Abstract
Objectives
We unexpectedly identified MRSA isolates carrying mecC (mecC-MRSA) from a Danish swine farm located in eastern Zealand. The objective of the present study was to investigate the origin of these isolates and their genetic relatedness to other mecC-MRSA isolates from Zealand.Methods
WGS was used to infer the phylogenetic relationship between 19 identified mecC-MRSA isolates from the swine farm and 34 additional epidemiologically unrelated human isolates from the same geographical region of Denmark. Variations in the accessory genome were investigated by bioinformatics tools, and antibiotic susceptibility profiles were assessed by MIC determination.Results
mecC-MRSA was isolated from a domestic swine farm, but not from cattle reared at the same farm. Phylogenetic analysis revealed that all mecC-MRSA isolates from both farm animals and workers formed a separate cluster, whereas human isolates from the same municipality belonged to a closely related cluster. Analysis of the accessory genome supported this relationship.Conclusions
To the best of our knowledge, this is the first report of mecC-MRSA isolated from domestic swine. The investigation strongly indicates that transmission of mecC-MRSA has taken place on the swine farm between the farmers and swine. The close clustering of farm isolates and isolates from the same municipality suggests a local transmission of mecC-MRSA.Free full text

Report of mecC-carrying MRSA in domestic swine
Abstract
Objectives
We unexpectedly identified MRSA isolates carrying mecC (mecC-MRSA) from a Danish swine farm located in eastern Zealand. The objective of the present study was to investigate the origin of these isolates and their genetic relatedness to other mecC-MRSA isolates from Zealand.
Methods
WGS was used to infer the phylogenetic relationship between 19 identified mecC-MRSA isolates from the swine farm and 34 additional epidemiologically unrelated human isolates from the same geographical region of Denmark. Variations in the accessory genome were investigated by bioinformatics tools, and antibiotic susceptibility profiles were assessed by MIC determination.
Results
mecC-MRSA was isolated from a domestic swine farm, but not from cattle reared at the same farm. Phylogenetic analysis revealed that all mecC-MRSA isolates from both farm animals and workers formed a separate cluster, whereas human isolates from the same municipality belonged to a closely related cluster. Analysis of the accessory genome supported this relationship.
Conclusions
To the best of our knowledge, this is the first report of mecC-MRSA isolated from domestic swine. The investigation strongly indicates that transmission of mecC-MRSA has taken place on the swine farm between the farmers and swine. The close clustering of farm isolates and isolates from the same municipality suggests a local transmission of mecC-MRSA.
Introduction
MRSA carrying a new variant of the mecA gene, now named mecC, was first described in 2011 from humans and cattle in the UK, Ireland and Denmark.1,2 García-Álvarez et al.1 demonstrated that the mecC gene is encoded by the type XI staphylococcal cassette chromosome mec (SCCmec) element, which is known to be present in at least four Staphylococcus aureus lineages: CC130, CC325, CC705 and CC1943. mecC was retrospectively detected in a human MRSA isolate from Denmark isolated in 1975.3
Petersen et al.4 found that mecC-MRSA CC130 and CC2361 account for ~2% of the annual number of MRSA cases in Denmark. They also found that some human mecC-MRSA CC130 isolates with spa type t843 were related to isolates from sheep and cows. Farm transmission of mecC-MRSA between humans and ruminants has also been demonstrated previously by WGS.5 mecC-MRSA has been isolated from a number of livestock species, such as cattle, sheep and horses, as well as from companion animals and a large number of wild animal species, including a single report from wild boar in Spain.6,7 To the best of our knowledge, this is the first report of mecC-MRSA isolated from domestic swine.
Materials and methods
Nasal swabs were initially collected from 10 weaners in a Danish swine farm in connection with an ongoing MRSA study. The farm was situated in eastern Zealand and contained ~600 sows and 100 beef cattle. The cattle were occasionally located in the same buildings as the swine and occasionally in pens that had been used by swine. The workers at the farm did not change clothes or boots when moving between the buildings or animal species.
The samples were cultivated on Brilliance MRSA 2 agar plates (Oxoid) with and without pre-enrichment in tryptic soy broth (Sigma–Aldrich) containing 6.5% NaCl, and denim blue colonies were subcultured for verification. Two out of 10 weaner samples were found to be positive for MRSA. The isolates were found to be positive for mecC by PCR.3 As this was, to the best of our knowledge, the first identification of mecC-MRSA from domestic swine, additional samples were collected. Nasal samples were collected from 10 sows in the farrowing unit, 10 piglets in the weaning unit and 10 slaughter pigs. In addition, nasal swabs from 20 beef cattle, 2 workers and the farm dog were tested. Approval from the National Committee on Health Research Ethics was not necessary for this investigation (Decision No. 16035690).
