Abstract
Free full text

Rb1 and Trp53 cooperate to suppress prostate cancer lineage plasticity, metastasis, and antiandrogen resistance
Associated Data
Abstract
Prostate cancer relapsing from antiandrogen therapies can exhibit variant histology with altered lineage marker expression, suggesting that lineage plasticity facilitates therapeutic resistance. The mechanisms underlying prostate cancer lineage plasticity are incompletely understood. Studying mouse models, we demonstrate that Rb1 loss facilitates lineage plasticity and metastasis of prostate adenocarcinoma initiated by Pten mutation. Additional loss of Trp53 causes resistance to antiandrogen therapy. Gene expression profiling indicates that mouse tumors resemble human prostate cancer neuroendocrine variants; both mouse and human tumors exhibit increased expression of epigenetic reprogramming factors such as Ezh2 and Sox2. Clinically relevant Ezh2 inhibitors restore androgen receptor expression and sensitivity to antiandrogen therapy. These findings uncover genetic mutations that enable prostate cancer progression; identify mouse models for studying prostate cancer lineage plasticity; and suggest an epigenetic approach for extending clinical responses to antiandrogen therapy.
As molecularly targeted cancer therapy improves, lineage plasticity is increasingly appreciated as a potential mechanism underlying therapeutic resistance. Lineage plasticity facilitates conversion of a cancer cell that is dependent on the therapeutic target to one that is indifferent to its function. For example, relapse of EGFR (epidermal growth factor receptor) mutant lung adenocarcinomas after EGFR-targeted therapy is associated with the appearance of histologically distinct variants that lack EGFR expression but express neuroendocrine lineage markers such as SYP (1, 2). Likewise, prostate adenocarcinoma (PADC) relapsing from antiandrogen therapies (ADTs) is associated with histological variants exhibiting altered histology, reduced androgen receptor (AR) levels, and expression of neuroendocrine markers (3–5). These neuroendocrine prostate cancer variants (NEPCs) emerge from PADC because they share clonal origin (5–8). The identification of effective therapies for NEPCs has been hindered by incomplete understanding of the mechanisms driving lineage plasticity and the lack of relevant experimental models.
The retinoblastoma tumor suppressor gene RB1 is more commonly mutated in metastatic and ADT-recurrent prostate cancer—NEPC variants in particular—than it is in primary tumors (5, 9–12). This suggests that there is selective pressure for RB1 loss during tumor evolution and that loss of this gene might drive PADC progression and lineage plasticity. To test this hypothesis, we engineered Rb1 deletion in a previously characterized mouse model of PADC initiated by Pten mutation (13). In the original model, the PBCre4 transgene (14) is used to delete floxed Pten alleles specifically in prostate epithelium (fig. S1). PBCre4:Ptenf/f mice, where f designates a floxed allele, develop prostatic intraepithelial neoplasia (PIN) by 6 weeks of age and invasive PADC by 9 weeks, but these cancers rarely progress to metastatic disease (13, 15–17). Prostate cancer in PBCre4:Ptenf/f:Rb1f/+ mice is similar, so both genotypes are used interchangeably here and are referred to as single knockout (SKO). Rb1 mutation alone is insufficient to initiate prostate cancer development in the mouse because PBCre4:Rb1f/f mice do not develop prostate cancer (18, 19). The combination of these mutations in PBCre4:Ptenf/f:Rb1f/f (DKO) mice leads to prostate cancer development, and the mice had a significantly shorter median survival of 38 weeks compared with 48 weeks for SKO mice (Fig. 1A). Rb1 loss did not affect end-stage tumor cell proliferation significantly, but similar to the loss of the tumor suppressor gene Trp53 (17), Rb1 loss abrogated the cellular senescence that occurs in Pten-deficient, premalignant prostate epithelium (fig. S2).
