Abstract
Free full text

GRIL-Seq, a method for identifying direct targets of bacterial small regulatory RNA by in vivo proximity ligation
Abstract
The first step in the post-transcriptional regulatory function of most bacterial small non-coding RNAs (sRNAs) is base-pairing with partially complementary sequences of targeted transcripts. We present a simple method for identifying sRNA targets in vivo and defining processing sites of the regulated transcripts. The technique (referred to as GRIL-Seq) is based on preferential ligation of sRNAs to ends of base-paired targets in bacteria co-expressing T4 RNA ligase, followed by sequencing to identify the chimeras. In addition to the RNA chaperone Hfq, the GRIL-Seq method depends on the activity of the pyrophosphorylase RppH. Using PrrF1, an iron-regulated sRNA in Pseudomonas aeruginosa, we demonstrate that direct regulatory targets of this sRNA can be readily identified. Therefore, GRIL-Seq represents a powerful tool not only for identifying direct targets of sRNAs in a variety of environments, but can also result in uncovering novel roles for sRNAs and their targets in complex regulatory networks.
Introduction
Post-transcriptional regulation mechanisms, primarily through the activities of regulatory small RNAs (sRNAs), play an important role in the bacterial stress response, metabolism, quorum sensing and virulence1–6. The sharp increase in the description of this abundant class of RNA regulators during the past decade is the direct result of transcriptome analyses using next generation RNA sequencing (RNA-seq) and the development of in silico methods to identify non-protein coding transcripts7. In bacteria, regulation of gene expression by sRNAs can be divided into two mechanistically distinct categories. One class of sRNAs function by modifying the activities of regulatory proteins8. Other sRNAs regulate gene expression by base-pairing with mRNAs; this process is accelerated by the RNA chaperone Hfq9. The targets of base-pairing sRNAs can range from a few to as many as 1% of total cellular transcripts10. Transcripts positively controlled by these sRNAs form secondary structures near their ribosome binding sites (RBSs) and can be disrupted by alternative base-pairing with sRNAs, allowing translational initiation11, 12. Negative regulation depends on base-pairing of sRNAs near the translation initiation regions, and downstream protein coding regions, which can lead to degradation of the transripts13.
In spite of the relative ease in identifying regulatory RNAs, their targets are less well defined. The main difficulty in predicting sRNA targets is the limited and non-contiguous base-pairing regions with frequent internal secondary structures and existence of multiple targets with different base-pairing configurations. Consequently, although several different computational algorithms have been developed, their performance in predicting direct regulatory targets of sRNAs is highly variable7. A number of experimental approaches have also been developed to facilitate the identification of direct targets of regulatory RNAs. Several methods (CLASH14 and iPAR-CLIP15) have been used to identify targets of eukaryotic micro RNAs (miRNAs) based on immunoprecipitation of transcripts crosslinked to Argonaut proteins. Crosslinking followed by in vitro ligation and sequencing was utilized to define the targets of non-coding RNAs in human cells15. In bacteria, transcriptome analysis following brief expression of sRNAs can be used to predict likely targets using translational reporters or ribosome profiling16, 17. However, these methods are often unable to distinguish between direct and indirect effects of sRNA regulation. A variation of these methods was applied to the identification of Hfq-bound mRNAs and sRNAs in E. coli and Salmonella, however, the assignment of a direct regulatory relationship could not be made18, 19. Another approach relies on fusing the MS2 coat protein-binding hairpin sequence to the sRNA and capturing the sRNA/mRNA complex with the MS2 bacteriophage coat protein20.
In order to facilitate the analysis of global effects of bacterial sRNA-target mRNA interactions, we have developed a robust yet simple method for identifying targets of sRNAs. We named this method Global sRNA Target Identification by Ligation and Sequencing (GRIL-Seq). The method takes advantage of the proximity of the sRNA and mRNA target sites in a complex that is likely to be stabilized by the Hfq protein. This arrangement facilitates a preferential ligation of the 3′ and 5′ ends by bacteriophage T4 RNA ligase, co-expressed in the same cell and the detection of the chimeric RNAs by sequencing. The GRIL-Seq method is an easy, readily accessible approach towards defining post-transcriptional regulatory networks controlled by sRNAs. Conceivably, this method could be applicable to the analyses of miRNA directed silencing of eukaryotic mRNAs as well.
Results
Small RNA-target RNA ligation by T4 RNA ligase in the cell
We exploited the ability of bacteriophage T4 RNA ligase to link two base-paired RNA molecules expressed in the same cell. In order to ligate two RNAs, the 5′ terminal donor sequence must be monophosphorylated. While the majority of bacterial small RNAs are primary transcripts with 5′ triphosphoryl termini, a recent study demonstrated that a fraction of Salmonella sRNAs carries 5′ monophosphate and their enrichment was observed in Hfq-coIP21. Taking advantage of the proximity of a base-paired sRNA to its target RNA, we reasoned that, in T4 RNA ligase expressing cells, the 5′ monophosphates of either sRNAs or mRNAs would preferentially ligate to the 3′ hydroxyl groups of mRNAs or sRNAs, respectively, creating chimeras between these two molecules (Fig. 1a).
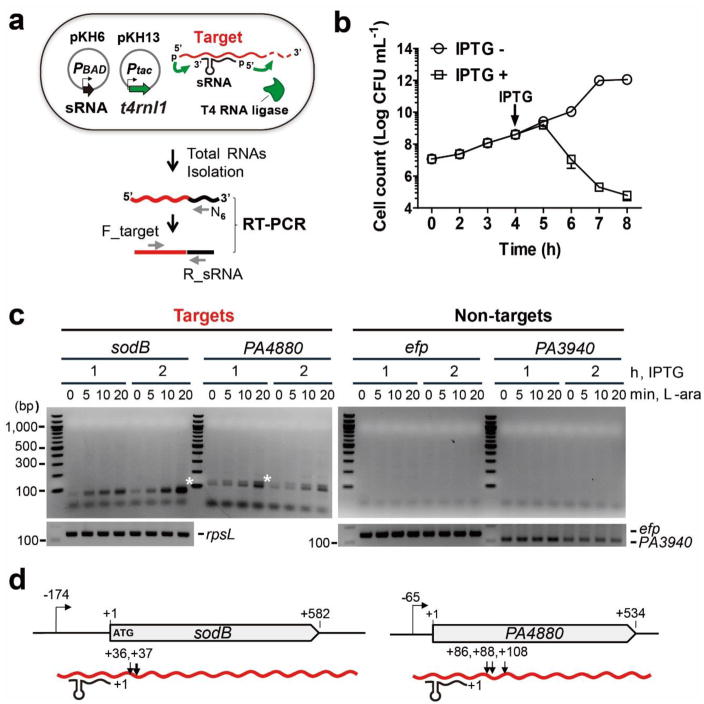
(a) Schematic of sRNA-target formation in cells expressing T4 RNA ligase and an sRNA. Also shown is the RT-PCR based strategy for the detection of chimeras between an sRNA and specific target mRNAs. (b) Effect of expression of T4 RNA ligase based on P. aeruginosa cell growth. The T4 RNA ligase was induced in P. aeruginosa (pKH13) when it reached an OD600 ~0.5 (indicated by the arrow) and cell viability was monitored for an additional 4 hours. Error bars represent the standard deviation of the average of three biological replicates (c) Determination of optimal induction time for the detection of PrrF1-containing chimeras (white star). Amplicons were detected only, when during the PCR step, primers for the targets (sodB and PA4880, white stars), but not for non-targets (efp and PA3940) were used. PCR amplification of cDNA for a housekeeping gene, rpsL and two non-targets (efp and PA3940) was carried out to ensure the presence of equal amounts of cDNA of two non-target genes in all samples. Results are representative of duplicate experiments. (d) Sites of ligation between PrrF1 and sodB and PA4880. Arrows indicate the location of each junction (relative to the start of translation) based on the sequence of the amplicons indicated by the white stars in 1c.
