Abstract
Free full text

Non-cell-autonomous Effects of Autophagy Inhibition in Tumor Cells Promote Growth of Drug-resistant Cells
Abstract
Autophagy, the mechanism by which cells deliver material to the lysosome, has been associated with resistance to anticancer drugs, leading autophagy inhibition to be widely studied as a potential chemosensitization strategy for cancer cells. This strategy is based on the idea that inhibition of autophagy will increase drug sensitivity and kill more cancer cells. Here we report an unintended negative effect of this strategy. When modeling the effect of drug resistance in a heterogeneous cancer cell population, we found that autophagy inhibition in drug-sensitive tumor cells causes increased growth of drug-resistant cells in the population through a mechanism involving caspase activation and prostaglandin E2 signaling. These results emphasize the importance of understanding how autophagy manipulation in a tumor cell can have both cell-autonomous and nonautonomous effects and suggest that attempts to chemosensitize by inhibiting autophagy could be enhanced by adopting methods aimed at reducing tumor repopulation.
Introduction
Macroautophagy (hereafter referred to as autophagy) is a mechanism whereby cellular material is engulfed in double membrane vesicles that fuse with lysosomes, resulting in the degradation of the engulfed material and recycling of macromolecular precursors. Autophagy has been widely studied in cancer where it is thought to have context-dependent roles that sometimes promote and sometimes suppress cancer (White, 2012; Galluzzi et al., 2015). Autophagy manipulation (induced or inhibited autophagy) is of potential value in many diseases (Rubinsztein et al., 2012). However, most current clinical studies that aim to target autophagy are in cancer and focus on autophagy inhibition (Thorburn et al., 2014a; Kroemer, 2015). These studies all use lysosomal inhibitors of autophagy, chloroquine or hydroxychloroquine, in combination with another anticancer drug (Barnard et al., 2014; Mahalingam et al., 2014; Rangwala et al., 2014a,b; Rosenfeld et al., 2014; Vogl et al., 2014). The rationale for this approach is that autophagy inhibition will increase drug sensitivity in the tumor cells. This idea is based on in vitro and preclinical data showing chemosensitization effects by autophagy inhibition for many different classes of cancer drugs (Thorburn et al., 2014a). Caution is warranted with this interpretation when only pharmacological approaches are used to inhibit autophagy, because chloroquine can chemosensitize and have anticancer effects by autophagy-independent mechanisms as well (Maycotte et al., 2012; Eng et al., 2016). Nevertheless, a wealth of evidence using genetic, as well as pharmacological inhibition, of autophagy supports the idea that autophagy inhibition can increase cancer cell sensitivity to toxic insults and specifically anticancer drugs.
Intrinsic or acquired drug resistance is a major problem in cancer therapy (Holohan et al., 2013), but the mechanisms that control growth of drug-resistant tumor cells are poorly understood. It is known that tumor cells that are killed by an anticancer treatment can affect tumor repopulation by drug-resistant cells that were not killed by the treatment. For example, apoptotic cells can promote growth of neighboring cells to promote tissue regeneration (Li et al., 2010). This pathway, which involves caspase-3 activation, leads to increased prostaglandin E2 (PGE2) signaling and can promote tumor repopulation by cancer cells that were not killed by anticancer treatments (Huang et al., 2011). Similarly, secretome-dependent signals from drug-treated tumor cells can promote drug resistance and tumor progression/metastasis (Obenauf et al., 2015) and PGE2-dependent signaling from dying tumor cells can stimulate cancer stem cell-mediated tumor repopulation (Kurtova et al., 2015). Senescence-associated secretion also leads to non-cell-autonomous effects on neighboring cells and is linked with autophagy (Gewirtz, 2014), and recent studies show that non-cell autonomous effects of autophagy in tumor stroma promotes growth of pancreas tumors through autophagic secretion of alanine (Sousa et al., 2016). This raises an important question—does autophagy inhibition that is aimed at increasing sensitivity to anticancer drugs have effects on drug-resistant cells in the population through non-cell-autonomous mechanisms? To address this question, we modeled the effects of autophagy inhibition in drug-sensitive tumor cells in a mixed population of drug-resistant and drug-sensitive tumor cells and followed the effects on growth of the resistant cells. We found that selective inhibition of autophagy in drug-sensitive cells can increase the growth of drug-resistant cells through caspase and PGE2 signaling.
Materials and Methods
Cell Culture and Reagents.
Mouse glioblastoma cell line GL261 was obtained from National Cancer Institute (Frederick, MD) and maintained at 37°C under 5% CO2 in Dulbecco’s modified Eagle medium (Corning, Corning, NY) supplemented with 10% fetal bovine serum. Where indicated doxycycline was obtained from Clontech Laboratories (Mountain View, CA, cat. no. 631311) and used at a final concentration of 2 µg/ml and replenished every 24 hours. Human breast cancer cell line MDA-MB-231 was maintained at 37°C under 5% CO2 in Dulbecco’s modified Eagle medium:nutrient mixture F-12 (Ham) (Gibco, cat. no.11320082) supplemented with 10% fetal bovine serum. Human BJAB, an EBV-negative B-cell lymphoma line cell line, was previously provided by Marcus Peter. The cells were maintained at 37°C under 5% CO2 in RPMI1640 medium (Corning, cat. no. 10-040-CV) supplemented with 10% fetal bovine serum.
Matched Drug-sensitive and -resistant Cell Lines.