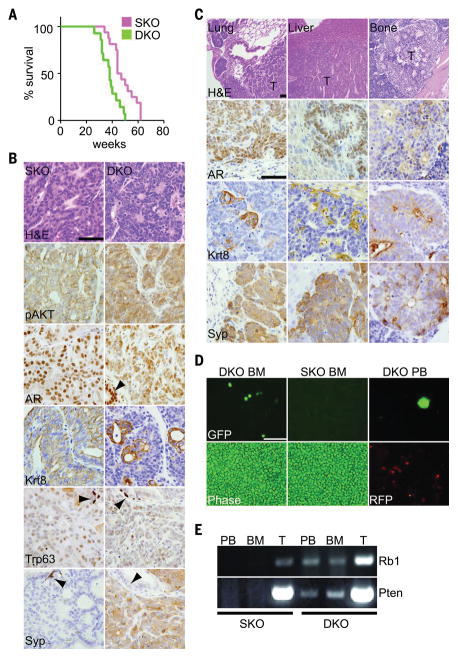
(A) Survival plot showing a significant difference in survival of SKO (n = 16) and DKO (n = 14) mice (log rank P = 0.0013). (B) End-stage tumor sections stained with hematoxylin and eosin (H&E) or antibodies against the indicated proteins. Arrowheads indicate uninvolved prostate epithelium. Scale bars, 100 μm. (C) Sections of DKO metastases from indicated tissues stained and presented as in (B). (D) Bone marrow (BM) or peripheral blood (PB) from SKO and DKO mice was imaged under phase or fluorescent microscopy. Cancer cells were genetically marked with green fluorescent protein (GFP), and normal cells were marked with red fluorescent protein (RFP). Scale bar, 100 μm. (E) Polymerase chain reaction (PCR) was used to detect Cre-deleted alleles in PB, BM, or tumor DNA (T).
End-stage SKO PADC showed expression of phosphorylated AKT (pAKT), nuclear AR, and the luminal epithelial marker Krt8 (Fig. 1B). Expression of the basal epithelial marker Trp63 was low, and expression of the neuroendocrine marker Syp was undetectable. DKO PADC also showed expression of pAKT, but Krt8 and AR levels were heterogeneous between cells and regionally within contiguous tumors (Fig. 1B and fig. S3A). DKO PADCs also contained cells expressing Syp. Cells surrounding acini were Krt8high:Syplow, whereas cells interspersed between acini were Krt8low:Syphigh (fig. S3B), suggesting the presence of at least two molecularly distinct cell populations within these tumors.
Metastasis was not detected in SKO mice, which is consistent with previous reports (15–17). In contrast, distant metastasis was detected in all DKO mice examined to date (Fig. 1C). Common metastatic sites were lymph node, lung, and liver. Bone metastasis was detected in 2 of 10 mice; this is likely an underestimate because we examined only a tibia and femur. All metastases recapitulated the heterogeneous Syp and Krt8 expression pattern of the primary tumors. Metastases disseminated through the vasculature because DKO cancer cells marked by green fluorescent protein (GFP) in PBCre4:Ptenf/f:Rb1f/f:RosamT/mG mice (20) were detected in both peripheral blood and bone marrow (Fig. 1, D and E). These observations suggest that Rb1 suppresses metastatic dissemination of PADC initiated by Pten loss.
The existence of both luminal-like Krt8high: Syplow cells and neuroendocrine-like Krt8low: Syphigh cells within DKO primary and metastatic tumors suggests that these cancers exhibit lineage plasticity, but other explanations are possible. To explore whether molecular heterogeneity is a consequence of polyclonal tumors, we incorporated the Brainbow 2.1 lineage tracing allele (21) into DKO mice. All end-stage tumors were monocolor, with one and sometimes two independent primary tumors per mouse (fig. S3C). Thus, DKO tumors, both primary and metastatic, were likely derived from a single neoplastic cell clone. Mosaic Cre-mediated gene deletion within tumor clones may also contribute to molecular heterogeneity. RNA-sequencing (RNA-seq) analysis of tumor specimens revealed a few reads mapping to Cre-deleted exons (fig. S3D); these reads are likely contributed by non-epithelial cells that contaminate the bulk tumor specimens. Immunostaining of tumor tissue sections also failed to detect cells expressing protein encoded by the deleted gene (fig. S3E). Thus, it is unlikely that mosaic Cre deletion accounts for the molecular heterogeneity observed.