To test this hypothesis, we engineered a P. aeruginosa strain containing two compatible plasmids. In the plasmid pKH6, the gene for an sRNA was cloned with +1 transcription start site (TSS) downstream of a pBAD promoter while the t4rnl1 gene, coding for the RNA ligase, was cloned in the plasmid pKH13 where its expression is controlled by the IPTG-inducible tac promoter (Fig 1a, Supplementary Fig. 1, Methods). Before introducing the sRNA expressing plasmid into the cell, we first assessed the effect of expression of T4 RNA ligase on growth (Fig. 1b). Bacteria continued dividing for at least one hour, followed by a decline in viability. For each experiment, induction of the T4 RNA ligase expression was kept under one hour.
To evaluate the utility of the proximity ligation method for identification of unknown targets of known sRNAs, we selected the P. aeruginosa sRNA PrrF1, one of the two small RNAs controlled by the iron-responsive Fur repressor22. PrrF1, together with a second sRNA PrrF2, is the functional orthologue of the E. coli RyhB sRNA23. To identify candidate targets of PrrF1 and suitable non-target controls, we created an arabinose inducible PrrF1 expression vector (pKH6-PrrF1) and introduced it into a P. aeruginosa Δprrf1Δprrf2 double mutant. Using RNA-seq, we compared the transcript levels in cells grown in the absence of the sRNAs (carrying the empty vector) to those recovered from bacteria expressing PrrF1 over a 20-minute period of induction (Supplementary Table 1). We selected two candidate target genes (sodB and PA4880) based on their likely regulation by PrrF1, and two genes whose transcript levels were not altered in response to PrrF1 overexpression (efp and PA3940). The sodB and PA4880 genes were also identified in several transcriptome studies evaluating the effect of iron, the Fur repressor, and the sRNAs22, 24. We then fused in-frame lacZ to their 5′-UTRs and the coding sequence encompassing 20 codons, and compared β-galactosidase expression in cells also carrying pKH6-PrrF1 or the empty vector (Supplementary Fig. 2a). As shown in Supplementary Fig. 2b, PrrF1-dependent repression of β-galactosidase was seen in cells expressing the target::lacZ fusions, but not when lacZ was fused to the 5′ regions of non-target genes.
We also used these two pairs of genes to test whether specific ligation between PrrF1 and the target mRNAs is observed in the same cell. T4 RNA ligase was expressed for one or two hours, followed by induction of expression of the sRNA for up to 20 min. To detect the ligated chimeric RNAs, we carried out RT-PCR using the individual gene-specific forward primers and a PrrF1-specific reverse primer as described in Fig. 1a. The individual gene-specific forward primers were designed to anneal to the Shine-Dalgarno/AUG regions of each mRNA. Unique amplicons were detected when the target gene-specific primers were used, but none when the non-target gene-specific primers were used (Fig. 1c).
We analyzed the ligated products between PrrF1 and the target mRNAs by sequencing the specific amplicons identified in Fig. 1c. The junction sequences and sites between the 5′ end of PrrF1 and 3′-OH of target genes are shown in Supplementary Fig. 3a and Fig. 1d, respectively. PrrF1 was ligated to two sites on sodB mRNA (36 and 37 nts downstream from the start codon) and to three sites on PA4880 mRNA (86, 88, and 108 nts downstream from the start codon), which explains the double bands seen in Fig. 1c. It is likely that the preferential ligation of the 5′ phosphate end of PrrF1 to different sites within the target transcripts represents the locations of RNase cleavage sites, creating multiple 3′ hydroxyl end substrates for the formation of the covalent linkage. In addition, we observed two additional adenosines at the junction of PA4880-PrrF1 chimeric RNA, suggesting that the ligation appears to occur following poly(A) addition, commonly seen in the bacterial RNA degradation pathway25 and can be observed during sRNA induced mRNA decay26.
Factors influencing the specific ligation of sRNA to target mRNA
The specificity of the ligation reaction between PrrF1 and sodB was further tested by generating single nucleotide substitutions in the region of the sRNA that is predicted to base-pair with the target22 and engineering a defective T4 RNA ligase27 (Fig. 2a and 2b). Three mutations (M1-M3) were created and tested for an effect on sodB transcript levels (Fig. 2c). Unlike wild-type and mutant PrrF1-M1, whose mutation is located at the end of the seed region, mutants PrrF1-M2 and M3 failed to negatively impact the concentration of sodB mRNA (Fig. 2c) suggesting that they lost their regulatory activity due to insufficient base-pairing with the target. Consequently, no ligation products were detected in cells expressing PrrF1-M2 or M3 (Fig. 2d, lanes 4 and 5) implying that the base-pairing between two RNAs plays an important role for ligation between sRNA and mRNA. In addition, the ligation between PrrF1 and sodB required active T4 RNA ligase, since a substitution mutation K99N failed to catalyze the formation of a covalent linkage between sodB (Fig 2d, lane 3) as well as PA4480 (Fig 2e, lane 3). These studies show that formation of an sRNA-mRNA chimera requires base-pairing between the two RNAs and active T4 RNA ligase.
