Abstract
Objective
The clinical presentation and course of Crohn's disease (CD) is highly variable. We sought to better understand the cellular and molecular mechanisms that guide this heterogeneity, and characterise the cellular processes associated with disease phenotypes.Design
We examined both gene expression and gene regulation (chromatin accessibility) in non-inflamed colon tissue from a cohort of adult patients with CD and control patients. To support the generality of our findings, we analysed previously published expression data from a large cohort of treatment-naïve paediatric CD and control ileum.Results
We found that adult patients with CD clearly segregated into two classes based on colon tissue gene expression-one that largely resembled the normal colon and one where certain genes showed expression patterns normally specific to the ileum. These classes were supported by changes in gene regulatory profiles observed at the level of chromatin accessibility, reflective of a fundamental shift in underlying molecular phenotypes. Furthermore, gene expression from the ilea of a treatment-naïve cohort of paediatric patients with CD could be similarly subdivided into colon-like and ileum-like classes. Finally, expression patterns within these CD subclasses highlight large-scale differences in the immune response and aspects of cellular metabolism, and were associated with multiple clinical phenotypes describing disease behaviour, including rectal disease and need for colectomy.Conclusions
Our results strongly suggest that these molecular signatures define two clinically relevant forms of CD irrespective of tissue sampling location, patient age or treatment status.Free full text

Molecular classification of Crohn’s disease reveals two clinically relevant subtypes
Abstract
Objective
The clinical presentation and course of Crohn’s disease (CD) is highly variable. We sought to better understand the cellular and molecular mechanisms that guide this heterogeneity, and characterize the cellular processes associated with disease phenotypes.
Design
We examined both gene expression and gene regulation (chromatin accessibility) in non-inflamed colon tissue from a cohort of adult CD and control patients. To support the generality of our findings, we analyzed previously published expression data from a large cohort of treatment-naïve pediatric CD and control ileum.
Results
We found that adult CD patients clearly segregated into two classes based on colon tissue gene expression—one that largely resembled the normal colon and one where certain genes showed expression patterns normally specific to the ileum. These classes were supported by changes in gene regulatory profiles observed at the level of chromatin accessibility, reflective of a fundamental shift in underlying molecular phenotypes. Further, gene expression from the ilea of the treatment-naïve pediatric CD patient cohort could be similarly subdivided into colon- and ileum-like classes. Finally, expression patterns within these CD subclasses highlight large-scale differences in the immune response and aspects of cellular metabolism, and were associated with multiple clinical phenotypes describing disease behavior, including rectal disease and need for colectomy.
Conclusion
Our results strongly suggest that these molecular signatures define two clinically relevant forms of CD irrespective of tissue sampling location, patient age or treatment status.
INTRODUCTION
Crohn’s disease (CD) is a chronic heterogeneous inflammatory disorder with distinct patterns of clinical behavior. CD may present or evolve with time into a more complex phenotype with patients developing strictures, fistulae, and/or abscesses, and many patients experience highly variable response to therapies. Genetic associations[1, 2] and a recently defined lipid metabolism-based gene expression signature predictive of disease involvement[3] suggest that molecular or genetic factors are associated with and may contribute to disease heterogeneity, but precise mechanisms are poorly understood. Molecular subtypes defined by gene expression that impact clinical phenotypes have also been documented in other complex diseases, especially cancers[4, 5, 6]. Whether adult CD can be similarly separated into two or more subgroups and whether these molecular classes can explain disease phenotypes remains largely unknown.
Understanding how genes are regulated provides complementary information to gene expression. We and others have studied gene regulation by focusing on accessible chromatin, which allows transcriptional regulators to bind to the otherwise highly condensed nuclear genome. Chromatin accessibility has been associated with promoters, enhancers, silencers, and insulators[7], and changes as cellular identity is established through differentiation and development[8, 9] or in response to cellular stresses. Chromatin profiling provides a mechanism as to why expression is changing, and whether observed changes may be transient or persistent. We have shown that chromatin accessibility can differentiate disease subtypes[10] and helps to describe genetic and environmental contributors to disease[11]. We therefore sought to determine whether multiple clinically distinct subclasses of adult CD exist by examining both gene expression, using RNA-seq, and chromatin accessibility, using Formaldehyde-Assisted Isolation of Regulatory Elements (FAIRE-seq)[12], in CD and non-IBD patient samples of unaffected colon mucosa.