To characterize the origin of Syphigh cells, we performed a longitudinal study of DKO PADC development. PIN lesions and early invasive PADC were apparent in DKO mice by 12 weeks of age, but these neoplastic lesions lacked Syphigh cells (fig. S3F). Small foci of Syphigh cells were readily detectable by 20 to 25 weeks of age. These early Syphigh foci are Krt8low but still express AR (fig. S3G). These observations are consistent with the derivation of Krt8low:Syphigh cells from preexisting Krt8high:Syplow neoplasia. We cannot, however, exclude the possibility that Krt8low:Syphigh cells arise independently, perhaps from a different cell type of origin, and they subsequently give rise to Krt8high:Syplow cells in tumors. Either scenario is consistent with the hypothesis that Rb1 loss enhances lineage plasticity of prostate neoplasia initiated by Pten loss.
Previous work has shown that PBCre4:Rb1f/f: Trp53f/f mice develop ARlow, castration-resistant, NEPC-like tumors exclusively from within the stem cell–rich proximal region of the prostate (18, 22). DKO tumors, in contrast, appeared in the anatomically distinct distal region of the prostate (fig. S4A). Whereas DKO PIN lesions were detected within the proximal prostate, they exhibited an ARhigh:Syplow phenotype; in contrast, the earliest PIN lesions detected in PBCre4:Rb1f/f:Trp53f/f mice were ARlow:Syphigh (fig. S4B). The AR and Krt8 immunostaining patterns were different in DKO and PBCre4:Rb1f/f: Trp53f/f tumors (fig. S4C). These observations suggest that the cell of origin is different in DKO and PBCre4:Rb1f/f:Trp53f/f prostate cancers.
PBCre4:Rb1f/f:Trp53f/f tumors are ADT-resistant de novo because castration does not extend survival (18). However, the AR levels in DKO tumors are higher (fig. S4D), suggesting that they may be ADT sensitive. To test this, we surgically castrated 30-week-old, tumor-bearing DKO mice and monitored their survival. Castration extended median survival of DKO mice from 38 to 48 weeks (Fig. 2A), but all mice eventually died from prostate cancer by 67 weeks. The response of individual mice varied considerably, with postcastration survival ranging from 3 to 37 weeks, but survival did not correlate consistently with precastration total prostate volume (Fig. 2, B and C). DKO cancer that relapsed after castration retained heterogeneous Krt8 and Syp expression but showed markedly reduced AR levels (Fig. 2D). Bone metastasis was detected in three of eight mice with postcastration recurrent disease. We found that two of five postcastration recurrent cancers analyzed with RNA-seq had acquired spontaneous Trp53 mutations (Fig. 2E). The Trp53 mutations are analogous to loss-of-function R282Q and V173M mutants found commonly in human cancers. Trp53 mutations were not detected in eight DKO tumors from noncastrated mice. This implies that Trp53 mutation cooperates with Rb1 loss to confer an ARlow, ADT-resistant phenotype.
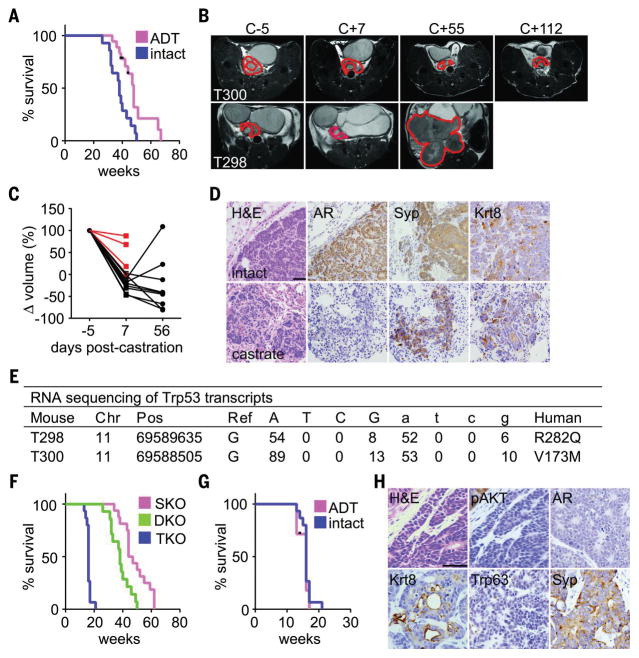
(A) Survival plot for DKO mice, either intact (n = 14) or castrated at 30 weeks (n = 18, log rank P = 0.003). Black tic marks represent mice alive at the end of the study. (B) Axial T2-weighted MR images from two mice acquired at indicated times relative to castration. The prostate and resulting tumors are outlined in red. T300 initial prostate volume (68 mm3) was larger than T298 (21 mm3), but T300 survived longer after castration (37 weeks) than did T298 (12 weeks). (C) Prostate volumes of DKO mice were measured with magnetic