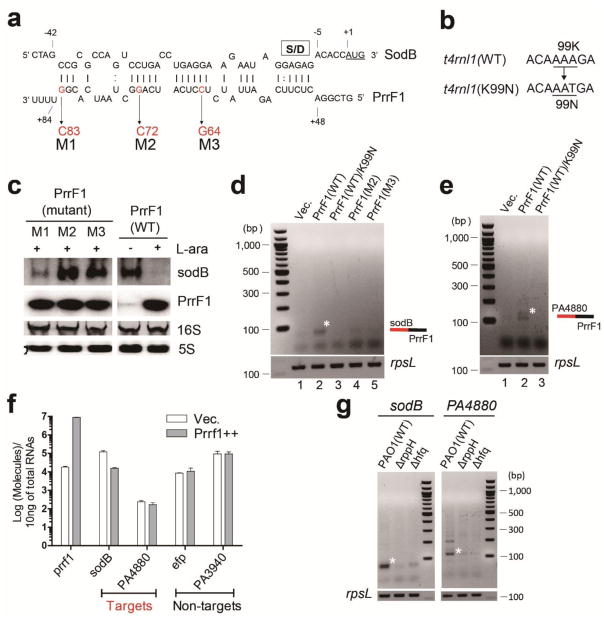
(a) Effect of mutations in PrrF1, indicated as M1(G83C), M2(G72C) and M3(C64G), predicted to be in its base-pairing region with sodB mRNA. (b) Requirement for a functional T4 RNA ligase in generating sRNA-mRNA chimeras. A mutation in the t4rnl1 gene was engineered, in the codon for lysine (K) position 99 changing it asparagine (N), creating a catalytically inactive T4 RNA ligase. (c) Northern blot analysis of the effects of mutations in the PrrF1 base-pairing region. The M2 or M3 mutations are unable to induce sodB mRNA degradation, while a smaller defect was seen with the M1 mutant of PrrF1. PCR amplification of cDNA for rpsL was carried out to ensure the presence of equal amount of cDNA. (d) Effect of PrrF1 M2 and M3 mutants and catalytically-inactive T4 RNA ligase on ligation of the sRNA to sodB. Ligation between sodB mRNA and PrrF1 (wt, wild type) was monitored by RT-PCR between sodB and wt PrrF1 and M2 and M3 mutants (lanes 2, 4 and 5, respectively) and the inactive (K99N) T4 RNA ligase (lane 3). (e) Requirement for an enzymatically active T4 RNA ligase for the detection of a PA4880 mRNA and PrrF1 chimeric product. (f) Expression levels of target and non-target mRNA in total RNA isolated from in vivo RNA ligated samples in three biological replicates. Error bars represent the standard deviation. Transcript levels were determined by quantitative qRT-PCR after induction of PrrF1 for 20 min when the cultures were at OD600 ~1.5. (g) Requirement for the RppH and Hfq proteins for efficient ligation between PrrF1 and sodB (or PA4880), monitored by RT-PCR. All results are representative of at least duplicate experiments.
We next examined whether the abundance of the target mRNA influences the covalent linkage of PrrF1 to its targets. Following induction of PrrF1 for 20 minutes, we assessed the mRNA levels of target and non-target by qRT-PCR and found that the levels of sodB and PA4480 varied. Following induction of PrrF1, a strong (8 fold) decrease in sodB and a modest (1.5 fold) decrease in PA4480 were observed (Fig. 2f). No significant change in the non-target transcript levels was observed. The extent of ligation between the sRNA and its targets, shown in Fig. 1c, does not seem to correlate with the transcript levels prior to or after PrrF1 overexpression. The ligation to non-target mRNAs was not seen in spite of their expression levels significantly exceeding that of PA4480. Therefore, it is not the transcript levels but other factors such as base-pairing between sRNA and the target as well as mRNA processing (creating ligatable 3′-OH termini) that are the main determinants in creation of chimeras in the cell.
The main impediment to the ligation between RNAs with 5′ phosphate and 3′ hydroxyl ends is the presence of the 5′ terminal triphosphate groups in primary transcripts of both sRNAs and mRNAs. However, it is likely that a fraction of sRNA molecules bound by Hfq as well as mRNAs in a degradation pathway contain monophosphorylated 5′ ends22, 28. The removal of pyrophosphate from the 5′ ends of primary transcripts is catalyzed by the pyrophosphohydrolase RppH28. Therefore, we examined whether the RNA-ligase catalyzed formation of the chimeras requires the activity of RppH as well as Hfq. We analyzed the formation of PrrF1 and sodB or PA4480 chimeras in wild-type P. aeruginosa and in rppH and hfq mutants (Fig. 2g). Notably, the ligation of both mRNAs to PrrF1 was strongly reduced in the two mutants.
These results demonstrate that the interaction of the PrrF1 sRNA with any one of its predicted targets gives rise to a complex which, following the removal of pyrophosphate from the 5′ end of the sRNA by RppH, can be covalently linked to one of several 3′ OH groups created by endonucleolytic processing of the mRNAs. Moreover, using the E. coli RyhB sRNA, we demonstrated T4 RNA ligase-mediated formation of chimeras between the sRNA and both positively and negatively regulated targets (Supplementary Note 1).
Identification of multiple RNA targets directly interacting with a single sRNA by GRIL-Seq
We expanded the ability of the RNA proximity ligation method to detect targets of sRNAs within the entire transcriptome. We developed an optimized pipeline shown schematically in Fig. 3 and we refer to this method as “GRIL-Seq: Global sRNA Target Identification by Ligation and Sequencing”. A key feature of this procedure is the enrichment step of sRNA-target chimeras following in vivo ligation and RNA recovery. The denatured total RNAs containing the ligated RNAs are annealed to poly(A) tailed oligonucleotides complementary to the sRNA, followed by capture of the sRNAs (including the chimera) on oligo-dT magnetic beads. The location of the annealing region in PrrF1 and sequences of the captured oligonucleotides are shown in Supplementary Fig. 5a. These enriched RNAs are then reverse transcribed using random primers and the cDNAs are used for library construction and Illumina sequencing.
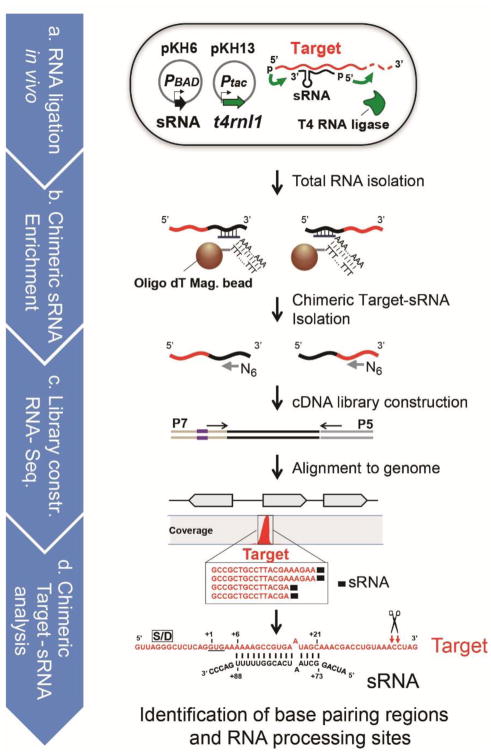
(a) Ligation of two RNAs is carried out in cells carrying two compatible expression plasmids: pKH6, where the expression of an sRNA (indicated in black) is under the control of the arabinose-inducible PBAD promoter, while in plasmid pKH13, the expression of the t4rnl1, coding for the T4 RNA ligase (indicated in green) is regulated by IPTG. For optimal ligation, T4 RNA ligase is induced first by addition of 1mM IPTG for 1hr and then the sRNA is expressed by the addition of 0.2% L-arabinose for 20 min. (b) Enrichment for transcripts containing the sRNA chimeras. Magnetic beads with attached oligo dT and poly(A)-tailed sRNA binding oligomer, target (red)-sRNA (black) chimeric RNAs are immobilized. The chimeric RNAs are recovered following DNase treatment. (c) The DNA library for Illumina sequencing is constructed using a commercially available kit (New England BioLab, NEBNext® Ultra™ RNA Library Prep Kit). (d) The sequences of chimeric target-sRNA are collected and aligned to the reference genome. For the identification of targets and processing sites, coverage of the chimeric RNAs is determined using CLC Genome Workbench (ver. 6.0.1).