RESULTS
Whole genome interrogation of the colonic transcriptional and chromatin landscape reveals two distinct molecular classes in Crohn’s disease
To determine, in an unbiased manner, whether gene expression levels separated samples into distinct molecular groups, we performed a principal components analysis (PCA) using gene expression profiles from a combined set of 21 CD and 11 non-IBD patients. A striking clustering pattern emerged whereby the CD individuals were divided into two distinct, expression-based subclasses, one of which clustered with the non-IBD controls (Figure 1A). To specifically interrogate these two CD subclasses, we identified genes differentially expressed between these two groups of CD patients (849 genes at FDR<0.05; Figure 1B, see Supplementary Table 1 for top 20 differentially expressed genes in each CD subclass). Surprisingly, when looking at the top 25 differentially expressed genes regardless of direction, most had tissue-specific expression patterns that discriminated colon from the small intestine (ileum), including NXPE4, CWH43, and CA2 (colon-specific), as well as RBP2, TM6SF2, APOB, MTTP, CREB3L3, and CPS1 (ileum-specific)[13]. CD samples similar to non-IBD controls (Figure 1A) exhibited abundant expression of the above colon-specific genes, whereas the other CD subclass showed expression patterns more consistent with ileum despite being sampled from the colon. To explore this more globally, we compared all these differentially expressed genes to 947 genes with known significant differential expression between colon and ileum (Figure 1C)[14]. We found that 34% of the genes more highly expressed in the colon-like CD samples were indeed markers of normal colon, and 44% of ileum marker genes were more highly expressed in the ileum-like CD samples (p<1×10−95, hypergeometric test). To validate these expression differences, we performed RT-qPCR on 18 CD samples of unaffected colon mucosa (9 colon-like, 9 ileum-like) using CEACAM7 and APOA1 as a proxy for colon-like and ileum-like expression patterns (Supplementary Figure 1A). In agreement with the RNA-seq data, CEACAM7 was significantly more abundant in colon-like CD samples (p=0.017, one-sided t-test), whereas APOA1 was significantly more abundant in ileum-like CD samples (p=0.020, one-sided t-test).
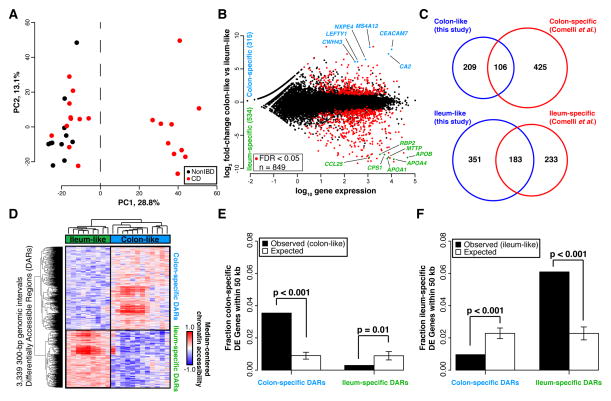
(A) PCA analysis of RNA-seq data from colon tissue from 21 CD and 11 non-IBD patients shows two distinct clusters. (B) 849 genes are differentially expressed between the two CD subclasses (adjusted p<0.05, DEseq), defined as colon-like and ileum-like. Known markers of colon and ileum are highlighted. (C) Genes upregulated in colon-like (top) and ileum-like (bottom) CD subclasses overlap previously defined colon-specific and ileum-specific genes, respectively. (D) Differentially accessible regions (DARs) identified using FAIRE-seq (p<0.05, two-sided t-test, normalized read counts, 300-bp windows) show distinct profiles in colon-like and ileum-like CD samples. (E) Colon-like-associated DARs are enriched and ileum-like-associated DARs are depleted near genes upregulated in colon-like samples (p≤0.01, permutation test). (F) Ileum-like-associated DARs are enriched and colon-like-associated DARs are depleted near genes upregulated in ileum-like samples (p<0.001, permutation test).
