.05, ** P<0.01, *** P<0.001
The expression changes observed in PDAC are not unique to this model, as the related KPC mouse model (26), which is initiated by a point mutation in Trp53, showed similar changes in gene expression (fig. S7A). Furthermore, these decreases in gene expression do not appear to be a consequence of the relative decrease in cancer cellularity of PDAC tumors (7), as sorted pancreatic cancer cells showed similar expression of genes involved in proximal BCAA catabolism relative to whole tumor extracts (fig. S7B). In further agreement with neither lung nor pancreatic cancers showing evidence of downstream BCAA-carbon oxidation, the expression of enzymes from this pathway was not markedly different in either of these cancers (figs. S7, C and D). In contrast, glycolytic gene expression was increased in both tumor types (figs. S7, E and F), which is consistent with known increases in glycolysis in each tumor type (27–29). Finally, to relate these data to tissue-of-origin, we performed principal component and clustering analyses, which demonstrated segregation of each tumor with the normal tissue from which it arose (figs. S7, G and H).
To ascertain whether similar changes in gene expression were also found in human cancers, we examined expression of BCAA catabolic enzymes in NSCLC and PDAC relative to their respective normal tissues in publically available data sets (30). Consistent with our observations in mice, human NSCLC had increased expression of SLC7A5, BCAT, and BCKDH, while expression of BCAA catabolism pathway enzymes was decreased in human PDAC (P < 0.0001 for the pathway) (Fig. 3E and tables S1 and S2). The distinct expression patterns for each tumor type were highly correlated across multiple data sets (fig. S7I and tables S3–6). Interestingly, the similarity between human NSCLC and the mouse model of NSCLC was observed despite KRAS and TP53 mutations occurring in less than 50% of human tumors (8) and similar expression patterns were also seen in squamous cell lung cancer (fig. S7I and table S6), further supporting the notion that tissue-of-origin can dictate metabolic phenotype.
The increased contribution of plasma BCAAs to biomass in NSCLC tumors suggests that these tumors may rely on BCAA metabolism for growth. To test this possibility, we used CRISPR-Cas9 mediated genome editing to disrupt exon sequences present in both the Bcat1 (cytosolic) and the Bcat2 (mitochondrial) isoforms (fig. S8A) in cancer cell lines derived from KP mice with NSCLC (Bcat null Clones A and B) or PDAC (Bcat null) (fig. S8B). Expression analysis and 15N-leucine tracing studies confirmed functional deletion of Bcat in both the NSCLC and PDAC cancer cells (figs. S8, C–F). Despite loss of both Bcat isoforms, these cells proliferate at a rate that is similar to the parental and vector control infected cell lines in vitro (Figs. 4, A and B). When Bcat null NSCLC cells were implanted subcutaneously in vivo, however, the ability of these cells to form tumors was significantly impaired, and one clone failed to produce tumors (Fig. 4C and fig. S8G). In contrast, Bcat null PDAC cells implanted subcutaneously generated tumors (Fig. 4D and fig. S8H). Additionally, orthotopic transplantation of NSCLC Bcat null cells failed to form lung tumors (Fig. 4E), while PDAC Bcat null cells formed tumors in the pancreas (Fig. 4F). Unlike subcutaneously implanted PDAC Bcat knockout cells, these cells formed smaller tumors in the pancreas than control cells (fig. S8I). Taken together, these data suggest that while KP lung tumors require Bcat activity for growth, this enzyme activity is dispensable for KP pancreas tumor formation, although PDAC tumor growth may be aided by Bcat activity in some tissue environments.