In biological duplicate experiments, we obtained 6,811,939 and 6,473,670 PrrF1-containing chimeras, representing 14 and 18% of total reads, respectively (Supplementary Fig. 5b). The duplicate samples show high reproducibility, with Pearson correlation coefficient of 0.99 (Supplementary Fig. 5c). The P. aeruginosa sequences within the chimeras were mapped to the genome and the number of reads at each peak was averaged for the two samples. Transcripts corresponding to 3505 P. aeruginosa genes were identified as chimeras with PrrF1; they were ranked according to the maximal coverage and are listed in Supplementary Table 2 and 3. Among the top 40, the gene products of 19 are enriched for iron containing proteins and 15 contain recognizable iron sulfur clusters. The list of top 36 mRNA transcripts includes 12 mRNAs that were predicted to bind PrrF1 by the CopraRNA algorithm29 (Supplementary Table 3).
To assess the relationship between abundance of a transcript and its presence in chimera with PrrF1, we generated a scatter plot using the data from RNA-Seq and the genes detected by GRIL-Seq. As shown in the Supplementary Fig. 6, there is poor correlation (R2 = 0.005) between RNA abundance detection of ligation products with PrrF1. This analysis indicates that the chimera identified by GRIL-Seq represent a specific class of transcripts and are not generated by random ligation based on their abundance.
We also mapped and quantified the PrrF-containing chimeras relative to the genomic locations of the corresponding mRNAs and non-coding RNAs (Fig. 4a). Interestingly, the mapping of the chimeric reads for the top-ranked genes was remarkably enriched at a specific location within the individual transcript (Supplementary Fig. 7). In the examples shown in Fig. 4b and 4c, a limited number of sites account for the majority of the reads. Over 50% of chimeric sequences containing sodB are the result of ligation to three proximal sites. Similarly, the majority of PrrF1 and gloA1 chimeras are separated by only one nucleotide. The model for base-pairing of the sRNAs in the vicinity of the ligation sites can be readily predicted using the IntaRNA algorithm30 (Fig. 4d). This strong preference for specific locations reflects the proximity of the annealed sRNA to the available 3′ hydroxyl sites created by RNase cleavage; ligation to 3′ hydroxyl termini from naturally terminated transcripts is seen infrequently. We also observed the addition of one or more adenosines at the 3′ OH of mRNA at the junction sites in a few PrrF1 chimeras in both cases (Supplementary Fig. 8), suggesting that a fraction of the ligations appear to occur following polyadenylation of mRNA that may be triggered by sRNA-mediated mRNA degradation.
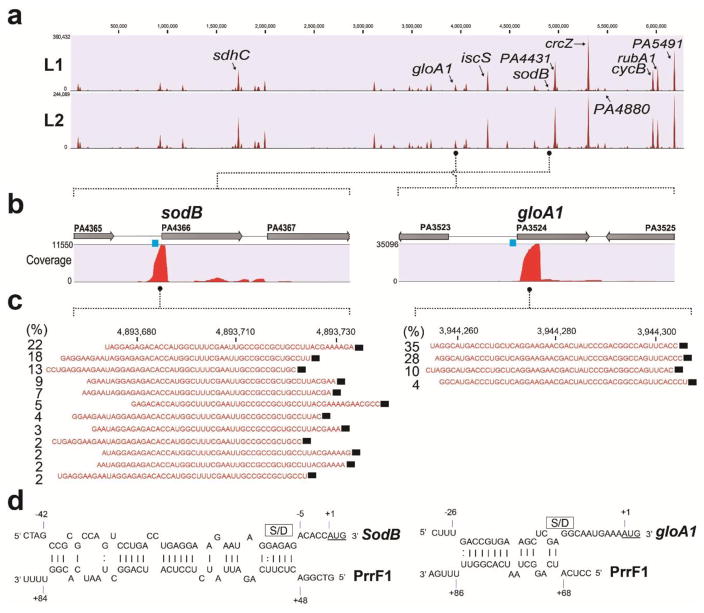
(a) Coverage profiling of enriched chimeric PrrF1-target RNAs following mapping to PAO1Δprrf1Δprrf2 genome. Putative targets of PrrF1 sRNA are shown in red color peaks. Peak height represents the value of maximum coverage. Duplicate GRIL-Seq experiments (L1 and L2) are shown. (b) Enrichment of the chimeric RNAs containing sodB and gloA1. The coverage of sequenced chimeric reads corresponding to sodB or gloA1 are shown as red peaks. Data is from the L2 experiment. (c) Sequencing reads corresponding to chimeras between PrrF1 and sodB or gloA1 mRNA. For each sequencing read, the location of the junction between the ligated sRNA and mRNA, at a single nucleotide resolution, was identified. The percentage of each read in the various chimeras was also determined. The sequences shown in red are the most common sodB or gloA1 sequences found in the chimeras, while the PrrF1 sequence is shown as a black rectangle. (d) The predicted base-pairing region between PrrF1 and sodB or gloA1 generated using the IntaRNA algorithm. The Shine-Dalgarno (S/D) sequence and the AUG start codon are indicated as a box and red letters, respectively.
The list of RNAs ligated to PrrF1 very likely contains several groups of transcripts. These include mRNAs regulated by the sRNA, affecting their translation and/or stability. Some can also act as “sponges” sequestering sRNAs from their targets. To differentiate between these possibilities, we compared the list of genes identified by GRIL-Seq to those whose transcript levels were affected by overexpression of PrrF1 (Supplementary Table 1). As shown in Fig. 5, the transcripts of 17 genes are significantly reduced (2–10 fold) among the top 40 genes identified by the GRIL-Seq procedure. The transcripts of these genes (Table 1) likely represent direct targets of PrrF1, where the interaction with the sRNA results in their enhanced degradation.
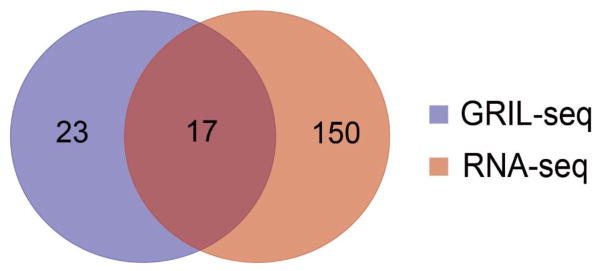
In the Venn diagram, the blue set represents the top 40 targets of PrrF1 as determined by their coverage following application of the GRIL-Seq method. The red set represents the top 167 differentially expressed genes (q-value < 2.0e-09) based on the relative abundance of transcripts in P. aeruginosa PAO1Δprrf1Δprrf2 and the same strain over-expressing PrrF1 by RNA-seq. Seventeen genes intersect both sets. Results are representative of duplicate experiments.