Construction of the GL261-inducible shATG12 cell line was previously described (Macintosh et al., 2012). GL261 cells do not express the epidermal growth factor receptor (EGFR) and are therefore insensitive to diphtheria toxin-EGF (DT-EGF)-induced cell killing (Liu et al., 2003; Thorburn et al., 2009). To create DT-EGF-sensitive GL261 cells, pBABE-puro human EGFR was constructed using SalI and SnaBI double digestion. Retrovirus particles were produced by cotransfecting GP2-293 cells (Clontech Laboratories) pBABE-puro human EGFR and vesicular stomatitis G protein using TransIT-LT1 (Mirus Bio LLC, Madison, WI) as previously described (Thorburn et al., 2014b). The GL261inducible shATG12 cells were transduced with retrovirus using 8 µg/ml polybrene and maintained with 0.5 µg/ml puromycin. To label DT-EGF-resistant cells, the parental GL261 cell line (DT-EGF resistant) was transduced with pLJM1-GFP-3xNLS-blasticidin lentivirus using 8 µg/ml polybrene and selected with 20 µg/ml blasticidin. After selection, the cells were flow-sorted for the brightest cells (top 15%). MDA-MB-231-wt [sensitive to human tumor necrosis factor-related apoptosis-inducing ligand (hTRAIL)] and MDA-MB-231TRAIL-R (resistant to hTRAIL) matched cell lines were developed through long-term exposure to increasing concentrations of hTRAIL (L. Dimberg et.al., submitted manuscript). The MDA-MB-231TRAIL-R cells were transduced with pLJM1-GFP-3xNLS-puromycin lentivirus using 8 µg/ml polybrene and selected with 1 µg/ml puromycin. After selection, the cells were flow-sorted for the brightest cells (top 15%). The parental BJAB cell line (sensitive to hTRAIL) and the BJAB LexR cell line (resistant to hTRAIL) were previously described (Menke et al., 2011). For quantitation of the resistant cells, BJAB LexR cells were transduced with pMSCV luciferase PGK-hygromycin retrovirus produced as described above using 8 µg/ml polybrene and selected with 400 µg/ml hygromycin. After selection, the cells were used as a polyclonal population. (BJABLexR-luc cell line).
Chemicals and Reagents.
hTRAIL/TNFSF10 was obtained from R & D Systems (Minneapolis, MN) and used at a final concentration of 150 ng/ml. DT-EGF was synthesized by Dr. A. Frankel and used at the following final concentrations: 0, 50, 100, 150 ng/ml. Rabbit anti-ATG5 (D5F5U) (cat. no. 12994; 1:1000), rabbit anti-ATG7 (cat. no. 2631; 1:1000), and horseradish peroxidase (HRP)-conjugated anti-mouse- and anti-rabbit-HRP secondary antibodies (cat. no. 7076 and cat. no. 7074 respectively; 1:1000) were obtained from Cell Signaling (Beverley, MO). Rabbit anti-LC3B/MAP1LC3B (cat. no. NB100-2220; 1:2500) antibody was obtained from Novus Biologicals, Littleton, CO. Mouse anti-β-actin (cat. no. A5441; 1:10,000) was obtained from Sigma Aldrich, St. Louis, MO. Chloroquine (cat. no. 193919; 40 µM final concentration) was obtained from MP Biomedicals, Solon, OH. QVD-OPh (cat. no. A1901; 20 µM final concentration) was obtained from ApexBio, Houston, TX. Prostaglandin E2 (2B5) monoclonal antibody (cat. no. 10009814; 100 nM final concentration) was obtained from Cayman Chemical, Ann Arbor, MI. Molecular Probes CellEvent Caspase 3/7 Green Detection Reagent (cat. no. C10423; 5 µM final concentration) was obtained through ThermoFisher Scientific (Waltham, MA). BD BBL Gram Crystal Violet (cat. no. B12525) was obtained through Fisher Scientific, Denver, CO. Plasmid pMSCV luciferase PGK-hygro (cat. no. 187820) was obtained from Addgene, Cambridge, MA. Hexadimethrine bromide (Polybrene) (cat. no. H9268) was obtained from (Sigma Aldrich). One-Glo Luciferase Assay Reagent (cat. no. E6110) was obtained from Promega (Madison, WI).
shRNA Knockdowns and IncuCyte Cell Growth Assays.
MDA-MB-231-wt cells (TRAIL sensitive) were plated at ~60,000 cells per 6 cm dish in 4 ml of complete medium. Twenty- to twenty-four hours postplating, the medium was changed for fresh medium containing polybrene at a final concentration of 8 µg/ml. The cells were incubated for 1–2 hours. For shRNA knockdowns, pLKO.1 shRNA lentivirus constructs were obtained from the University of Colorado Cancer Center Functional Genomics Shared Resource: pLKO.1 human shATG5: Target sequence: CCTTTCATTCAGAAGCTGTTT; ID TRCN0000151474. pLKO.1 human shATG7: Target sequence: CCCAGCTATTGGAACACTGTA; ID TRCN0000007587. Lentiviral particles were produced by co-transfecting HEK293FT cells (Clontech, Mountain View, CA) with the structural plasmids Rev, RRE, and vesicular stomatitis G protein along with pKLO.1 shRNA lentivirus, pLJMGFP-3xNLS-blasticidin, and pLJMGFP-3xNLS-puromycin constructs using TransIT-LT1. Media containing viral particles were collected at 48 and 72 hours posttransfection, pooled, and spun down, and supernatant was stored at −80°C. One milliliter of lentivirus targeting ATG5 or ATG7 (plus polybrene at a final concentration of 8 µg/ml) was added drop-wise to the appropriate dish and gently mixed and cells incubated overnight. After removing the virus-containing medium, the cells were rinsed with 1× PBS, and fresh medium added with puromycin at a concentration of 1 µg/ml for selection. Forty-eight hours posttransduction, the cells were replated in 96-well plates (Corning, cat. no. 3585) at 4000 cells per well either alone or together with 100 GFP-labeled, MDA-MB-231-TRAIL-R cells in 100 µl of medium containing puromycin at a final concentration of 0.5 µg/ml. Eighteen hours later, the medium was changed for fresh medium without puromycin (90 µl). hTRAIL at a final concentration of 150 ng/ml was delivered in 10 µl of medium to the appropriate wells. Alternatively, 10 µl of medium was added to the appropriate wells without hTRAIL. Immediately after treatment, the GFP-expressing resistant cells were imaged in an IncuCyte Zoom at time 0 and then every 12 hours using a 4× objective. Imaging continued for up to 192 hours. Each experiment shown was performed with six technical replicates and repeated at least three times (biologic replicates) with similar results.