To determine whether these expression changes represented a fundamental shift in the functional cellular identity of these tissues, we investigated chromatin accessibility by performing FAIRE-seq[12] on the same samples from both CD subclasses. Supporting a fundamental shift in underlying molecular phenotypes, we identified 3,339 300-bp regions with significantly differential chromatin accessibility between colon-like and ileum-like CD samples (Figure 1D; p<0.05, two-sided t-test), hereafter referred to as Differentially Accessible Regions (DARs). These DARs could be divided into two classes based on greater accessibility in colon-like or ileum-like CD samples, and further, an unsupervised PCA of FAIRE-seq data nearly separated ileum-like from colon-like CD subclasses (Supplementary Figure 1C). Subclass-specific changes in the chromatin landscape corresponded strongly to differences in nearby (within 50 kb) gene expression (Figure 1E–F; p≤0.01, permutation). Additionally, both colon-specific and ileum-specific DARs exhibited a significant enrichment for CD GWAS SNP loci[15] compared to what was expected due to random chance (colon-specific p=0.018, ileum-specific p=0.006; permutation), suggestive that changes in chromatin accessibility occur at disease relevant regions of the genome.
We next sought to annotate these DARs based on tissue-specific gene regulatory information. Post-translational modifications on histone proteins serve to compartmentalize the genome and demarcate putative function of regulatory elements[16]. Using ChIP-seq data from the Roadmap Epigenomics Project[17], we assessed the enrichment of seven histone modifications reflective of underlying regulatory activity (active: H3K4me1, H3K4me3, H3K27ac, H3K36me3; repressive: H3K27me3, H3K9me3) around colon-specific and ileum-specific DARs. We found that colon-specific DARs were demarcated by H3K27ac and H3K4me1 modifications present in colon but not ileum (Supplementary Figure 1B), suggesting these DARs function as active regulatory regions only in the normal colon. In contrast, ileum-specific DARs demonstrated positive H3K27ac and H3K4me1 enrichment found only in normal small intestine, despite these samples originating from colon tissue. These suggest that regulatory activity in DARs contribute to the colon-like and ileum-like expression levels. To confirm regulatory activity, we cloned three DARs (two with colon-specific and one with ileum-specific chromatin accessibility) into luciferase vectors upstream of a minimal promoter in both orientations using THP-1 monocytes (Supplementary Figure 1D). Relative to empty vector controls, two DARs (associated with SATB2-AS1 and DEPDC7) exhibited a significant increase (p<0.01, one-sided t-test) in luciferase activity in both orientations, strongly suggestive of enhancer function. The third DAR (associated with SLC16A9) also enhanced luciferase activity significantly (p=8.9×10−5, one-sided t-test), however only in the reverse orientation.
Together, these data support the existence of two molecularly distinct subclasses of CD. Further, chromatin accessibility data suggests these subclasses exist due to stable molecular transformations of the genomic architecture in colon tissue cells, and not transient differences due to external cellular signaling.
Whole genome RNA-seq analysis reveals colon-like and ileum-like subclasses in treatment-naïve pediatric Crohn’s disease patients
Gene expression profiles in adult CD patients may vary due to patient treatment histories. Therefore, we sought to determine whether treatment-naïve pediatric CD patients also segregated into similar molecular classes. We performed PCA on previously published RNA-seq data from ileal biopsies in age-matched pediatric CD (n=201) and non-IBD (n=40) patients generated within the Pediatric Risk Stratification Study[3]. Though a clustering as distinct as with the adult samples was not observed, non-IBD ileum samples clustered with some CD samples along the first principal component, whereas the other CD samples were separate (Supplementary Figure 2). To determine whether this pattern was related to the adult CD molecular subtypes, we performed PCA on combined adult colon and pediatric ileum expression data (Figure 2A). Unsurprisingly, samples predominantly separated by tissue of origin (colon vs. ileum; first principal component). However, a separation indicative of two molecular subclasses was evident along the second principal component, and correlated nearly exactly with the first principal components in single cohort PCAs (Figure 1A, Supplementary Figure 2). Further, pediatric CD samples fell on a spectrum highly correlated with expression of APOA1, a marker gene of the ileum and indicator of disease outcome in the pediatric cohort[3]. This pattern aligned well with the ileum-like (APOA1-high) and colon-like (APOA1-low) subclasses we identified in the adult CD colon.