Table 1
Also shown are the values for the GRIL-Seq maximum coverage and their expression levels from RNA-seq in the two strain backgrounds.
| Locus tag | Name | Product Name | Max coverage (GRIL-Seq) | qValue (RNA-seq) | Expression (Δprrf1 Δprrf 2) | |
|---|---|---|---|---|---|---|
|
| ||||||
| Vec. | PrrF1++ | |||||
| PA0849 | trxB2 | Thioredoxin reductase | 37,196 | 1.25e-10 | 164 | 80 |
| PA1554 | ccoN1 | Cytochrome c oxidase, cbb3-type, CcoN subunit | 24,260 | 6.80e-13 | 146 | 73 |
| PA1787 | acnB | aconitate hydratase 2 | 25,808 | 1.17e-16 | 324 | 159 |
| PA1838 | cysI | sulfite reductase | 80,581 | 2.87e-122 | 278 | 64 |
| PA2953 | electron transfer flavoprotein-ubiquinone oxidoreductase | 19,407 | 1.53e-53 | 230 | 75 | |
| PA3094.2 | Asp tRNA | 40,509 | 1.78e-10 | 6,810 | 3,781 | |
| PA3262.1 | Asp tRNA | 41,289 | 1.45e-09 | 3,945 | 2,185 | |
| PA3524 | gloA1 | lactoylglutathione lyase | 46,443 | 9.43e-77 | 409 | 102 |
| PA3621 | fdxA | ferredoxin I | 56,776 | 1.08e-83 | 1,004 | 247 |
| PA4236 | katA | catalase | 44,545 | 9.71e-54 | 226 | 71 |
| PA4366 | sodB | superoxide dismutase | 14,027 | 0.00e+00 | 2,197 | 250 |
| PA4431 | iron-sulfur protein | 204,548 | 5.94e-16 | 345 | 162 | |
| PA4880 | bacterioferritin | 22,422 | 1.82e-53 | 82 | 21 | |
| PA5300 | cycB | cytochrome C5 | 154,313 | 5.38e-236 | 1,348 | 238 |
| PA5351 | rubA1 | rubredoxin | 158,050 | 3.48e-116 | 1,121 | 241 |
| PA4812 | fdnG | formate dehydrogenase-O, major subunit | 13,169 | 4.16e-80 | 61 | 17 |
| PA5490 | cc4 | cytochrome c4 precursor | 14,265 | 1.38e-64 | 659 | 198 |
The targets identified by GRIL-Seq were subjected to experimental validation using two independent methods. Briefly, we created in-frame fusions to the predicted 5′-UTR and an additional 20 codons to a lacZ reporter and also expressed full-length proteins carrying a C-terminal 6His tag (Supplementary Fig. 9a). Induction of PrrF1 expression caused a reduction in β–galactosidase levels and protein expression detected by Western immunoblot analysis, in 11 out of 12 predicted targets (Supplementary Fig. 9b and c). For each target, we were also able to predict the most likely region base paired with PrrF1 using the IntaRNA algorithm. We selected two targets (sodB and gloA1) and confirmed experimentally that mutations of sRNA in the base-pairing region abolish its regulatory activity and these can be reverted by compensatory mutations in the target sequence (Supplementary Note 2, Supplementary Fig. 10).
The majority of predicted base-pairing of PrrF1 with its targets could be localized to the 5′ ends of mRNAs, in agreement with the expected role of sRNAs in regulating translation or mRNA stability. However, on several occasions, ligation of PrrF1 was seen near 3′ ends of mRNAs. We investigated one such interaction, between PrrF1 and the katA mRNA, leading to the discovery of another sRNA sequestration (sponge) mechanism for modulating gene expression. Details of this work are described in the Supplemental Note 3.
Non-coding RNAs
Using the GRIL-Seq approach we also identified ligation products to several non-coding RNAs. Among these, the CrcZ sRNA is one of the regulators of carbon catabolite repression in P. aeruginosa31, 32. Two PrrF1 regions base-pairing can be predicted within CrcZ and two sites (at position +227 and +242) are mainly linked to PrrF1 in the chimeric reads (Supplementary Fig. 7-#1). Moreover, 3′ OH of fragmented PrrF1 (mainly at position +74) was linked to 5′ of the fragmented form (+242) of CrcZ, which suggests that the ligation of PrrF1 to CrcZ is a result of direct interactions, concurrent with the cleavage of both RNAs. We also identified several tRNAs ligated to PrrF1, including two tRNA-vals and three tRNA-asps. Detailed analysis of the interactions between PrrF1 and the tRNAs can be found in Supplementary Note 4.
Discussion
The GRIL-Seq method described here allows identification of transcripts recognized by sRNAs following the base-pairing between segments of the complementary RNAs and proximity ligation of the 3′ hydroxyl and 5′ phosphate ends by T4 RNA ligase, expressed in live bacteria. GRIL-Seq can therefore be used to identify multiple targets of sRNAs based on their ability to form transient complexes with mRNAs as well as other transcripts including RNA traps, anti-sRNAs, and sponges20, 33–35.
An important feature of GRIL-Seq is that it can be carried out in live bacteria, without modification of an sRNA. Moreover, transcripts of negatively regulated genes are usually degraded following their continuous interaction with sRNAs, as it occurs under natural steady state conditions. Consequently, regulated induction of the sRNA provides the ability to identify putative targets under conditions when they are highly abundant and before they are degraded.
We applied the GRIL-Seq method using the PrrF1 sRNA and were able to show its interaction with a group of transcripts, 17 of which were also de-stabilized following overexpression of the sRNA (Fig. 5). Examination of these PrrF1 targets showed that the ligation reaction in the majority (12) of mRNAs occurred at sites near the sequence specifying the amino terminus of each translated protein, presumably reflecting the location of a preferred endonucleolytic cleavage site. Predicted base-pairing interactions, generated using the IntaRNA algorithm, were in all cases located 5′ to the ligation sites and often (but not always) contained the sites of initiation of translation (Shine-Dalgarno sequence, S/D) and the start codon. These results are consistent with the generally accepted model for sRNA regulation, where base-pairing with an mRNA inhibits initiation of translation, inducing endonucleolytic cleavage followed by exonucleolytic degradation of the transcript3, 13, 25. However, several targets formed chimeras in GRIL-Seq with PrrF1 at their 3′ ends and some of them have predictable secondary regions capable of base-pairing with PrrF1 near the sites of initiation of translation. Although the consequences of ligation of an sRNA at 3′ end of mRNA remain to be explored further, we have shown that a 3′ fragment of katA mRNA, which base pairs with PrrF1, relieves the negative effect of this sRNA at the 5′end of the full-length mRNA (Supplementary Note 3).
In addition to identification of putative base-pairing sites of PrrF1-targets, we found that PrrF1 can be linked to two different non-coding RNAs (CrcZ and two aspartyl tRNAs), implying additional uncharacterized roles of PrrF1. CrcZ is an sRNA decoy capable of relieving Hfq-mediated translational repression. It contains A-rich sequences capable of binding to the distal surface of Hfq, previously shown to be the site of mRNA interaction32, while PrrF1 can bind Hfq presumably with 3′ poly U sequences at the proximal surface36, potentially allowing for ligation of the two RNAs in the same Hfq oligomer. Given that they also appear to have sequences to form a stable duplex (hybridization energy, −34.5 kcal/mol) and their ligation appears to occur following the cleavage of both RNAs, they likely interact by base-pairing. However, our RNA-seq results did not show significant changes in CrcZ levels during 20 min of ectopic overexpression of PrrF1 (Supplementary Table 1). It is conceivable that CrcZ functions as an sRNA sponge, sequestering PrrF1. GRIL-Seq also identified ligations of PrrF1 to the 3′ end of three precursor aspartyl tRNAs. Given that Hfq binds to several tRNAs and participates in tRNA processing in E. coli18, 37, this finding suggests that sRNAs (such as PrrF1) may also participate in some unknown aspects of tRNA function.