GL261ind shATG12-hEGFR cells (drug sensitive) were plated at ~300,000 cells in 10-cm dishes in medium supplemented with 10% Tet System Approved FBS obtained from Clontech (cat. no. 631106) and 2 µg/ml doxycycline. Doxycycline was refreshed every 24 hours. Sixty to seventy-two hours postplating, the cells were replated into 96-well plates (Corning, cat. no. 3585) at 4000 cells per well either alone or together with 100 GL261-GFP cells (drug resistant) in 100 µl of conditioned medium. Eighteen to twenty-four hours later, the medium was changed for fresh conditioned medium (90 µl). Doses of DT-EGF were delivered in 10 µl of conditioned medium. Alternatively, where appropriate, DMSO, QVD-OPh, and anti-PGE2 antibody were delivered in 5 µl of conditioned medium and incubated for ~0.5 hours before the addition of doses of DT-EGF also in 5 µl of conditioned medium. Caspase 3/7 activity assays using CellEvent Caspase 3/7 were performed following the manufacturer’s instructions. Immediately after treatment, the cells were imaged in an IncuCyte Zoom (Essen Bioscience, Ann Arbor, MI) to monitor growth rate of drug-resistant cells at time 0 and then every 12 hours using a 4× objective. After 24 hours of imaging doxycycline and where appropriate, DMSO, caspase inhibitor, QVD-OPh, and anti-PGE2 antibody were refreshed. Imaging continued for up to 192 hours. Live cell content was determined by using the custom parameters for measuring fluorescence. Each IncuCyte experiment shown was performed with six technical replicates (data points shown are mean ± S.D.) and repeated at least three times (biologic replicates) that gave similar results.
BJAB wt cells (TRAIL sensitive) were plated at ~50,000 cells per 6 cm dish in 1.5 ml of medium containing polybrene at a final concentration of 8 µg/ml. The cells were incubated for 1–2 hours. One milliliter of lentivirus plus polybrene (at a final concentration of 8 µg/ml) was added drop-wise to the appropriate dish and gently mixed. The cells were incubated overnight. Early in the morning, the cells were transferred to 1–3 10-cm dishes containing fresh medium with puromycin at a concentration of 0.5 µg/ml for selection. The cells were selected for a minimum of 7 days. The cells were plated in 12-well plates (Corning, cat. no. 3512) at 40,000 cells per well (98.75%) either alone or together with 500 BJABLexR-luc cells (TRAIL-resistant cells) (1.25%) in 800 µl of medium without puromycin. hTRAIL at a final concentration of 150 ng/ml was delivered in 200 μl of medium to the appropriate wells. The cells were allowed to grow between 10 and 12 days, and a luciferase assay was performed. Each experiment shown was performed with three technical replicates and repeated at least three times (biologic replicates) with similar results.
Clonogenic Growth Assays.
After doxycycline-induced shATG12 knockdown, described above, the GL261-inducible shATG12-hEGFR cells (drug-sensitive cells) were also replated into 12-well plates (Corning cat. no. 3512) at 20,000 cells per well either alone or together with 500 GL261-GFP cells (drug-resistant cells) in 1 ml of conditioned medium. Eighteen to twenty-four hours later the medium was changed for fresh conditioned medium (1 ml). Doses of DT-EGF were delivered in 1 µl of conditioned medium. Doxycycline was refreshed after 24 hours post-drug treatment. Seven days posttreatment, the cells were fixed with 10% MeOH, 10% glacial acetic acid in dH2O, and stained with crystal violet. The crystal violet was resolubilized in 30% glacial acetic acid and analyzed at 540 nM using a Benchmark Plus microplate reader (Bio-Rad Laboratories, Inc., Hercules, CA). Experiments were performed in triplicate and repeated at least three times.
Autophagic Flux Assays.
The GL261ind shATG12-hEGFR cells and the MDA-MB-231-wt cells were plated into 6-well plates (Corning, cat. no. 3506; and Greiner Bio-one, cat. no. 657160) at 120,000 cells per well in 2 ml of conditioned medium. Eighteen to twenty-four hours later, the medium was changed for fresh conditioned medium. The cells were treated -/+ chloroquine as indicated at a final concentration of 40 µM for 2.5 hours to block autophagic flux and then harvested for Western blot analysis to confirm ATG gene knockdown and a reduction of LC3 II processing. Total cell lysates were prepared in stringent RIPA buffer plus Roche Protease Inhibitor Cocktail purchased through Sigma Aldrich. Proteins were separated on 10% and 12% SDS-PAGE gels and transferred onto PVDF membranes. The membranes were probed with antibodies that recognize ATG5, ATG7, LC3, and β-actin. Proteins were visualized using EMD Millipore Immobilon Western Chemiluminescent HRP Substrate (ECL) purchased through Fisher Scientific, Denver, CO (cat. no. WBKLS0500) and exposed to X-ray film or analyzed on an Odyssey Fc imaging system (Li-Cor, Lincoln, NE).
Luciferase Assays.
Cell suspensions were collected in Eppendorf tubes, spun down for 5 minutes at 1500 rpm, and the cell pellet resuspended in 500 µl of medium. The luciferase assay was performed as per the manufacturer's instructions using a Turner Biosystems Microplate Reader.
Results
Growth of drug-resistance subsets of tumor cells after killing of drug-sensitive tumor cells can explain how tumors become resistant to treatment. We were interested to test if autophagy inhibition, which is being tested in the clinic as a chemosensitization strategy as a means to improve cancer therapy, could affect the behavior of drug-resistant cells. Because cancer cells can show both growth inhibitory and stimulatory effects to autophagy inhibition (Guo et al., 2011; Maycotte et al., 2014), treatment with pharmacological autophagy inhibitors that would affect both drug sensitive and resistant cells equally is unable to test rigorously for non-cell-autonomous effects of autophagy in a heterogeneous tumor cell population. We previously reported that autophagy inhibition sensitizes cancer cells to TRAIL-induced apoptosis by making it easier for the cancer cells to activate caspases after mitochondrial membrane disruption (Thorburn et al., 2014b). Therefore, to model non-cell-autonomous effects of autophagy inhibition intended to chemosensitize tumor cells, we mixed isogenic tumor necrosis factor-related apoptosis-inducing ligand (TRAIL)-sensitive and TRAIL-resistant MDA-MB-231 breast tumor cells (Fig. 1) so that the great majority (97.5%) of cells were sensitive to TRAIL-induced apoptosis with only a small minority of cells that were previously derived to be resistant to the drug and that were also stably expressing nuclear GFP. We blocked autophagy in the TRAIL-sensitive cells (but not the resistant cells) using shRNAs targeted to two essential autophagy regulators (ATG5 and ATG7), treated with doses of TRAIL sufficient to kill all the sensitive cells (Fig. 1A), and monitored growth of the GFP-labeled resistant cells over several days (Fig. 1B). This strategy allows detection of non-cell-autonomous effects that specifically affect the resistant cells that were due to autophagy inhibition in the drug-sensitive cells. Figure 2, A–C shows that knockdown of ATG5 or ATG7 in the TRAIL-sensitive cells inhibited autophagic flux and caused an increase in the growth rate of the resistant cells. Experiments using a different isogenic TRAIL-sensitive and TRAIL-resistant BJAB cell line pair (Menke et al., 2011) showed a similar growth promoting effect in resistant cells when autophagy was inhibited by ATG knockdown in sensitive cells (Fig. 2D).