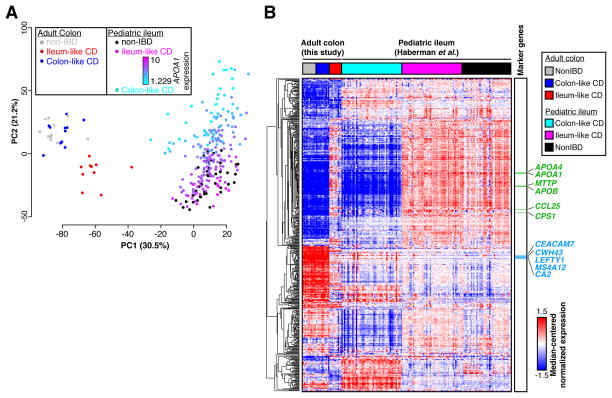
(A) PCA analysis of combined RNA-seq data from adult colon tissue and pediatric ileum tissue from CD and non-IBD patients shows separation of tissue types (PC1) and replicates ileum-like and colon-like clusters (PC2). Expression of APOA1 (blue-pink, low-high) in pediatric samples aligns well with subclasses. (B) Hierarchical clustering of RNA-seq data using 500 colon- and ileum-specific genes show clusters of genes associated with ileum-like and colon-like samples across both the adult colon and pediatric ileum cohorts, as well as genes associated with tissue of origin.
To closely examine the relationship between the two CD subclasses across the adult and pediatric cohorts, we assessed gene expression patterns across the 500 most-variably-expressed known colon and ileum marker genes[14] using hierarchical clustering (Figure 2B). To focus this analysis, we selected the 50 pediatric ileum samples each that were most colon-like and most ileum-like based on the PCA (Figure 2A, second PC). Many of the colon and ileum representative genes described above (e.g. APOA1, CEACAM7, MTTP, LEFTY1, and CA2) exhibited highly consistent expression patterns across all samples in a defined molecular subclass, regardless of cohort. Interestingly, for these 500 genes, expression patterns were extremely consistent between colon-like CD and non-IBD colon samples, as well as between ileum-like CD and non-IBD ileum samples. Importantly, we note that a subset of genes differentiate all colon tissues from all ileum tissues indicating that tissue-of-origin-specific expression is not completely lost. Together, these data strongly suggest that the colon-like and ileum-like molecular signatures define two forms of CD present regardless of tissue sampling location, patient age, or treatment status.
Metabolic and immune activation gene expression profiles characterize distinct molecular phenotypes in adult and pediatric CD
Gene expression differences between colon-like and ileum-like subclasses in the adult and pediatric CD cohorts went beyond the marker genes described above (Figure 1B–C). To evaluate this more broadly, we computed and compared pathway-level expression patterns of both CD subclasses and non-IBD controls in both patient cohorts[18, 19]. We then grouped significantly altered pathways based on similarity in both gene composition and direction of expression difference (Figure 3, Supplementary Figures 3 and 4). First, numerous pathways related to interferon signaling, GPCR signaling, and antigen processing were significantly upregulated in CD patients as a whole relative to non-IBD controls in both cohorts (Supplementary Figure 3, “CD-enriched”, red). Given that the adult patient cohort consisted of disease-unaffected tissue, and the pediatric cohort was treatment-naïve, this suggests a basal activation of the immune system in CD. In contrast, many pathways related to the RNA processing, translation, and transcription were downregulated in CD in the pediatric patients (Supplementary Figure 3, “CD-depleted”, blue). These results strongly corroborate studies in mice and humans linking overall defects in cellular protein processing in CD to immune activation and unabated inflammation[20].
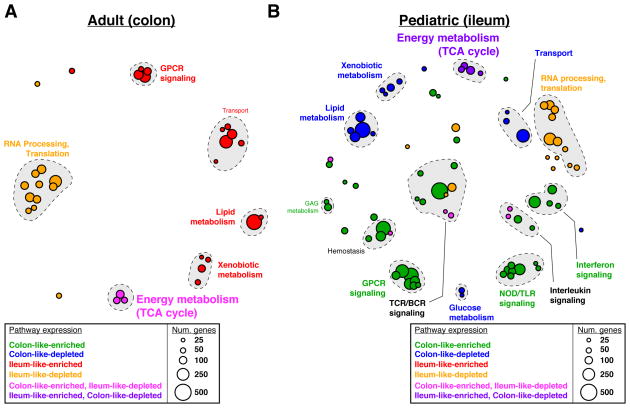
Pathway enrichments were determined using GSAA (FDR<0.1, permutation test) for all pairwise comparisons between all CD, colon-like CD, ileum-like CD, and non-IBD samples. Separate analyses in (A) adult colon and (B) pediatric ileum show similar and unique pathways associated with CD phenotypes. Each circle represents Reactome-defined pathway with the size reflecting the number of genes in the pathway. Pathways were grouped based on similarity in gene membership, and labels describing multi-pathway clusters are shown. See Supplementary Figures 3–4 for CD vs non-IBD comparisons and full list of pathways.