As illustrated with PrrF1, the GRIL-Seq method leads to the detection of a large number of chimeras, some of which could represent products of random ligations, particularly to highly abundant RNAs. However, our analysis of the distribution of chimeras revealed that the ligation junctions are limited to a small number of internal RNase cleavage sites, and these are independent of transcript abundance. Moreover, base-pairing between the sRNA and a transcript is essential for their ligation. Therefore, GRIL-Seq is a sensitive method for detecting base-paired RNA molecules and, in the case of sRNAs, these can have biological consequences such as enhanced degradation of targeted transcripts. Alternatively, an sRNA may block translation without significantly affecting mRNA stability; translational repression, in the absence of mRNA degradation was shown to be sufficient for several sRNAs to exert their full regulatory effect38, 39. Finally, the list of ligation products between any specific sRNA and various transcripts could also include potential “false positives”. These chimeras represent RNAs capable of base-pairing with an sRNA to form sufficiently stable structures that could be preferentially ligated to transcripts at sites created by endo- or exo-nuclease cleavage during normal RNA turnover25.
The GRIL-Seq method constitutes a simple and highly sensitive method for detecting interactions between various partially complementary transcripts in vivo, a key stage in the regulatory mechanism of non-coding sRNAs. When combined with computational sRNA target prediction algorithms, it appears to be a valuable tool to predict base-pairing regions between sRNAs and target transcripts. Conceptually, GRIL-Seq is analogous to the ChIP-seq method where the binding sites for regulatory proteins are mapped to a genome, with one major difference being that the site on the target ligated to the sRNA is usually generated by an endonucleolytic cleavage and can be located at a substantial distance from the site of base pairing between the two transcripts. GRIL-Seq is easily scalable, with the use of multiple oligonucleotides for the enrichment step; this could be applicable to multiplexing dozens of sRNAs for a single sequencing reaction. Moreover, given the performance of modern sequencing platforms, studies of sRNA regulation without the need for overexpression may soon be possible, allowing mapping of all sRNA targets under a specific set of conditions in a single experiment.
During the revision of this manuscript, another group published a method (RIL-seq) for the identification of Hfq-bound pairs of small RNAs and their targets40. It relies on capturing crosslinked sRNAs/Hfq complexes with antibody, subsequent RNase A/T1 digestion, ligation, protease treatment and sequencing. GRIL-Seq differs from RIL-seq not only in its simplicity and technical details (mainly the generation of the chimeras occurs in vivo) but it can also identify non-Hfq dependent targets of sRNAs that may represent a substantial fraction of sRNA targets41. Nevertheless, both of these techniques represent an important addition to the molecular toolbox and should greatly facilitate sRNA research.
Methods
Oligonucleotides and plasmids
The oligonucleotides and plasmids used in this study are listed in Supplementary Table 4 and 5, respectively. Most subclonings were carried out using an in-fusion cloning kit (Clontech) and when required, restriction enzyme treatment and T4 DNA ligation were performed. For engineering chromosomal mutations in P. aeruginosa and fusion of genes, the SOEing PCR method was employed and plasmid pEXG2 was used as described previously42. To construct the plasmid pKH13, the plasmid pPSV40 was used as a backbone of the pKH13 with some modifications. The two restriction sites, HindIII and SmaI in pPSV40, were digested and then removed by generating blunt ends using Klenow polymerase and re-ligated to generate pPSV40-1. The plasmid pPSV40-1 was linearized by EcoRI and XhoI. The Lac repressor gene (lacIq), promoter (Ptac) and rrnB terminator (rrnB T12) were amplified from plasmid pMMB67EH by using oligonucleotides (F_XhoI_pKH5/R_ERI_rrnBT) and cloned into the linearized pPSV40-1 to generate the plasmid pKH7. To replace the GenR marker (aacC1) in pKH7 with the CarbR resistance gene (bla) of pMMB67EH was amplified using oligonucleotides (F_Bla_gsn/R_Bla_gsn) and cloned into the pKH7 plasmid backbone amplified with oligonucleotides (F_pKH11_bla vec_gs/R_pKH11_bla vec_gs) to generate plasmid pKH11. To obtain tight control of the Ptac promoter, the promoter sequence of plasmid pBTK27, containing two lac repressor gene operators (lacO), was amplified using oligonucleotides (F_pspOMI_lacIq/R_pKH13_pBTK27) and then cloned to generate pKH13. The plasmid pKH13-t4rnl1 was constructed via PCR amplification from the original plasmid, pET16b-t4rnl1 (generously provided by Ushati Das, Stewart Shuman’s lab) using oligonucleotides (F_xbI_SD_T4RL_pKH13/R_Hnd3_T4RL_pKH11, Supplementary Fig. 1b and 15). The T4 RNA ligase mutant (K99N) was generated by SOEing PCR using oligonucleotides (F_t4rlK99N/R_ t4rlK99N).
The sRNA overexpression vector, pKH6, was constructed by modifying the PBAD promoter region of pJN105 to allow for the transcription of a cloned gene to start at the same 5′ end as the endogenous RNA. Briefly, the plasmid pJN105 was linearized with MluI and XbaI and a modified transcription site was introduced by cloning the DNA fragments amplified using oligonucleotides (F_MluI_pJN150/R_ERI_XbI_pJN150). The terminator sequence of rrnB (rrnB T12) amplified from pMMB67EH using oligonucleotides (F_PstI_rrnBT/R_sacI-rrnBT) was inserted between the PstI and SacI sites of the pJN105. To remove the additional lac promoter site located downstream of araC in pKH6, the vector was amplified with oligonucleotides (R_phos_notI_pKH6/F_phos_aac65_pKH6) and re-joined using T4 DNA ligase. To construct plasmids pKH6-PrrF1 and pKH6-RyhB, the prrf1 gene from PAO1 and the ryhB gene from MG1655 (Supplementary Fig. 15), containing transcription start site (in the case of RyhB, an adenosine was added) and its native terminator was amplified using oligonucleotides (F_XbaI_prrf1+1/R_Hd3_prrf1+135 and F_XbaI_ryhb+1A/R_Hd3_ryhb+113) and inserted between the XbaI and HindIII sites of pKH6 (Supplementary Fig. 1a). The mutations in PrrF1 (M1 to M3) were engineered by SOEing PCR with each oligonucleotides pair listed on Supplementary Table 5.
Bacterial strains
A list of bacterial strains created for this study is provided in Supplementary Table 4. Pseudomonas aeruginosa PAO1 is referred to as the wild-type strain and was used as a recipient for plasmids or mutant construction. To construct the P. aeruginosa strain KT4P1 containing two plasmids, pKH13(CarbR) and pKH6(GenR) (or their derivatives), sequential mating with the plasmid-bearing E. coli donor was carried out. Briefly, the plasmid pKH13 was first introduced into E. coli strain SM10λpir and then it was used as a donor for transfer to PAO1 wild type (or mutant) strain by mating. This PAO1 (pKH13) strain was then used, in a conjugation experiment, as a recipient for plasmid pKH6 or its derivatives.