The mecC-MRSA were analysed by WGS. For comparative purposes we reviewed the national MRSA database to identify and extract information on persons colonized or infected with mecC-MRSA between 2013 and 2016. A total of 34 mecC positive isolates from Zealand were included, of which five originated in the same municipality as the swine farm. Samples and isolates included in the study can be found in Table 1. The methods used for WGS, phylogenetic analysis of the core genome, analysis of the accessory genome, MLST, SCCmec typing and searching for resistance genes are available as Supplementary data at JAC Online. All sequences can be obtained using the ENA Study accession number PRJEB15105.
Table 1.
Samples and isolates included in the study
| Sampling site | Sampling date | Number of samples | mecC-positive samples | spa types (number of isolates) |
|---|---|---|---|---|
| Farm isolates (H1a–c, F1–9, W1–7) | ||||
 weaning unit weaning unit | Sep 2015 | 10 (W1–2) | 2 | t843 (2) |
 farrowing unit farrowing unit | Oct 2015 | 10 (F1–9) | 9 | t843 (8), t11205 (1) |
 weaning unit weaning unit | Nov 2015 | 10 (W3–7) | 5 | t843 (5) |
 slaughter unit slaughter unit | Nov 2015 | 10 | 0 | — |
 cattle cattle | Nov 2015 | 20 | 0 | — |
 dog dog | Jan 2016 | 1 | 0 | — |
 human workers human workers | Jan 2016 | 2 (H1a–c) | 1 (3 isolates) | t1535 (3) |
| Isolates from Zealand (H2–35) | ||||
 human isolates human isolates | 2013 | 5 | t843 (5) | |
| 2014 | 9 | t843 (9) | ||
| 2015 | 19 | t843 (16), t1535 (3) | ||
| 2016 | 1 | t15667 (1) | ||
Antibiotic susceptibility testing was performed by MIC determination using a custom-made panel (DKSSP2, TREK Diagnostics). Antimicrobials tested, including ranges and breakpoints, are described in Table S1 (available as Supplementary data at JAC Online). spa typing was performed as described previously.3
Results
From the farrowing unit, 9 out of 10 samples were found to be mecC-MRSA positive. In the weaning unit, five samples were found to be mecC-MRSA positive, in addition to the two isolates from the initial screening. All slaughter pigs and beef cattle as well as the farm dog were found to be MRSA negative. One MRSA isolate from each pig was further characterized. Fifteen swine isolates belonged to spa type t843 (repeat succession: 04-82-17-25-17-25-25-16-17), whereas the remaining swine isolate harboured spa type t11205 (repeat succession: 04-16-17). One of the two farm workers was MRSA positive, and in order to assess the genomic variation present within this individual, three isolates from this sample were whole-genome sequenced and spa typed; all three isolates belonged to spa type t1535 (repeat succession: 04-82-17-25-17-25-16-17).
All 19 mecC-MRSA isolates were phenotypically resistant to penicillin and cefoxitin, but susceptible to all other antimicrobials tested. All 19 mecC-MRSA isolates belonged to CC130 and did not carry mecA or any other resistance genes. The isolates were carrying the mecC gene on the mobile element SCCmec XI. All isolates carried the entire SCCmec XI element2 except strain H35, which did not contain three hypothetical genes downstream from the ars operon.
The phylogenetic analysis was based on 2530 variable SNPs identified within 84% of the reference genome and found to be conserved within the strain collection, and showed that the 19 farm isolates (16 swine and 3 human isolates) were found on a separate branch on the maximum-likelihood tree (Figure 1) with a pairwise distance between 0 and 30 SNPs (mean =
= 18). The three isolates from the farm worker (H1a–c) were found to branch out from the basal part of the group and differed by 5–12 SNPs. In comparison, between 11 and 24 SNP differences were observed between the human and swine isolates (W1–6 and F1–9). There were no canonical SNPs separating the human and swine farm isolates. The most closely related human isolates from the MRSA database originated from five persons living in the same municipality as the swine farm (H2–6) and all had spa type t843. The remaining human isolates originated from other parts of Zealand and were found to be diverse and only distantly related to the farm and municipality isolates.