resonance imaging (MRI) 5 days before castration at 30 weeks of age, or 1 and 8 weeks after castration. The plot shows relative prostate volumes for individual mice over time normalized to their precastration measurement. Red lines indicate mice that died before the 8-week MRI time point. (D) Tumor sections from intact or postcastration recurrent DKO tumors stained with H&E or antibodies directed against the indicated proteins. AR levels decline in postcastration recurrent tumors. Scale bar, 100 μm. (E) RNA-seq data from postcastration recurrent DKO tumors indicate that 88% of reads mapping to Trp53 have mutations analogous to loss-of-function mutations commonly found in human cancer. Single-letter abbreviations for the amino acid residues are as follows: A, Ala; C, Cys; D, Asp; E, Glu; F, Phe; G, Gly; H, His; I, Ile; K, Lys; L, Leu; M, Met; N, Asn; P, Pro; Q, Gln; R, Arg; S, Ser; T, Thr; V, Val; W, Trp; and Y, Tyr. In the mutants, other amino acids were substituted at certain locations; for example, R282Q indicates that arginine at position 282 was replaced by glutamine. (F) Survival plot showing significantly different survival of SKO (n = 16), DKO (n = 14), and TKO (n = 15) mice (log rank P < 0.0001). (G) Survival plot of TKO mice, either intact (n = 15) or castrated at 10 weeks (n = 7). Castration does not affect survival significantly (log rank P = 0.46). Black tic marks represent mice alive at the end of the study. (H) Tumor sections from TKO mice were stained with H&E or antibodies against the indicated proteins. Scale bar, 100 μm.
To test this hypothesis, we bred PBCre4:Ptenf/f: Rb1f/f:Trp53f/f (TKO) mice. TKO mice developed aggressive prostate cancer, limiting median survival to 16 weeks (Fig. 2F). Metastasis to the lung, liver, and bone (one of four mice examined) was detected in TKO mice despite their short life span (fig. S5A). Castration of TKO mice at 10 weeks did not extend survival (Fig. 2G), demonstrating that TKO PADC was ADT-resistant de novo. Primary TKO tumors showed patchy Krt8 and Syp expression like DKO tumors but had very low AR levels comparable with that of postcastration recurrent DKO tumors (Fig. 2H). Surprisingly, pAKT levels were also reduced in TKO tumors despite uniform Pten loss (fig. S5B). Thus, TKO tumors have reduced dependence on both AKT and AR signaling. We also examined young PBCre4:Ptenf/f:Rb1f/f:Trp53f/+ mice retaining one wild-type copy of Trp53. In these mice, the appearance of Syp+ neoplastic cells occurred earlier than in DKO mice and was coincident with reduced levels of the Trp53 target gene Cdkn1a (fig. S5C). This suggests that spontaneous loss of the remaining wild-type Trp53 allele facilitates the onset of Syp expression. These observations support the conclusion that Rb1 and Trp53 cooperate to suppress prostate cancer lineage plasticity underlying the development of ADT-resistant NEPC variants.
We next compared the gene expression patterns of SKO, DKO, and TKO tumors with that of normal prostate tissue. Gene expression patterns clustered primarily by genotype (fig. S6A). DKO and TKO tumors exhibited widespread changes in gene expression, encompassing ~15% of mouse genes (Fig. 3A). DKO and TKO tumors shared 85 to 90% of their differentially expressed genes. This similarity in gene expression between DKO and TKO tumors was not due to spontaneous silencing of Trp53 expression in DKO tumors (fig. S6B). This implies that the primary driver of gene expression reprogramming in DKO and TKO tumors is Rb1 loss.