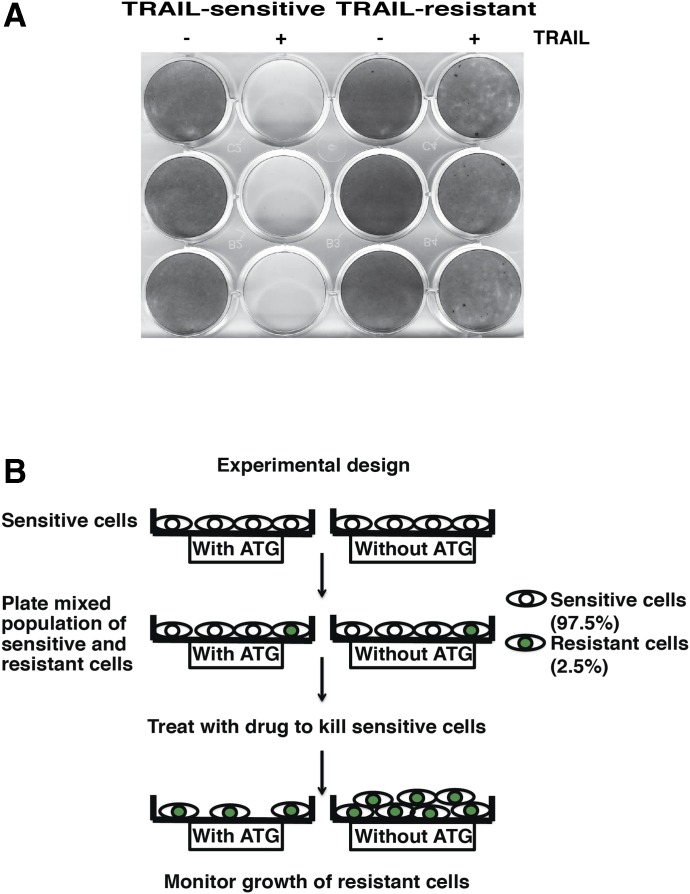
Characterization of matched TRAIL-sensitive and TRAIL-resistant MDA-MB-231 human breast cancer cells. (A) Clonogenic survival assay comparing MDA-MB-231-wt cells (TRAIL sensitive) and MDA-MB-231-TRAIL-R-GFP cells (TRAIL resistant) after TRAIL treatment. (B) Experimental design of resistant growth experiments. Drug-sensitive cells were plated at a density of 97.5% and the drug-resistant cells at a density of 2.5%, unless otherwise stated.

shATG5 and shATG7 knockdown in TRAIL-sensitive cells leads to increased growth of TRAIL-resistant cells. (A) Cell growth experiment in MDA-MB-231-TRAILR cells (TRAIL resistant) with control and ATG knockdown in MDA-MB-231 wt cells (TRAIL sensitive) after TRAIL treatment performed using the IncuCyte Zoom; knockdown of either ATG5 or ATG7 in TRAIL-sensitive cells promotes increased growth of the resistant cells in each of the experiments. Error bars represent S.D. (B) Western blot analysis of ATG gene knockdown. (C) Western blot analysis of LC3 II processing. (D) Cell growth experiment in BJABLexR-luc cells (TRAIL resistant) with control and ATG knockdown in BJAB wt cells (TRAIL sensitive) after TRAIL treatment performed using a luciferase-based assay system. Western blot analysis of ATG gene knockdown. Error bars represent S.D.
Caspase-mediated signaling through PGE2 can promote tumor repopulation after radiotherapy or chemotherapy (Huang et al., 2011). Because this mechanism is due to caspase activation and we previously found that autophagy inhibition can increase caspase activation in response to apoptotic stimuli like TRAIL (Thorburn et al., 2014b), we hypothesized that this mechanism is induced when autophagy is inhibited in drug-sensitive tumor cells. A test of this hypothesis requires that caspases be inhibited and effects on the growth of drug-resistant cells then determined. This cannot be done using TRAIL as an antitumor agent because the drug-sensitive cells would not die with caspase inhibition, and thus the experiment would be unable to discriminate between an effect that was caused by caspase signaling itself or by the death of the cell. Therefore, to test if caspase activity is responsible for the autophagy inhibition-mediated stimulation of drug-resistant cell growth, we need to kill drug-sensitive tumor cells using an agent that can activate caspase-dependent apoptosis that is stimulated by autophagy inhibition but will still result in drug-induced tumor cell death even if caspases are blocked. To do this, we used a diphtheria toxin-EGF fusion protein (DT-EGF) that kills epidermal growth factor receptor (EGFR)-expressing cells (Liu et al., 2003). We previously found that DT-EGF can kill glioma cells in both a caspase-dependent and caspase-independent manner. When glioma cells are treated with DT-EGF, they die primarily by a caspase-independent mechanism. However, upon autophagy inhibition, caspase activation is increased and the preferred mode of death is by caspase-dependent apoptosis (Thorburn et al., 2009). DT-EGF-sensitive, EGFR-expressing GL261 cells were killed irrespective of whether autophagy was inhibited by inducible knockdown of ATG12 and in the presence or absence of caspase inhibitor (Fig. 3A). As expected based on previous studies (Thorburn et al., 2009), autophagy inhibition caused an increase in caspase activity in response to DT-EGF (Fig. 3B). If a tumor repopulation mechanism like that previously described (Huang et al., 2011) is responsible for growth stimulation of resistant cells, caspase activity associated with autophagy inhibition in the DT-EGF-sensitive cells should promote growth of the DT-EGF-resistant cells; but because cell death occurs even if caspases are blocked (Fig. 3A), the caspase dependency of the growth stimulation as opposed to a growth effect caused by cell death itself can be determined. We therefore designed a similar mixing experiment with DT-EGF-sensitive cells where autophagy was inhibited and determined effects of autophagy inhibition on the growth of a small proportion of GFP-tagged DT-EGF-resistant GL261 cells. Figure 4 shows that as with the TRAIL-treated cells, autophagy inhibition in DT-EGF-sensitive cells, which causes increased caspase activity (Fig. 3B), also caused increased growth of the subpopulation of DT-EGF-resistant cells. Figures 5 and and66 show that both caspase inhibition and an anti-PGE2 blocking antibody that was previously shown to ablate PGE2-mediated cancer stem cell-mediated tumor growth (Kurtova et al., 2015) reduced the stimulation of drug-resistant cell growth caused by autophagy inhibition in drug-sensitive cells. In each case, Western blotting analysis of LC3-II levels from a sample of the same cells used for the growth assay showed that the ATG knockdown inhibited autophagic flux as measured by reduced accumulation of LC3-II in the presence of the lysosomal inhibitor chloroquine. These data indicate that it is not cell death itself that is responsible for the growth stimulation of the tumor cells that were not killed by the drug but rather the caspase activation and subsequent PGE2 signaling. Thus, although autophagy inhibition can enhance the apoptosis response to an anticancer agent, this can lead to increased growth of any drug-resistant cancer cells in the population through a caspase-dependent and PGE2-dependent mechanism similar to that previously identified (Huang et al., 2011).