Media and growth condition
Bacterial cells were grown aerobically with shaking at 37 °C, in LB (Luria–Bertani) broth with antibiotics as required (for P. aeruginosa, 150 μg/mL carbenicillin, 75 μg/mL gentamicin, 25μg/mL irgasan, 75 μg/mL tetracycline: for E. coli, 50 μg/mL carbenicillin, 15 μg/mL gentamicin). For the overnight seed culture of P. aeruginosa, strain KT4P1 carrying two plasmids, pKH13-t4rnl1 and pKH6-PrrF1, was cultured in LB with gentamicin and carbenicillin and then diluted into the same media to an optical density at 600 nm (OD600) of 0.01. For determination of cell viability following induction of the T4RNA ligase expression, IPTG (final conc., 1mM) was added when the culture of P. aeruginosa PAO1 (pKH13-t4rnl1) reached an OD600 ~ 0.5. At each time point, aliquots were withdrawn and serially diluted with LB medium. The duplicate cultures from the serial dilution were spread on LB plates containing carbenicillin (150 μg/mL) and incubated for 18 h at 37°C. The number the viable cells was determined and averaged from three independent experiments.
In silico prediction of base-pairing between sRNA and target RNAs
IntaRNA software30 was used to predict interactions between sRNA and target RNAs. For the prediction of interactions between PrrF1 and sodB (or PA4880), the previously published prediction22 was used. The regions of interaction between RyhB and targets (sodB, sdhD and shiA) in E. coli were also based on previously published work23, 43, 44.
Preparation of cells for GRIL-Seq and RNA-seq
For GRIL-Seq, the details are described in Supplementary Note 5. Briefly, overnight cultures of P. aeruginosa KT4P1 carrying two plasmids, pKH13-t4rnl1 and pKH6-PrrF1, was cultured in LB with gentamicin and carbenicillin and then diluted into the same media to OD600 of 0.01. The T4 RNA ligase was induced when a culture reached an OD600 of ~ 0.5 by addition of IPTG (final concentration, 1mM). After 1h, PrrF1 was induced for 20 min by addition of L-arabinose to 0.2%. The cells were harvested by centrifugation at 13,000g for 1 min and the pellets were frozen in liquid nitrogen for the subsequent RNA isolation. The same protocol of was followed for RNA-seq using strain PAO1Δprrf1Δprrf2.
RNA isolation
Total RNA was isolated using the Direct-zol™ RNA MiniPrep (Zymo), following the manufacturer’s instructions with some modifications. Cells from a 1.6 mL volume of culture (equivalent to 5 OD600 units) were collected by centrifugation (13,000g, 40 s) and the supernatant was discarded. To stop additional transcription, the pellets were immediately snap-frozen using liquid nitrogen. 700 μL of TRI Reagent® was added to the frozen pellet in a 1.5 mL centrifuge tube and cells were immediately lysed by vigorous vortexing (2 min). Cell debris was removed by centrifugation (13,000g, 1 min) and 650 μL of the lysate was transferred into a fresh tube containing the same volume of 100% ethanol. The ethanol mixture was transferred into Zymo-Spin™ IIC Column in a collection tube and centrifuged. The flow-through was discarded and this was repeated until all of the mixture was transferred. The column with bound RNAs was washed according to the manufacturer’s protocol. For RNA elution, 40 μL nuclease-free water was used.
Northern blot
For PrrF1 and 5S RNA, total RNA (5 μg) was mixed with RNA loading buffer II (Ambion) and denaturated at 95 °C for 3 min. The RNA was fractionated on 5% polyacrylamide/7 M urea gel in 1X TBE. For sodB mRNA, total RNA (10 μg) was mixed with RNA loading buffer (62% formaldehyde, 8% glycerol, 0.02% bromophenol blue and 0.02% xylene cyanol) and denaturated at 95 °C for 3 min. The RNA was separated by 1.5% agarose gel containing EtBr in 1× MOPS buffer. The fractionated RNAs were electrotransferred to a Hybond N+ membrane (GE Healthcare) and crosslinked on the membrane using the Stratalinker UV crosslinker on the Autocrosslink setting (2 times, 120,000 μJ/cm2). After prehybridization with Rapid-hyb buffer (GE Healthcare), a 32P-labeled oligonucleotide probe was hybridized at 43 °C and the membrane was washed with 2× SSC/0.1% SDS and 0.5× SSC/0.1% SDS buffer. The signal was visualized on Typhoon FLA 7000 (GE Healthcare). To detect PrrF1, 5S RNA and sodB mRNA, oligonucleotides listed in Supplementary Table 5 (R_PrrF1+46, R_5S+90 and R_sodB+24) were used for 5′ end labeling with [32P]γ –ATP, respectively.
Quantitative Real-Time PCR
For RNA quantification, One-Step qRT-PCR was carried out as previously described 36. The primers used for qRT-PCR are listed in Supplementary Table 5.
Enrichment of chimeric sRNA
1.6 mL of cells (5OD600) collected by centrifugation and liquid nitrogen frozen cells were broken with 700 μL of TRI Reagent®. DNA was removed on a column with 8 μL of DNase I (2 U/μL) at 37 °C for 30 min. Total RNA was precipitated with two volumes of 100% ethanol at −80 °C and 30 μL of nuclease free water was used for RNA recovery. Total RNAs (10 μg) with RIN numbers of more than 7 were used for the enrichment of sRNA. To enrich the samples for the chimeras containing the sRNA, the MICROBExpress™ Kit (Life Technologies) was used with some modification. As the Capture Oligo Mix, 5′ poly dA-tailed (18 mer) PrrF1 binding complementary oligonucleotides were designed based on the proper melting temperature (55–61 °C, 23 mer) and GC contents (48–65%) at the hybridization regions of sRNA (Supplementary Table 5). 1 μL of Capture Oligo Mix (20 μM) was mixed with the total RNA (10 μg, 14 μL) and the Binding Buffer (200 μL) was added as recommended by the manufacturer. Denaturation of secondary structure of RNAs and the Capture Oligo was carried out at 70 °C for 15 min and then the RNA/Capture Oligo mixture was incubated at 37 °C for 1 h for the specific annealing of the Capture Oligo to the sRNA. The preparation of oligo-dT containing magnetic beads (Oligo MagBeads) was carried out according to the manufacturer’s instructions. For “sandwich” hybridization of sRNA-Capture Oligo and Oligo MagBeads, 50 μL of the pre-equilibriated (37 °C) Oligo MagBeads was added to the RNA/Capture Oligo mixture and incubated 37 °C for 15 min. Wash Solution prepared as described previously45 was pre-equilibriated at 37 °C for 15 min before washing the beads. After magnetic capture with the Oligo MagBeads, they were washed three times by pipetting 300 μL of Wash Solution and discarding the supernatant from the collected Oligo MagBeads. RNA bound to the Oligo MagBeads was eluted by resuspending the beads in 50 μL of TURBO DNase I (3 U) and incubating at 37 °C for 25 min. The eluted RNA (in 47 μL) was collected and nuclease-free water (153 μL) was added to reach 200 μL. The enriched RNA was precipitated overnight at −80 °C, with 500 μL of 100% ethanol (2.5 volume), 20 μL of 0.1 M sodium (0.1 volume) acetate and 4 μL of glycogen (5 μg/μL).