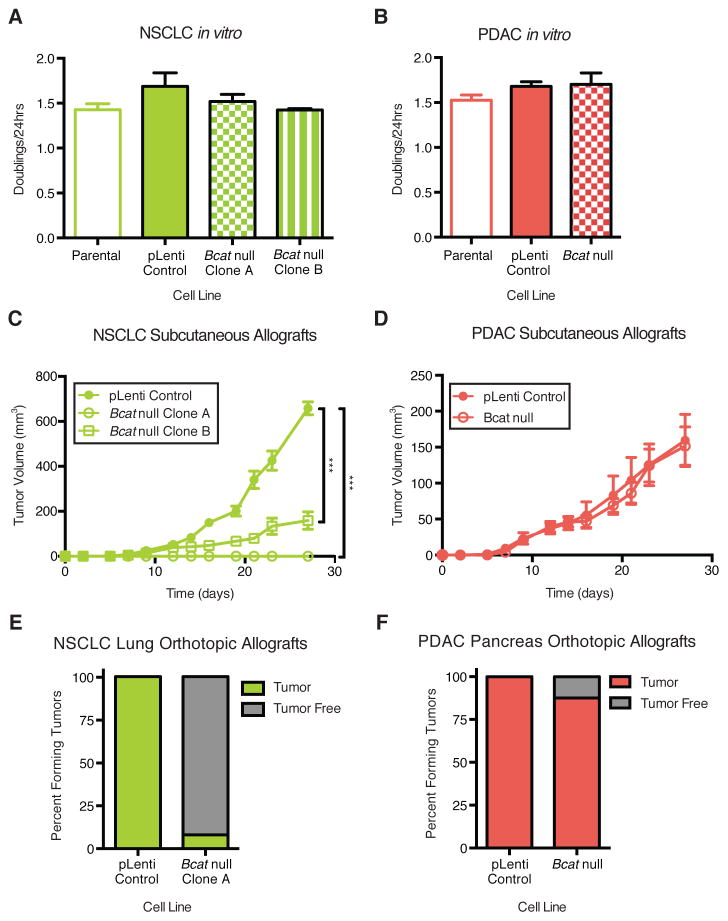
(A) Doubling time of parental, control CRISPR-Cas9 vector infected (pLenti), and NSCLC Bcat null cell lines in vitro. Data are presented as mean ± SEM. N = 3 per group. Representative experiment from ≥ 2 repeats. (B) Doubling time of parental, control CRISPR-Cas9 vector infected and PDAC Bcat null cell lines in vitro. Data are presented as mean ± SEM. N = 3 per group. Representative experiment from ≥ 2 repeats. (C) Estimated tumor volume (mm3) of subcutaneous allograft of control infected and Bcat null syngenic NSCLC cell lines into C57BL/6J mice. Data are presented as mean ± SEM. N = 6 per group. Two-way repeated measures ANOVA used for comparison between groups. (D) Estimated tumor volume (mm3) of subcutaneous allograft of control infected and Bcat null syngenic PDAC cell lines into C57BL/6J mice. Data are presented as mean ± SEM. N = 5 pLenti control and N = 6 Bcat null. Two-way repeated measures ANOVA used for comparison between groups. (E) Lung orthotopic allograft of control infected and Bcat null syngenic NSCLC cell lines into C57BL/6J mice. N = 23 vector control and N = 13 Bcat null Clone A. (F) Pancreatic orthotopic allograft of control infected and Bcat null syngenic PDAC cell lines into C57BL/6J mice. N = 8 per group. * P<0.05, ** P<0.01, *** P<0.001.
Proliferating cells need to acquire amino acids, both to make protein and as source of nitrogen for nucleotide and non-essential amino acid synthesis. Prior work has shown that macropinocytosis plays a role in filling this requirement in mutant RAS-driven PDAC tumors and cells (12, 14, 31). The data presented here argue that this process might be less active in mutant Ras transformed NSCLC tumors that acquire nitrogen in part from free BCAAs. Indeed, we observed less macropinocytosis in cells derived from mouse NSCLC relative to mouse PDAC cells (fig. S9). The decreased reliance of PDAC on free BCAAs however, does not necessarily imply that uptake of these amino acids would be toxic for this cancer. Overexpressing Slc7a5 in PDAC cells is sufficient to increase leucine uptake (figs. S10, A and B), but has minimal effects on proliferation in vitro (fig. S10C) or tumor growth in vivo (figs. S10, D and E).
A role for free BCAAs in supplying nitrogen to lung cancers is intriguing in light of recent studies in glioblastoma and NSCLC indicating that glutamine, which is the most abundant plasma amino acid and serves as the major free amino acid substrate for nitrogen and carbon in culture (32), contributes less to tumor metabolism in vivo (33, 34). Indeed, glucose-tracing studies in humans and mice demonstrate that glutamine is net synthesized from glucose (15, 33–37), and alternative sources of nitrogen are required to support glutamine production. Thus, in these contexts, extraction of nitrogen from BCAAs for de novo amino acid and nucleotide biosynthesis in vivo may explain how lung tumors satisfy their nitrogen requirements. Consistent with this possibility, BCAT1 expression is known to be important for glioblastoma growth (38), suggesting that tumors arising in tissues other than the lung may also utilize BCAAs as a source of nitrogen. Multiple factors including local environment, tumor cell-of-origin, and genetic mutations can lead to convergent metabolic adaptations in disparate tumor types.
Elevations in plasma BCAA levels are associated with early PDAC and result from increased tissue protein breakdown (18). The finding that PDAC tumors have decreased utilization of circulating BCAAs contributes to this phenotype as well. In contrast, NSCLC tumors actively utilize BCAAs leading to plasma BCAA depletions, particularly since the liver does not regulate levels of these amino acids (23). Many patients with PDAC and NSCLC tumors develop cachexia with end-stage disease (39). Our findings suggest that differential use of amino acids by tumors and the resulting impact on whole body metabolism might play a role in the initiation and natural history of cachexia. In addition, as personalized medicine plays a larger role in the clinical management of cancer, it will be critical to understand how cell-of-origin and tissue environment interact with genetic events to influence metabolic dependencies of tumors and select the right treatment approaches for patients.