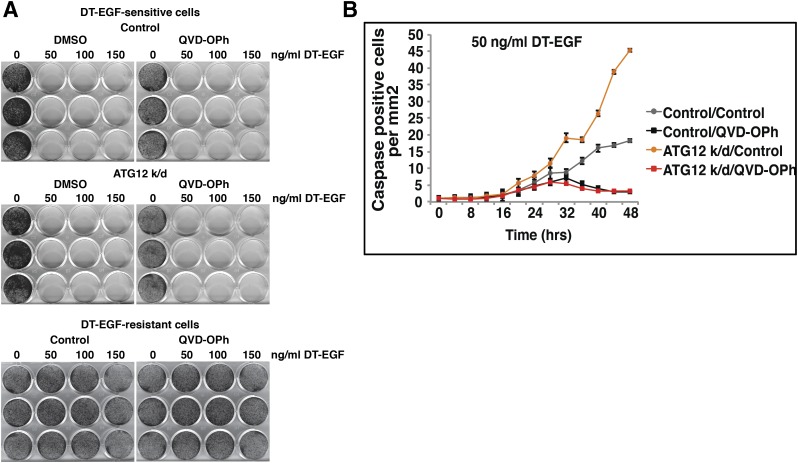
Characterization of matched DT-EGF-sensitive and DT-EGF-resistant GL261 glioblastoma cells. (A) Clonogenic survival assay comparing GL261inducible shATG12-hEGFR cells (DT-EGF sensitive) and GL261-GFP cells (DT-EGF resistant) in the presence and absence of QVD-OPh treated with DT-EGF. The DT-EGF-sensitive cells die in the presence or absence of the pan-caspase inhibitor QVD-OPh. GL261-GFP cells were DT-EGF-resistant. (B) Caspase 3/7 activity assays. shATG12 knockdown in the DT-EGF-sensitive cells leads to increased caspase 3/7 activity after DT-EGF treatment that is inhibited by the pan-caspase inhibitor QVD-OPh. Error bars represent S.D.
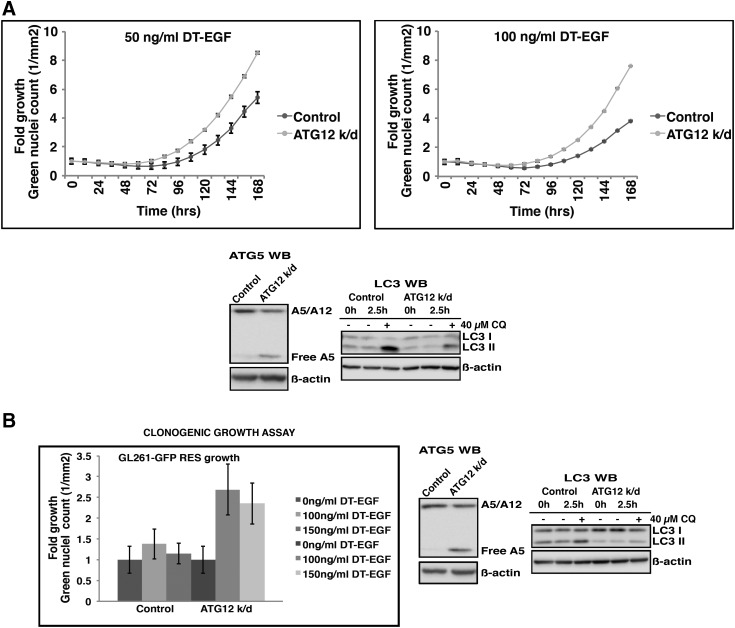
ATG12 knockdown in DT-EGF-sensitive leads to increased growth of the DT-EGF-resistant cells. (A) Cell growth experiment in GL261-GFP cells (DT-EGF resistant) with and without ATG12 knockdown in the GL261inducible shATG12-hEGFR cells (DT-EGF-sensitive) performed using the IncuCyte Zoom showing increased growth when autophagy is inhibited. Error bars represent S.D. Lower panel shows Western blot analysis of ATG gene knockdown and LC3 II processing, indicating reduced ATG5-ATG12 conjugate and inhibition of autophagic flux by doxycycline-induced ATG12 knockdown. (B) Clonogenic growth assay (left panel) . shATG12 knockdown in the GL261inducible shATG12-hEGFR cells (DT-EGF sensitive) leads to increased growth of the GL261-GFP cells (DT-EGF resistant) as in the IncuCyte experiments. Combined data from three separate experiments. Error bars represent S.D. Representative Western blot analysis (right panel) of ATG gene knockdown and LC3 II processing indicating reduced ATG5-ATG12 conjugate and inhibition of autophagic flux by doxycycline-induced ATG12 knockdown.