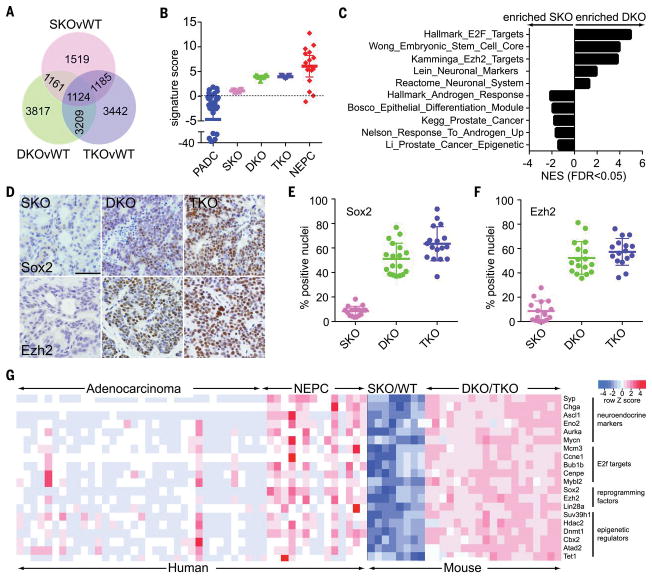
(A) Venn diagrams showing the number of differentially expressed genes between the indicated genotypes (wild type, WT; n = 4 or 5 mice per genotype). (B) Plot showing the signature scores for mouse (SKO, DKO, and TKO) and human (PADC and NEPC) prostate cancer by using the Beltran et al. (23) weighted gene expression signature. Dots represent individual patients. Bars represent the mean and interquartile range. (C) Selected gene sets enriched in DKO versus SKO tumors, with the x axis representing normalized enrichment score (NES). (D) Tumor sections stained with antibodies directed against indicated proteins. Scale bar, 100 μm. (E) Quantitation of Sox2 immunostaining in tumor sections of the indicated genotypes. Each dot represents one analyzed image taken from three different mice for each genotype, with bars representing the mean and standard deviation. Sox2 immunostaining in DKO tumors is greater than in SKO tumors (t test P < 0.0001) and greater in TKO tumors than in DKO tumors (P = 0.01). (F) Quantitation of Ezh2 immunostaining as in (E). Ezh2 immunostaining is greater in DKO tumors than in SKO tumors (P < 0.0001), but immunostaining in DKO and TKO tumors is not significantly different (P = 0.25). (G) A heat map comparing gene expression data from human (5, 23) and the indicated mouse specimens. The select genes deregulated in DKO and TKO tumors are similarly deregulated in human NEPC.
Consistent with immunostaining results, DKO and TKO tumors exhibited decreased levels of AR RNA and elevated levels of neuroendocrine lineage marker RNA (Figs. 1B and and2H2H and fig. S6C). These changes are similar to those observed in human NEPC variants. To compare prostate cancer in these mouse models with the human disease, we used a weighted gene expression signature developed by Beltran et al. (5, 23) to distinguish human adenocarcinoma from NEPC variants. We found that SKO cancer was similar to human PADC, whereas DKO and TKO cancers were similar to human NEPC (Fig. 3B). The Beltran signature accurately distinguishes SKO from DKO/TKO tumors through hierarchical clustering (fig. S6D).
Gene set enrichment analysis revealed that DKO and TKO tumors have altered expression of E2F target genes and neuroendocrine lineage genes (Fig. 3C). We also noted altered expression of gene sets related to stem cells and epigenetic reprogramming, including increased expression of Sox2 [SRY (sex determining region Y)-box 2] and Ezh2 (histone methyltransferase enhancer of zeste homolog 2) (Fig. 3, D to F). Gene expression changes corresponding to stem cell reprogramming factors and neuroendocrine lineage markers were associated with a switch in posttranslational histone H3 modifications from the repressive H3K27me3 mark to the active H3K4me3 mark (fig. S7). Human NEPC also exhibited increased expression of E2F target genes, neuroendocrine markers, stem cell reprogramming factors, and epigenetic regulators (Fig. 3G). Thus, DKO and TKO prostate cancers mimic the molecular phenotype of human NEPC variants, and both human and mouse NEPC variants exhibit deregulation of stem cell reprogramming factors.
Rb1/E2f protein complexes can directly repress expression of Sox2 and Ezh2 (24, 25). We hypothesized that Rb1 loss in prostate cancer derepresses these genes, enabling epigenetic reprogramming toward a stem cell–like state. Lineage plasticity inherent in this state would facilitate metastasis, NEPC transformation, and ADT resistance. This hypothesis predicts that ADT resistance may be reversible through appropriate epigenetic modulation.