18). The three isolates from the farm worker (H1a–c) were found to branch out from the basal part of the group and differed by 5–12 SNPs. In comparison, between 11 and 24 SNP differences were observed between the human and swine isolates (W1–6 and F1–9). There were no canonical SNPs separating the human and swine farm isolates. The most closely related human isolates from the MRSA database originated from five persons living in the same municipality as the swine farm (H2–6) and all had spa type t843. The remaining human isolates originated from other parts of Zealand and were found to be diverse and only distantly related to the farm and municipality isolates.
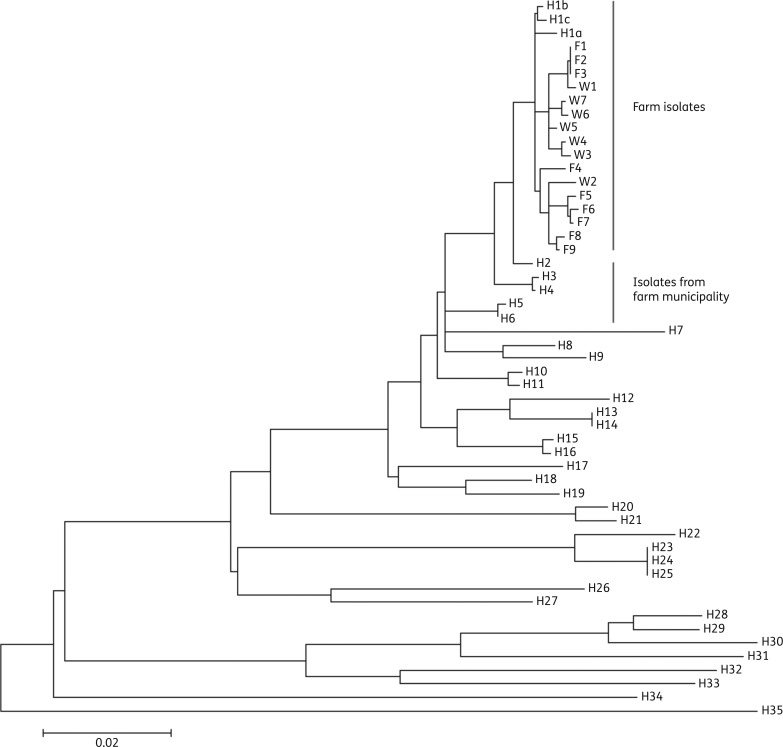
Phylogenetic analysis of mecC-carrying S. aureus from Zealand. The phylogenetic relationship of 53 isolates was inferred using maximum likelihood based on 2530 SNPs, contained within 84% of the reference chromosome (2.35 Mb). W1–7, isolates from weaning pigs; F1–9, isolates from farrowing unit; H1a–c, isolates from farm worker; H2–6, human isolates from the same municipality as the farm; H7–35, human isolates from other parts of Zealand. The bar indicates substitutions per site, and is equivalent to ~50 SNPs.
Analysis of the accessory genome confirmed a very close affiliation between the farm isolates and the five human isolates from the same municipality (Figure S1, available as Supplementary data at JAC Online). A cluster of 48 genes was consistently present in this group compared with most of the other human isolates investigated. Most genes encoded hypothetical proteins or were phage-related ORFs, but none of the genes represented known virulence or host adaptation factors. Based on the Illumina sequencing data, it was not possible to decide whether these genes comprise a novel entire or partial phage; however, using PHAST (http://phast.wishartlab.com), some of the identified genes are associated to an incomplete phage found in S. aureus strain LGA251 (data not shown).
Discussion
To the best of our knowledge, this is the first report of mecC-carrying S. aureus isolated from domestic swine, emphasizing that this variant of S. aureus has a broad host range.7 This report also underlines the importance of characterizing MRSA isolates for the presence of both mecA and mecC. In the herd, the genome sequences of the pig isolates did not support any clear differentiation between isolates from the weaning unit (W1–7) and the farrowing unit (F1–9). This was not unexpected due to the lack of restriction in moving animals between the different units. The observed SNP variation in the core genome also supports that the isolates have been present in the farm for an extended period of time. Previous reports have indicated that mecC-carrying MRSA show a preference for cattle.1,8 Surprisingly, at this farm no mecC-MRSA was detected in the cattle. The mecC strains did not carry any tetracycline resistance genes, unlike the more commonly occurring mecA-MRSA isolates circulating in Danish swine herds.9 Tetracycline was used regularly on this farm for individual treatment of sows and nursery pigs; however, the total antibiotic usage was lower than the Danish average for swine herds. Therefore, antibiotic usage alone cannot explain the occurrence of mecC-carrying MRSA in the case herd. The introduction and survival of new MRSA clones such as this mecC-carrying MRSA into Danish pig production may complicate attempts to lower MRSA frequencies by decreasing the use of antimicrobials such as tetracycline.