Increased growth of GL261-GFP cells (DT-EGF-resistant) induced by shATG12 knockdown in the GL261inducible shATG12-hEGFR cells (DT-EGF sensitive) is caspase-dependent. The pan-caspase inhibitor QVD-OPh inhibits the increased growth of DT-EGF-resistant cells induced by ATG12 knockdown. Error bars represent S.D. Lower panel shows Western blot analysis of ATG gene knockdown and LC3 II processing, demonstrating doxycycline-induced knockdown of ATG12 leading to reduced levels of ATG5-ATG12 conjugate and inhibition of autophagic flux.

Increased growth of GL261-GFP cells (DT-EGF resistant) induced by shATG12 knockdown in the GL261inducible shATG12-hEGFR cells (DT-EGF sensitive) is PGE2 dependent. Anti-PGE2 blocking antibody inhibits the increased growth. Error bars represent S.D. Lower panel shows Western blot analysis of ATG gene knockdown and LC3 II processing, demonstrating doxycycline-induced knockdown of ATG12 leading to reduced levels of ATG5-ATG12 conjugate and inhibition of autophagic flux.
Discussion
A large number of studies have demonstrated that inhibition of autophagy can sensitize tumor cells to anticancer drugs (Levy and Thorburn, 2011; Thorburn et al., 2014a; Rebecca and Amaravadi, 2016). This has led to dozens of clinical trials where autophagy inhibitors (so far, invariably chloroquine or hydroxychloroquine) are combined with other agents. An underlying rationale for these trials is that autophagy protects against tumor cell apoptosis. For example, autophagy inhibition causes an increase in the proapoptotic BH3 protein PUMA, which sensitizes tumor cells to TRAIL-induced apoptosis (Thorburn et al., 2014b). Our studies identify an unexpected negative effect of this approach because autophagy inhibition in drug-sensitive cells also causes more rapid growth of neighboring drug-resistant cells. This occurs through a mechanism of tumor repopulation, which is mediated by caspase activation and PGE2 signaling from the dying apoptotic cells. Thus, although autophagy inhibition may cause more effective tumor cell apoptosis, this might not always be a good thing in the long run if the end result is more rapid tumor repopulation from drug-resistant cells that were unable to be killed. Our results indicate that these effects are due to caspase activation [and PGE2 signaling, which can be stimulated by caspases (Li et al., 2010)] rather than cell death itself. Therefore, if one kills tumor cells by a mechanism other than caspase-dependent apoptosis, it may be feasible to circumvent this growth stimulatory response.
Our experiments were done with mixed populations of isogenic tumor cells where only the resistant cells were marked with a fluorescent protein, allowing them to be followed specifically. This allowed a rigorous assessment of non-cell-autonomous effects of autophagy inhibition in the dying cells at drug doses that killed all the drug-sensitive cells, but also identifies a potential problem in studies intended to test if autophagy alters cancer cell chemosensitivity. Such studies usually involve inhibition of autophagy in a homogeneous population of drug-sensitive tumor cells followed by analysis of effects on tumor cell survival and growth. Because it is impossible to detect a sensitization or resistance effect of autophagy manipulation using drug doses that kill all the target cells, these experiments must be done at doses that are only partially effective. However, the mechanism identified here may lead to increased growth of cells that were not killed, resulting in autophagy inhibition-mediated sensitization to the drug being underestimated or, especially with fast growing cells, even an apparent increase in overall number of drug-treated cells when autophagy is blocked. Such an effect might cause an investigator to underestimate the chemosensitization effects of autophagy inhibition or mistakenly conclude that autophagy was necessary for tumor cell killing. This may explain some cases in the literature where autophagic cell death has been proposed to mediate chemotherapy effects (Thorburn et al., 2014a).
In summary, the work described here identifies a potential problem with autophagy inhibition strategies designed to promote chemosensitization; improved drug-induced tumor cell apoptosis achieved by autophagy inhibition may inadvertently also increase growth of drug-resistant tumor cells. This emphasizes the fact that autophagy can have non-cell-autonomous effects that affect tumor cell behavior and suggest that efforts to minimize tumor repopulation from drug-resistant cells could further enhance the efficacy of autophagy inhibition. For some anticancer agents (e.g., the BRAF inhibitor vemurafenib), autophagy inhibitors cannot only synergize with the drug but can also overcome resistance to that drug (Ma et al., 2014), and there are indications of clinical benefit in overcoming acquired resistance to vemurafenib when it is combined with chloroquine (Levy et al., 2014). The work described here suggests that autophagy inhibition in combination with another agent may be most effective in this type of situation where overcoming resistance, as well as achieving synergy in sensitive cells, is feasible because autophagy inhibition would enhance killing of the drug-sensitive tumor cells while simultaneously targeting drug-resistance as well. Additionally finding ways to kill cancer cells by methods other than induction of apoptosis could lead to more effective long-term treatment by avoiding this mechanism.
Abbreviations
| DT-EGF | diphtheria toxin-epidermal growth factor fusion protein |
| EGFR | epidermal growth factor receptor |
| GFP | green fluorescent protein |
| HRP | horseradish peroxidase |
| PGE2 | prostaglandin E2 |
| TRAIL | tumor necrosis factor-related apoptosis-inducing ligand |
Authorship Contributions
Participated in research design: J. Thorburn, A. Thorburn.
Conducted experiments: J. Thorburn, Staskiewicz.
Performed data analysis: J. Thorburn, Goodall.
Contributed new reagents or analytic tools: Dimberg, Frankel, Ford.
Wrote or contributed to the writing of the manuscript: J. Thorburn, A. Thorburn.
Footnotes
This work was supported by National Institutes of Health National Cancer Institute [Grants RO1CA150925;, RO1CA190170;, P30CA046934].