Ezh2 is the enzymatic component of the Polycomb Repressive complex 2 that methylates the lysine 27 residue of histone H3. Ezh2 inhibitors (Ezh2i) are being evaluated as cancer therapies in clinical trials. We isolated cell lines from mouse prostate cancers to test whether Ezh2i can restore ADT sensitivity in vitro. The cell lines behave like the tumors from which they were derived. For example, SKO, DKO, postcastration recurrent DKO (DKOCr), and TKO cell lines exhibit differences in AR expression, AR activity, neuroendocrine lineage marker expression, Ezh2 and Sox2 expression, cell proliferation, and motility analogous to the behavior of their corresponding tumors in vivo (fig. S8, A to G). Enzalutamide sensitivity was highest in SKO cells, reduced in DKO cells, and lowest in cells from postcastration recurrent DKO (DKOCr) and TKO tumors (fig. S8H).
We tested two Ezh2 inhibitors (Ezh2i), GSK126 and EPZ6438, and found that they both sensitized TKO and DKOCr cells to enzalutamide when used at concentrations sufficient to inhibit histone H3K27 methylation but without significant single-agent cell growth inhibitory activity (Fig. 4, A to C, and fig. S9, A and B). Ezh2i plus enzalutamide also had a greater effect on DKO cells than either drug as single agents (fig. S9C), and Ezh2i pretreatment inhibited DKO cell motility (fig. S9E). Ezh2i did not significantly alter the enzalutamide sensitivity of SKO cells (fig. S9D), suggesting that its effects are specific for NEPC variants. We also evaluated the effects of Ezh2i in vivo. We propagated a primary DKOCr tumor by transplantation into SCID mice and then treated the mice with Ezh2i and enzalutamide, alone or in combination. Enzalutamide plus Ezh2i treatment slowed tumor growth significantly compared with treatment with enzalutamide [two-way analysis of variance (ANOVA) P = 0.037] or Ezh2i (P = 0.027) alone (Fig. 4D). Enzalutamide treatment alone did not slow tumor growth relative to vehicle control (P = 0.43), confirming that the DKOCr tumor was ADT-resistant.
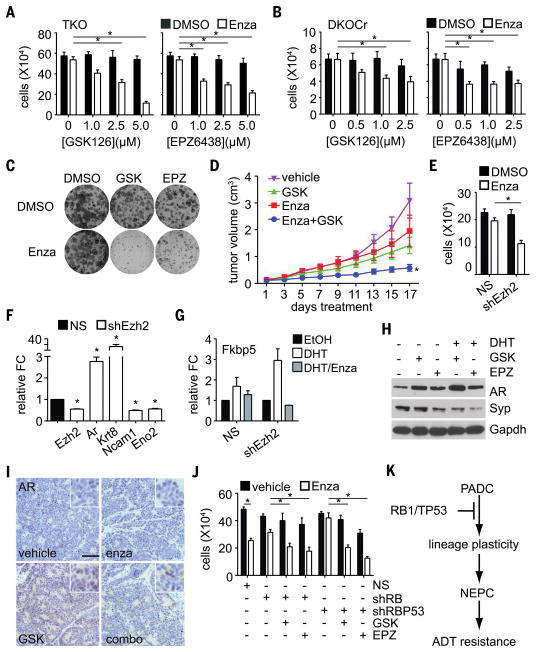
(A) A TKO cell line was treated with enzalutamide or dimethyl sulfoxide (DMSO), with or without Ezh2i, at the indicated concentrations, and the viable cells were then counted. Mean cell number and standard error are shown for three experiments. Asterisks indicate significant differences (P < 0.05). (B) A DKOCr cell line was treated and analyzed as in (A). (C) A DKOCr cell line was plated at low density, then treated as indicated. Resulting colonies were stained 10 days later. A representative result is shown (quantitation is provided in fig. S8A). (D) A DKOCr tumor was transplanted into a cohort of mice, and the mice were treated with GSK503 (GSK) and/or enzalutamide (Enza) as indicated. Tumor volume for each mouse (n = 7 or 8 for each treatment) was recorded every other day. The mean and standard error for all mice are shown. Asterisk indicates significantly slower growth than any of the other treatments (ANOVA, P < 0.05). (E) Ezh2-targeted shRNA (shEzh2), or nonsilencing control (NS), were expressed in DKOCr cells. The cells were then treated with enzalutamide or DMSO, and cell number was measured as in (A). The mean and standard error for three experiments are shown. (F) RNA was extracted from DKOCr cells in (E) and analyzed by means of real-time PCR for the indicated genes. The mean and standard error of fold change (FC) relative to the NS control are shown for two experiments in duplicate. (G) DKOCr cells silenced for Ezh2 as in (C) were