Preparation of cDNA library for RNA-Sequencing
The precipitated enriched chimeric sRNA was recovered in 15 μL of nuclease free water. The enriched RNAs (100 ng) were used for cDNA library preparation. Library construction for Illumina sequencing was carried out using the NEB Next® Ultra™ Directional RNA Library Prep Kit for Illumina (New England Biolab) following manufacturer’s instructions (Supplementary Notes 5) The Index primers used in this study are listed in Supplementary Table 5. To enrich the libraries, 16 cycles of PCR reaction were carried out at the last enrichment step. To obtain high purity of the library from the adaptor dimer, the bead purification was carried out two times at the last step.
Detection of sRNA –target chimeras by RT-PCR
To remove residual DNA from total RNAs, 10 μg of RNA sample in a 50 μL reaction volume was treated with TURBO DNase (4 U) for 25 min, followed by its inactivation for 5 min using TURBO DNA-free Kit (Thermo Fisher Scientific). Total RNA was recovered (45 μL) by centrifuge at 10,000g for 2 min and the total RNA (1 μg) was converted to cDNA using SuperScript III First-Strand Synthesis system (Invitrogen) with random hexamer. When required, 5 pmol of gene specific primer was used for the gene specific cDNA synthesis. Reverse transcription was carried out at 50 °C for 1 h and terminated at 85 °C for 10 min. The residual RNA was removed using 2 μL of an enzyme mixture containing RNase H (2.5 U, New England Biolab) and Riboshredder (0.5 U, Epicentre) for each reaction. Approximately 10% of the reaction was used as the template for PCR amplification using GoTaq Green Master Mix (Promega) and the primer pairs listed in Supplementary Table 5. Cycling conditions were: 95 °C/3 min; 30 cycles of 94 °C/25 sec, 58 °C/25 sec, 72 °C/60 sec and a final 72 °C/5 min. The PCR products were separated by 2% agarose gel electrophoresis; DNA bands were eluted and cloned into pJET1.2 vector (Thermo Fisher Scientific) as recommended by the manufacturer’s instructions and the inserts were sequenced using the pJET1.2 reverse primer.
RNA-seq data analysis
Sequencing reads were aligned to the PAO1 genome using the Rockhopper system46. The two biological replicate RNA-seq experiments corresponding to the PrrF1-PrrF2 deletion strain resulted in 7,520,964 sequencing reads and 8,655,351 sequencing reads, of which 87% mapped to the reference genome. The two biological replicate RNA-seq experiments corresponding to PrrF1 overexpression resulted in 5,687,213 sequencing reads and 9,884,669 sequencing reads, of which 88% mapped to the reference genome. Sequencing read data was normalized using upper quartile normalization47. Differential expression in the two conditions was tested using the approach of DESeq248, where p-values were computed to indicate the probability of observing each gene’s two expression levels, in the two conditions, by chance. Because multiple tests were performed across the set of genes, p-values were corrected to q-values in order to control the false discovery rate at less than 1%49.
Data availability
The sequencing data have been deposited in NCBI-SRA under accession no. SAMN05933141, SAMN05933142, SAMN05933143, SAMN05933144, SAMN05933145, SAMN05933146.
Acknowledgments
We thank Ushati Das and Stewart Shuman for the gift of a recombinant plasmid with the T4 RNA ligase gene and William Robins for help with Illumina sequencing. We thank Thomas Dougherty for critical reading of the manuscript. This work was supported by NIH Grant R37 AI021451 to SL and R15 GM102755 to BT.
Footnotes
Author contributions
S.L. and K.H. conceived of the approach. K.H. carried out all of the experiments. S.L., K.H., and B.T. analyzed the expression as well as GRIL-Seq data. S.L., K.H., and B.T. wrote the paper.Supplementary information is available for this paper.
Competing Financial Interests
The authors declare no conflict of interest.
References
Full text links
Read article at publisher's site: https://doi.org/10.1038/nmicrobiol.2016.239
Read article for free, from open access legal sources, via Unpaywall:
https://www.ncbi.nlm.nih.gov/pmc/articles/PMC5567838
Citations & impact
Impact metrics
Citations of article over time
Alternative metrics
Smart citations by scite.ai
Explore citation contexts and check if this article has been
supported or disputed.
https://scite.ai/reports/10.1038/nmicrobiol.2016.239
Article citations
Global analysis of the RNA-RNA interactome in Acinetobacter baumannii AB5075 uncovers a small regulatory RNA repressing the virulence-related outer membrane protein CarO.
Nucleic Acids Res, 52(18):11283-11300, 01 Oct 2024
Cited by: 2 articles | PMID: 39149883 | PMCID: PMC11472050
RNA interactome of hypervirulent Klebsiella pneumoniae reveals a small RNA inhibitor of capsular mucoviscosity and virulence.
Nat Commun, 15(1):6946, 13 Aug 2024
Cited by: 0 articles | PMID: 39138169 | PMCID: PMC11322559
Hfq-Antisense RNA I Binding Regulates RNase E-Dependent RNA Stability and ColE1 Plasmid Copy Number.
Int J Mol Sci, 25(7):3955, 02 Apr 2024
Cited by: 0 articles | PMID: 38612765 | PMCID: PMC11012335
ChimericFragments: computation, analysis and visualization of global RNA networks.
NAR Genom Bioinform, 6(2):lqae035, 17 Apr 2024
Cited by: 0 articles | PMID: 38633425 | PMCID: PMC11023125
Agnodice: indexing experimentally supported bacterial sRNA-RNA interactions.
mBio, 15(3):e0301023, 06 Feb 2024
Cited by: 0 articles | PMID: 38319109
Go to all (53) article citations
Data
Data behind the article
This data has been text mined from the article, or deposited into data resources.
BioStudies: supplemental material and supporting data
Similar Articles
To arrive at the top five similar articles we use a word-weighted algorithm to compare words from the Title and Abstract of each citation.
Toward a Comprehensive Analysis of Posttranscriptional Regulatory Networks: a New Tool for the Identification of Small RNA Regulators of Specific mRNAs.
mBio, 12(1):e03608-20, 23 Feb 2021
Cited by: 8 articles | PMID: 33622723 | PMCID: PMC8545128
Mapping the small RNA interactome in bacteria using RIL-seq.
Nat Protoc, 13(1):1-33, 07 Dec 2017
Cited by: 38 articles | PMID: 29215635
Conditional Hfq Association with Small Noncoding RNAs in Pseudomonas aeruginosa Revealed through Comparative UV Cross-Linking Immunoprecipitation Followed by High-Throughput Sequencing.
mSystems, 4(6):e00590-19, 03 Dec 2019
Cited by: 14 articles | PMID: 31796567 | PMCID: PMC6890931
Target activation by regulatory RNAs in bacteria.
FEMS Microbiol Rev, 39(3):362-378, 30 Apr 2015
Cited by: 123 articles | PMID: 25934124 | PMCID: PMC4542691
Review Free full text in Europe PMC
Funding
Funders who supported this work.
NIAID NIH HHS (2)
Grant ID: R37 AI021451
Grant ID: R21 AI125972
NIGMS NIH HHS (1)
Grant ID: R15 GM102755