References
- Barnard RA, Wittenburg LA, Amaravadi RK, Gustafson DL, Thorburn A, Thamm DH. (2014) Phase I clinical trial and pharmacodynamic evaluation of combination hydroxychloroquine and doxorubicin treatment in pet dogs treated for spontaneously occurring lymphoma. Autophagy 10:1415–1425. [Europe PMC free article] [Abstract] [Google Scholar]
- Eng CH, Wang Z, Tkach D, Toral-Barza L, Ugwonali S, Liu S, Fitzgerald SL, George E, Frias E, Cochran N, et al. (2016) Macroautophagy is dispensable for growth of KRAS mutant tumors and chloroquine efficacy. Proc Natl Acad Sci USA 113:182–187. [Europe PMC free article] [Abstract] [Google Scholar]
- Galluzzi L, Pietrocola F, Bravo-San Pedro JM, Amaravadi RK, Baehrecke EH, Cecconi F, Codogno P, Debnath J, Gewirtz DA, Karantza V, et al. (2015) Autophagy in malignant transformation and cancer progression. EMBO J 34:856–880. [Europe PMC free article] [Abstract] [Google Scholar]
- Gewirtz DA. (2014) Autophagy and senescence in cancer therapy. J Cell Physiol 229:6–9. [Abstract] [Google Scholar]
- Guo JY, Chen H-Y, Mathew R, Fan J, Strohecker AM, Karsli-Uzunbas G, Kamphorst JJ, Chen G, Lemons JMS, Karantza V, et al. (2011) Activated Ras requires autophagy to maintain oxidative metabolism and tumorigenesis. Genes Dev 25:460–470. [Europe PMC free article] [Abstract] [Google Scholar]
- Holohan C, Van Schaeybroeck S, Longley DB, Johnston PG. (2013) Cancer drug resistance: an evolving paradigm. Nat Rev Cancer 13:714–726. [Abstract] [Google Scholar]
- Huang Q, Li F, Liu X, Li W, Shi W, Liu F-F, O’Sullivan B, He Z, Peng Y, Tan A-C, et al. (2011) Caspase 3-mediated stimulation of tumor cell repopulation during cancer radiotherapy. Nat Med 17:860–866. [Europe PMC free article] [Abstract] [Google Scholar]
- Kroemer G. (2015) Autophagy: a druggable process that is deregulated in aging and human disease. J Clin Invest 125:1–4. [Europe PMC free article] [Abstract] [Google Scholar]
- Kurtova AV, Xiao J, Mo Q, Pazhanisamy S, Krasnow R, Lerner SP, Chen F, Roh TT, Lay E, Ho PL, et al. (2015) Blocking PGE2-induced tumour repopulation abrogates bladder cancer chemoresistance. Nature 517:209–213. [Europe PMC free article] [Abstract] [Google Scholar]
- Levy JM, Thorburn A. (2011) Targeting autophagy during cancer therapy to improve clinical outcomes. Pharmacol Ther 131:130–141. [Europe PMC free article] [Abstract] [Google Scholar]
- Levy JMM, Thompson JC, Griesinger AM, Amani V, Donson AM, Birks DK, Morgan MJ, Mirsky DM, Handler MH, Foreman NK, et al. (2014) Autophagy inhibition improves chemosensitivity in BRAF(V600E) brain tumors. Cancer Discov 4:773–780. [Europe PMC free article] [Abstract] [Google Scholar]
- Li F, Huang Q, Chen J, Peng Y, Roop DR, Bedford JS, Li C-Y. (2010) Apoptotic cells activate the “phoenix rising” pathway to promote wound healing and tissue regeneration. Sci Signal 3:ra13. [Europe PMC free article] [Abstract] [Google Scholar]
- Liu TF, Cohen KA, Ramage JG, Willingham MC, Thorburn AM, Frankel AE. (2003) A diphtheria toxin-epidermal growth factor fusion protein is cytotoxic to human glioblastoma multiforme cells. Cancer Res 63:1834–1837. [Abstract] [Google Scholar]
- Ma X-H, Piao S-F, Dey S, McAfee Q, Karakousis G, Villanueva J, Hart LS, Levi S, Hu J, Zhang G, et al. (2014) Targeting ER stress-induced autophagy overcomes BRAF inhibitor resistance in melanoma. J Clin Invest 124:1406–1417. [Europe PMC free article] [Abstract] [Google Scholar]
- Macintosh RL, Timpson P, Thorburn J, Anderson KI, Thorburn A, Ryan KM. (2012) Inhibition of autophagy impairs tumor cell invasion in an organotypic model. Cell Cycle 11:2022–2029. [Europe PMC free article] [Abstract] [Google Scholar]
- Mahalingam D, Mita M, Sarantopoulos J, Wood L, Amaravadi RK, Davis LE, Mita AC, Curiel TJ, Espitia CM, Nawrocki ST, et al. (2014) Combined autophagy and HDAC inhibition: a phase I safety, tolerability, pharmacokinetic, and pharmacodynamic analysis of hydroxychloroquine in combination with the HDAC inhibitor vorinostat in patients with advanced solid tumors. Autophagy 10:1403–1414. [Europe PMC free article] [Abstract] [Google Scholar]
- Maycotte P, Aryal S, Cummings CT, Thorburn J, Morgan MJ, Thorburn A. (2012) Chloroquine sensitizes breast cancer cells to chemotherapy independent of autophagy. Autophagy 8:200–212. [Europe PMC free article] [Abstract] [Google Scholar]
- Maycotte P, Gearheart CM, Barnard R, Aryal S, Mulcahy Levy JM, Fosmire SP, Hansen RJ, Morgan MJ, Porter CC, Gustafson DL, et al. (2014) STAT3-mediated autophagy dependence identifies subtypes of breast cancer where autophagy inhibition can be efficacious. Cancer Res 74:2579–2590. [Europe PMC free article] [Abstract] [Google Scholar]
- Menke C, Bin L, Thorburn J, Behbakht K, Ford HL, Thorburn A. (2011) Distinct TRAIL resistance mechanisms can be overcome by proteasome inhibition but not generally by synergizing agents. Cancer Res 71:1883–1892. [Europe PMC free article] [Abstract] [Google Scholar]
- Obenauf AC, Zou Y, Ji AL, Vanharanta S, Shu W, Shi H, Kong X, Bosenberg MC, Wiesner T, Rosen N, et al. (2015) Therapy-induced tumour secretomes promote resistance and tumour progression. Nature 520:368–372. [Europe PMC free article] [Abstract] [Google Scholar]
- Rangwala R, Chang YC, Hu J, Algazy KM, Evans TL, Fecher LA, Schuchter LM, Torigian DA, Panosian JT, Troxel AB, et al. (2014a) Combined MTOR and autophagy inhibition: phase I trial of hydroxychloroquine and temsirolimus in patients with advanced solid tumors and melanoma. Autophagy 10:1391–1402. [Europe PMC free article] [Abstract] [Google Scholar]