treated with AR ligand R1881 (DHT) and/or enzalutamide (Enza), RNA was extracted, and the expression of AR target gene Fkbp5 was assayed by means of real-time PCR. Mean and standard error of FC relative to the NS control are shown for two experiments in duplicate. (H) DKO cells were treated as indicated, and protein extracts were analyzed by means of Western blot for the listed proteins. Gapdh serves as loading control. (I) Tumors dissected from transplanted mice in (D) after 17 days of the indicated treatment were sectioned and immunostained for AR. Inset image is magnified so as to highlight nuclear staining. Scale bar, 100 μm. Ezh2i treatment restores patchy AR expression. (J) LNCaP-AR cells stably expressing RB1 (shRB) or RB1/TP53 shRNA (shRBP53) were treated as indicated, and viable cells were counted as in (A). The mean and standard error of three experiments are shown. (K) A model summarizing the proposed role of Rb1 and Trp53 in suppressing lineage plasticity, neuroendocrine lineage transformation, and ADT resistance.
To genetically verify Ezh2 as the relevant target of Ezh2i, we silenced Ezh2 expression using short hairpin RNA (shRNA). Ezh2 silencing sensitized DKOCr cells to enzalutamide (Fig. 4, E and F). Silencing Ezh2 also increased AR expression, augmented AR activity, increased expression of the luminal lineage marker Krt8, and decreased expression of neuroendocrine lineage markers (Fig. 4, F and G). Ezh2i treatment also increased AR expression and decreased Syp expression in vitro (Fig. 4H). Ezh2i-treated transplanted DKOCr tumors showed evidence of increased AR expression in vivo (Fig. 4I), although immunostaining was patchy with both ARhigh and ARlow regions present. These findings suggest that Ezh2i sensitizes NEPC variants to enzalutamide by reversing or suppressing lineage transformation.
To investigate whether Ezh2i has similar effects in human prostate cancer, we used LNCaP-AR cells from Mu et al. (26) in which RB1 and TP53 expression is stably silenced. Because LNCaP-AR cells were PTEN-null (27), RB1- and RB1/TP53–silenced derivatives were analogous to DKO and TKO mouse cells, respectively. As in mouse cells, RB1 and RB1/TP53 silencing reduced AR levels and enzalutamide sensitivity, but enzalutamide sensitivity was restored by Ezh2i (Fig. 4J and fig. S9F). Thus, enzalutamide resistance is reversible in both human and mouse NEPC variants.
Rb1 and Trp53 repress epigenetic reprogramming factors such as Ezh2 and Sox2, which are important in generating induced pluripotent stem cells (24, 28, 29). The data presented here support a hypothesis in which RB1 and TP53 loss in prostate cancer derepresses these same factors, creating a stem cell–like epigenetic environment permissive for lineage plasticity (Fig. 4K). Lineage plasticity is proposed to drive prostate cancer progression by enabling adaptation to selective pressures experienced during metastasis and ADT. Because the mouse models characterized in this study develop metastatic PADC reminiscent of human NEPC variants, they will be useful for testing this hypothesis and identifying molecular mechanisms underlying cancer lineage plasticity. Our results also suggest a therapeutic approach to treat NEPC variants—that is, epigenetic modulation may reverse or delay lineage transformation, extending the durability of clinically beneficial ADT responses. As lineage plasticity is increasingly appreciated in other types of human cancers relapsing from molecularly targeted therapy, this approach may be generally applicable.