Next, we identified significantly altered pathways that described how the ileum-like and colon-like CD subclasses differed. Among the most pronounced effects in both cohorts reflected strong differences in metabolic activity including pathways involved in lipid metabolism and metabolism of foreign (xenobiotic) agents (Figure 3A; “Ileum-like-enriched”, red; Figure 3B; “Colon-like-depleted”; blue). Interestingly, energy production by way of the TCA cycle was significantly affected in opposing ways: in adults, it was upregulated in the colon-like class and simultaneously downregulated in the ileum-like class in colon tissue (Figure 3A; “Colon-like-enriched, Ileum-like-depleted”, pink); whereas in pediatric patients it was downregulated in the colon-like class and upregulated in the ileum-like class in ileal tissue (Figure 3B; “Ileum-like-enriched, Colon-like-depleted”, purple). This suggests that energy production increases in patients where the subtype is more similar to the tissue of origin, and decreases when gene expression adopts patterns of the opposite tissue. In addition, several pathways related to G-protein coupled receptor (GPCR) signaling were upregulated in the ileum-like subclass in colon tissue (Figure 3A; “Ileum-like-enriched”, red) and upregulated in the colon-like subclass in ileum tissue (Figure 3B; “Colon-like-enriched”, green). GPCRs are highly expressed in monocytes and macrophages central to the development and progression of inflammation in CD, mainly through migration and accumulation within the inflamed tissues[21].
Taken together, dysregulation of metabolic pathways may represent defining features of CD subtypes. Although dysregulation of lipid metabolism has been previously described in CD[3], our data suggest these alterations may be specific only to patients within a certain subclass and dependent on the tissue being assayed. Further, these data indicate that despite a striking similarity in expression of key ileum and colon marker genes (Figure 2B), there are key differences in pathway-level expression patterns between adult and pediatric CD patients, such as the immune response (e.g. NOD/TLR signaling, interleukin signaling), that point toward clinically relevant phenotypes and characteristics of each subclass.
Molecular subclasses are associated with clinical outcomes
To understand the impact of molecular phenotype on clinical disease, we studied the clinical characteristics of treatment-naïve pediatric patients with respect to molecular subclassification. We again used the 50 ileum-like and 50 colon-like pediatric samples defined above. We found that significantly more colon-like CD patients displayed both colon and ileum involvement (p=0.0014), had deep ulcers (p=0.0002), and showed macroscopic inflammation (p=0.0156), whereas more ileum-like patients showed no inflammation (p=0.0122) and a strong skew in prevalence towards colon-only involvement (p=0.0528; Table 1).
Table 1
Pediatric and adult patient phenotypes, segregated by colon-like and ileum-like classifications, were compared using Fisher’s Exact Tests (for categorical data) or two-sided t-tests (for continuous data). Significant associations (p<0.05) are bolded. See Supplementary Tables 2–3 for data on individual patients.