- Rangwala R, Leone R, Chang YC, Fecher LA, Schuchter LM, Kramer A, Tan K-S, Heitjan DF, Rodgers G, Gallagher M, et al. (2014b) Phase I trial of hydroxychloroquine with dose-intense temozolomide in patients with advanced solid tumors and melanoma. Autophagy 10:1369–1379. [Europe PMC free article] [Abstract] [Google Scholar]
- Rebecca VW, Amaravadi RK. (2016) Emerging strategies to effectively target autophagy in cancer. Oncogene 35:1–11. [Europe PMC free article] [Abstract] [Google Scholar]
- Rosenfeld MR, Ye X, Supko JG, Desideri S, Grossman SA, Brem S, Mikkelson T, Wang D, Chang YC, Hu J, et al. (2014) A phase I/II trial of hydroxychloroquine in conjunction with radiation therapy and concurrent and adjuvant temozolomide in patients with newly diagnosed glioblastoma multiforme. Autophagy 10:1359–1368. [Europe PMC free article] [Abstract] [Google Scholar]
- Rubinsztein DC, Codogno P, Levine B. (2012) Autophagy modulation as a potential therapeutic target for diverse diseases. Nat Rev Drug Discov 11:709–730. [Europe PMC free article] [Abstract] [Google Scholar]
- Sousa CM, Biancur DE, Wang X, Halbrook CJ, Sherman MH, Zhang L, Kremer D, Hwang RF, Witkiewicz AK, Ying H, et al. (2016) Pancreatic stellate cells support tumour metabolism through autophagic alanine secretion. Nature 536:479–483. [Europe PMC free article] [Abstract] [Google Scholar]
- Thorburn A, Thamm DH, Gustafson DL. (2014a) Autophagy and cancer therapy. Mol Pharmacol 85:830–838. [Europe PMC free article] [Abstract] [Google Scholar]
- Thorburn J, Andrysik Z, Staskiewicz L, Gump J, Maycotte P, Oberst A, Green DR, Espinosa JM, Thorburn A. (2014b) Autophagy controls the kinetics and extent of mitochondrial apoptosis by regulating PUMA levels. Cell Reports 7:45–52. [Europe PMC free article] [Abstract] [Google Scholar]
- Thorburn J, Horita H, Redzic J, Hansen K, Frankel AE, Thorburn A. (2009) Autophagy regulates selective HMGB1 release in tumor cells that are destined to die. Cell Death Differ 16:175–183. [Europe PMC free article] [Abstract] [Google Scholar]
- Vogl DT, Stadtmauer EA, Tan K-S, Heitjan DF, Davis LE, Pontiggia L, Rangwala R, Piao S, Chang YC, Scott EC, et al. (2014) Combined autophagy and proteasome inhibition: a phase 1 trial of hydroxychloroquine and bortezomib in patients with relapsed/refractory myeloma. Autophagy 10:1380–1390. [Europe PMC free article] [Abstract] [Google Scholar]
- White E. (2012) Deconvoluting the context-dependent role for autophagy in cancer. Nat Rev Cancer 12:401–410. [Europe PMC free article] [Abstract] [Google Scholar]
Articles from Molecular Pharmacology are provided here courtesy of American Society for Pharmacology and Experimental Therapeutics
Full text links
Read article at publisher's site: https://doi.org/10.1124/mol.116.106070
Read article for free, from open access legal sources, via Unpaywall:
https://molpharm.aspetjournals.org/content/molpharm/91/1/58.full.pdf
Citations & impact
Impact metrics
Citations of article over time
Smart citations by scite.ai
Explore citation contexts and check if this article has been
supported or disputed.
https://scite.ai/reports/10.1124/mol.116.106070
Article citations
Nanoplatform-Mediated Autophagy Regulation and Combined Anti-Tumor Therapy for Resistant Tumors.
Int J Nanomedicine, 19:917-944, 26 Jan 2024
Cited by: 0 articles | PMID: 38293604 | PMCID: PMC10826716
Review Free full text in Europe PMC
Multifaceted role of redox pattern in the tumor immune microenvironment regarding autophagy and apoptosis.
Mol Cancer, 22(1):130, 10 Aug 2023
Cited by: 9 articles | PMID: 37563639 | PMCID: PMC10413697
Review Free full text in Europe PMC
Mitochondrial-derived vesicles compensate for loss of LC3-mediated mitophagy.
Dev Cell, 56(14):2029-2042.e5, 24 Jun 2021
Cited by: 62 articles | PMID: 34171288 | PMCID: PMC8319140
Molecular Mechanisms Underlying Autophagy-Mediated Treatment Resistance in Cancer.
Cancers (Basel), 11(11):E1775, 11 Nov 2019
Cited by: 49 articles | PMID: 31717997 | PMCID: PMC6896088
Review Free full text in Europe PMC
Cancer Cells Upregulate NRF2 Signaling to Adapt to Autophagy Inhibition.
Dev Cell, 50(6):690-703.e6, 01 Aug 2019
Cited by: 53 articles | PMID: 31378590 | PMCID: PMC7233142
Go to all (8) article citations
Similar Articles
To arrive at the top five similar articles we use a word-weighted algorithm to compare words from the Title and Abstract of each citation.
Downregulation of ATG14 by EGR1-MIR152 sensitizes ovarian cancer cells to cisplatin-induced apoptosis by inhibiting cyto-protective autophagy.
Autophagy, 11(2):373-384, 01 Jan 2015
Cited by: 101 articles | PMID: 25650716 | PMCID: PMC4502709
Enhanced caspase-8 recruitment to and activation at the DISC is critical for sensitisation of human hepatocellular carcinoma cells to TRAIL-induced apoptosis by chemotherapeutic drugs.
Cell Death Differ, 11 Suppl 1:S86-96, 01 Jul 2004
Cited by: 128 articles | PMID: 15105837
Concurrent Autophagy Inhibition Overcomes the Resistance of Epidermal Growth Factor Receptor Tyrosine Kinase Inhibitors in Human Bladder Cancer Cells.
Int J Mol Sci, 18(2):E321, 04 Feb 2017
Cited by: 20 articles | PMID: 28165387 | PMCID: PMC5343857
Autophagy is a major metabolic regulator involved in cancer therapy resistance.
Cell Rep, 36(7):109528, 01 Aug 2021
Cited by: 34 articles | PMID: 34407408
Review
Funding
Funders who supported this work.
NCI NIH HHS (3)
Grant ID: R01 CA190170
Grant ID: R01 CA150925
Grant ID: P30 CA046934