Acknowledgments
RNA-seq data generated in this study are deposited in the Gene Expression Omnibus (accession no. GSE90891). We acknowledge the RPCI writing group for critical reading of the manuscript and helpful comments. We thank H. Baumann for important assistance with multispectral imaging. The work was supported by funding from the National Cancer Institute (NCI) (grants R21 CA179907 and R01 CA70292 to D.W.G.; R01 CA155169, R01 CA19387, and P50 CA092629 to C.L.S.). P.M. was supported by a Congressionally Directed Medical Research Programs Prostate Cancer Research Program Postdoctoral Prostate Cancer Training Award (PC141607). L.E. and D.P.L. were supported by Prostate Cancer Foundation Young Investigator Awards. D.W.G. and M.S. were supported by the Roswell Park Alliance Foundation. D.P.L. is a recipient of a Scholarship for the Next Generation of Scientists from the Cancer Research Society and a Canadian Institute of Health Research Fellowship. C.L.S. was supported by the Howard Hughes Medical Institute (SU2C/AACR DT0712). The study was supported by the NCI RPCI Cancer Center Support Grant (P30 CA016056) and the NCI MSKCC Cancer Center Support Grant (P30 CA008748, P3 CA008748). C.L.S. is a co-inventor of enzalutamide and may be entitled to royalties. Enzalutamide is commercially available from Selleck Chemicals. C.L.S. serves on the board of directors of Novartis and is a paid consultant to ORIC Pharmaceuticals.
Footnotes
REFERENCES AND NOTES
Full text links
Read article at publisher's site: https://doi.org/10.1126/science.aah4199
Read article for free, from open access legal sources, via Unpaywall:
https://europepmc.org/articles/pmc5367887?pdf=render
Citations & impact
Impact metrics
Citations of article over time
Alternative metrics
Article citations
Integrated single-cell transcriptomic analyses identify a novel lineage plasticity-related cancer cell type involved in prostate cancer progression.
EBioMedicine, 109:105398, 16 Oct 2024
Cited by: 0 articles | PMID: 39418984 | PMCID: PMC11530610
IMPA1-derived inositol maintains stemness in castration-resistant prostate cancer via IMPDH2 activation.
J Exp Med, 221(11):e20231832, 29 Oct 2024
Cited by: 0 articles | PMID: 39470689
The neuroendocrine transition in prostate cancer is dynamic and dependent on ASCL1.
Nat Cancer, 11 Oct 2024
Cited by: 0 articles | PMID: 39394434
Epigenomic heterogeneity as a source of tumour evolution.
Nat Rev Cancer, 16 Oct 2024
Cited by: 0 articles | PMID: 39414948
Review
Castration-resistant prostate cancer monitoring by cell-free circulating biomarkers.
Front Oncol, 14:1394292, 10 Sep 2024
Cited by: 0 articles | PMID: 39319053 | PMCID: PMC11420116
Review Free full text in Europe PMC
Go to all (590) article citations
Data
Data behind the article
This data has been text mined from the article, or deposited into data resources.
BioStudies: supplemental material and supporting data
GEO - Gene Expression Omnibus
- (1 citation) GEO - GSE90891
Similar Articles
To arrive at the top five similar articles we use a word-weighted algorithm to compare words from the Title and Abstract of each citation.
TMPRSS2-ERG Controls Luminal Epithelial Lineage and Antiandrogen Sensitivity in PTEN and TP53-Mutated Prostate Cancer.
Clin Cancer Res, 24(18):4551-4565, 29 May 2018
Cited by: 36 articles | PMID: 29844131 | PMCID: PMC6139075
SOX2 promotes lineage plasticity and antiandrogen resistance in TP53- and RB1-deficient prostate cancer.
Science, 355(6320):84-88, 01 Jan 2017
Cited by: 598 articles | PMID: 28059768 | PMCID: PMC5247742
Lineage plasticity in cancer: a shared pathway of therapeutic resistance.
Nat Rev Clin Oncol, 17(6):360-371, 09 Mar 2020
Cited by: 237 articles | PMID: 32152485 | PMCID: PMC7397755
Review Free full text in Europe PMC
Resistance to androgen receptor signaling inhibition does not necessitate development of neuroendocrine prostate cancer.
JCI Insight, 6(8):146827, 22 Apr 2021
Cited by: 27 articles | PMID: 33724955 | PMCID: PMC8119192
Funding
Funders who supported this work.
NCI NIH HHS (8)
Grant ID: P30 CA016056
Grant ID: R01 CA155169
Grant ID: R21 CA179907
Grant ID: R01 CA193837
Grant ID: R01 CA207757
Grant ID: P50 CA092629
Grant ID: R01 CA070292
Grant ID: P30 CA008748
NIDCD NIH HHS (1)
Grant ID: R01 DC019387
NIGMS NIH HHS (1)
Grant ID: R25 GM095459