Acknowledgments
JRM acknowledges support from F30CA183474 and T32GM007753. ANL is a Robert Black Fellow of the Damon Runyon Cancer Research Foundation, DRG-2241-15. MGVH acknowledges support from P30CA1405141, R01CA168653, R01CA201276, the Burroughs Wellcome Fund, the Eisen and Chang Families, the Ludwig Center at MIT, SU2C, and the Lustgarten Foundation.
Footnotes
References
Full text links
Read article at publisher's site: https://doi.org/10.1126/science.aaf5171
Read article for free, from open access legal sources, via Unpaywall:
https://science.sciencemag.org/content/sci/353/6304/1161.full.pdf
Citations & impact
Impact metrics
Citations of article over time
Alternative metrics
Smart citations by scite.ai
Explore citation contexts and check if this article has been
supported or disputed.
https://scite.ai/reports/10.1126/science.aaf5171
Article citations
IMPA1-derived inositol maintains stemness in castration-resistant prostate cancer via IMPDH2 activation.
J Exp Med, 221(11):e20231832, 29 Oct 2024
Cited by: 0 articles | PMID: 39470689
Tryptophan Metabolism Disorder-Triggered Diseases, Mechanisms, and Therapeutic Strategies: A Scientometric Review.
Nutrients, 16(19):3380, 04 Oct 2024
Cited by: 0 articles | PMID: 39408347 | PMCID: PMC11478743
Review Free full text in Europe PMC
Multivariate analysis of metabolic state vulnerabilities across diverse cancer contexts reveals synthetically lethal associations.
Cell Rep, 43(10):114775, 20 Sep 2024
Cited by: 0 articles | PMID: 39305483 | PMCID: PMC11511630
Identification of New Single Nucleotide Polymorphisms Potentially Related to Small Ruminant Lentivirus Infection Susceptibility in Goats Based on Data Selected from High-Throughput Sequencing.
Pathogens, 13(10):830, 25 Sep 2024
Cited by: 0 articles | PMID: 39452702 | PMCID: PMC11510762
Branched-chain amino acids and risk of lung cancer: insights from mendelian randomization and NHANES III.
J Thorac Dis, 16(8):5248-5261, 17 Aug 2024
Cited by: 0 articles | PMID: 39268127 | PMCID: PMC11388241
Go to all (333) article citations
Data
Data behind the article
This data has been text mined from the article, or deposited into data resources.
BioStudies: supplemental material and supporting data
Similar Articles
To arrive at the top five similar articles we use a word-weighted algorithm to compare words from the Title and Abstract of each citation.
BCAT2-mediated BCAA catabolism is critical for development of pancreatic ductal adenocarcinoma.
Nat Cell Biol, 22(2):167-174, 06 Feb 2020
Cited by: 93 articles | PMID: 32029896
Tumour-reprogrammed stromal BCAT1 fuels branched-chain ketoacid dependency in stromal-rich PDAC tumours.
Nat Metab, 2(8):775-792, 06 Jul 2020
Cited by: 82 articles | PMID: 32694827 | PMCID: PMC7438275
GEO data mining and TCGA analysis reveal altered branched chain amino acid metabolism in pancreatic cancer patients.
Aging (Albany NY), 13(8):11907-11918, 21 Apr 2021
Cited by: 7 articles | PMID: 33882453 | PMCID: PMC8109144
Targeting metabolic reprogramming in KRAS-driven cancers.
Int J Clin Oncol, 22(4):651-659, 24 Jun 2017
Cited by: 66 articles | PMID: 28647837
Review
Funding
Funders who supported this work.
Burroughs Wellcome Fund
Damon Runyon Cancer Research Foundation (1)
Grant ID: DRG-2241-15
Eisen and Chang Families
Ludwig Center at Massachusetts Institute of Technology
Lustgarten Foundation
NCI (3)
Grant ID: P30CA1405141
Grant ID: R01CA168653
Grant ID: R01CA201276
NCI NIH HHS (5)
Grant ID: U01 CA210171
Grant ID: P30 CA014051
Grant ID: R01 CA168653
Grant ID: R01 CA201276
Grant ID: F30 CA183474
NIGMS NIH HHS (1)
Grant ID: T32 GM007753
NIH National Cancer Institute (NCI) (1)
Grant ID: F30CA183474
National Institute of General Medical Sciences (1)
Grant ID: T32GM007753