Three different spa types were found among the isolates from the swine farm. The spa types t843 and t1535 differ by only a single repeat, emphasizing the close affiliation between the isolates from the farmer and the pigs (Table 1). One of the pig isolates belonged to spa type t11205, which differs from t843 by a loss of a group of six adjacent repeats and most probably represents a single genetic event.
The swine isolates were closely affiliated to the human isolates from the same farm, indicating a transmission between the farmer and the swine. Zoonotic transmission of mecC-carrying MRSA has also been previously reported.4,5 Concluding on clonal transmission based on single isolates can, however, be problematic due to the diversity present among MRSA isolates in a single host.10 Genetic diversity was also observed among the three isolates obtained from the farm worker. The clustering of the multiple isolates from the farmer suggests that the farmer could have been the source of introduction into the herd. This is further supported by the adjacent clustering of human isolates recently obtained from the same geographical area, based on analysis of both the core genomes and the accessory genomes. A high diversity was observed between the human mecC-carrying isolates from Zealand in 2013–16, indicating multiple introductions or a genetic diversity that has been accumulating over many years. This corresponds well with the fact that mecC-carrying MRSA has been present in Denmark consistently from 2003 onwards.1,3
References
Articles from Journal of Antimicrobial Chemotherapy are provided here courtesy of Oxford University Press
Full text links
Read article at publisher's site: https://doi.org/10.1093/jac/dkw389
Read article for free, from open access legal sources, via Unpaywall:
https://academic.oup.com/jac/article-pdf/72/1/60/8497248/dkw389.pdf
Citations & impact
Impact metrics
Citations of article over time
Alternative metrics
Smart citations by scite.ai
Explore citation contexts and check if this article has been
supported or disputed.
https://scite.ai/reports/10.1093/jac/dkw389
Article citations
Genomic epidemiology of mecC-carrying Staphylococcus aureus isolates from human clinical cases in New Zealand.
Access Microbiol, 6(9):000849.v2, 05 Sep 2024
Cited by: 0 articles | PMID: 39239568 | PMCID: PMC11376224
Prevalence, antibiotic resistance and molecular characterization of Staphylococcus aureus in ready-to-eat fruits and vegetables in Shanghai, China.
Curr Res Food Sci, 8:100669, 25 Dec 2023
Cited by: 3 articles | PMID: 38226140 | PMCID: PMC10788225
Genomic Evidence for Direct Transmission of mecC-MRSA between a Horse and Its Veterinarian.
Antibiotics (Basel), 12(2):408, 17 Feb 2023
Cited by: 3 articles | PMID: 36830318 | PMCID: PMC9952710
mecC MRSA in Israel-genomic analysis, prevalence and global perspective.
JAC Antimicrob Resist, 4(4):dlac085, 27 Aug 2022
Cited by: 0 articles | PMID: 36042980 | PMCID: PMC9418563
The European Union Summary Report on Antimicrobial Resistance in zoonotic and indicator bacteria from humans, animals and food in 2019-2020.
EFSA J, 20(3):e07209, 29 Mar 2022
Cited by: 106 articles | PMID: 35382452 | PMCID: PMC8961508
Go to all (15) article citations
Data
Data behind the article
This data has been text mined from the article, or deposited into data resources.
BioStudies: supplemental material and supporting data
BioProject
- (1 citation) BioProject - PRJEB15105
Similar Articles
To arrive at the top five similar articles we use a word-weighted algorithm to compare words from the Title and Abstract of each citation.
Transmission of MRSA between humans and animals on duck and turkey farms.
J Antimicrob Chemother, 71(1):58-62, 21 Oct 2015
Cited by: 17 articles | PMID: 26490016
Prevalence, risk factors and genetic diversity of methicillin-resistant Staphylococcus aureus carried by humans and animals across livestock production sectors.
J Antimicrob Chemother, 68(7):1510-1516, 20 Feb 2013
Cited by: 44 articles | PMID: 23429641
Isolation and characterization of methicillin-resistant Staphylococcus aureus from swine and workers in China.
J Antimicrob Chemother, 64(4):680-683, 14 Aug 2009
Cited by: 106 articles | PMID: 19684078
The emergence of mecC methicillin-resistant Staphylococcus aureus.
Trends Microbiol, 22(1):42-47, 09 Dec 2013
Cited by: 201 articles | PMID: 24331435 | PMCID: PMC3989053
Review Free full text in Europe PMC
Funding
Funders who supported this work.