| Phenotype | Pediatric (ileum) | Adult (colon) | ||||
|---|---|---|---|---|---|---|
|
| ||||||
| Ileum-like (n = 50) | Colon-like (n = 50) | p-value | Ileum-like (n = 10) | Colon-like (n = 11) | p-value | |
| Location | ||||||
|
| ||||||
| Ileum-only | 10 | 10 | 1 | 2 | 0 | 0.21 |
| Colon-only | 21 | 11 | 0.0528 | 1 | 6 | 0.06 |
| Ileum+Colon | 19 | 29 | 0.0014 | 7 | 5 | 0.39 |
|
| ||||||
| Patient characteristics | ||||||
|
| ||||||
| Mean age | 12.1 | 12.4 | 0.6193 | 35.1 | 35.3 | 0.98 |
| Male | 28 | 29 | 1 | 3 | 3 | 1 |
| Female | 22 | 21 | 1 | 7 | 8 | 1 |
| Smoker | NA | NA | NA | 5 | 4 | 0.67 |
|
| ||||||
| Inflammation | ||||||
|
| ||||||
| Macroscopic | 29 | 41 | 0.0156 | NA | NA | NA |
| Microscopic | 8 | 6 | 0.7742 | NA | NA | NA |
| None | 13 | 3 | 0.0122 | NA | NA | NA |
|
| ||||||
| Phenotypes and involvement | ||||||
|
| ||||||
| Deep ulcers | 10 | 29 | 0.0002 | NA | NA | NA |
| Perianal | NA | NA | NA | 0 | 4 | 0.09 |
| Sigmoid | NA | NA | NA | 2 | 7 | 0.08 |
| Rectal | NA | NA | NA | 0 | 9 | <0.01 |
| Ileal Disease | 29 | 39 | 0.0528 | 9 | 5 | 0.06 |
 Inflammatory Inflammatory | NA | NA | NA | 1 | 4 | 0.31 |
 Stricturing Stricturing | NA | NA | NA | 7 | 4 | 0.2 |
 Penetrating Penetrating | NA | NA | NA | 5 | 4 | 0.67 |
|
| ||||||
| Pre-operative treatment history | ||||||
|
| ||||||
| Steroids | NA | NA | NA | 8 | 10 | 0.59 |
| 5-ASA | NA | NA | NA | 8 | 10 | 0.59 |
| Immunomodulation | NA | NA | NA | 8 | 8 | 1 |
| Anti-TNF | NA | NA | NA | 7 | 10 | 0.31 |
| Non-anti-TNF biologic | NA | NA | NA | 0 | 2 | 0.48 |
|
| ||||||
| Post-operative outcome | ||||||
|
| ||||||
| Post-operative disease recurrence | NA | NA | NA | =2/8 | =0/10 | 0.18 |
| Post-operative biologic use | NA | NA | NA | =6/9 | =1/11 | 0.02 |
| Colectomy | NA | NA | NA | 0 | 6 | 0.01 |
| Second resection | NA | NA | NA | 3 | 6 | 0.39 |
| Median Time to first resection (years) | NA | NA | NA | 3 | 5 | 0.91 |
| Median Time from first to second resection (years, if applicable) | NA | NA | NA | 3.5 | 2 | 0.33 |
We next performed an in-depth retrospective chart review and a prospective follow-up of all adult CD patients (Table 1; Supplementary Table 1) to see if a similar clinical impact of molecular subclass was observed. Patients with colon-like CD had greater rectal disease involvement (p<0.01) and were more likely to eventually need a colectomy (p=0.01). Patients with ileum-like CD tended to have ileal disease (p=0.06) and required the use of post-operative biological therapy (p=0.02). Though our sample size is small, these data suggest that molecular subtypes of CD can stratify patients into clinically distinct and relevant subgroups, and may prospectively identify those more likely to require intensive medical therapy.
DISCUSSION
We identified two distinct molecular phenotypes in colon tissue obtained from adult CD patients. When we applied the same analytical approach to pediatric RNA-seq data from ileum, two molecular phenotypes also emerged, albeit the clinical consequences in both circumstances were relevant to the population of study and limited to the data collected for each population in terms of clinical phenotype. Although the adult patient sample size was limited, in only the colon-like CD subclass were there patients with rectal disease, or that required a colectomy. Rectal CD is particularly difficult to manage, and though it may represent a unique CD phenotype, its underlying molecular mechanisms are unknown[22, 23, 24]. Pediatric patients with colon-like disease were more likely to have macroscopic inflammation, deep ulcers, and involvement of both the ileum and colon. These data emphasize the need to continue and expand these studies over time to incorporate the evolving clinical phenotype in both adult and pediatric patients, and the need to study both tissues in the same patient. Only through such studies will we uncover whether the tissue of origin dictates potential clinical phenotype similarly.
The ileum-like CD subclass was primarily characterized by an upregulation of pathways involved in lipid and xenobiotic metabolism. The mechanistic link between these clinical phenotypes and the lipid metabolic signature defined here is yet to be explored, however the implication that lipid metabolism may be involved is interesting for several reasons. First, lipid metabolism and altered levels of certain lipids have been previously associated with IBD (reviewed in [25]), and may be linked to inflammation state and immune signaling (reviewed in [26]), in addition to diet and intestinal microbiota composition[27]. Lipid levels may also be a cause or result of increases in intestinal oxidative stress, which has also been shown to be elevated in CD patients (reviewed in [28]). Notably, two of the top differentially expressed genes between CD subclasses were in fact GSTA1 and GSTA2, key mediators of the oxidative stress response[29]. Furthermore, diet, cholesterol, and microbiota composition have each been studied either themselves as potential therapies or how their levels change as a result of surgery or biologic use (reviewed in [25]).
The diversity in cellular state demonstrated at both the transcriptional and chromatin levels has important potential therapeutic implications. Our results suggest that additional testing of CD patients for particular molecular signatures, especially metabolic pathways, may determine potential therapeutic subgroups. This approach has already increased our understanding of human cancer biology. Patient sub-classification in various forms of cancer, most notably invasive breast tumors[4, 5], has led to numerous associations with clinical outcome and helped to shape future treatment strategies. There is growing consensus that subtypes exist in CD as well, each with its own presentation, genetic makeup, and prognosis. As a first step, molecular stratifications of archived patient tissue and serum from major clinical trials could be performed in the context of response to biologic and microbial therapies for CD[30].
Our current studies did not allow for the investigation of both intestinal regions from the same individual, but they provide significant motivation for future exploration of these molecular subclasses longitudinally in larger cohorts of matched colon and ileum tissue of the same patient. New larger studies should allow for a more complete understanding of the molecular effects of host-environment interactions on disease, and its utility in guiding clinical decisions. In addition, our sample numbers here are too limited to study the effects of genetic variation on identified changes in chromatin accessibility and gene expression. Our chromatin accessibility data strongly suggested that changes occurred at genomic loci previously associated with disease through genetic variation. Future studies with larger sample sizes will help identify specific relationships between genetic disease predisposition, regulatory activity, gene expression levels and clinical phenotypes and to better characterize individual-level disease sub-phenotypes within this very heterogeneous disease and tissue. Genomic studies on composite cell types (e.g. immune and epithelial cells) may also become necessary to study cell specific mechanisms driving phenotype specific disease pathogenesis.
MATERIALS AND METHODS
For more detailed information, see the Supplemental Methods. All processed sequencing data are available in GEO under accession GSE85499 with raw sequence data available through dbGaP. Full data tables are also hosted at http://fureylab.web.unc.edu/datasets/crohns-disease-molecular-subtypes/.
Statistics
Differential gene expression was detected using DEseq with an adjusted p-value threshold of 0.05. Differential chromatin accessibility was detected using a 2-sided t-test with p-value threshold of 0.05. Pathway enrichments were determined using GSAA (FDR<0.1, permutation test) for all pairwise comparisons between all CD, colon-like CD, ileum-like CD, and non-IBD samples based on differential gene expression p-values calculated using 2-sided t-tests. Clinical phenotype associations were tested with 2-sided t-tests or Fisher’s Exact Tests.
Study Approval
All procedures were approved under University of North Carolina IRB protocols 10-0355, 14-2445 and 11-0359.
Supplementary Material
Sup Fig 1
Sup Fig 2
Sup Fig 3
Sup Fig 4
Sup Table 1
Sup Table 2
Acknowledgments
We thank B. Aronow and L. Denson for assistance with pediatric patient data (CCFA RISK cohort). This work was supported by American Gastroenterological Association Research Scholar Award (SZS), Broad Medical Research Program (SZS), Crohn’s and Colitis Foundation of America’s Career Development Award (SZS), and Microbiome Consortium (RBS, DPBM), R01-ES024983 (SZS, TSF), UNC Team Translational Science Award (SZS, TSF), R01-DK094779 (RBS, SZS), P30-DK034987 (RBS, SZS), U01-DK062413, P01-DK046763 (DPBM), Helmsley Trust SHARE 2, Project 3 (SZS, RBS, DPBM), and T32-DK007634 (BK).
Abbreviations
| CD | Crohn’s disease |
| PCA | principal components analysis |
| FAIRE | formaldehyde-assisted isolation of regulatory elements |
| DARs | differentially accessible regions |
Footnotes
COMPETING INTERESTS
The authors have declared that no competing interests exist
AUTHOR CONTRIBUTIONS
MW, JMS, BK, AT, AR, GRG, MSS, and DPBM acquired data. MW, JMS, and JWI analyzed and interpreted data. MW and JMS prepared figures, drafted and revised the manuscript. RBS, HHH, RR, TS and MJK provided help with tissue acquisition and patient phenotyping. SZS and TSF designed and supervised the study, acquired, analyzed and interpreted the data, drafted and revised the manuscript, and obtained funding. SZS conceptualized the study and acted as study sponsor. All authors uphold the integrity of the work, approved the manuscript in its entirety, and are accountable for all aspects of the work.References
Full text links
Read article at publisher's site: https://doi.org/10.1136/gutjnl-2016-312518
Read article for free, from open access legal sources, via Unpaywall:
https://gut.bmj.com/content/gutjnl/67/1/36.full.pdf
Citations & impact
Impact metrics
Citations of article over time
Alternative metrics
Smart citations by scite.ai
Explore citation contexts and check if this article has been
supported or disputed.
https://scite.ai/reports/10.1136/gutjnl-2016-312518
Article citations
Ileal Crohn's Disease Exhibits Reduced Activity of Phospholipase C-β3-Dependent Wnt/β-Catenin Signaling Pathway.
Cells, 13(11):986, 05 Jun 2024
Cited by: 0 articles | PMID: 38891118 | PMCID: PMC11171731
Identification of Key Disulfidptosis-Related Genes and Their Association with Gene Expression Subtypes in Crohn's Disease.
J Inflamm Res, 17:3655-3670, 07 Jun 2024
Cited by: 0 articles | PMID: 38863903
Catalase inhibition can modulate the ability of peripheral blood T cells to undergo apoptosis in Crohn's disease.
Clin Exp Immunol, 217(1):45-56, 01 Jun 2024
Cited by: 1 article | PMID: 38247555
The molecular subtypes of autoimmune diseases.
Comput Struct Biotechnol J, 23:1348-1363, 28 Mar 2024
Cited by: 0 articles | PMID: 38596313 | PMCID: PMC11001648
Review Free full text in Europe PMC
A MTA2-SATB2 chromatin complex restrains colonic plasticity toward small intestine by retaining HNF4A at colonic chromatin.
Nat Commun, 15(1):3595, 27 Apr 2024
Cited by: 2 articles | PMID: 38678016 | PMCID: PMC11055869
Go to all (58) article citations
Data
Data behind the article
This data has been text mined from the article, or deposited into data resources.
BioStudies: supplemental material and supporting data
GEO - Gene Expression Omnibus
- (1 citation) GEO - GSE85499
Similar Articles
To arrive at the top five similar articles we use a word-weighted algorithm to compare words from the Title and Abstract of each citation.
Colonic epithelial miR-31 associates with the development of Crohn's phenotypes.
JCI Insight, 3(19):122788, 04 Oct 2018
Cited by: 14 articles | PMID: 30282822 | PMCID: PMC6237459
An ileal Crohn's disease gene signature based on whole human genome expression profiles of disease unaffected ileal mucosal biopsies.
PLoS One, 7(5):e37139, 14 May 2012
Cited by: 27 articles | PMID: 22606341 | PMCID: PMC3351422
Differential miRNA Expression in Ileal and Colonic Tissues Reveals an Altered Immunoregulatory Molecular Profile in Individuals With Crohn's Disease versus Healthy Subjects.
J Crohns Colitis, 13(11):1459-1469, 01 Oct 2019
Cited by: 17 articles | PMID: 31001642 | PMCID: PMC6821350
Redefining the IBDs using genome-scale molecular phenotyping.
Nat Rev Gastroenterol Hepatol, 16(5):296-311, 01 May 2019
Cited by: 38 articles | PMID: 30787446 | PMCID: PMC8311467
Review Free full text in Europe PMC
Funding
Funders who supported this work.
NCI NIH HHS (1)
Grant ID: P30 CA016086
NIDDK NIH HHS (6)
Grant ID: P30 DK034987
Grant ID: P01 DK046763
Grant ID: R01 DK104828
Grant ID: T32 DK007634
Grant ID: U01 DK062413
Grant ID: P01 DK094779
NIEHS NIH HHS (1)
Grant ID: R01 ES024983
National Institute of Diabetes and Digestive and Kidney Diseases (5)
Grant ID: P30-DK034987
Grant ID: R01-DK094779
Grant ID: T32-DK007634
Grant ID: U01-DK062413
Grant ID: P01-DK046763
National Institute of Environmental Health Sciences (1)
Grant ID: R01-ES024983





